Abstract
OBJECTIVE:
The aim of this study was to evaluate the prevalence and spectrum of Nkx2.5 mutations associated with idiopathic atrial fibrillation (AF).
METHODS:
A cohort of 136 unrelated patients with idiopathic atrial fibrillation and 200 unrelated, ethnically matched healthy controls were enrolled. The coding exons and splice junctions of the Nkx2.5 gene were sequenced in 136 atrial fibrillation patients, and the available relatives of mutation carriers and 200 controls were subsequently genotyped for the identified mutations. The functional characteristics of the mutated Nkx2.5 gene were analyzed using a dual-luciferase reporter assay system.
RESULTS:
Two novel heterozygous Nkx2.5 mutations (p.N19D and p.F186S) were identified in 2 of the 136 unrelated atrial fibrillation cases, with a mutational prevalence of approximately 1.47%. These missense mutations co-segregated with atrial fibrillation in the families and were absent in the 400 control chromosomes. Notably, 2 mutation carriers also had congenital atrial septal defects and atrioventricular block. Multiple alignments of the Nkx2.5 protein sequences across various species revealed that the altered amino acids were completely conserved evolutionarily. Functional analysis demonstrated that the mutant Nkx2.5 proteins were associated with significantly reduced transcriptional activity compared to their wild-type counterpart.
CONCLUSION:
These findings associate the Nkx2.5 loss-of-function mutation with atrial fibrillation and atrioventricular block and provide novel insights into the molecular mechanism involved in the pathogenesis of atrial fibrillation. These results also have potential implications for early prophylaxis and allele-specific therapy of this common arrhythmia.
Keywords: Atrial Fibrillation, Transcriptional Factor, Nkx2.5, Genetics, Reporter Gene
INTRODUCTION
Atrial fibrillation (AF) is the most common form of sustained cardiac arrhythmia, accounting for approximately one-third of hospital admissions for miscellaneous cardiac rhythm conditions (1). The prevalence of AF is approximately 1% in the general population and increases with age, rising from less than 1% in individuals aged under 60 years to nearly 10% of those over 80 years of age (1). Among people older than 40 years, the lifetime risk of developing AF is approximately 25% (2). In the United States, the number of persons with AF is currently estimated at 2.3 million and is projected to reach 15.9 million by 2050 (3). This prevalent dysrhythmia is associated with decreased quality of life, reduced exercise tolerance, impaired cognition, tachycardiomyopathy, thromboembolism, congestive heart failure, and even death (1,4,5). Patients with AF have a 5-fold increased risk of ischemic stroke (6), and the mortality rate of patients with AF is approximately 2-fold greater than that of individuals with a normal sinus rhythm (7). Therefore, AF confers a substantial economic burden on both affected individuals and society (8). Despite the high prevalence and clinical significance, the molecular etiology of AF remains largely elusive.
Traditionally, AF has been thought of as a complication secondary to various structural heart diseases or systemic disorders, including ischemic heart disease, rheumatic heart disease, congenital heart disease, cardiomyopathy, congestive heart failure, essential hypertension, and hyperthyroidism (1,9,10). However, in 30% to 45% of patients, AF occurs separately in the absence of underlying diseases or precipitating factors, which is a condition defined as idiopathic or lone AF; at least 15% of these patients have a positive family history, termed familial AF (11). Increasing epidemiological evidence has demonstrated the familial aggregation of AF and the enhanced susceptibility to AF in close relatives of patients with AF, strongly suggesting that genetic components play pivotal roles in the initiation and maintenance of AF in a subset of cases (12). Genome-wide linkage analyses have mapped specific susceptibility loci for AF on human chromosomes 10q22, 6q14-16, 11p15.5, 5p15, 10p11-q21, and 5p13; in particular, AF-causative mutations in 2 genes, namely KCNQ1 on chromosome 11p15.5 and NUP155 on chromosome 5p13, were identified and functionally characterized (13). Direct scans of candidate genes revealed a long list of AF-associated genes, including KCNE2, KCNH2, KCNJ2, KCNA5, SCN5A, ANP, and GJA5 (13). Nevertheless, AF has substantial genetic heterogeneity, and the genetic determinants for AF in most cases remain to be identified.
It is well established that focal triggered activity is involved in both the initiation and maintenance of AF and that the myocardial sleeves surrounding the pulmonary veins at their junctions with the atrial chambers are the predominant source of ectopic activity (14). Recent studies have further highlighted the critical role for several transcription factors, including Nkx2.5, GATA4, GATA5, and GATA6, in the normal embryonic development of the cardiovascular system (15-17); in addition, variations in GATA4, GATA5, and GATA6 have been causally linked to the pathogenesis of AF (18-26). Cardiac-specific Nkx2.5 is a member of the NK2 family of homeodomain-containing transcription factors, and its expression and functions overlap with those of GATA4, GATA5, and GATA6 during cardiovascular genesis, especially in the regulation of target gene expression synergistically with GATA4 (27), which justifies screening for Nkx2.5 as a prime candidate gene for idiopathic AF.
