Abstract
Connective tissue growth factor (CTGF) has different roles in different types of cancer. However, the involvement and molecular basis of CTGF in tumor progression and prognosis of human nasopharyngeal carcinoma (NPC) have almost never been reported. In this study, we observed that downregulated CTGF expression was significantly associated with NPC progression and poor prognosis. Knockdown of CTGF markedly elevated the ability of cell proliferation in vivo and in vitro. Subsequently, we discovered that the reduction of CTGF increased the expression of miR-18b, an oncomir-promoting cell proliferation. Further, we discovered that attenuated CTGF-mediated upregulation of miR-18b was dependent on the increased binding of transcription factors Jun proto-oncogene (C-Jun) and v-Myc myelocytomatosis viral oncogene homolog (C-Myc) to miR-18b promoter region via phosphoinositide 3-kinase (PI3K)/AKT pathway. Finally, we further found that miR-18b directly suppressed the expression of CTGF in NPC. In clinical fresh specimens, miR-18b was widely overexpressed and inversely correlated with CTGF expression in NPC. Our studies are the first to demonstrate that reduced CTGF as an unfavorable prognosis factor mediates the activation of miR-18b, an oncomir directly suppresses CTGF expression, by PI3K/AKT/C-Jun and C-Myc and promotes cell growth of NPC.
Keywords: CTGF, NPC, miR-18b, PI3K/AKT
NPC (nasopharyngeal carcinoma) is one of the commonest carcinomas with high degree of malignant phenotype and geographical pattern characteristics in Southern China and Southeast Asia. It is easy to find typical cervical lymph node metastases frequently in its early stage. Synergetic effects of viral infections, genetic alterations, and environmental factors are thought to drive abnormal gene expression that contributes to the initiation and development of NPC.1, 2, 3, 4, 5 In our previous investigation, we used complementary DNA (cDNA) microarray to examine differentially expressed genes among NPC tissues and noncancerous nasopharyngeal tissues (NP). By means of the analysis of BRB-array tools, the expression of connective tissue growth factor (CTGF), a member of CCN family, was shown to be markedly decreased in NPC tissues, suggesting a possible role of CTGF in suppressing the pathogenesis of NPC.1
CTGF, a cysteine-rich, matrix-associated, heparin-binding protein, is widely expressed in various human tissues and organs, such as connective tissue, pancreas, placenta, lung, and so on, and related to tumor cell proliferation, adhesion, and angiogenesis.6, 7 Its abnormal expression is shown and serves as prognostic markers in many types of human cancer.8, 9 Interestingly, CTGF has different roles in different types of cancer; in pancreatic cancer, prostate cancer, liver cancer, breast cancer, and sarcoma, CTGF was shown to be an oncogenic factor in promoting the progression of these tumors.6, 10, 11, 12, 13 On the contrary, CTGF was taken as a tumor suppressor participating in the pathogenesis of lung cancer, ovarian cancer, and oral squamous cell cancer and so on.14, 15, 16 However, the expression pattern and functional mechanism of CTGF in NPC have been unclear yet. The present study investigated its expression pattern in NPC, the correlation of its protein expression with clinicopathologialc features in NPC patients, and its effects on NPC cell growth and invasion. Our studies demonstrated that reduced CTGF as a tumor suppressor could activate miR-18b, an oncomir in turn targeting CTGF directly, through regulating phosphoinositide 3-kinase (PI3K)/AKT/Jun proto-oncogene (C-Jun) and v-Myc myelocytomatosis viral oncogene homolog (C-Myc) signaling, and finally stimulating NPC progression and poor prognosis.
Results
CTGF is lowly expressed in NPC
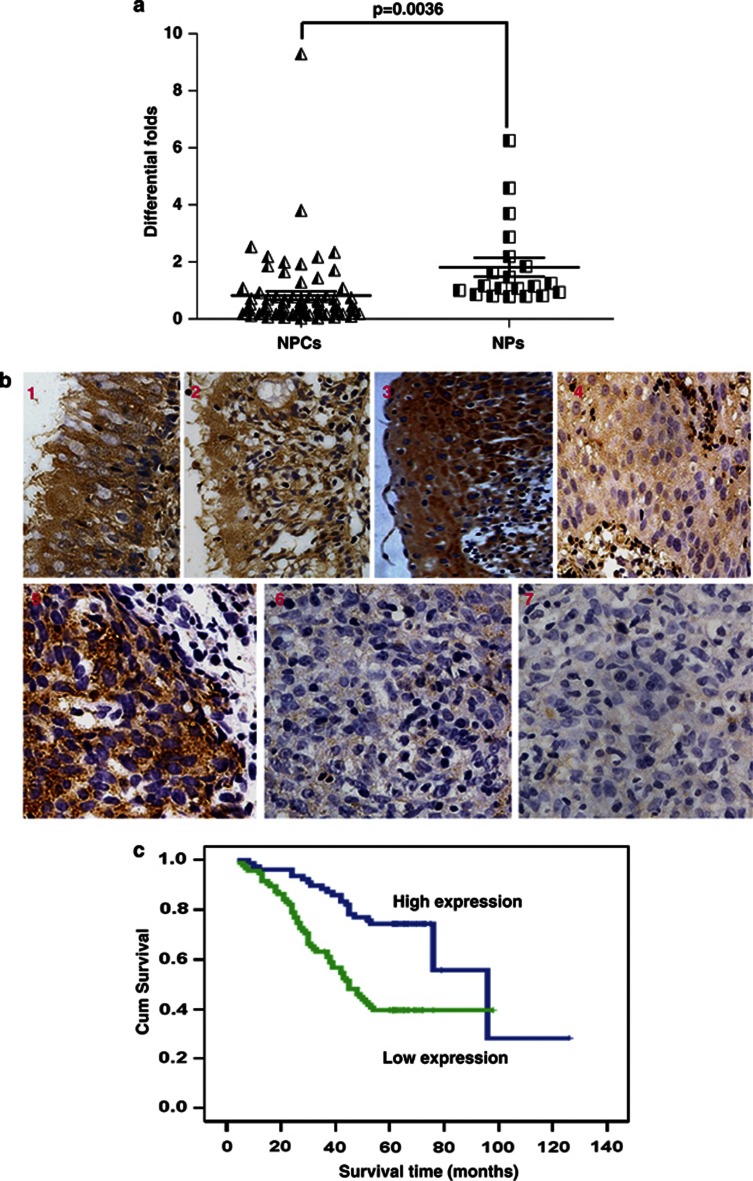
QPCR analysis indicated that downregulation of CTGF mRNA was showed in NPC tissues compared with NP tissues (P=0.0036; Figure 1a). Further, we measured the expression levels of CTGF protein in archived paraffin-embedded NPC samples and noncancerous nasopharynx specimens using immunohistochemical staining (Figure 1b). Specific CTGF protein staining was found in the cytoplasm of noncancerous and malignant nasopharynx tissues. CTGF protein was highly expressed in normal nasopharynx epithelium compared with atypical hyperplasia (P=0.025) and NPC samples (P<0.001). Furthermore, CTGF expression was relatively higher in atypical hyperplasia samples than that in NPC samples (P<0.001) (Table 1). However, no significant difference was found between normal and squamous epithelium samples (P=0.155).
