Abstract
We previously demonstrated that non-small cell lung cancer (NSCLC) cells and primary human lung tumors aberrantly express the vitamin D3-catabolizing enzyme, CYP24, and that CYP24 restricts transcriptional regulation and growth control by 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) in NSCLC cells. To ascertain the basis for CYP24 dysregulation, we assembled a panel of cell lines that represent distinct molecular classes of lung cancer: Cell lines were selected which harbored mutually exclusive mutations in either the K-ras or the Epidermal Growth Factor Receptor (EGFR) genes. We observed that K-ras mutant lines displayed a basal vitamin D receptor (VDR)lowCYP24high phenotype, whereas EGFR mutant lines had a VDRhighCYP24low phenotype. A mutation-associated difference in CYP24 expression was also observed in clinical specimens. Specifically, K-ras mutation was associated with a median 4.2-fold increase in CYP24 mRNA expression (p = 4.8 × 10−7) compared to EGFR mutation in a series of 147 primary lung adenocarcinoma cases. Because of their differential basal expression of VDR and CYP24, we hypothesized that NSCLC cells with an EGFR mutation would be more responsive to 1,25(OH)2D3 treatment than those with a K-ras mutation. To test this, we measured the ability of 1,25(OH)2D3 to increase reporter gene activity, induce transcription of endogenous target genes, and suppress colony formation. In each assay, the extent of 1,25(OH)2D3 response was greater in EGFR mutation-positive HCC827 and H1975 cells than in K-ras mutation-positive A549 and 128.88T cells. We subsequently examined the effect of combining 1,25(OH)2D3 with erlotinib, which is used clinically in the treatment of EGFR mutation-positive NSCLC. 1,25(OH)2D3/erlotinib combination resulted in significantly greater growth inhibition than either single agent in both the erlotinib-sensitive HCC827 cell line and the erlotinib-resistant H1975 cell line. These data are the first to suggest that EGFR mutations may identify a lung cancer subset which remains responsive to and is likely to benefit from 1,25(OH)2D3 administration.
Keywords: non-small cell lung cancer; vitamin D receptor; 1,25-dihydroxyvitamin D3; epidermal growth factor receptor; K-ras
Introduction
Pre-clinical models support the idea that the active metabolite of vitamin D3, 1,25-dihydroxyvitamin D3(1,25(OH)2D3) inhibits lung cancer growth [1, 2]. Anti-proliferative effects of 1,25(OH)2D3 are mediated by binding to the vitamin D receptor (VDR) [3]. Upon ligand binding, VDR forms a heterodimer with the retinoid-X-receptor (RXR) and regulates the expression of genes whose promoters contain vitamin D response elements (VDREs). Transcriptional targets of 1,25(OH)2D3 include genes that regulate cell cycle arrest and apoptosis [4–7]. To explore the potential role of vitamin D signaling in clinical disease control, the relationship between serum 25-hydroxyvitamin D3 (25(OH)D3) levels or tumor VDR expression and non-small cell lung cancer (NSCLC) survival was determined. Early-stage NSCLC patients who had 25(OH)D3 levels ≥ 21.6 ng/mL experienced a significant improvement in survival as compared to patients with 25(OH)D3 levels ≤ 10.2 ng/mL [8]. With regard to VDR status, 5 year overall survival rates were 59% for patients with high nuclear VDR expression versus 27% for low nuclear VDR expression [9]. In light of these results, mechanisms that decrease VDR expression and/or vitamin D levels in tumor cells would be predicted to adversely affect lung cancer outcomes.
1α,25-dihydroxyvitamin D3 24-hydroxylase (CYP24) is the primary enzyme responsible for the catabolic inactivation of 1,25(OH)2D3 and is considered a candidate oncogene [10, 11]. CYP24 is frequently over-expressed in primary lung tumors [12–14], and its expression is independently prognostic of poor survival [15]. In prior mechanistic studies by us, the selective CYP24 inhibitor CTA091 suppressed 1,25(OH)2D3 catabolism, preserved 1,25(OH)2D3 regulation of gene expression through a VDR-dependent process, and reinforced its growth inhibitory effects in NSCLC cells [7]. These data support the hypothesis that CYP24 expression promotes tumor growth by enabling NSCLC cells to bypass growth regulation by 1,25(OH)2D3.
To dissect the mechanisms contributing to aberrant CYP24 expression in lung cancer, we assembled a panel of NSCLC cell lines that harbored mutually exclusive mutations in either the epidermal growth factor receptor (EGFR) or K-ras genes. These were selected because they represent independent oncogenic pathways in lung cancer. Mutations within the EGFR gene occur in approximately 10% of all lung adenocarcinomas and are observed most commonly in the subset of patients who have never smoked [16]. Patients whose tumors harbor activating EGFR mutations show nearly 80% response rates to EGFR tyrosine kinase inhibitors (TKIs) [17, 18]. K-ras mutations occur in approximately 25% of lung adenocarcinomas and are associated with a history of cigarette use and resistance to EGFR TKIs [19]. Our analysis of NSCLC cell lines revealed that K-ras mutation-positive cells have a basal VDRlowCYP24high phenotype that is associated with limited response to 1,25(OH)2D3. Conversely, NSCLC cells that harbor EGFR mutations have a VDRhighCYP24low, 1,25(OH)2D3-sensitive phenotype. Differential CYP24 expression in the K-ras and EGFR mutation-positive subsets of lung adenocarcinomas was confirmed in a clinical case series. To the best of our knowledge, these data are the first to identify mutation-related differences in CYP24 expression and the response of NSCLC cells to 1,25(OH)2D3 and to suggest that vitamin D supplementation may be most effective in the management of lung cancers that harbor EGFR mutations.