MATERIALS AND METHODS
Ethics statement
This investigation was conducted in compliance with the ethical principles of the revised Declaration of Helsinki (Somerset West, Republic of South Africa, 1996). The research protocol was reviewed and approved by the local institutional ethics committee, and written informed consent was obtained from all of the participants prior to the study.
Study subjects
A cohort of 136 unrelated patients with idiopathic AF was recruited from the Han Chinese population. The available relatives of the patients harboring the identified Nkx2.5 mutations were also included. In total, 200 ethnically matched, unrelated healthy individuals were enrolled as controls. Peripheral venous blood samples were prepared, and clinical data including medical records, electrocardiograms, and echocardiography reports were collected. The study subjects were clinically classified using a consistently applied set of definitions (11,23).
Mutational scan
Genomic DNA was extracted from the peripheral venous blood lymphocytes of all participants using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). The coding exons and exon-intron boundaries of Nkx2.5 were sequenced in 136 unrelated patients with idiopathic AF. Subsequently, genotyping of Nkx2.5 in the available relatives of mutation carriers and 200 control individuals was performed to detect the presence of mutations that were identified in the probands. The referential genomic DNA sequence of Nkx2.5 was derived from GenBank (accession No. NT_023133) at the National Center for Biotechnical Information (NCBI; http://www.ncbi.nlm.nih.gov/). With the aid of Primer 3 software (http://frodo.wi.mit.edu), the primer pairs used to amplify the entire coding region and the splice junction sites of Nkx2.5 by polymerase chain reaction (PCR) were designed as previously described (28). PCR was performed using HotStar Taq DNA Polymerase (Qiagen, Hilden, Germany) with a Veriti Thermal Cycler (Applied Biosystems, Foster City, CA, USA), and standard conditions and reagent concentrations were used. The amplified products were analyzed on 1% agarose gels stained with ethidium bromide and purified using a QIAquick Gel Extraction Kit (Qiagen). Both strands of each amplicon were sequenced with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI PRISM 3130 XL DNA Analyzer (Applied Biosystems). The sequencing primers were the same as those previously designed for specific region amplification. The DNA sequences were then analyzed using DNA Sequencing Analysis Software v5.1 (Applied Biosystems). Each variant was validated by re-sequencing an independent PCR-generated amplicon from the subject and met the quality control thresholds, with a call rate that was greater than 99%. In addition, the novelty of each identified sequence variant was confirmed by querying the Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS) and NCBI's single nucleotide polymorphism (SNP; http://www.ncbi.nlm.nih.gov/SNP) online databases.
Alignment of multiple Nkx2.5 protein sequences
Multiple Nkx2.5 protein sequences across various species were aligned using the program MUSCLE version 3.6 (http://www.ncbi.nlm.nih.gov/).
Prediction of the disease-causing potential of an Nkx2.5 sequence variation
The causative potential of an Nkx2.5 sequence variation was predicted using MutationTaster (available at http://www.mutationtaster.org), which automatically assigned a probability for the variation to be either a pathogenic mutation or a benign polymorphism. Notably, the P value used was the probability of the correct prediction rather than the probability of error, as is used in t-test statistics (i.e., a value close to 1 indicated high accuracy of the prediction).
Expression plasmids and site-directed mutagenesis
The recombinant expression plasmid Nkx2.5-pEFSA and the atrial natriuretic factor (ANF)-luciferase reporter plasmid, which contains the 2,600-bp 5'-flanking region of the ANF gene, referred to as ANF(-2600)-Luc, were provided by Dr. Ichiro Shiojima from the Department of Cardiovascular Science and Medicine, Chiba University Graduate School of Medicine, Chuo-ku, Chiba, Japan. Each of the identified mutations was introduced into wild-type Nkx2.5 using a QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) with a complementary pair of primers. The mutants were sequenced to confirm the desired mutations and to exclude any other sequence variations.