Figure 1.
Reduced expression of CTGF was correlated with poor prognosis in NPC. (a) Using qPCR examination, CTGF mRNA expression was markedly decreased in 62 NPC tissues compared with 20 nasopharynx tissues (P=0.0036). (b) CTGF protein expression was progressively decreased in 37 atypical hyperplasia and 213 NPC samples compared with 49 normal nasopharynx tissues and 21 squamous epithelium by immunohistochemistry assay. (1) CTGF expression in normal epithelium, 1–2: strong expression; (2) CTGF expression in squamous epithelium, 3: strong expression; (3) CTGF expression in atypical hyperplasia tissue, 4: weak expression; (4) CTGF expression in NPC, 5: strong expression; 6: weak expression; 7: negative expression. (c) Lower expression levels of CTGF reduced the overall survival time of 173 NPC patients with clinical prognosis information.
Table 1. Gradual downregulation of CTGF protein in NPC and atypical hyperplasia samples compared with squamous and normal nasopharynx epithelium tissues.
| Group |
Protein expression (n) |
P-value |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Low | High | S/N/A/C | S/N | A/N | C/N | A/C | S/C | |
| Squamous epithelium | 21 | 3 | 18 | ||||||
| Normal epithelium | 49 | 2 | 47 | ||||||
| Atypical hyperplasia | 37 | 9 | 28 | ||||||
| Cancer | 213 | 124 | 89 | 0.000a | 0.155b | 0.025b | 0.000b | 0.000b | 0.000b |
Abbreviations: A, atypical; C, cancer; N, normal; n, patient case; S, squamous.
Kruskal Wallis test
χ2 test
Downregulated protein expression of CTGF is associated with NPC progression and poor prognosis
We analyzed the relationship between clinicopathological characteristics and CTGF expression levels in individuals with NPC (Table 2). We did not find a significant association of CTGF expression levels with patient's age, sex, smoking status, family tumor history, disease recurrence, or distant metastasis (M classification) in 173 NPC cases. However, we observed that the reduced expression level of CTGF was significantly correlated with tumor size (T classification; P=0.020), lymph node metastasis (N classification; N0–N1 versus N2–N3; P=0.014), and clinical stage (I–II versus III–IV; P=0.006) in NPC patients. Further, we investigate the prognostic value of CTGF expression for NPC patients using Kaplan–Meier analysis with the log-rank test. In 173 NPC cases, we observed that the level of CTGF protein expression was significantly correlated with overall survival, as patients with lower CTGF expression had worse survival (Figure 1c) than those with higher CTGF expression (P<0.001). Furthermore, we found that higher level of CTGF expression resulted in better prognosis in NPC patients, regardless of disease stage and TNM classification (Supplementary Figure S1). Univariate analysis displayed that radiotherapy, T, N, and M classification, and clinical stage were also significantly correlated with patients' survival (P<0.001, P<0.001, P=0.006, P<0.001, and P<0.001, respectively). To determine whether CTGF is an independent prognostic factor for NPC, we performed multivariate analysis of CTGF protein expression levels adjusted for radiotherapy, T, N, and M classification, and clinical stage of NPC patients using Cox proportional-hazards model. The results indicated that decreased CTGF expression was a poor independent prognostic factor for NPC patients (P<0.001) (Supplementary Table S8).
Table 2. Correlation between the clinicopathological characteristics and expression of CTGF protein in NPC.
| Characteristics | n |
CTGF (%) |
P | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Gender | ||||
| Male | 117 | 48(41.0) | 69 (59.0) | |
| Female | 56 | 30(53.6) | 26 (46.4) | 0.143 |
| Age (years) | ||||
| ≥50 | 79 | 32 (40.5) | 47 (59.5) | |
| <50 | 94 | 46 (48.9) | 48(51.1) | 0.286 |
| Smoking | ||||
| Yes | 31 | 9 (29.0) | 22 (71.0) | |
| No | 142 | 69 (48.6) | 73 (51.4) | 0.072 |
| Family tumor history | ||||