Materials and Methods
Cells
A549, HCC827, H1650, and H1975 cells were obtained from the American Type Culture Collection (Manassas, VA). 128.88T cells were generously provided by Dr. Jill Siegfried (University of Pittsburgh, Pittsburgh PA). HCC827, H1650, and H1975 cells were maintained in RPMI 1640 (Mediatech, Manassa, VA). A549 and 128.88T cells were maintained in BME (Life Technologies, Grand Island, NY). To prepare complete growth medium, RPMI or BME was supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Inc., Logan, UT), 2 mM L-glutamine, and 100 U/ml penicillin-streptomycin. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. The presence of a K-ras codon 12 mutation was confirmed in A549 and 128.88T cells using the method described by Mitchell et al. [20]. HCC827, H1650, and H1975 cells were authenticated by RADIL prior to use in these studies.
Chemicals
Erlotinib was purchased as a powder from ChemieTek (Indianapolis, IN). Stock solutions were prepared at a final concentration of 10 mM in dimethylsulfoxide (DMSO) and stored at −20°C. On the day of use, stocks were diluted in tissue culture medium to achieve the desired final concentration.
Preparation of whole cell extracts and immunoblot analysis
Procedures for preparation of whole cell extracts and immunoblot detection of VDR and CYP24 were the same as those described by us previously [7].
Construction of a CYP24 promoter-luciferase reporter construct
Genomic DNA was isolated from 128.88T cells using the ChargeSwitch gDNA kit from Life Technologies. A 533 bp fragment of the CYP24 promoter corresponding to nucleotides 725 to 1257 of GenBank entry HSU60669 was amplified from 100 ng of genomic DNA by 32 cycles of PCR using primers modified to contain either an Asp 718 or Bgl II site. The PCR product was gel-purified, digested with Asp 718 and Bgl II, and ligated into the Asp 718/Bgl II sites of the firefly luciferase reporter plasmid, pGL2 (Promega Corporation). Candidate clones were identified by restriction digestion and verified to contain human CYP24 promoter sequences by automated DNA sequencing and nucleotide sequence alignment using NCBI BLAST.
Assay of CYP24 promoter activity
Cells were plated in six-well plates at 4 × 105 cells per well in complete growth medium lacking antibiotics. When cells achieved 50–70% confluence, they were transiently transfected using Fugene 6 (Roche Applied Science, Mannheim, Germany). Each reaction contained 100 ng of the CYP24 promoter-luciferase reporter, 50 ng of CMV-β-galactosidase (β-gal), and pBluescript II (to adjust total DNA content to 1 μg per reaction). After 5 h, transfection medium was removed and cells were treated with either vehicle or 100 nM 1,25(OH)2D3. Treatments were done in fresh medium containing 10% charcoal-stripped serum (CSS). Cells were harvested 24 h post-treatment. Firefly luciferase activity was assayed using the Promega Luciferase Assay System. Luminescence was read for 60 sec using an AutoLumat LB953 luminometer (Berthold, Pforzheim, Germany). Reporter activity was calculated as the ratio of firefly luciferase activity to β-gal activity. For each cell line, the ratio of luciferase/ β-gal activity for vehicle-treated cells was assigned a value of 1.0.
RT-PCR Assay
Cells were seeded into 6-well plates in complete growth medium. When still sub-confluent, the cells were treated with vehicle (control) or 1,25(OH)2D3 (100 nM) for 17 h. Treatments were done in fresh medium containing 10% CSS. RNA was extracted using the PerfectPure RNA Cultured Cell Kit (5 Prime, Gaithersburg, MD, USA) in accordance with the manufacturer’s instructions. The RNA concentrations were determined by NanoDrop. RNA (250–500 ng) was converted into cDNA using the First Strand cDNA Synthesis kit from Origene (Rockville, MD). A fixed volume of cDNA (2.0 μL) was used for each PCR reaction. Reactions were run using Hot Star Taq Plus Master Mix (Qiagen, Valencia, CA) on a BioRad C1000 Thermal Cycler. VDR, CYP24, CAMP, and G3PDH primer sequences were described previously [7]. PCR products were resolved on 1.2% agarose gels and visualized by staining with ethidium bromide. Gels were imaged using the BioRad Gel Doc XR instrument.