Luciferase reporter gene assay
COS-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The ANF(-2600)-Luc reporter construct and an internal control reporter plasmid pGL4.75 (hRluc/CMV, Promega) were used in transient transfection assays to explore the transactivational activity of the Nkx2.5 mutants. The COS-7 cells were transfected with 0.4 μg of wild-type or mutant Nkx2.5-pEFSA expression vector or empty vector pEFSA, 1.0 μg of ANF(-2600)-Luc reporter construct, and 0.04 μg of pGL4.75 control reporter vector using the Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). For the co-transfection experiments, 0.2 μg of wild-type Nkx2.5-pEFSA, 0.2 μg of mutant Nkx2.5-pEFSA or empty vector pEFSA, 1.0 μg of ANF(-2600)-Luc, and 0.04 μg of pGL4.75 were used. Firefly luciferase and Renilla luciferase activities were measured using the Dual-Glo luciferase assay system (Promega) 48 h after transfection. The activity of the ANF promoter was expressed as the fold activation of Firefly luciferase relative to Renilla luciferase. A minimum of 3 independent experiments was performed for wild-type or mutant Nkx2.5.
Statistical analysis
The data are expressed as the means ± SD. Continuous variables were tested for their normality of distribution, and the Student′s unpaired t-test was used to compare numeric variables between 2 groups. The categorical variables were compared between 2 groups using Pearson's chi-squared test or Fisher′s exact test, when appropriate. A 2-sided P-value less than 0.05 was considered statistically significant.
RESULTS
Clinical characteristics of the study population
A cohort of 136 unrelated patients with idiopathic AF was enlisted, clinically evaluated, and compared to 200 ethnically matched, unrelated healthy individuals. None of the subjects had traditional risk factors for AF. There were no significant differences between the patient and control groups regarding the baseline characteristics, including age, gender, body mass index, blood pressure, fasting blood glucose, serum lipids, left atrial dimension, left ventricular ejection fraction, heart rate at rest, and life style (data not shown). The baseline clinical characteristics of the 136 patients with idiopathic AF are summarized in Table 1.
Table 1.
Baseline clinical characteristics of the 136 unrelated patients with idiopathic AF.
| Parameter | Number or mean | Percentage or range |
| Male | 71 | 52 |
| Age at the initial diagnosis of AF (years) | 51.8 | 20–60 |
| Age at the time of the present study (years) | 56.7 | 22–65 |
| Type of AF at presentation | ||
| Paroxysmal AF | 88 | 65 |
| Persistent AF | 27 | 20 |
| Long-lasting persistent AF | 21 | 15 |
| Positive family history of AF | 48 | 35 |
| History of cardioversion | 112 | 82 |
| History of pacemaker | 7 | 5 |
| Resting heart rate (beats per minute) | 76.5 | 58-160 |
| Systolic blood pressure (mmHg) | 126.4 | 90-136 |
| Diastolic blood pressure (mmHg) | 87.2 | 62-89 |
| Body mass index (kg/m2) | 22.4 | 20-24 |
| Left atrial dimension (mm) | 38 | 30-40 |
| Left ventricular ejection fraction | 0.6 | 0.5-0.7 |
| Fasting blood glucose (mmol/L) | 4.6 | 3.6-5.8 |
| Total cholesterol (mmol/L) | 4.0 | 3.0-5.6 |
| Triglycerides (mmol/L) | 1.5 | 0.8-1.7 |
| Medications | ||
| Amiodarone | 101 | 74 |
| Warfarin | 29 | 21 |
| Digoxin | 23 | 17 |
| Beta-blocker | 16 | 12 |