| Yes | 5 | 3(60.0) | 2(40.0) | |
| No | 168 | 75(44.6) | 93(55.4) | 0.659 |
| Recurrence | ||||
| Yes | 46 | 22(47.8) | 24(52.2) | |
| No | 127 | 56(44.1) | 71(55.9) | 0.730 |
| T classification | ||||
| T1–T2 | 122 | 62(50.8) | 60 (49.2) | |
| T3–T4 | 51 | 16 (31.4) | 35(68.6) | 0.020 |
| N classification | ||||
| N0–N1 | 95 | 51 (53.7) | 44(46.3) | |
| N2–N3 | 78 | 27(34.6) | 51 (65.4) | 0.014 |
| Distant metastasis | ||||
| Yes | 11 | 4(36.4) | 7(63.6) | |
| No | 162 | 74(45.7) | 88(54.3) | 0.756 |
| TNM Clinical stage | ||||
| I∼II | 58 | 35 (60.3) | 23 (39.7) | |
| III∼IV | 115 | 43(37.4) | 72(62.6) | 0.006 |
Stably downregulated or transiently inhibited CTGF expression stimulates cell cycle progression and cell proliferation in NPC
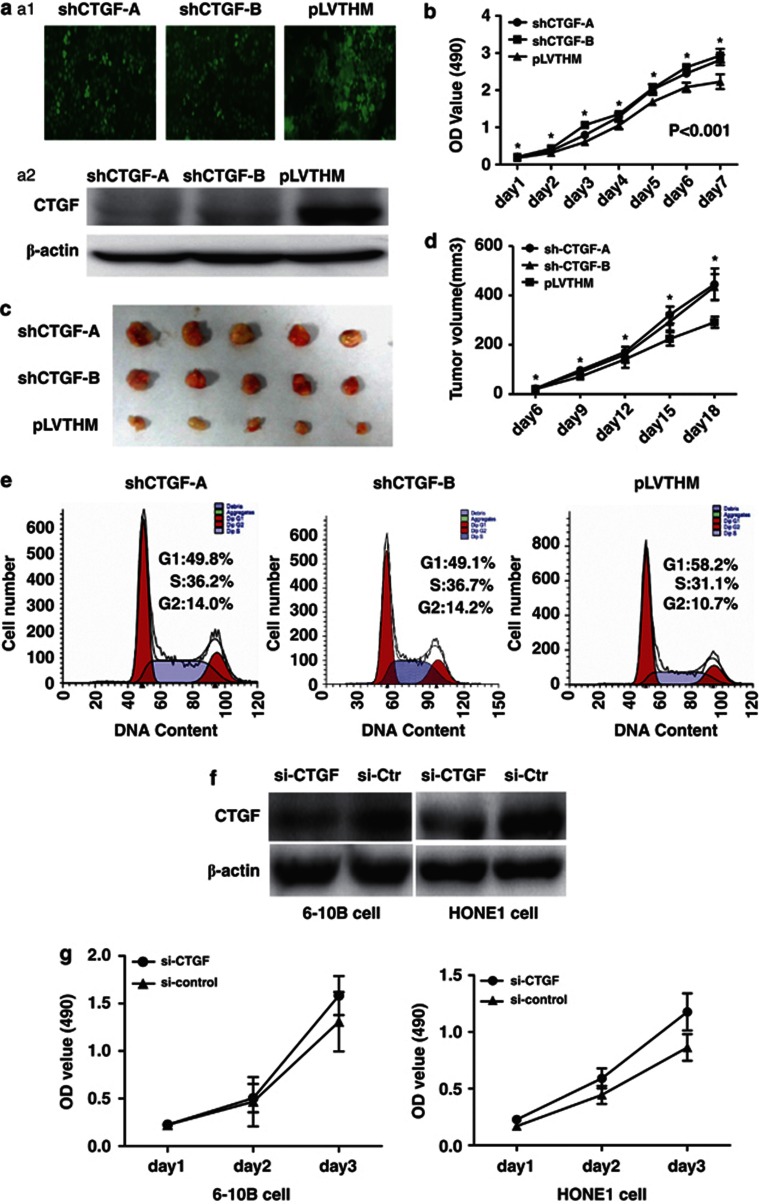
We used a lentiviral short-hairpin RNA (shRNA) vector specifically targeting CTGF to stably knock down the expression of CTGF in 6-10B cell line that showed the highest expression level of CTGF (data not shown). Stably decreased expression of CTGF protein was confirmed by western blot in shRNA-1024 and 1047 cells compared with PLV-Ctr cells (Figures 2a1 and a2). Subsequently, we examined the effect of decreased CTGF expression on NPC cell growth in vitro and in vivo. The growth curves determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed that suppressed CTGF significantly elevated cell proliferation relative to PLV-Ctr cells (Figure 2b). The results from in vivo tumorigenesis study inoculating shRNA-CTGF-1024 and 1047 cells into nude mice showed that the average weight and volume were significantly elevated in shRNA-CTGF xenograft mice compared with PLV-Ctr xenograft mice (Figures 2c and d). Immunohistochemistry analysis indicated that CTGF protein expression was markedly lower in shRNA-CTGF xenograft tumor specimens than in PLV-Ctr group (Supplementary Figure S2). Furthermore, suppression of CTGF markedly triggered cell cycle transition from G1 to S in shRNA-CTGF cells compared with PLV-Ctr cells (Figure 2e). Interestingly, similar results were observed in small-interfering RNA (siRNA)-mediated suppression of CTGF in NPC cells (Figure 2f). We found that knocking down endogenous CTGF expression speeded up the cell growth (Figure 2g). These results suggested an inhibitory effect of CTGF on tumorigenesis in vitro and in vivo.
Figure 2.
Stable suppression of CTGF expression stimulated the proliferation of NPC cells and speeded up the transition of cell cycle from G1 to S. (a) (A1) Polyclonal cells of lentivirus-mediated shRNA-CTGF-A and B, and PLV-Ctr were screened by GFP using FACS cytometry assay. (A2) Expression of CTGF was suppressed in shRNA-CTGF-A and B compared with PLV-Ctr cells by western blot. (b) In vitro proliferative ability of NPC cells was significantly restored in CTGF-suppressed cells compared with PLV-Ctr cells by MTT assay. (c) When compared with PLV-Ctr cells, tumor weight of shRNA-CTGF-A and B cells was markedly increased in vivo. (d) When compared with PLV-Ctr cells, tumor volume of shRNA-CTGF-A and B cells was markedly increased in vivo. (e) Downregulated CTGF expression stimulated cell cycle progression from G1 to S in shRNA-CTGF-A and B cells (P<0.001). (f) Specific siRNA was used to suppress the expression of CTGF in NPC 6-10B and HONE1 cells by western blot examination. (g) Cell growth ability was enhanced in si-CTGF-treated NPC cells (*P<0.05)
CTGF modulates the expression of miR-18b in NPC
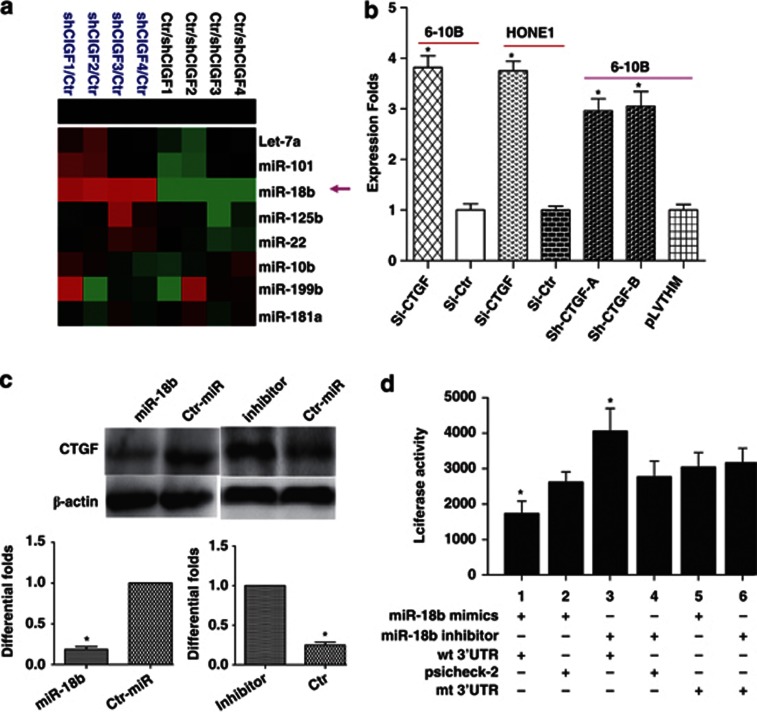
To investigate the effect of CTGF on microRNAs (miRNAs) in NPC, we used miRNA chip to compare the differential miRNA expression between SiRNA-CTGF and siRNA-Ctr in 6-10B cells. We found that miR-18b was notably upregulated in SiRNA-CTGF-treated 6-10B NPC cells (Figure 3a). Further, we used real-time PCR to confirm our finding in SiRNA-CTGF-treated HONE1, 6-10B, and shRNA-6-10B-treated cells (Figure 3b).
Figure 3.