Clonogenic Assay
Cells were seeded in triplicate in 6-well plates in complete growth medium. The next day, cells were treated with fresh medium (controls) or medium containing the indicated concentrations of 1,25(OH)2D3 ± erlotinib. Treatments were repeated every 3 days. After 7 d, colonies were fixed with methanol and stained with crystal violet. Grids were scored onto the back of each plate. Colonies in each section of the grid were inspected using a microscope, and those containing ≥ 30 cells were counted. The percent remaining colonies was calculated using the equation: % Colonies Remaining = 100 × [number colonies for treatment group/average number colonies for control group].
Results
VDR and CYP24 are differentially expressed in K-ras and EGFR mutation-positive lung cancers
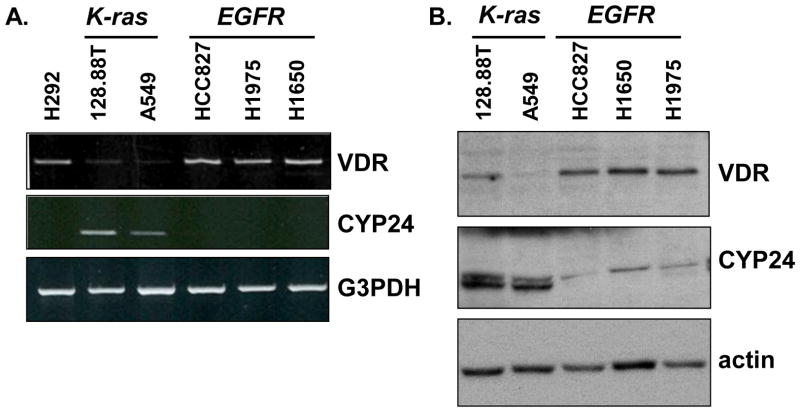
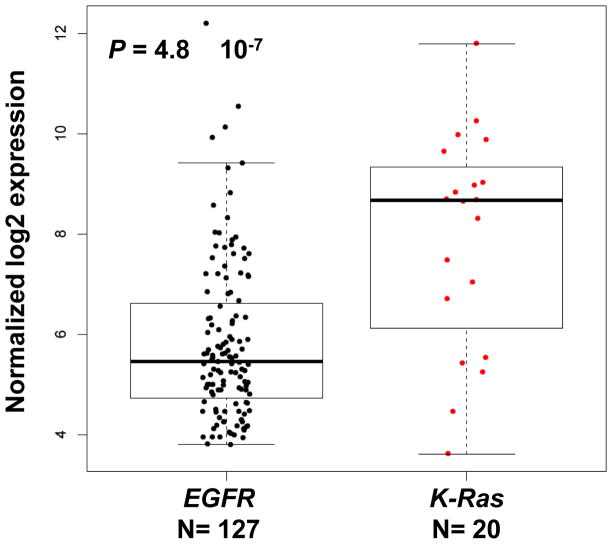
To identify factors that may contribute to aberrant basal expression of CYP24 in NSCLC, we assembled a panel of cell lines that harbored distinct mutations in either the K-ras (A549, 128.88T) or EGFR genes (HCC827, H1650, and H1975). The VDR and CYP24 expression profile of each line was established following growth in complete medium. Cells that harbor an activating K-ras mutation had lower levels of VDR mRNA and higher levels of CYP24 mRNA than cells with an EGFR mutation (Fig. 1A). The differential expression profiles were also observed when VDR and CYP24 protein levels were evaluated by immunoblot (Fig. 1B). To determine whether the observed mutation-associated differences in CYP24 expression had any clinical relevance, we performed a secondary analysis of a publically available microarray dataset that included 127 EGFR and 20 K-ras mutation–positive lung adenocarcinomas [21]. Consistent with the cell line data, we observed a statistically significant 4-fold increase in CYP24 expression in the K-ras mutation-positive subset of lung cancers (Fig. 2).
Figure 1. EGFR and K-ras mutation positive NSCLC cells differ in their basal expression of VDR and CYP24.
(A) mRNA expression levels of VDR and CYP24 were evaluated in the indicated cell lines by semi-quantitative RT-PCR. (B) Whole cell extracts were prepared from the indicated cell lines. Equivalent amounts of protein were analyzed by immunoblot for VDR and CYP24. Blots were reprobed for actin as a control for protein quantitation and loading. Results are representative of 3 independent experiments.
Figure 2. CYP24 is differentially expressed in primary human lung adenocarcinomas that harbor EGFR or K-ras mutations.
Gene expression values were abstracted for the 20 K-ras mutation-positive lung adenocarcinomas and 127 EGFR mutation-positive lung adenocarcinomas included in the whole genome, gene expression profiling study of Okayama et al., (GSE31210) [21]. Each point represents an individual case. Data were analyzed using the Bioconductor packages in the R statistical computing environment microarray data processing. Specifically, the RMA function was used to generate expression summary values through convolution background correction, quantile normalization, and a summarization based on a multi-array model fit using the medial polish algorithm. The Limma program was used to calculate the level of differential expression based on the log2 transformed gene expression value. Multiple testing was corrected using Benjamin and Hochberg’s algorithm. Box edges indicate the 25%-ile and 75%-ile of CYP24 expression for each population. Horizontal bars indicate the median CYP24 expression value.