| Calcium channel blocker | 11 | 8 |
Nkx2.5 mutations
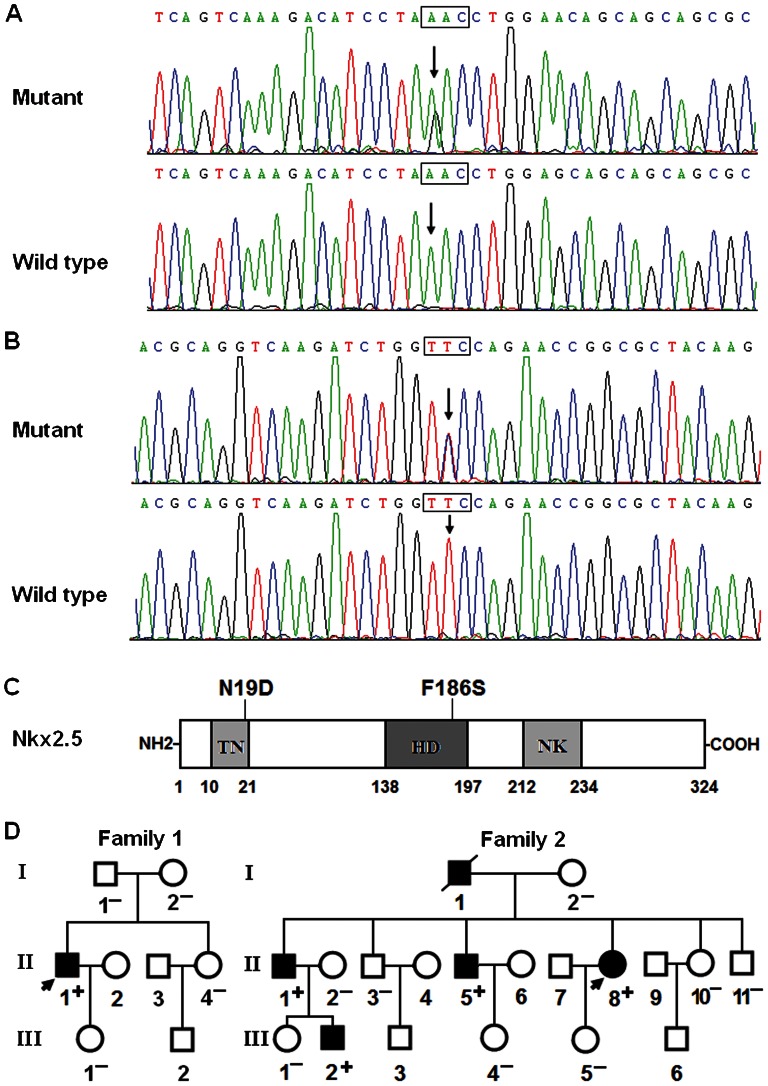
Two heterozygous missense mutations in Nkx2.5 were identified in 2 of the 136 unrelated patients with idiopathic AF. The total population prevalence of Nkx2.5 mutations, based on the patient cohort, was approximately 1.47%. Specifically, the substitution of guanine (G) for adenine (A) in the first nucleotide of codon 19 (c.55A>G), which predicted the transition of asparagine (N) into aspartic acid (D) at amino acid 19 (p.N19D), was identified in the index patient from family 1. In addition, the change of thymine (T) to cytosine (C) in the second nucleotide of codon 186 (c.557T>C), corresponding to the transversion of phenylalanine (F) into serine (S) at amino acid 186 (p.F186S), was found in the proband from family 2. The sequence chromatograms displaying the detected heterozygous Nkx2.5 mutations of c.55A>G and c.557T>C in contrast to the control sequences are presented in Figure 1A and Figure 1B, respectively. A schematic diagram of Nkx2.5 depicting the putative structural domains and location of the mutations identified in the AF patients is presented in Figure 1C. The missense mutations were neither observed in the control population nor reported in the EVS and NCBI SNP databases. Genetic scans of the available family members of the mutation carriers revealed that, in each family, the mutation was present in all of the living, affected family members but was absent in the unaffected family members examined. Analysis of the pedigrees demonstrated that, in each family, the mutation that co-segregated with AF was transmitted in an autosomal dominant pattern with complete penetrance. The pedigree structures of the 2 families are illustrated in Figure 1D. The phenotypic characteristics and the results of genetic screening of the affected family members are listed in Table 2.
Figure 1.
Nkx2.5 mutations associated with idiopathic AF. A, Sequence electropherograms showing the c.55A>G mutation of Nkx2.5 in contrast to the control. The arrow indicates the heterozygous nucleotides of A/G in the index patient from family 1 (mutant) or the homozygous nucleotides of A/A in the corresponding control individual (wild-type). The rectangle designates the nucleotides comprising a codon of Nkx2.5. B, Sequence electropherograms showing the c.557T>C mutation of Nkx2.5 in contrast to the control. The arrow denotes the heterozygous nucleotides of T/C in the proband from family 2 (mutant) or the homozygous nucleotides of T/T in the corresponding control individual (wild-type). C, Schematic representation of the Nkx2.5 protein structure, with the AF-related mutations indicated. The mutations identified in patients with idiopathic AF are shown above the structural domains. NH2, amino-terminus; TN, transcriptional activation domain; HD, homeodomain; NK, NK2-specific domain; and COOH, carboxyl-terminus. D, Pedigree structures of the families with AF. The families are designated as family 1 and family 2, and the family members are identified by generations and numbers. Squares indicate male family members; circles, female members; a symbol with a slash, a deceased member; closed symbols, affected members; open symbols, unaffected members; arrows, probands; “+”, carriers of the heterozygous mutations; and “−”, non-carriers.
Table 2.