CTGF inhibited the expression of miR-18b, and miR-18b directly targets CTGF in NPC. (a) MiR-18b was notably upregulated in SiRNA-CTGF-treated 6-10B NPC cells by miRNA arrays analysis. (b) Real-time PCR confirmed the reliability of miRNA arrays in SiRNA-CTGF-treated HONE1, 6-10B and shRNA-6-10B-treated cells. (c) CTGF expression was repressed or elevated with the transfection of miR-18b mimics and inhibitor, respectively, at 100 nm in NPC 6-10B cells. (d) Luciferase reporter assay was used to determine whether miR-18b could directly target the 3′UTR of CTGF in NPC cells. Lanes 1 and 2: a significant decrease of luciferase activity was indicated in miR group. Lanes 5 and 6: the activity of mt 3′UTR vector was unaffected by a simultaneous transfection with miR-18b. Lanes 3 and 4: co-transfection with miR-18b inhibitor and wt 3′UTR vector led to a marked increase of luciferase activity. Data are presented as mean±S.D. for three independent experiments (*P<0.05)
MiR-18b directly targets CTGF in NPC
miR-18b was predicted to directly target CTGF. We transfected 6-10B cells with miR-18b mimics and inhibitor at 100 nm and then examined CTGF expression levels. As shown in Figure 3c, miR-18b mimics induced a decreased CTGF protein level. Moreover, inhibition of endogenous miR-18b by its inhibitor resulted in upregulated expression of CTGF. Attributed to the negative regulation of miR-18b to CTGF, we further performed luciferase reporter assay to determine whether miR-18b could directly target the 3′ untranslated regions (UTR) of CTGF in NPC cells. The target sequence of CTGF 3′UTR (wild-type (wt) 3′UTR) or the mutant sequence (mt 3′UTR) was cloned into a luciferase reporter vector. 6-10B cells were then transfected with wt or mt 3′UTR vector and miR-18b mimics. The results showed a significant decrease of luciferase activity with treatment of wt 3′UTR vector when compared with miR control (Figure 3d, lanes 1 and 2; P=0.008). The activity of mt 3′UTR vector was unaffected by a simultaneous transfection with miR-18b (Figure 3d, lanes 5 and 6, P=0.85). Moreover, co-transfection with miR-18b inhibitor and wt 3′UTR vector in 6-10B cells led to a marked increase of luciferase activity (Figure 3d, lanes 3 and 4; P= 0.015). Taken together, all these results strongly suggested that CTGF was a direct target of miR-18b in NPC cells.
CTGF inhibits miR-18b expression by modulating PI3K/AKT/C-Jun and C-Myc pathway
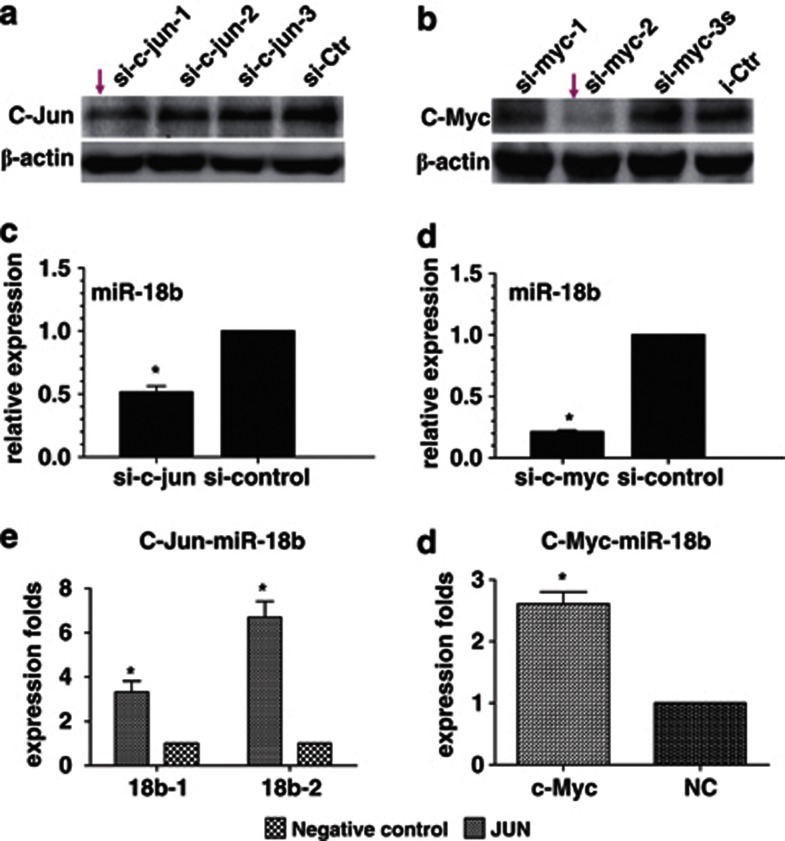
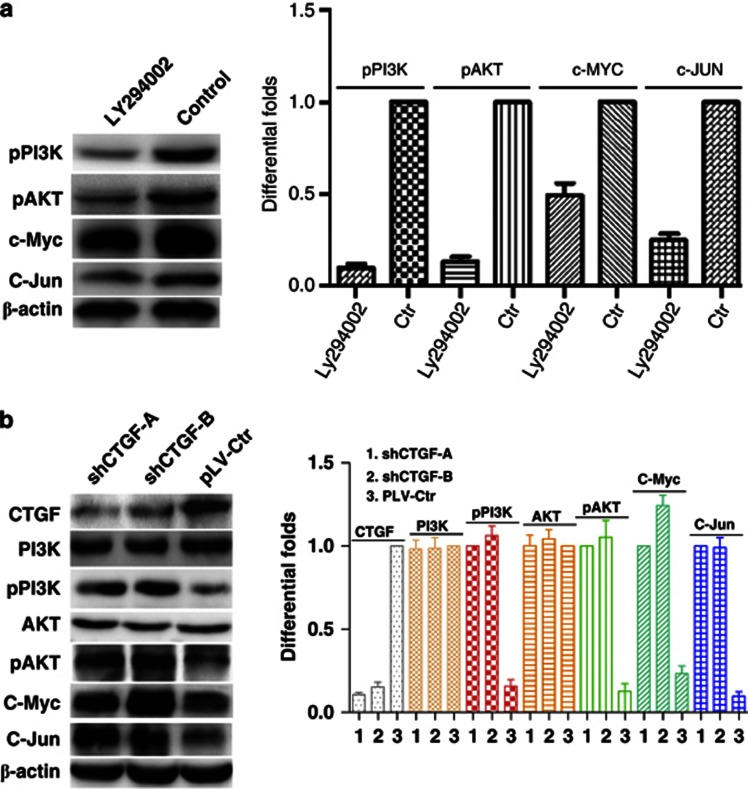
To investigate the effect of CTGF on miR-18b in NPC, bioinformatic assay was used to predict and find that there were binding sites of C-Jun (AP1) and C-Myc, two oncogenic transcription factors also regulated by PI3K/AKT pathway, in putative promoter region of miR-18b. We speculated that CTGF might participate in regulating the expression of miR-18b through the PI3K/AKT/C-Jun and C-Myc pathway. To examine the role of C-Jun and C-Myc in regulating miR-18b expression, we first used siRNAs to suppress the expression of C-Jun and C-Myc in NPC 6-10B cells (Figures 4a and b). QPCR analysis indicated that miR-18b expression was markedly reduced in 6-10B cells with the knockdown of C-Jun and C-Myc (Figures 4c and d). In subsequent investigation, we used chromatin immunoprecipitation combined with qPCR analysis to confirm that C-Jun and C-Myc could bind miR-18b promoter and activate its expression in NPC (Figures 4e and f). Furthermore, we used specific inhibitor of PI3K to suppress the expression of PI3K and observed that the protein expression of C-Jun and C-Myc was decreased in NPC 6-10B cells (Figure 5a). Finally, we observed that reduced CTGF significantly increased the expression of phos-PI3K and phos-AKT, but not their total protein levels (Figure 5b). Our results demonstrated that CTGF suppressed the expression of miR-18b through PI3K/AKT/C-Jun and C-Myc pathway.