NSCLC cells with an EGFR mutation respond preferentially to 1,25(OH)2D3 treatment
Given that VDR mediates transcriptional regulation by 1,25(OH)2D3, and CYP24 negatively regulates 1,25(OH)2D3 levels and activity, we predicted that NSCLC cells that have an EGFR mutation and a VDRhigh/CYP24low phenotype would be more responsive to 1,25(OH)2D3 than cells with a K-ras mutation and a VDRlow/CYP24high phenotype. To test this, 1,25(OH)2D3 transcriptional activation was measured. Cells were transfected with a CYP24 promoter-luciferase reporter vector and CMV-β-gal then treated 5h later with either vehicle (0.004% EtOH) or 100 nM 1,25(OH)2D3. When bound by 1,25(OH)2D3, the VDR increases CYP24 transcription through a series of VDREs located in the CYP24 promoter [22, 23]. HCC827 and H1975 cells displayed robust transcriptional responses to 1,25(OH)2D3 administration, with average increases in reporter activity of 10-fold (range 5.7–14.5) and 9-fold (range 5.7–16.6), respectively (Fig. 3A). 128.88T and A549 cells also responded to treatment, but the average 1,25(OH)2D3-mediated increase in reporter activity in these cells was 3-fold. When the reporter activity at 100 nM 1,25(OH)2D3 was compared among the cell lines, significant differences were observed between HCC827 and A549 (p = 0.009); HCC827 and 128.88T (p =0.023); and H1975 and A549 (p=0.05). To confirm these differential transcriptional responses, the effect of 1,25(OH)2D3 treatment on endogenous gene expression was examined (Fig. 3B). To do this, we analyzed expression of the CYP24, CD14, and CAMP genes, whose promoters contain VDREs [24]. In both HCC827 and H1975 cells, 1,25(OH)2D3 treatment was associated with a discernible increase in the expression of all 3 genes. In contrast, expression of the 1,25(OH)2D3 target genes was essentially unchanged upon treatment in 128.88T and A549 cells. Cumulatively, these data indicate that transcriptional responses to 1,25(OH)2D3 are preferentially retained in NSCLC cells that harbor an EGFR mutation.
Figure 3. EGFR and K-ras mutation-positive NSCLC cells differ in their transcriptional responses to 1,25(OH)2D3.
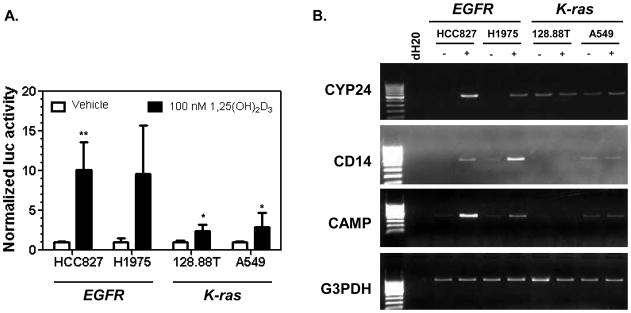
(A) Cells were co-transfected with a CYP24 promoter-luciferase reporter plasmid and CMV β-galactosidase (to control for transfection efficiency). Transfected cells were treated with vehicle or 100 nM 1,25(OH)2D3. Luciferase and β-galactosidase activities were measured after 17 h. Normalized activity values (luc/β-gal) were calculated for each treatment group. For each cell line, the control (vehicle) value was set to a value of 1.0. Values represent the mean ± SD for 3–6 determinations. The difference between the reporter activity in the control group versus the 100 nM 1,25(OH)2D3 treatment group was assessed using one-sided t-tests. * p <0.05; ** p < 0.001. (B) The indicated cell lines were treated with vehicle (-) or 100 nM 1,25(OH)2D3 (+). RNA was isolated 17h later, and the expression of endogenous 1,25(OH)2D3 target genes CYP24, CD14, and CAMP was measured by semi-quantitative PCR. G3PDH was used as a control for RNA quantitation and cDNA synthesis. Similar results were obtained in a second, independent study.
