Abstract
The casein kinase 1 (CK1) family, a major intracellular serine/threonine kinase, is implicated in multiple pathways; however, understanding its regulation has proven challenging. A recent study published in Science identifying allosteric activation of CK1 by the DEAD-box RNA helicase DDX3 expands our understanding of the control of this abundant kinase family.
The human CK1 protein kinase family is encoded by six genes (α, γ1, γ2, γ3, δ, and ε) and regulates diverse biochemical processes including hedgehog signaling, circadian rhythms and the p53 tumor suppression [reviewed in 1]. In the Wnt/β-catenin pathway, all CK1 family members are involved, each with a distinct role. To carry out their functions, CK1 family members achieve specificity by several mechanisms, but how their kinase activity is regulated has been less clear. Here, we discuss the findings of the Niehrs lab2 in the context of what is known about CK1 control in the Wnt pathway.
CK1γ proteins are membrane bound due to C-terminal S-palmitoylation and phosphorylate the Wnt co-receptor LRP5/6 in the presence of Wnts and Disheveled to activate the pathway3,4. One mechanism of activation may be via 'priming' by upstream phosphorylation of LRP5/6, a common characteristic of CK1 substrate recognition5. CK1δ and CK1ε bind to and phosphorylate Disheveled, an activity regulated by Wnt signaling and protein phosphatases6,7. CK1α interacts with and phosphorylates APC, Axin and Ser45 of β-catenin in an apparently unregulated reaction. The CK1α-catalyzed phosphorylation primes β-catenin for further phosphorylation by GSK3 and subsequent degradation. How does CK1 accomplish so many different jobs in the Wnt pathway and how is it controlled?
A key mechanism for regulation is CK1s' differential interaction with scaffolds and membranes. CK1δ and CK1ε bind to substrates including Disheveled, Period and NFAT1; CK1α interacts with Axin, and CK1γ localizes to membranes where it phosphorylates LRP6. These interactions take place at protein motifs distinct from the phosphorylation sites. However, binding and co-localization alone are probably not sufficient for precise biological control.
Each CK1 isoform is likely to be regulated differently. CK1α is the smallest member of the family (∼38 kDa), and has been thought to be constitutively active. CK1δ and CK1ε have closely-related C-terminal domains (148-184 aa) that are actively autophosphorylated, resulting in a kinase-phosphotail interaction that restricts access of protein substrates to the active site of the kinase. CK1δ and CK1ε can be relieved of this auto-inhibition by the action of protein phosphatases that in turn can be stimulated by extracellular signals such as glutaminergic and Wnt signaling1,6. The regulation of CK1γ is not well understood.
Although the kinase domains between CK1s are highly conserved, subtle differences govern their binding to scaffolds. For example, two key residues determine the differential binding of CK1α and CK1ε to Disheveled and Period8. Motifs on the scaffolds also facilitate binding to CK1. CK1ε binds to an F-X-X-X-F motif on PER2 and NFAT1 that is quite distal from the phosphorylation sites9. The F-X-X-X-F motif is also present on additional CK1 partners including DDX3, although its importance has not yet been tested. The presence of kinase-binding motifs can greatly enhance the phosphorylation of the substrate. Thus, regulating the affinity of CK1 for scaffold-binding sites can have profound effects on rates of phosphorylation.
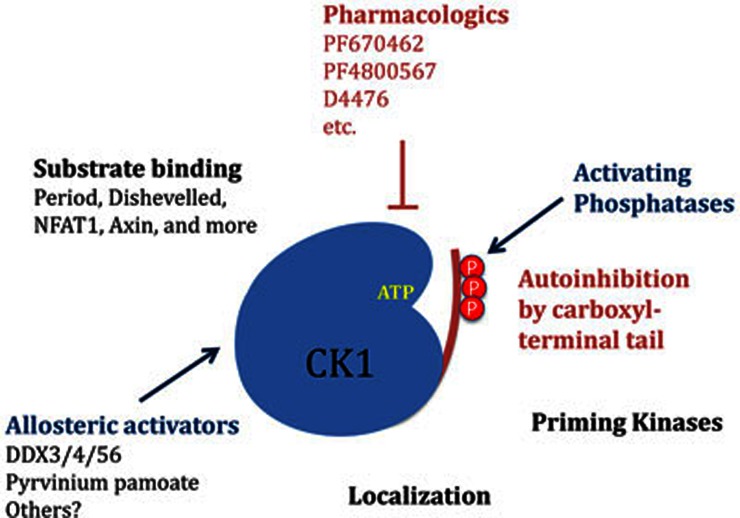
Protein kinase activity can be controlled by diverse mechanisms, the most commonly studied being phosphorylation, addition or removal of regulatory subunits, and targeting to scaffolds (Figure 1). An additional, under-explored mechanism is allosteric regulation. While allostery has a proud history in enzymology, there are only a few examples (e.g., AMP-kinase, phosphorylase kinase) of small-molecule allosteric regulation of protein kinases [reviewed in 10]. Notably, a recent screen for inhibitors of the Wnt/β-catenin pathway identified the drug pyrvinium pamoate as an allosteric activator of CK1α11. As a clue to mechanism, pyrvinium bound to but did not activate other CK1 isoforms. However, it could activate CK1δ lacking its C-terminal regulatory domain. This suggests that there is a conserved site in the CK1 family to which pyrvinium binds that allosterically activates the kinases. Additional inhibitory mechanisms, such as the C-terminal phosphodomains of CK1δ and CK1ε, may be able to override the small-molecule activation. The finding of allosteric activation by pyrvinium suggests that endogenous allosteric regulators of the CK1 family may also exist.