Figure 4.
C-Jun and C-Myc directly positively modulated miR-18b expression in NPC. (a) siRNA (fragment 1) was used to suppress the expression of C-Jun in NPC 6-10B cells. (b) The expression of C-Myc was suppressed by its specific siRNA (fragment 2) in NPC 6-10B cells. (c) Knocking down C-Jun downregulated the expression of miR-18b in NPC 6-10B cells. (d) Reduced C-Myc expression downregulated the expression of miR-18b in NPC 6-10B cells. (e and f) Crosslinked chromatin preparation from mock and pcDNA3.1-C-Myc or pcDNA3.1-C-Jun-transfected 6-10B cells was immunoprecipitated with anti-C-Myc, anti-C-Jun, or a normal rabbit IgG. The AP1 binding sites on the immunoprecipitated DNA was determined by quantitative RT-PCR. Amplification of input chromatin (input) before immunoprecipitation was served as positive controls for chromatin extraction and PCR amplification. Chromatin immunoprecipitation using a nonspecific antibody (normal human IgG) served as negative controls. QPCR analysis indicated that C-Jun and C-Myc could bind more miR-18b promoter region than that in control group in NPC 6-10B cells (*P<0.05)
Figure 5.
Reduced CTGF activated PI3K/AKT-mediated expression of C-Jun and C-Myc in NPC. (a) Suppressing the expression of PI3K by its specific inhibitor Ly294002 reduced C-Jun and C-Myc expression by inactivating pPI3K/AKT in NPC 6-10B cells. (b) Reduced CTGF stimulated the expression of C-Jun and C-Myc by activating phosphorylated PI3K/AKT, but not their total proteins in NPC 6-10B cells
MiR-18b promotes cell proliferation in NPC
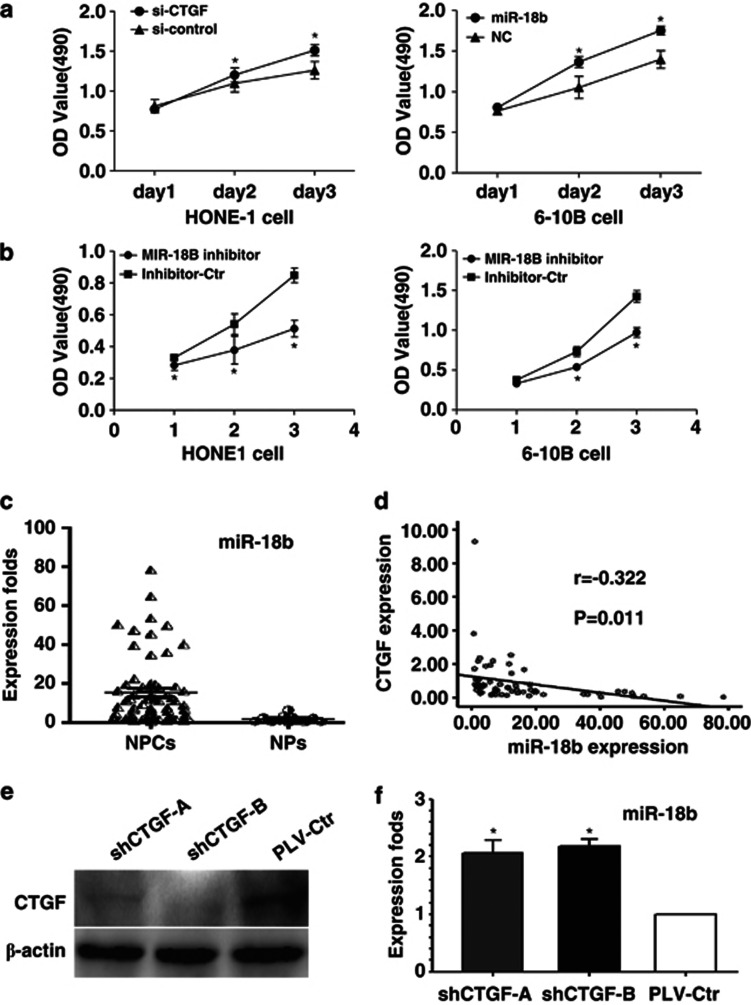
To study the biological functions of miR-18b, we introduced miR-18b mimics or its inhibitor into NPC HONE1 and 6-10B cell lines. Compared with their negative controls, we found that miR-18b mimics and inhibitor could promote and inhibit cell growth in NPC cells, respectively, according to MTT assays (Figures 6a and b).
Figure 6.
High expression of miR-18b promotes cell proliferation and is negatively correlated with the expression of CTGF in NPC. The ability of cell proliferation was markedly elevated after increasing miR-18b mimics expression in NPC. (b) Transduction of miR-18b inhibitor suppressed cell growth in NPC cells. (c) miR-18b expression was dramatically elevated in NPC tissues and NPC cells compared with nasopharynx tissues. (d) miR-18b was negatively correlated with the expression of CTGF in NPC samples. (e) CTGF protein expression was suppressed in xenograft tumor specimens after stably knocking down CTGF expression. (f) Reduced CTGF expression induced the miR-18b expression in xenograft tumor specimens. Data are presented as mean±S.D. for three independent experiments (*P<0.05)
MiR-18b is highly expressed and negatively correlated with the expression of CTGF in NPC
We further measured the expression of miR-18b in NPC specimens and normal nasopharyngeal epithelial tissues. The results showed that the average expression level of miR-18b was significantly higher in NPC specimens than in normal NPs (Figure 6c; P<0.001). Furthermore, a significantly inverse correlation was observed between CTGF mRNA and miR-18b expression (Figure 6d; P=0.005) in NPC tissues. Similarly, we observed that the expression of miR-18b was upregulated in xenograft tumor specimens after knocking down CTGF expression (Figures 6e and f).
Discussion
CTGF has dual roles as oncogene or tumor suppressor in different cancer types,6, 7, 8, 9, 10, 11, 12, 13, 14 which may attribute to tissue-specific patterns of expression in different tissues and organs in tumourigenesis. However, its roles and molecular mechanisms linking the initiation and development of NPC are seldom reported.1
In previous study, we have found that CTGF was less expressed in NPC tissues than in NP tissues by microarray examination.1 In this study, we first confirmed CTGF mRNA was lowly expressed in NPC compared with NP by qPCR assay. Further, we used immunohistochemistry to examine the expression level of CTGF protein in NPC tissues and noncancerous NP tissues. We observed that CTGF was an expressed factor of cytoplasm in NPC and noncancerous NP tissues. The expression of CTGF was progressively decreased in atypical hyperplasia and cancer tissues compared with normal and squamous epithelium. These results were not only consistent with mRNA expression of CTGF, but also hinted that loss of CTGF expression was involved in the stage of initiation and precancerous lesion of NPC.