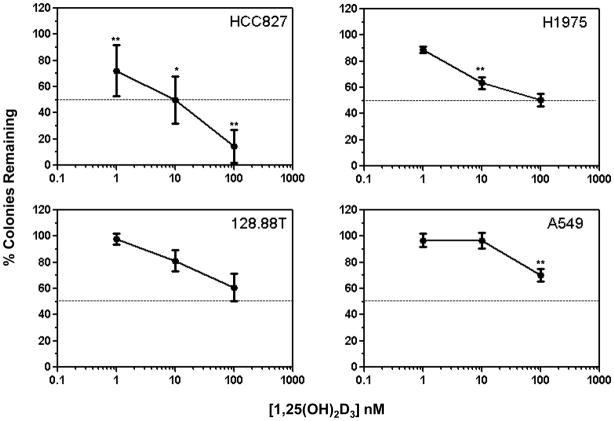
To determine whether the observed differential transcriptional responses were indicative of differential sensitivity to 1,25(OH)2D3 –mediated growth inhibition, clonogenic assays were conducted. Cells were allowed to attach and then were treated every 3 days with vehicle (control) or increasing concentrations of 1,25(OH)2D3. Clonogenic survival was measured after 7 days. Although treatment resulted in dose-dependent growth inhibition in all 4 cell lines, the preferential sensitivity of EGFR mutation-positive cells to 1,25(OH)2D3 was supported by two findings (Fig. 4). First, HCC827 and H1975 cells but not 128.88T or A549 cells displayed significant increases in growth inhibition between 1 and 10 nM 1,25(OH)2D3. Second, at the highest concentration of 1,25(OH)2D3 tested (100 nM), the average percent growth inhibition for HCC827 (86%) and H1975 (50%) cells was greater than that which was observed for 128.88T (39%) and A549 (30%) cells.
Figure 4. EGFR and K-ras mutation-positive NSCLC cells are differentially sensitive to 1,25(OH)2D3-mediated growth inhibition.
The indicated cells were seeded into 6 well plates (3 wells per treatment group). The next day, cells were treated with fresh medium containing vehicle (control) or varying concentrations of 1,25(OH)2D3. Treatments were repeated every 3 days. Crystal violet staining was used to assess colony formation as described in Methods. Values represent the mean ± SD for 6–9 determinations, pooled from 2–3 independent experiments per cell line. The association between cell growth inhibition and treatment was assessed using a one-way ANOVA. The ordered pair-wise comparisons between successive dose-levels were made using t-tests (* p<0.05, ** p≤ 0.001).
1,25(OH)2D3/erlotinib combination results in increased growth suppression of EGFR mutation-positive lung cancer cells
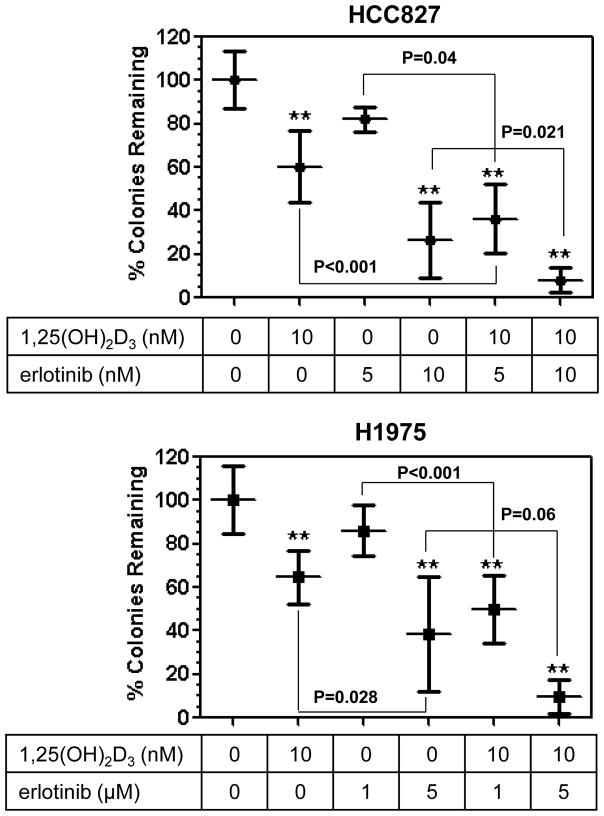
The EGFR tyrosine kinase inhibitor (EGFR TKI), erlotinib, is effective in the treatment of individuals with advanced NSCLC whose tumors harbor an activating mutation in the EGFR gene (reviewed in [17]). Because we found that NSCLC cells with an EGFR mutation are also likely to respond to 1,25(OH)2D3, we sought to examine the effect of combining 1,25(OH)2D3 with erlotinib. For this purpose, we used both HCC827 and H1975 cells. HCC827 cells have an exon 19 deletion (delE746-A750) in the EGFR gene and are sensitive to EGFR TKIs [7, 25]. H1975 cells are refractory to EGFR TKIs due to the presence of the T790M mutation in EGFR [26]. After overnight attachment, the cells were treated with vehicle, 1,25(OH)2D3 alone, erlotinib alone, or 1,25(OH)2D3 plus erlotinib. Consistent with the results presented above, 1,25(OH)2D3 treatment resulted in significant inhibition of clonogenic survival in both cell lines (Fig. 5). Erlotinib inhibited the growth of both cell lines in a dose-dependent manner and, as expected, the concentration that significantly inhibited H1975 survival (5 μM) was greater than that required for HCC827 cells (10 nM). In both cell lines, a 1,25(OH)2D3/erlotinib combination was identified which resulted in significantly greater growth inhibition than either single agent. These results suggest that 1,25(OH)2D3 treatment may represent an effective approach to increase the responsiveness of erlotinib-sensitive and erlotinib-resistant EGFR-mutant NSCLCs to EGFR TKIs.