Phenotypic characteristics and the Nkx2.5 mutation status of the affected pedigree members.
| Subject Information | Phenotype | Electrocardiogram | Echocardiogram | Genotype | |||||
| Identity | Gender | Age at time of study (years) | Age at first diagnosis of AF (years) | AF (classification) | QRS interval (ms) | QT/QTc | LAD (mm) | LVEF (%) | Nkx2.5 mutations |
| Family 1 | N19D | ||||||||
| II-1 | M | 52 | 45 | Long-lasting persistent | 98 | 396/465 | 38 | 65 | +/– |
| Family 2 | F186S | ||||||||
| I-1 | M | 65a | 46 | Long-lasting persistent | 120 | 408/427 | 46 | 58 | N/A |
| II-1 | M | 46 | 32 | Persistent | 110 | 388/450 | 42 | 65 | +/– |
| II-5 | M | 40 | 35 | Paroxysmal | 104 | 438/482 | 40 | 62 | +/– |
| II-8 | F | 38 | 38 | Paroxysmal | 88 | 418/424 | 37 | 68 | +/– |
| III-2 | M | 22 | 22 | Paroxysmal | 86 | 382/424 | 32 | 64 | +/– |
Note: AF = atrial fibrillation; F = female; M = male; N/A = not available or not applicable; LAD = left atrial dimension; LVEF = left ventricular ejection fraction; QT = QT interval; QTc = corrected QT interval. + indicates present and – denotes absent. a Age at death.
Interestingly, congenital atrial septal defect and progressive atrioventricular block were confirmed in individual I-1 and individual II-1 from family 2 according to previous echocardiograms and electrocardiograms, as well as their medical history. Individual I-1 had an atrial ostium secundum septal defect, paroxysmal AF, a second-degree Wenchebach-type atrioventricular block, and an incomplete right bundle branch block at the age of 35 years. He subsequently developed a third-degree atrioventricular block and a complete right bundle branch block, and a pacemaker was implanted at age 49. Individual II-1 had a small atrial ostium secundum septal defect, paroxysmal AF, and a first-degree atrioventricular block at age 38. A pacemaker was implanted at age 46 after several syncopal episodes that were ascribed to third-degree atrioventricular block. One young individual (III-2) was an apparently asymptomatic mutation carrier. However, Holter monitoring revealed paroxysmal AF and an intermittent first-degree atrioventricular block. No sinus node dysfunction was observed in the studied individuals from these 2 families.
In addition, 2 previously reported Nkx2.5 sequence polymorphisms, namely c.63A>G and c.606C>G, were observed in the AF patients and control individuals. However, there was no significant difference in either of the 2 allele frequencies between the AF patients and the control groups. All of the sequence variants and their allele frequencies are listed in Table 3.
Table 3.
Nkx2.5 sequence variations identified in this study.
| Location | Nucleotide | Amino acid | Allele frequency | |
| Patients | Controls | |||
| Exon 1 | c.55A>G | p.N19D | (0.004) 1/272 | (0.000) 0/400 |
| Exon 1 | c.63A>G | p.E21E | (0.246) 67/272 | (0.238) 95/400 |
| Exon 2 | c.557T>C | p.F186S | (0.004) 1/272 | (0.000) 0/400 |
| Exon 2 | c. 606C>G | p.L202L | (0.022) 6/272 | (0.020) 8/400 |
Alignments of multiple Nkx2.5 protein sequences across species
A cross-species alignment of the Nkx2.5 protein sequences revealed that the altered amino acids were completely conserved evolutionarily (Figure 2).
Figure 2.
Alignment of the multiple Nkx2.5 protein sequences across species. The altered amino acids of p.N19 and p.F186 are completely conserved evolutionarily among the various species.
Disease-causing potential of the Nkx2.5 variation
The Nkx2.5 sequence variations of c.55A>G and c.557T>C were both automatically predicted to be disease-causing mutations, with p-values of 0.977377 and 0.999997, respectively. No SNPs in the altered regions were found in the MutationTaster database.
Transcriptional activity of the Nkx2.5 mutants
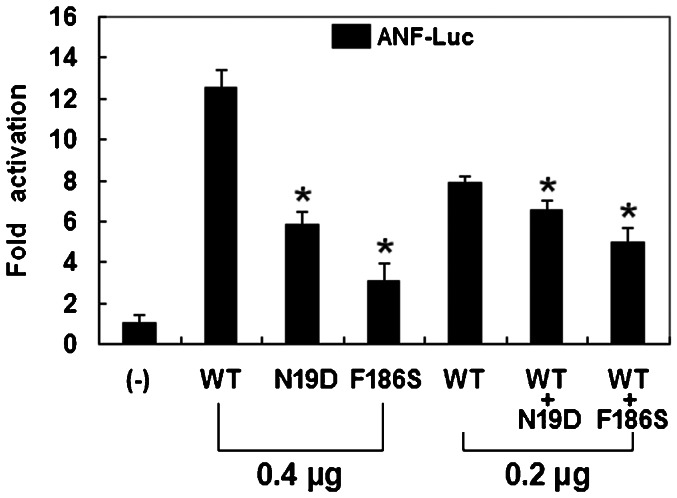
The same amounts of wild-type (0.4 μg), N19D-mutant (0.4 μg), and F186S-mutant (0.4 μg) Nkx2.5 activated the ANF promoter by ∼13-fold, ∼6-fold, and ∼3-fold increase, respectively, when compared with the empty plasmid. When the same amount of wild-type Nkx2.5 (0.2 μg) was cotransfected with N19D-mutant Nkx2.5 (0.2 μg) or F186S-mutant Nkx2.5 (0.2 μg), the induced activation of the ANF promoter was ∼7-fold and ∼5-fold increase, respectively, in comparison with the empty plasmid (Figure 3).
