Abstract
FBW7 tumor suppressor suppresses the growth and survival of tumor cells by promoting the degradation of several oncoproteins, and induces endothelial differentiation by modulating the NF1/RAS axis. A recent study in this issue of Cell Research showed that FBW7 further regulates endothelial functions via degrading transcription factor KLF2.
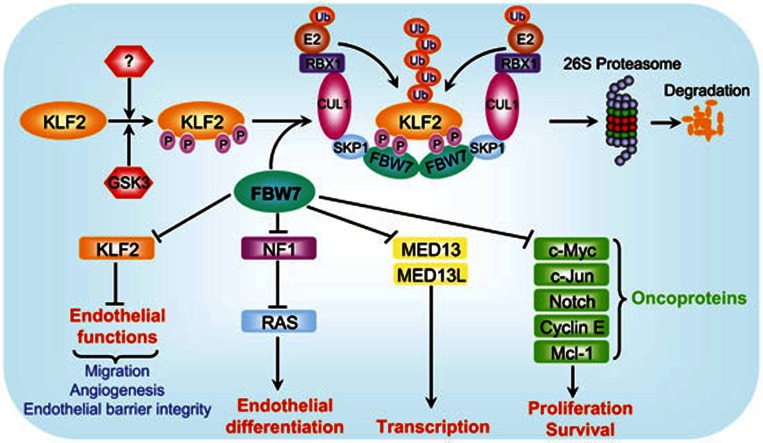
The SCFFBW7 E3 ubiquitin ligase, consisting of SKP1 adaptor protein, CUL1 scaffold protein, RBX1 or RBX2 (also known as SAG) RING protein, and FBW7 substrate recognizing protein, regulates various cellular processes, including cell proliferation and survival, endothelial differentiation, and tumorigenesis by promoting a timely ubiquitylation and degradation of its substrates1 (Figure 1). As a substrate receptor, FBW7 binds to its substrates upon phosphorylation by a specific kinase(s) through conserved CDC4 phospho-degron (CPD) consensus motifs (I/L-I/L/P-pT-P)1. Previous studies showed that SCFFBW7-RBX1 promotes the degradation of oncoproteins, including c-Myc, c-Jun, cyclin E, Notch and Mcl-1 to inhibit proliferation and survival of tumor cells1,2, whereas SCFFBW7-RBX2/SAG promotes the degradation of NF1, thus activating RAS, to induce endothelial differentiation3. Consistently, Fbw7-deficient embryos die in utero around embryonic day 10.5 (E10.5) with remarkable abnormalities in vascular development, indicating that FBW7 is a physiological regulator of endothelial functions4,5. Most recently, SCFFBW7-RBX1 was found to regulate basal transcription machinery by targeted degradation of the components of the Mediator complex, MED13 and MED13L6 (Figure 1). Given the fact that the loss-of-function mutations of FBW7 are found in many human cancers, FBW7 is generally considered as a tumor suppressor7.
Figure 1.
A model for SCFFBW7-mediated KLF2 degradation to regulate endothelial functions. Tumor suppressor FBW7 is a substrate-recognizing subunit of SCF E3 ubiquitin ligase and promotes degradation of a) oncoproteins to regulate proliferation and survival; b) NF1 to modulate endothelial differentiation; and c) MED13/MED13L to regulate transcription. A new study reported in this issue of Cell Research revealed that KLF2, a transcription factor that negatively regulates endothelial functions, is also a substrate of SCFFBW7 E3 ligase. Upon phosphorylation by GSK3 and likely other kinases under an undefined physiological trigger(s), KLF2 is recognized by FBW7 via two CPD motifs. Dimerization of SCFFBW7 E3 ligase promotes KLF2 ubiquitylation and subsequent degradation by the 26S proteasome. Thus, by targeted degradation of KLF2, SCFFBW7 activates endothelial functions, including migration, angiogenesis and the maintenance of endothelial barrier integrity.