Figure 5. Combination of 1,25(OH)2D3 with erotinib results in increased growth inhibition of EGFR mutation-positive NSCLC cells.
(A) HCC827 and H1975 cells were seeded into 6 well plates (3 wells per treatment group). The next day, the cells were treated with vehicle (control), 1,25(OH)2D3 alone, erlotinib alone, or the combination. Treatments were repeated every 3 days. Colony formation was assessed as described in Fig. 4. Values represent the mean ± SD for 3 pooled experiments per cell line. The association between treatment and cell growth for each cell line was assessed using a one-way ANOVA test. One-sided post-hoc t-tests, conducted with Dunnett’s adjustment for multiple tests, were used to compare the control group to all other treatments (** p<0.001). Within each cell line, the combination treatments were subsequently compared to the individual treatments using one-sided t-tests with a Bonferroni adjustment for multiple tests (p values for each comparison are shown adjacent to brackets).
Discussion
Lung cancer remains the primary cause of cancer-related death in the United States. However, significant improvements in outcomes have been realized in recent years. Advances have resulted from the application of molecularly targeted therapies to specific subsets of lung cancer patients [17]. Based on the results presented in this manuscript, we suggest that EGFR mutations may identify a lung cancer subset which is likely to respond to and benefit from 1,25(OH)2D3 administration. Specifically, we find that EGFR mutation-positive lung cancer cells express relatively high levels of the VDR, have an intact 1,25(OH)2D3 signaling axis, and are significantly inhibited by 1,25(OH)2D3 administration. Because erlotinib is effective in the treatment of individuals with advanced NSCLC whose tumors harbor an activating mutation in the EGFR gene, we examined the potential impact of 1,25(OH)2D3 on its activity. We discovered that combination of 1,25(OH)2D3 with erlotinib resulted in significant inhibition of both HCC827 cells (a model of erlotinib-sensitive lung cancer) and H1975 cells (a model of acquired resistance to erlotinib). These results suggest that 1,25(OH)2D3 may be useful in preventing the outgrowth of erlotinib-resistant disease. Pertinent to the potential clinical application of these findings, results from a prior Phase 1 clinical trial demonstrate that 1,25(OH)2D3 (calcitriol) can be safely co-administered with an EGFR tyrosine kinase inhibitor in the oncology setting [27].
We observe an inverse relationship between VDR and CYP24 expression in NSCLC cells (Fig 1). A statistically significant inverse association between these molecules was also observed in a large series of primary lung adenocarcinomas [15]. One possible explanation for these findings is that VDR functions as a repressor of basal CYP24 transcription in NSCLC. The existence of such a regulatory mechanism is supported by prior studies in kidney and breast cancer cells [28, 29]. Using VDR over-expression and VDR siRNA approaches, Alimirah et al. were able to demonstrate the repressive action of unliganded VDR on CYP24 expression in multiple breast cancer cell lines [29]. It was hypothesized by these investigators that CYP24 repression occurs when VDR recruits the nuclear co-repressors, NCoR and SMRT, to the CYP24 promoter. Once bound, the VDR/corepressor complex recruits other proteins that mediate transcriptional repression via epigenetic processes. If this model also applies to lung cancer, then it is expected that in K-ras mutation-positive NSCLC cells that have low VDR levels, NCoR/SMRT are no longer recruited to the promoter, CYP24 repression is relieved and basal transcription increases. In constructing such models, it is important to consider the possibility that VDR/NCoR complexes may also mediate basal repression of CYP24 through elements other than the promoter proximal VDREs. VDR and NCoR both bind to a VDRE located 50 kb downstream of the CYP24 gene in human colonic LS180 cells in the absence of 1,25(OH)2D3 [30]. In 3-dimensional space, this region comes into proximity with the CYP24 transcription start site [30]. In this configuration, VDR/NCoR complexes bound to the +50 kb element may contribute to basal repression of the CYP24 promoter.
A549 and 128.88T cells, which harbor a K-ras mutation, display a VDRlow/CYP24high, vitamin D-resistant phenotype. Might the K-ras mutation itself contribute to CYP24 upregulation? Ras signaling activates Ets transcription factors, and there is a functional Ets-binding site (EBS) located adjacent to the proximal VDRE within the CYP24 promoter [31]. Ras-activated Ets transcription factors cooperate with VDR in the 1,25(OH)2D3-mediated induction of CYP24 transcription [31]. Ets-1 also increases CYP24 transcription in the absence of 1,25(OH)2D3, but only when repressive VDR:RXR heterodimers cannot form at the VDRE within the CYP24 promoter [31]. Based on these data, we speculate that the low VDR levels observed in K-ras mutant NSCLC cells preclude the formation of repressive VDR:RXR complexes under basal growth conditions. Consequently, K-ras activated Ets-1 transcription factors are able to bind to the EBS, where they stimulate CYP24 transcription. Consistent with this idea, we find that mutation of the EBS within a CYP24 promoter-luciferase construct significantly decreases basal reporter activity in A549 and 128.88T cells (data not shown).