Figure 3.
Functional defects resulting from Nkx2.5 mutations. Activation of the atrial natriuretic factor promoter-driven luciferase reporter in COS-7 cells by wild-type (WT), N19D-mutant, or F186S-mutant Nkx2.5, alone or in combination, demonstrated significantly decreased transactivational activity of the mutant proteins. The experiments were performed in triplicate, and the mean and standard deviations are shown. *represents p<0.0005 when compared to wild-type Nkx2.5.
DISCUSSION
In the present study, 2 novel heterozygous Nkx2.5 mutations, p.N19D and p.F186S, were identified in 2 AF families. In each family, the mutant allele was present in all of the available, affected family members but was absent in the unaffected relatives examined and in the 400 control chromosomes. A cross-species alignment of the Nkx2.5 protein sequences revealed that the altered amino acids were completely conserved evolutionarily, and functional analysis demonstrated that the Nkx2.5 mutant proteins were associated with significantly decreased transcriptional activity. Therefore, it is highly probable that functionally impaired Nkx2.5 contributes to AF in these families.
The human Nkx2.5 gene maps to chromosome 5q34, which consists of 2 exons encoding a protein of 324 amino acids (29). Nkx2.5 is highly expressed in the early cardiac progenitor cells in both primary and secondary heart fields during embryogenesis and continues to be expressed at a high level in the heart throughout adulthood, thereby playing a crucial role in normal cardiovascular genesis and maturation (15). The functional and structural domains of Nkx2.5 are presumed to comprise the amino-terminal TN domain, the homeodomain that recognizes and binds to the specific consensus DNA sequence 5′-T[C/T]AAGTG-3′, and the carboxy terminal NK2-specific domain (Figure 1C). The TN and NK domains function to regulate the transcriptional activity of Nkx2.5, whereas the homeodomain is essential for nuclear translocation, DNA binding, and interactions with other transcription factors (30). The p.N19D and p.F186S mutations in Nkx2.5 identified in current study are located in the TN domain and in the homeodomain, respectively; thus, these mutations may be expected to exert influence on the transcriptional activity of Nkx2.5 by direct transcriptional inhibition or by interfering with its nuclear distribution or DNA-binding ability.
Previous studies have demonstrated that Nkx2.5 serves as an upstream regulator of multiple target genes that are expressed in the heart during embryogenesis, including the genes that encode ANF, brain natriuretic peptide, and α-cardiac actin (31). Hence, the functional effect of an Nkx2.5 mutation could be characterized by analysis of the transcriptional activity of the ANF promoter in cells expressing mutated Nkx2.5 in comparison to its wild-type counterpart. In the present study, the functional characteristics of the 2 novel Nkx2.5 mutations identified in AF patients were analyzed using transcriptional activity assays, and the results demonstrated significantly decreased transcriptional activity of a downstream gene. These findings imply that dysfunctional, mutated Nkx2.5 may serve as a potential alternative pathological mechanism of AF.
The findings that functionally compromised Nkx2.5 confers susceptibility to AF may be partially due to the abnormal development of the pulmonary venous myocardium. The pulmonary venous vessel is sheathed by a layer of myocardium known as the pulmonary myocardial sleeve, which has been implicated in the initiation and maintenance of AF by several potential electrophysiological mechanisms, encompassing enhanced intrinsic pacemaker activity, anisotropic arrangement of the myocardial fibers, and slowed conduction that facilitates reentry (32-35). Genetic labeling and lineage-tracing investigations have revealed that Nkx2.5 is abundantly expressed in the atria and the pulmonary myocardium but is indispensable for the localized formation of the sinoatrial node during embryogenesis. As a repressor of the sinoatrial node lineage gene program, Nkx2.5 may limit pacemaker activity to the sinoatrial and atrioventricular nodes. In a hypomorphic model demonstrating reduced Nkx2.5 protein expression, pulmonary cardiomyocytes were shown to transition into connexin40-negative, HCN4-positive cells and adopt a nodal-like phenotype with pacemaker activity (34). In Nkx2.5-null mouse embryos, HCN4 is activated along the entire embryonic heart tube, whereas connexin40 expression is suppressed, and ectopic pacemaker cells are present throughout the heart (35). Therefore, a Nkx2.5 loss-of-function mutation presumably predisposes an individual to formation of the pulmonary myocardium sleeve and switching of the pulmonary myocardium to a sinoatrial node-like phenotype, thereby creating an atrial arrhythmogenic substrate liable to AF.