In previous other studies, different expression pattern of CTGF in tumor cells had been shown to be associated with tumor progression, displaying a favorable or unfavorable prognostic significance in various tumor types. Elevated expression of CTGF was observed in melanoma, papillary thyroid carcinoma, esophageal squamous cell carcinoma, gastric cancer, and cervical tumors, and so on,17, 18, 19, 20, 21 in which CTGF was positively associated with tumor progression and poor prognosis. Conversely, in oral squamous cell carcinoma, ovarian cancer, and lung adenocarcinomas,22, 23, 24 reduced CTGF expression was favorable for their progression and poor prognosis. In this study, we evaluated the correlation of cytoplasmic CTGF expression with clinical parameters of NPC patients, and found that attenuated CTGF expression was negatively associated with T, N classification and clinical stages of NPC patients. Furthermore, we observed that suppressed CTGF expression was positively correlated with NPC patient's overall survival. Patients with lower expression of CTGF protein had shorter overall survival time. Multivariate analyses showed that decreased expression of CTGF protein was a significant independent predictor of poor prognosis for NPC patients. These above results suggested a tumor suppressive role for CTGF in NPC.
Subsequently, we presented strong evidence that CTGF was taken a tumor suppressor participating in NPC pathogenesis. We found that stably decreased expression of CTGF using CTGF-shRNA could convert 6-10B cells into more aggressive cells, which showed a high capability of cell growth in vitro and in vivo xenograft animal model and G1/S cell cycle transition in vitro compared with PLV-Ctr cells. Similar to the results of stably suppressed CTGF expression, we observed that suppression of CTGF expression by transfection of exogenous siRNA could also induce elevated ability of cell proliferation in 6-10B and HONE1 cells in vitro.
CTGF was previously reported to block FAK/PI3K/AKT or miR-504/FOXP1 signaling suppressing cell motility and invasion in oral squamous cell carcinoma.16, 22 In this study, we investigated the molecular mechanism of CTGF-mediated miRNAs expression in NPC cell growth. We did miRNA expression profile analysis of siRNA-CTGF and siRNA-Ctr-treated 6-10B cells. Interestingly, miR-18b, a miR-18 family member from paralog clusters of miR-17-92 cluster, was notably upregulated in NPC cells with knockdown of CTGF. In previous investigations, miR-18a, another miRNA belonging to miR-17-92 cluster, had been identified to directly target CTGF.25, 26, 27 In subsequent bioinformatic assay, we found that miR-18b has the same seed sequence with miR-18a, which therefore, we expected to regulate the same transcripts, including CTGF. In further exploration, we confirmed this conjecture and demonstrated that CTGF was also a direct target of miR-18b in NPC cells.
In a previous study, the expression of miR-18a controlled by C-Myc had been documented,28, 29 which hinted that expression of miR-18b might be regulated by transcription factors. In subsequent study, we employed bioinformatic assay to find that there were binding sites for C-Jun (AP1) and C-Myc, two oncogenic transcription factors regulated by PI3K/AKT pathway,16, 30, 31 in putative promoter region of miR-18b. We speculated that CTGF might participate in regulating the expression of miR-18b through the PI3K/AKT/C-Jun and C-Myc pathway. Knocking down C-Jun and C-Myc by siRNAs, we observed that miR-18b expression was significantly decreased. Chromatin immunoprecipitation combined with qPCR analysis also indicated that C-Jun and C-Myc could bind miR-18b promoter and activate its expression in NPC. Furthermore, we used specific PI3K inhibitor to suppress the expression of PI3K, and found that the expression of C-Jun and C-Myc was reduced accordingly. Finally, we found that inhibition of CTGF activated the PI3K/AKT/C-Jun and C-Myc pathway.
In addition to our first discovery of the oncogenic effect of miR-18b on NPC cell proliferation in vitro and in vivo, which was inversely correlated with CTGF role, we showed that miR-18b was naturally upregulated in clinical NPC specimens and inversely correlated with CTGF levels, suggesting an important role of miR-18b in NPC tumorigenesis in the presence of CTGF silence.
In summary, this study provides evidence that CTGF is a tumor suppressor in NPC; its reduced expression, as an unfavorable prognosis factor, can activate miR-18b, an oncomir directly suppressing CTGF expression, by regulating PI3K/AKT/C-Jun and C-Myc pathway, and thereby promote cell growth of NPC.
Materials and Methods
Cell culture and sample collection
Eight NPC cell lines 5-8F, 6-10B, CNE2, CNE1, C666-1, HONE1, HNE1 and SUNE1 were obtained from Cancer Research Institute of Southern Medical University, Guangzhou, China, and maintained in RPMI 1640 medium supplemented with 10% newborn calf serum (PAA Laboratories, Inc, Pasching, Austria). All of these cell lines were incubated in a humidified chamber with 5% CO2 at 37 °C. Sixty-two fresh NPC tissues, 20 fresh nasopharynx tissues, 213 paraffin-embedded undifferentiated NPC specimens (173 cases with clinical and prognosis information) and 107 paraffin-embedded noncancerous nasopharynx specimens (Table 1) were obtained at the time of diagnosis before initial therapy in People's Hospital in Zhongshan City (Guangdong, China). The clinical processes were approved by the Ethics Committees of People's Hospital of Zhongshan City. The patients were confirmed with the informed consents. The pathological stage of all specimens was confirmed according to the 1997 NPC staging system (WHO).
RNA isolation and qRT-PCR
RNA was extracted from the NPC cell lines, NPC tissues and NP using Trizol (Takara, Shiga, Japan). For miR-18b real-time quantitative reverse transcription PCR (qRT-PCR), RNA was transcribed into cDNA and amplified with sense primer 5′-CTAAGGTGCATCTAGTGCAGTTAG-3′ and antisense general primer using miRNA PrimeScript RT Enzyme Mix kit, according to the manufacturer's instructions (Ambion, Austin, TX, USA). For CTGF, C-Jun and C-Myc qRT-PCR, RNA was transcribed into cDNA and amplified with specific sense/antisense primers (Supplementary Table S1). The assays were performed in accordance with manufacturer's instructions (Takara). The PCR reaction for each gene was repeated three times. MiRNA and mRNA expression was normalized to U6 and ARF5, respectively, using the 2−ΔΔCt method.16
Immunohistochemistry and evaluation of staining
Immunohistochemistry and evaluation of staining of CTGF (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were performed in NPC and noncancerous NPs, according to the previous description.3
Western blot analysis
Western blot assays were carried out according to the previous description3 with rabbit polyclonal anti-CTGF antibody (1 : 200; Epitomics, Burlingame, CA, USA), anti-ACTB, C-Myc (1 : 400; Santa Cruz Biotechnology), C-Jun, AKT, p-Akt(Ser-473), PI3K, and pPI3K antibody(1 : 1000, Cell Signaling Technology, Danvers, MA, USA). HRP-conjugated anti-rabbit IgG antibody was used as the secondary antibody (Zhongshan, Beijing, China). Signals were detected using enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA). Quantification of western blots was performed using Quantity One Software (Bio-Rad, Foster City, CA, USA).