The Krüppel-like factors (KLFs) are a subclass of the zinc finger family of transcription factors involved in the regulation of cell growth, differentiation and tissue development8. Accumulated lines of evidence imply that several KLFs, including KLF2, 4, 6, 10 and 15, are involved in endothelial biology. Among them, KLF2 has emerged as being particularly important in the regulation of endothelial functions, including cell migration, angiogenesis and endothelial barrier integrity (Figure 1)9. Although the regulation of KLF2 has been well studied at the transcriptional level, it remains elusive whether KLF2 protein is also regulated at the posttranslational level in endothelial cells. In this issue of Cell Research, Wang and coworkers reported that KLF2 is a physiological substrate of FBW7 in endothelial cells10. Through targeting KLF2 for degradation, FBW7 regulates various endothelial functions.
Using RNA interference, the authors found that FBW7 knockdown in endothelial cells significantly inhibited tube formation, reduced vessel sprouting in vitro, and blocked VEGF-induced angiogenesis in vivo. FBW7 knockdown also markedly reduced migration of HUVEC cells. After a global protein database search for potential FBW7 substrates, the authors identified two putative CPD sites on KLF2, a well-established regulator of endothelial functions, suggesting that FBW7 may regulate endothelial functions through the destruction of KLF2. Indeed, siRNA-based FBW7 knockdown significantly increased the basal level of endogenous KLF2 and extended its protein half-life in HUVEC cells. Conversely, in a co-transfection experiment, overexpression of FBW7, but not other F-box proteins examined, remarkably reduced KLF2 levels and significantly shortened KLF2 protein half-life. Immunofluorescence staining showed that KLF2 co-localized with FBW7α in the nucleus. Co-immunoprecipitation assays further confirmed that endogenous FBW7 interacted with endogenous KLF2. Furthermore, the in vitro ubiquitination assay showed that FBW7 promoted ubiquitylation of KLF2. Taken together, these results support the notion that KLF2 is a substrate of SCFFBW7 E3 ligase, which promotes its ubiquitylation for targeted degradation.
Further biochemical characterization with site-directed mutagenesis revealed that FBW7-KLF2 binding and subsequent KLF2 degradation is mediated by two conserved CPDs and requires KLF2 phosphorylation. To define the involved kinase(s), the authors generated an anti-KLF2 antibody that specifically recognized phosphorylated T243 within the CPD motif. A limited kinase inhibitor screen identified LiCl, a GSK3 inhibitor, but not other kinase inhibitors tested, as an inhibitor of T243 phosphorylation, which stabilized KLF2. Consistently, siRNA-based GSK3β knockdown significantly increased the basal levels of endogenous KLF2 and extended KLF2 protein half-life in HUVECs. The in vitro kinase assay further confirmed that GSK3β indeed phosphorylated KLF2 on T243.
To determine the biological significance of KLF2 degradation by FBW7, the authors co-expressed FBW7 with wild-type KLF2 or a KLF2 mutant with CPD motifs deleted in HUVECs. They found that FBW7 significantly attenuated the inhibition of tube formation by wide-type KLF2, but not by its CPD-deletion mutant. Consistently, FBW7 knockdown-triggered abnormal endothelial functions, as assayed for tube formation, in vivo angiogenesis and endothelial barrier integrity, can be abrogated by simultaneous KLF2 knockdown, indicating that KLF2 plays a causal role. Using an in vivo embryonic stem (ES) cell differentiation model, the authors further demonstrated that FBW7-mediated KLF2 degradation was required for the development of blood vessels in teratomas. They showed that the Fbw7 mRNA level was dramatically increased during endothelial cell differentiation, and that Fbw7 knockdown markedly blocked teratoma growth and reduced the number of blood vessels, which was abrogated by simultaneous KLF2 knockdown. Finally, the authors demonstrated that the FBW7-KLF2 axis was required for vasculogenesis and angiogenesis during vertebrate embryogenesis in a zebrafish model.