Figure 1.
Regulation of the CK1 family. As described in the text, diverse mechanisms exist to regulate the activity of CK1.
Cruciat et al.2 now provide evidence for protein allosteric activators of CK1. In a cell-based RNAi screen for new regulators of the Wnt/β-catenin signaling, they identified the DEAD-box RNA helicase DDX3. DDX3 is required both for Wnt/β-catenin signaling in human cells as well as Wnt-dependent apical-posterior neural patterning of the central nervous system in Xenopus and neuroblast migration in C. elegans. Epistatic and biochemical analysis place DDX3 at the level of LRP6 and Disheveled phosphorylation. DDX3 cooperates with CK1ε in phosphorylating Disheveled, and physically interacts with CK1ε after Wnt stimulation. Kinetic analysis revealed that DDX3 is an allosteric activator of all CK1 family members tested.
The DDX genes encode a family of DEAD-box RNA helicases, so named for the “DEAD” (Asp-Glu-Ala-Asp) amino acid sequence in their motif II. The 37 members of the DDX family of proteins have the defining function of unwinding double-stranded RNA in an ATP-dependent manner. DDX proteins are multifunctional, playing roles in translation initiation, mRNA transport and splicesosome assembly12. Stimulating CK1 activity is a 'moonlighting' function of DDX3, as key helicase domains and the helicase activity of DDX3 are not required to stimulate Wnt signaling and activate CK1ε.
DDX3 and pyrvinium are similar in that both are allosteric activators of CK1. Whether the underlying mechanisms of activation are similar remains to be determined. The C-terminal regulatory domain of CK1δ blocks pyrvinium action11, whereas DDX3 is able to activate full-length CK1δ. Pyrvinium and DDX3 might interact with the same region on CK1, but the protein activators may function more efficiently than small molecules. Future structural studies may provide us with more definitive answers.
While DDX3 was identified as a Wnt/β-catenin activator via CK1ε, it also activates CK1α and CK1γ, raising the question of why one activity predominates over the other in the cell- and animal-based assays used by Cruciat et al. Furthermore, the authors found that a handful of other DDX proteins activate CK1. While this suggests redundancy, they found strong phenotypes with loss-of-function DDX3 alone. This may be due to tissue-specific expression. It is also possible that other binding partners may modulate the interaction of DDX proteins with CK1.
As always, more questions remain. Does DDX3 regulation of CK1 extend beyond Disheveled signaling? For example, could DDX proteins activate CK1δ/ε to phosphorylate Period proteins during circadian rhythms? How does Wnt signaling stimulate the interaction between CK1ε and DDX3? Is DDX3 also a substrate for Wnt-activated CK1ε, and what effect does that have on DDX3's other functions? How do all these regulatory mechanisms on the various CK1s work together temporally and spatially? Further work will illuminate what happens when the DEAD-box family brings CK1 to life.
References
- Cheong JK, Virshup DM. Int J Biochem Cell Biol. 2011. pp. 465–469. [DOI] [PubMed]
- Cruciat CM, Dolde C, de Groot RE, et al. Science. 2013. pp. 1436–1441. [DOI] [PubMed]
- Davidson G, Wu W, Shen J, et al. Nature. 2005. pp. 867–872. [DOI] [PubMed]
- Zeng X, Tamai K, Doble B, et al. Nature. 2005. pp. 873–877. [DOI] [PMC free article] [PubMed]
- Flotow H, Graves PR, Wang AQ, et al. J Biol Chem. 1990. pp. 14264–14269. [PubMed]
- Swiatek W, Tsai IC, Klimowski L, et al. J Biol Chem. 2004. pp. 13011–13017. [DOI] [PubMed]
- Gao ZH, Seeling JM, Hill V, et al. Proc Natl Acad Sci USA. 2002. pp. 1182–1187. [DOI] [PMC free article] [PubMed]
- Dahlberg CL, Nguyen EZ, Goodlett D, et al. PLoS One. 2009. p. e4766. [DOI] [PMC free article] [PubMed]
- Okamura H, Garcia-Rodriguez C, Martinson H, et al. Mol Cell Biol. 2004. pp. 4184–4195. [DOI] [PMC free article] [PubMed]
- Lindsley JE, Rutter J. Proc Natl Acad Sci USA. 2006. pp. 10533–10535. [DOI] [PMC free article] [PubMed]
- Thorne CA, Hanson AJ, Schneider J, et al. Nat Chem Biol. 2010. pp. 829–836. [DOI] [PMC free article] [PubMed]
- Schröder M. Biochem Pharmacol. 2010. pp. 297–306. [DOI] [PubMed]