Establishment of NPC 6-10B cell line with stable expression of CTGF-shRNA
The preparation of lentivirus expressing human CTGF-shRNA-1024,1047 (Supplementary Table S2) was performed using the pLVTHM-green fluorescent protein (GFP) lentiviral RNAi expression system.3, 13 NPC 6-10B cells were infected with lentiviral particles containing specific or negative control vectors, and the polyclonal cells with GFP signals were enriched for further experiments using FACS flow cytometer assay.
Transient transfection with siRNAs, miR-18b mimics or its inhibitor and PI3K inhibitor Ly294002
siRNA for CTGF, C-Myc, and C-Jun (Supplementary Tables S3 and S4), as well as miR-18b mimics (Supplementary Table S5) and its inhibitor (Patent, not published) were designed and synthesized by Guangzhou RiboBio (RiboBio Inc., Guangzhou City, China). The sequences of each gene and its control are shown in Supplementary Table S4. PI3K inhibitor Ly294002 was bought from Sigma-Aldrich, St Louis, MO, USA. Twenty-four hours before transfection, NPC cells 6-10B and HONE1 were plated onto a 6-well plate or a 96-well plate (Nest Biotech, Guangzhou City, China) at 30–50% confluence. They were then transfected into cells using TurboFect siRNA Transfection Reagent (Fermentas, Vilnius, Lithuania), according to the manufacturer's protocol. Cells were collected after 48–72 h for the further experiments.
Cell proliferation analysis
Cell proliferation was analyzed using MTT assay, as described previously.2, 3 For shRNA-CTGF, the cells were incubated for 1, 2, 3, 4, 5, 6, or 7 days. For siRNA-CTGF or miR-18b mimics, the cells were incubated for 1, 2, or 3 days.
In vivo tumorigenesis in nude mice
A total of 1 × 106 logarithmically growing 6-10B cells (CTGF-shRNA-1024, 1047, and Ctr-vector) in 0.1 ml RPMI 1640 medium were subcutaneously injected into the left flank of 4–6-week-old male BALB/c nu/nu mice. The mice were maintained in a barrier facility on HEPA-filtered racks and fed with an autoclaved laboratory rodent diet. All animal studies were conducted in accordance with the principles and procedures outlined in Southern Medical University Guide for the Care and Use of Animals under assurance number SCXK (Guangdong) 2008-0002. Tumor volume was monitored using a calliper in the process of tumor growth. After 18 days the mice were killed, and tumor tissues were excised and weighted.
Cell cycle assay
To evaluate cell cycle distributions, 6-10B cells with stable suppression of CTGF were fixed in 70% ice-cold ethanol for 48 h at 4 °C, stained by incubation with PBS containing 10 μg/ml propidium iodide and 0.5 mg/ml RNase A for 15 min at 37 °C, and analyzed for the DNA content of labeled cells by FACS Caliber Cytometry (BD Bioscience, Franklin Lakes, NJ, USA). Each experiment was done in triplicate.
MiRNA array for Si-CTGF
We used in-house two-channel oligo-arrays (CCDTM-miRNA850-V4px) from NIH including 713 human, mammalian, and viral mature antisense miRs plus two internal controls with seven serial dilutions,32 which were endowed by Professor Wang E and Marincola FM. Total RNA (6–7 μg) samples isolated respectively from each Si-CTGF and its control NPC 6-10B cells were labeled by Cy5-dCTP and Cy3-dCTP, respectively, in a reverse transcription reaction. The balanced mixture of Cy5- and Cy3-labeled targets was co-hybridized against miRNAs chip. The experiments were repeated four times and scanned images were converted to digital data by GenePixpro V6.0 (Molecular Devices Corporation, Sunnyvale, CA, USA). Finally, the data were analyzed using BRB ArrayToolsV3.7.2 software (National Cancer Institute, Bethesda, MD, USA).
MiRNA target validation
A 291-bp fragment of CTGF 3′UTR was amplified by PCR primers (Supplementary Table S6) and cloned downstream of the firefly luciferase gene in psiCHECK-2 vector. The vector was named wt 3′UTR. Site-directed mutagenesis of the miR-18b-binding site in CTGF 3′UTR was performed using GeneTailor Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA) and named mt 3′UTR. For reporter assays, wt or mt 3′UTR vector and the control vector psiCHECK-2 vector coding for Renilla luciferase were co-transfected into 6-10B cells with miR-18b mimics or inhibitor by Lipofectamine 2000. Luciferase activity was measured 48 h after transfection using the activity ratio between Firefly and Renila luciferase.
Chromatin immunoprecipitation assay
DNA–protein complexes were immunoprecipitated from 6-10B cells with the transfection of C-Myc or C-Jun cDNA by using the Chromatin Immunoprecipitation Kit (Millipore, Billerica, MA, USA), according to the manufacturer's protocol with the following polyclonal antibodies: anti-C-Jun 1 μg, C-Myc 1 μg, or normal mouse IgG (Millipore) 1 μg. The precipitated DNA was subjected to qPCR analysis by using specific primers (Supplementary Table S7) to amplify across the miR-18b promoter region. Finally, the data were analyzed using the 2−ΔΔCt method.33
Statistical analysis
All data were analyzed for statistical significance using SPSS 13.0 software (SPSS, Chicago, IL, USA). The Kruskal Wallis test was used to examine the differences of CTGF expression among squamous epithelium, normal epithelium, atypical hyperplasia, and cancer tissues of nasopharynx. The χ2 test was applied to the examination of relationship between CTGF expression levels and clinicopathological characteristics. Survival analysis was performed using Kaplan–Meier method. Multivariate Cox proportional-hazards method was used for analyzing the relationship between the variables and patient's survival time. The relationship between CTGF mRNA and miR-18b expression in NPC tissues was explored by Pearson's test. A two-tailed Student's t-test was used for comparisons of two independent groups. One-way ANOVA was used to determine the differences between groups for all in vitro analyses. A P-value of <0.05 was considered statistically significant.