It has been confirmed that certain downstream genes are transactivated by Nkx2.5, and mutations in multiple target genes have been linked to AF, including ANF, GATA4, GATA5, GATA6, connexin40, and connexin43 (13),. Therefore, mutated Nkx2.5 may enhance susceptibility to AF by down-regulating the expression of target genes.
Notably, congenital atrial septal defect and progressive atrioventricular conduction block were documented in some of the AF patients who carried the p.F186S mutation in Nkx2.5. Similarly, congenital cardiovascular malformations and progressive atrioventricular conduction block have been previously associated with diverse Nkx2.5 mutations, and the most common phenotypes caused by Nkx2.5 mutations include atrial septal defects and atrioventricular block (36). These observational results suggest that a proportion of AF cases may share a common genetic origin with congenital heart disease and atrioventricular block.
In conclusion, the present study links the Nkx2.5 loss-of-function mutation to AF and atrioventricular block. These results provide novel insight into the molecular mechanisms of AF and have potential implications for early prophylaxis and allele-specific treatment of this common arrhythmia.
ACKNOWLEDGMENTS
We are grateful to the individuals who participated in the present study. This work was supported in part by grants from the Personnel Development Foundation of Shanghai, China (2010019), the National Basic Research Program of China (2010CB912604), and the National Natural Science Fund of China (81070153, 81270161, and 30570768).
Footnotes
No potential conflict of interest was reported.
REFERENCES
- 1.Fuster V, Rydén LE, Cannom DS, Crijns HJ, Curtis AB, Ellenbogen KA, et al. 2011 ACCF/AHA/HRS Focused Updates Incorporated Into the ACC/AHA/ESC 2006 Guidelines for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;123(10):e269–367. doi: 10.1161/CIR.0b013e318214876d. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Wang TJ, Leip EP, Larson MG, Levy D, Vasan RS, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110(9):1042–6. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 3.Miyasaka Y, Barnes ME, Gersh BJ, Cha SS, Bailey KR, Abhayaratna WP, et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation. 2006;114(2):119–25. doi: 10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 4.Mansur Ade P, Takada JY, Avakian SD, Strunz CM. Warfarin doses for anticoagulation therapy in elderly patients with chronic atrial fibrillation. Clinics. 2012;67(6):543–6. doi: 10.6061/clinics/2012(06)01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.dos Santos AC, Nobre MR, Nussbacher A, Rodrigues GH, Gebara OC, Azul JB, et al. Predictors of the risk of falls among elderly with chronic atrial fibrillation. Clinics. 2012;67(4):305–11. doi: 10.6061/clinics/2012(04)02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham Study. Stroke. 1991;22:983–8. doi: 10.1161/01.str.22.8.983. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin EJ, Wolf PA, D'Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98(10):946–52. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 8.Wolowacz SE, Samuel M, Brennan VK, Jasso-Mosqueda JG, Van Gelder IC. The cost of illness of atrial fibrillation: a systematic review of the recent literature. Europace. 2011;13(10):1375–85. doi: 10.1093/europace/eur194. [DOI] [PubMed] [Google Scholar]
- 9.Saritas A, Kandis H, Baltaci D, Erdem I. Paroxysmal atrial fibrillation and intermittent left bundle branch block: an unusual electrocardiographic presentation of mad honey poisoning. Clinics. 2011;66(9):1651–3. doi: 10.1590/S1807-59322011000900025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armaganijan L, Lopes RD, Healey JS, Piccini JP, Nair GM, Morillo CA. Do omega-3 fatty acids prevent atrial fibrillation after open heart surgery? A meta-analysis of randomized controlled trials. Clinics. 2011;66(11):1923–8. doi: 10.1590/S1807-59322011001100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41(12):2185–92. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 12.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 2010;304(20):2263–9. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahida S, Lubitz SA, Rienstra M, Milan DJ, Ellinor PT. Monogenic atrial fibrillation as pathophysiological paradigms. Cardiovasc Res. 2011;89(4):692–700. doi: 10.1093/cvr/cvq381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nattel S. New ideas about atrial fibrillation 50 years on. Nature. 2002;415(6868):219–26. doi: 10.1038/415219a. [DOI] [PubMed] [Google Scholar]