Although K-ras mutation may be a determinant of the VDRlow/CYP24high phenotype in NSCLC, it is clearly not sufficient for phenotype generation. Some K-ras mutation-positive lung adenocarcinomas express low levels of CYP24 mRNA (Fig. 2), and some K-ras positive NSCLC cells (SK-LU-1) are VDRhigh/CYP24low and 1,25(OH)2D3-responsive [15, 32]. An alternative hypothesis is that: (1) The mutation-related differences in 1,25(OH)2D3 sensitivity that we observe occur because K-ras and EGFR mutations are differentially associated with smoking; and (2) Smoking drives epigenetic changes in K-ras mutation-positive cells that contribute to inactivation of the vitamin D pathway. This idea is supported by prior reports showing that VDR and CYP24 are subject to epigenetic regulation [33, 34], and that specific epigenetic alterations are differentially observed in K-ras and EGFR mutant lung cancers [35]. Ongoing studies in our laboratory are designed to establish the relationship between smoking, K-ras mutation, and acquisition of the VDRlow/CYP24high, vitamin D-resistant phenotype.
Highlights.
Aberrant CYP24 expression limits 1,25(OH)2D3 activity in NSCLC cells
CYP24 levels are increased in NSCLC cells and lung tumors with K-ras mutations
NSCLC cells and lung tumors with EGFR mutations have low basal CYP24 levels
K-ras and EGFR mutation-positive NSCLC cells respond differentially to 1,25(OH)2D3
EGFR mutations may identify a NSCLC subset likely to benefit from 1,25(OH)2D3
Acknowledgments
The authors wish to thank Drs. Brenda Diergaarde, Richard Steinman, and Jill Siegfried (University of Pittsburgh Cancer Institute), and Drs. Candace Johnson and Donald Trump (Roswell Park Cancer Institute) for helpful discussions during the course of this work.
Abbreviations used
- CSS
Charcoal-stripped serum
- DMSO
Dimethylsulfoxide
- EGFR
Epidermal growth factor receptor
- FBS
Fetal bovine serum
- 1, 25(OH)2D3
1,25-dihydroxyvitamin D3
- NSCLC
non-small cell lung cancer
- TKI
tyrosine kinase inhibitor
- VDRE
vitamin D response element
Footnotes
This work was supported by grant R01 CA132844.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakagawa K, Kawaura A, Kato S, Takeda E, Okano T. Metastatic growth of lung cancer cells is extremely reduced in Vitamin D receptor knockout mice. J Steroid Biochem Mol Biol. 2004;89–90(1–5):545–547. doi: 10.1016/j.jsbmb.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 2.Norton R, O’Connell M. Vitamin D: Potential in the Prevention and Treatment of Lung Cancer. Anticancer Res. 2012;32:211–222. [PubMed] [Google Scholar]
- 3.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1998;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swami S, Raghavachari N, Muller UR, Bao YP, Feldman D. Vitamin D growth inhibition of breast cancer cells: gene expression patterns assessed by cDNA microarray. Breast Cancer Res Treat. 2003;80:49–62. doi: 10.1023/A:1024487118457. [DOI] [PubMed] [Google Scholar]
- 5.Krishnan AV, Shinghal R, Raghavachari N, Brooks JD, Peehl DM, Feldman D. Analysis of Vitamin D-Regulated Genes in LNCaP Human Prostate Cancer Cells Using cDNA Microarrays. The Prostate. 2004;59:243–251. doi: 10.1002/pros.20006. [DOI] [PubMed] [Google Scholar]
- 6.White JH. Profiling 1,25-dihydroxyvitamin D3-regulated gene expression by microarray analysis. Steroid Biochem Mol Biol. 2004;89–90:239–244. doi: 10.1016/j.jsbmb.2004.03.074. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Kanterewicz B, Buch S, Petkovich M, Parise R, Beumer J, Lin Y, Diergaarde B, Hershberger PA. CYP24 inhibition preserves 1α,25-dihydroxyvitamin D3 anti-proliferative signaling in lung cancer cells. Mol Cell Endocrinol. 2012;355(1):153–161. doi: 10.1016/j.mce.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou W, Heist RS, Liu G, Asomaning K, Neuberg DS, Hollis BW, Wain JC, Lynch TJ, Giovannucci E, Su L, et al. Circulating 25-hydroxyvitamin D levels predict survival in early-stage non-small-cell lung cancer patients. J Clin Oncol. 2007;25(5):479–485. doi: 10.1200/JCO.2006.07.5358. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan M, Parwani AV, Hershberger PA, Lenzner DE, Weissfeld JL. Nuclear vitamin D receptor expression is associated with improved survival in non-small cell lung cancer. J Steroid Biochem Mol Biol. 2011;123(1–2):30–36. doi: 10.1016/j.jsbmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29(12):664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Albertson DG, Yistra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo W-L, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nature Genetics. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 12.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 13.Anderson MG, Nakane M, Ruan X, Kroeger PE, Wu-Wong JR. Expression of VDR and CYP24A1 mRNA in human tumors. Cancer Chemother Pharmacol. 2006;57:234–240. doi: 10.1007/s00280-005-0059-7. [DOI] [PubMed] [Google Scholar]