Acknowledgments
This study was supported by National Nature Science Fund of China (No. 81272268), Natural science fund of Guangdong Province (No. S2012010009583), Yangcheng scholar research projects from Universities of Guangzhou (No. 12A011D), New star plan of Pearl River Science and Technology from Guangzhou City (No. 2011J2200009), and Guangdong Province Breeding Project Plan (LYM11102). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- NPC
nasopharyngeal carcinoma
- CTGF
connective tissue growth factor
- NP
nasopharyngeal tissues
- PI3K
phosphoinositide 3-kinase
- AKT
v-akt murine thymoma viral oncogene homolog 1
- C-Jun
jun proto-oncogene
- C-Myc
v-myc myelocytomatosis viral oncogene homolog
- ACTB
actin beta
- qRT-PCR
real-time quantitative reverse transcription PCR
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- cDNA
complementary DNA
- miRNA
microRNA;
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Fang W, Li X, Jiang Q, Liu Z, Yang H, Wang S, et al. Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. J Transl Med. 2008;6:32. doi: 10.1186/1479-5876-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alajez NM, Shi W, Hui AB, Bruce J, Lenarduzzi M, Ito E, et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010;1:e85. doi: 10.1038/cddis.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Luo W, Zhou Y, Zhen Y, Yang H, Yu X, et al. Potential tumor suppressor NESG1 as an unfavorable prognosis factor in nasopharyngeal carcinoma. PLoS One. 2011;6:e27887. doi: 10.1371/journal.pone.0027887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fang W. Increased expression of hepatoma-derived growth factor correlates with poor prognosis in human nasopharyngeal carcinoma. Histopathology. 2011;58:217–224. doi: 10.1111/j.1365-2559.2010.03739.x. [DOI] [PubMed] [Google Scholar]
- Luo WR, Wu AB, Fang WY, Li SY, Yao KT. Nuclear expression of N-cadherin correlates with poor prognosis of nasopharyngeal carcinoma. Histopathology. 2012;61:237–246. doi: 10.1111/j.1365-2559.2012.04212.x. [DOI] [PubMed] [Google Scholar]
- Chien W, O'Kelly J, Lu D, Leiter A, Sohn J, Yin D, et al. Expression of connective tissue growth factor (CTGF/CCN2) in breast cancer cells is associated with increased migration and angiogenesis. Int J Oncol. 2011;38:1741–1747. doi: 10.3892/ijo.2011.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci. 2008;15:675–685. doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY, Zhang QY, et al. Expression and prognostic significance of THBS1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer. 2009;9:291. doi: 10.1186/1471-2407-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Z, Feng G, You W, Li J. Expression of connective tissue growth factor is in agreement with the expression of VEGF, VEGF-C, -D and associated with shorter survival in gastric cancer. Pathol Int. 2007;57:712–718. doi: 10.1111/j.1440-1827.2007.02162.x. [DOI] [PubMed] [Google Scholar]
- Bennewith KL, Huang X, Ham CM, Graves EE, Erler JT, Kambham N, et al. The role of tumor cell-derived connective tissue growth factor (CTGF/CCN2) in pancreatic tumor growth. Cancer Res. 2009;69:775–784. doi: 10.1158/0008-5472.CAN-08-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65:8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- Jia XQ, Cheng HQ, Li H, Zhu Y, Li YH, Feng ZQ, et al. Inhibition of connective tissue growth factor overexpression decreases growth of hepatocellular carcinoma cells in vitro and in vivo. Chin Med J (Engl) 2011;124:3794–3799. [PubMed] [Google Scholar]
- Croci S, Landuzzi L, Astolfi A, Nicoletti G, Rosolen A, Sartori F, et al. Inhibition of connective tissue growth factor (CTGF/CCN2) expression decreases the survival and myogenic differentiation of human rhabdomyosarcoma cells. Cancer Res. 2004;64:1730–1736. doi: 10.1158/0008-5472.can-3502-02. [DOI] [PubMed] [Google Scholar]
- Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, et al. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One. 2007;2:e534. doi: 10.1371/journal.pone.0000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbolina MV, Adley BP, Kelly DL, Shepard J, Fought AJ, Scholtens D, et al. Downregulation of connective tissue growth factor by three-dimensional matrix enhances ovarian carcinoma cell invasion. Int J Cancer. 2009;125:816–825. doi: 10.1002/ijc.24347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JY, Yang WY, Lai CH, Lin CD, Tsai MH, Tang CH. CTGF inhibits cell motility and COX-2 expression in oral cancer cells. Int Immunopharmacol. 2011;11:948–954. doi: 10.1016/j.intimp.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Braig S, Wallner S, Junglas B, Fuchshofer R, Bosserhoff AK. CTGF is overexpressed in malignant melanoma and promotes cell invasion and migration. Br J Cancer. 2011;105:231–238. doi: 10.1038/bjc.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Zhang Q, Mao Z, Chen J, Wang X, Qu J, et al. CTGF is overexpressed in papillary thyroid carcinoma and promotes the growth of papillary thyroid cancer cells. Tumour Biol. 2011;32:721–728. doi: 10.1007/s13277-011-0173-6. [DOI] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Wu JY, Shen ZY, Zhao Q, Du ZP, et al. Involvement of CYR61 and CTGF in the fascin-mediated proliferation and invasiveness of esophageal squamous cell carcinomas cells. Am J Pathol. 2010;176:939–951. doi: 10.2353/ajpath.2010.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CG, Lv L, Liu FR, Wang ZN, Liu FN, Li YS, et al. Downregulation of connective tissue growth factor inhibits the growth and invasion of gastric cancer cells and attenuates peritoneal dissemination. Mol Cancer. 2011;10:122. doi: 10.1186/1476-4598-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YF, Cheung TH, Tsao GS, Lo KW, Yim SF, Wang VW, et al. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int J Cancer. 2006;118:2461–2469. doi: 10.1002/ijc.21660. [DOI] [PubMed] [Google Scholar]
- Yang MH, Lin BR, Chang CH, Chen ST, Lin SK, Kuo MY, et al. Connective tissue growth factor modulates oral squamous cell carcinoma invasion by activating a miR-504/FOXP1 signalling. Oncogene. 2012;31:2401–2411. doi: 10.1038/onc.2011.423. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Tsuda H, Kanai Y, Kasamatsu T, Sengoku K, Hirohashi S, et al. Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Res. 2007;67:7095–7105. doi: 10.1158/0008-5472.CAN-06-4567. [DOI] [PubMed] [Google Scholar]
- Chang CC, Shih JY, Jeng YM, Su JL, Lin BZ, Chen ST, et al. Connective tissue growth factor and its role in lung adenocarcinoma invasion and metastasis. J Natl Cancer Inst. 2004;96:364–375. doi: 10.1093/jnci/djh059. [DOI] [PubMed] [Google Scholar]
- van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, et al. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29:3411–3422. doi: 10.1038/onc.2010.83. [DOI] [PubMed] [Google Scholar]
- Ohgawara T, Kubota S, Kawaki H, Kondo S, Eguchi T, Kurio N, et al. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583:1006–1010. doi: 10.1016/j.febslet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Salvatori B, Iosue I, Mangiavacchi A, Loddo G, Padula F, Chiaretti S, et al. The microRNA-26a target E2F7 sustains cell proliferation and inhibits monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis. 2012;3:e413. doi: 10.1038/cddis.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Reuss DE, Mucha J, Hagenlocher C, Ehemann V, Kluwe L, Mautner V, et al. Sensitivity of malignant peripheral nerve sheath tumor cells to TRAIL is augmented by loss of NF1 through modulation of MYC/MAD and is potentiated by curcumin through induction of ROS. PLoS One. 2013;8:e57152. doi: 10.1371/journal.pone.0057152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenamandra SP, Darekar SD, Kashuba V, Matskova L, Klein G, Kashuba E. Stem cell gene expression in MRPS18-2-immortalized rat embryonic fibroblasts. Cell Death Dis. 2012;3:e357. doi: 10.1038/cddis.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.