The study by Wang et al.10, mechanistically extended previous knowledge that Fbw7 is required for vasculogenesis during mouse embryogenesis4,5 and endothelial differentiation, which is mediated by modulating the NF1-RAS axis3. The authors showed that 1) FBW7 is highly expressed in endothelial cells, suggesting an important role in endothelial functions; 2) FBW7 depletion seriously impairs endothelial functions; 3) FBW7 promotes ubiquitylation and degradation of KLF2 after being phosphorylated by GSK3; and 4) FBW7-controlled endothelial functions are largely mediated via targeting KLF2 for degradation; and 5) the FBW7-KLF2 axis is required for zebrafish embryogenesis, suggesting that FBW7-mediated KLF2 degradation is an evolutionally conserved pathway.
A number of questions that warrant future investigation remain unanswered. First, how much do the other FBW7 substrates, known to regulate vasculogenesis (e.g. Notch and NF1), contribute to the regulation of endothelial functions described in this study? Second, can the death phenotype of Fbw7-deficient embryos be rescued by simultaneous deletion of Klf2, which would indicate that KLF2 is truly a physiological substrate of FBW7? Third, besides GSK3, are there any other kinases and their associated pathways that would trigger KLF2 phosphorylation and subsequent degradation? Fourth, under what physiological conditions is GSK3 activated to trigger KLF2 degradation? Fifth, does FBW7 regulate KLF2 functions in any other cell types? Finally, among KLF family members, it is known that KLF5 is also degraded by SCFFBW711; whereas KLF4 is destructed by SCFβTrCP ubiquitin ligase12. Do these degradations also contribute to the regulation of endothelial functions? Answers to these questions will advance our current knowledge of how SCF E3 ligases in general and SCFFBW7 E3 ligase in particular precisely modulate endothelial functions as well as other biological processes regulated by KLF family members.
In summary, the findings reported by Wang's group provide convincing evidence that the FBW7-KLF2 axis plays an essential role in regulation of endothelial functions and expands our current knowledge as to how SCFFBW7 modulates vascular biology. Since KLF2 negatively regulates angiogenesis, which is required for tumorigenesis, future work should be directed to study how to selectively target SCFFBW7 to cause KLF2 accumulation, thus blocking tumor angiogenesis.
References
- Welcker M, Clurman BE. Nat Rev Cancer. 2008. pp. 83–93. [DOI] [PubMed]
- Inuzuka H, Shaik S, Onoyama I, et al. Nature. 2011. pp. 104–109. [DOI] [PMC free article] [PubMed]
- Tan M, Zhao Y, Kim SJ, et al. Dev Cell. 2011. pp. 1062–1076. [DOI] [PMC free article] [PubMed]
- Tetzlaff MT, Yu W, Li M, et al. Proc Natl Acad Sci USA. 2004. pp. 3338–3345. [DOI] [PMC free article] [PubMed]
- Tsunematsu R, Nakayama K, Oike Y, et al. J Biol Chem. 2004. pp. 9417–9423. [DOI] [PubMed]
- Davis MA, Larimore EA, Fissel BM, et al. Genes Dev. 2013. pp. 151–156. [DOI] [PMC free article] [PubMed]
- Wang Z, Inuzuka H, Zhong J, et al. FEBS Lett. 2012. pp. 1409–1418. [DOI] [PMC free article] [PubMed]
- McConnell BB, Yang VW. Physiol Rev. 2010. pp. 1337–1381. [DOI] [PMC free article] [PubMed]
- Nayak L, Lin Z, Jain MK. Antioxid Redox Signal. 2011. pp. 1449–1461. [DOI] [PMC free article] [PubMed]
- Wang R, Wang Y, Liu N, et al. Cell Res. 2013. pp. 803–819. [DOI] [PMC free article] [PubMed]
- Liu N, Li H, Li S, et al. J Biol Chem. 2010. pp. 18858–18867. [DOI] [PMC free article] [PubMed]
- Kim MO, Kim SH, Cho YY, et al. Nat Struct Mol Biol. 2012. pp. 283–290. [DOI] [PubMed]