- 14.Parise RA, Egorin MJ, Kanterewicz B, Taimi M, Petkovich M, Lew AM, Chuang SS, Nichols M, El-Hefnawy T, Hershberger PA. CYP24, the enzyme that catabolizes the antiproliferative agent vitamin D, is increased in lung cancer. Int J Cancer. 2006;119(8):1819–1828. doi: 10.1002/ijc.22058. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Kim SH, King AN, Zhao L, Simpson RU, Christensen PJ, Wang Z, Thomas DG, Giordano TJ, Lin L, et al. CYP24A1 is an independent prognostic marker of survival in patients with lung adenocarcinoma. Clin Cancer Res. 2011;17(4):817–826. doi: 10.1158/1078-0432.CCR-10-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Schmid-Bindert G, Zhou C. Erlotinib in the treatment of advanced non-small cell lung cancer: an update for clinicians. Ther Adv Med Oncol. 2011;4(1):19–29. doi: 10.1177/1758834011427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15(16):5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell CE, Belinsky SA, Lechner JF. Detection and quantitation of mutant K-ras codon 12 restriction fragments by capillary electrophoresis. Anal Biochem. 1995;224(1):148–153. doi: 10.1006/abio.1995.1020. [DOI] [PubMed] [Google Scholar]
- 21.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 22.Chen K-S, DeLuca HF. Cloning of the human 1α,25-dihydroxyvitamin D3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta. 1995;1263:1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- 23.Ohyama Y, Ozono K, Uchida M, Yoshimura M, Shinki T, Suda T, Yamamoto O. Functional assessment of two vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J Biol Chem. 1996;271:30381–30385. doi: 10.1074/jbc.271.48.30381. [DOI] [PubMed] [Google Scholar]
- 24.Wang TT, Tavera-Mendoza LE, Laperriere D, Libby E, MacLeod NB, Nagai Y, Bourdeau V, Konstorum A, Lallemant B, Zhang R, et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol. 2005;19(11):2685–2695. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 25.Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, Halmos B, Pearlberg J, Tsuchihashi Z, Cantley LC, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97(16):1185–1194. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- 26.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski M, Kris M, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fakih MG, Trump DL, Muindi JR, Black JD, Bernardi RJ, Creaven PJ, Schwartz J, Brattain MG, Hutson A, French R, et al. A phase I pharmacokinetic and pharmacodynamic study of intravenous calcitriol in combination with oral gefitinib in patients with advanced solid tumors. Clin Cancer Res. 2007;13(4):1216–1223. doi: 10.1158/1078-0432.CCR-06-1165. [DOI] [PubMed] [Google Scholar]
- 28.Dwivedi PP, Muscat GE, Bailey PJ, Omdahl JL, May BK. Repression of basal transcription by vitamin D receptor: evidence for interaction of unliganded vitamin D receptor with two receptor interaction domains in RIP13delta1. J Mol Endocrinol. 1998;20(3):327–335. doi: 10.1677/jme.0.0200327. [DOI] [PubMed] [Google Scholar]
- 29.Alimirah F, Vaishnav A, McCormick M, Echchgadda I, Chatterjee B, Mehta RG, Peng X. Functionality of unliganded VDR in breast cancer cells: repressive action on CYP24 basal transcription. Mol Cell Biochem. 2010;342(1–2):143–150. doi: 10.1007/s11010-010-0478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1α,25-dihydroxyvitamin D3. J Biol Chem. 2010;285(20):15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dwivedi PP, Omdahl JL, Kola I, Hume DA, May BK. Regulation of rat cytochrome P450C24 (CYP24) gene expression. Evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D3-mediated induction. J Biol Chem. 2000;275(1):47–55. doi: 10.1074/jbc.275.1.47. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Chen G, King AN, Jeon CK, Christensen PJ, Zhao L, Simpson RU, Thomas DG, Giordano TJ, Brenner DE, et al. Characterization of vitamin D receptor (VDR) in lung adenocarcinoma. Lung Cancer. 2012;77(2):265–271. doi: 10.1016/j.lungcan.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung I, Karpf AR, Muindi JR, Conroy JM, Nowak NJ, Johnson CS, Trump DL. Epigenetic silencing of CYP24 in tumor-derived endothelial cells contributes to selective growth inhibition by calcitriol. J Biol Chem. 2007;282(12):8704–8714. doi: 10.1074/jbc.M608894200. [DOI] [PubMed] [Google Scholar]
- 34.Marik R, Fackler M, Gabrielson E, Zeiger MA, Sukumar S, Stearns V, Umbricht CB. DNA methylation-related vitamin D receptor insensitivity in breast cancer. Cancer Biol Ther. 2010;10(1):44–53. doi: 10.4161/cbt.10.1.11994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyooka S, Tokumo M, Shigematsu H, Matsuo K, Asano H, Tomii K, Ichihara S, Suzuki M, Aoe M, Date H, et al. Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res. 2006;66(3):1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]