- 15.Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol Ther. 2005;107(2):252–68. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63(2):196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Mommersteeg MT, Christoffels VM, Anderson RH, Moorman AF. Atrial fibrillation: a developmental point of view. Heart Rhythm. 2009;6(12):1818–24. doi: 10.1016/j.hrthm.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Yang YQ, Wang MY, Zhang XL, Tan HW, Shi HF, Jiang WF, et al. GATA4 loss-of-function mutations in familial atrial fibrillation. Clin Chim Acta. 2011;412(19-20):1825–30. doi: 10.1016/j.cca.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Jiang JQ, Shen FF, Fang WY, Liu X, Yang YQ. Novel GATA4 mutations in lone atrial fibrillation. Int J Mol Med. 2011;28(6):1025–32. doi: 10.3892/ijmm.2011.783. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Sun YM, Yang YQ. Mutation spectrum of the GATA4 gene in patients with idiopathic atrial fibrillation. Mol Biol Rep. 2012;39(8):8127–35. doi: 10.1007/s11033-012-1660-6. [DOI] [PubMed] [Google Scholar]
- 21.Posch MG, Boldt LH, Polotzki M, Richter S, Rolf S, Perrot A, et al. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur J Med Genet. 2010;53(4):201–3. doi: 10.1016/j.ejmg.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Yang YQ, Wang J, Wang XH, Wang Q, Tan HW, Zhang M, et al. Mutational spectrum of the GATA5 gene associated with familial atrial fibrillation. Int J Cardiol. 2012;157(2):305–7. doi: 10.1016/j.ijcard.2012.03.132. [DOI] [PubMed] [Google Scholar]
- 23.Gu JY, Xu JH, Yu H, Yang YQ. Novel GATA5 loss-of-function mutations underlie familial atrial fibrillation. Clinics. 2012;67(12):1393–9. doi: 10.6061/clinics/2012(12)08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang YQ, Wang XH, Tan HW, Jiang WF, Fang WY, Liu X. Prevalence and spectrum of GATA6 mutations associated with familial atrial fibrillation. Int J Cardiol. 2012;155(3):494–6. doi: 10.1016/j.ijcard.2011.12.091. [DOI] [PubMed] [Google Scholar]
- 25.Yang YQ, Li L, Wang J, Zhang XL, Li RG, Xu YJ, et al. GATA6 loss-of-function mutation in atrial fibrillation. Eur J Med Genet. 2012;55(10):520–6. doi: 10.1016/j.ejmg.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Liu WD, Yang ZL, Yang YQ. Novel GATA6 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2012;30(4):783–90. doi: 10.3892/ijmm.2012.1068. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Rath N, Hannenhalli S, Wang Z, Cappola T, Kimura S, et al. GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development. 2007;134(1):189–98. doi: 10.1242/dev.02720. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Xin YF, Liu XY, Liu ZM, Wang XZ, Yang YQ. A novel NKX2-5 mutation in familial ventricular septal defect. Int J Mol Med. 2011;27(3):369–75. doi: 10.3892/ijmm.2010.585. [DOI] [PubMed] [Google Scholar]
- 29.Shiojima I, Komuro I, Inazawa J, Nakahori Y, Matsushita I, Abe T, et al. Assignment of cardiac homeobox gene CSX to human chromosome 5q34. Genomics. 1995;27(1):204–6. doi: 10.1006/geno.1995.1027. [DOI] [PubMed] [Google Scholar]
- 30.Pradhan L, Genis C, Scone P, Weinberg EO, Kasahara H, Nam HJ. Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry. 2012;51(32):6312–9. doi: 10.1021/bi300849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126(6):1269–80. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 32.Haïssaguerre M, Jaïs P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339(10):659–66. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 33.Bittner A, Mönnig G, Vagt AJ, Zellerhoff S, Wasmer K, Köbe J, et al. Pulmonary vein variants predispose to atrial fibrillation: a case-control study using multislice contrast-enhanced computed tomography. Europace. 2011;13(10):1394–400. doi: 10.1093/europace/eur145. [DOI] [PubMed] [Google Scholar]
- 34.Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101(9):902–9. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 35.Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100(3):354–62. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 36.Reamon-Buettner SM, Borlak J. NKX2-5: an update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD) Hum Mutat. 2010;31(11):1185–94. doi: 10.1002/humu.21345. [DOI] [PubMed] [Google Scholar]