Abstract
Rab GTPases are emerging targets of diverse bacterial pathogens. Here, we perform biochemical and structural analyses of LepB, a Rab GTPase-activating protein (GAP) effector from Legionella pneumophila. We map LepB GAP domain to residues 313-618 and show that the GAP domain is Rab1 specific with a catalytic activity higher than the canonical eukaryotic TBC GAP and the newly identified VirA/EspG family of bacterial RabGAP effectors. Exhaustive mutation analyses identify Arg444 as the arginine finger, but no catalytically essential glutamine residues. Crystal structures of LepB313-618 alone and the GAP domain of Legionella drancourtii LepB in complex with Rab1-GDP-AlF3 support the catalytic role of Arg444, and also further reveal a 3D architecture and a GTPase-binding mode distinct from all known GAPs. Glu449, structurally equivalent to TBC RabGAP glutamine finger in apo-LepB, undergoes a drastic movement upon Rab1 binding, which induces Rab1 Gln70 side-chain flipping towards GDP-AlF3 through a strong ionic interaction. This conformationally rearranged Gln70 acts as the catalytic cis-glutamine, therefore uncovering an unexpected RasGAP-like catalytic mechanism for LepB. Our studies highlight an extraordinary structural and catalytic diversity of RabGAPs, particularly those from bacterial pathogens.
Keywords: Legionella, Rab GTPases, GTPase-activating protein (GAP), type IV secretion system, TBC GAP, membrane trafficking
Introduction
Rab GTPases constitute the largest family of small GTP-binding proteins that act as molecular switches in regulating vesicular transport in eukaryotic cells1,2. Rab GTPases are lipid-modified at their C termini and localized to distinct membrane-bound compartments to ensure corrected delivery of different cargos3. Like other small GTPases, Rabs cycle between the GDP-bound ''off'' and GTP-bound ''on'' states, the latter of which can interact with downstream effector proteins for appropriate functioning. The conformational states and activities of Rab GTPases are subjected to precise regulation by several classes of Rab modulators4,5. GTPase-activating proteins (GAPs) catalyze GTP hydrolysis and thereby inactivate Rabs on the membrane. Guanine nucleotide dissociation inhibitors (GDIs) extract GDP-bound Rab from the membrane and complex with them in the cytosol. For activation, Rabs are released from RabGDI by the GDI-displacement factor to allow for membrane re-association and subsequent exchange of GDP for GTP, catalyzed by guanine nucleotide exchange factors (GEFs).
The importance of modulating Rab function has also been appreciated by bacterial pathogens. Recent intensive studies on Legionella pneumophila, a successful intracellular bacterial pathogen responsible for Legionnaires' disease, have uncovered a series of stepwise manipulations of Rab1 function by Legionella Dot/Icm type IV secretion system effectors. L. pneumophila establishes a replication niche called Legionella-containing vacuole (LCV) in host macrophages, which requires recruitment of endoplasmic reticulum (ER)-derived vesicles6,7,8. In line with the fact that the Dot/Icm type IV secretion system is required for maturation and intracellular trafficking of the LCV, several Legionella type IV effectors are endowed with activities of manipulating Rab1 cycle and function. SidM/DrrA, one of the Dot/Icm-secreted effectors, recruits Rab1 from the cytosol to the LCV and uses its unique GEF activity to activate Rab1 from the GDI complex9,10,11,12,13,14. The N-terminal domain of SidM bears an additional adenylyl transferase domain that adenylylates Rab1-GTP on a switch II tyrosine residue, which prevents GAP-catalyzed GTP hydrolysis15 and prolongs the duration of Rab1-GTP for activating LidA, a Legionella-supplied Rab effector. SidM-induced adenylylation is enzymatically reversed by another Dot/Icm effector called SidD16,17. This de-adenylylation modification allows for Rab1 GTP hydrolysis and thereby recycling back to the cytosol, the former of which is catalyzed by LepB, a Legionella-encoded RabGAP effector whose mechanism remains unknown18.
Very recently we discovered a Rab1-specific GAP activity of the VirA and EspG family of type III secretion system effectors from non-vacuole pathogens Shigella flexneri and enteropathogenic E. coli (EPEC)19. These effectors function to counteract autophagy and cytokine secretion-mediated host defenses during S. flexneri and EPEC infection, respectively. Notably, the VirA/EspG family of bacterial GAPs adopts a completely different three-dimensional architecture from the host TBC GAPs for Rab GTPases20, but employs a TBC-like arginine and glutamine dual-finger catalytic mechanism19. Inspired by this study, here we perform biochemical and structural analyses of LepB RabGAP activity. We map LepB GAP domain to residues 313-618 and identify Arg444 as the only arginine residue required for LepB313-618 GAP activity. Crystal structures of LepB313-618 alone and the GAP domain of L. drancourtii LepB in complex with Rab1-GDP-AlF3 confirm Arg444 being the catalytic arginine finger. More importantly, the structures reveal a 3D fold and a GTPase-recognition mode distinct from those of all known GAPs. Glu449 undergoes a drastic movement and interacts strongly with Rab1 Gln70 in the complex structure. This interaction induces a side-chain flipping of Rab1 Gln70, which acts as the catalytic cis-glutamine, a situation similar to that in the RasGAP-Ras complex. Thus, LepB is a structurally novel GAP that employs a RasGAP-like catalytic mechanism to promote GTP hydrolysis in Rab1. Our study highlights an extraordinary structural and catalytic diversity of RabGAPs, particularly those from bacterial pathogens.
Results
Identification of LepB313-618 as the minimal RabGAP domain and Arg444 as the candidate arginine finger
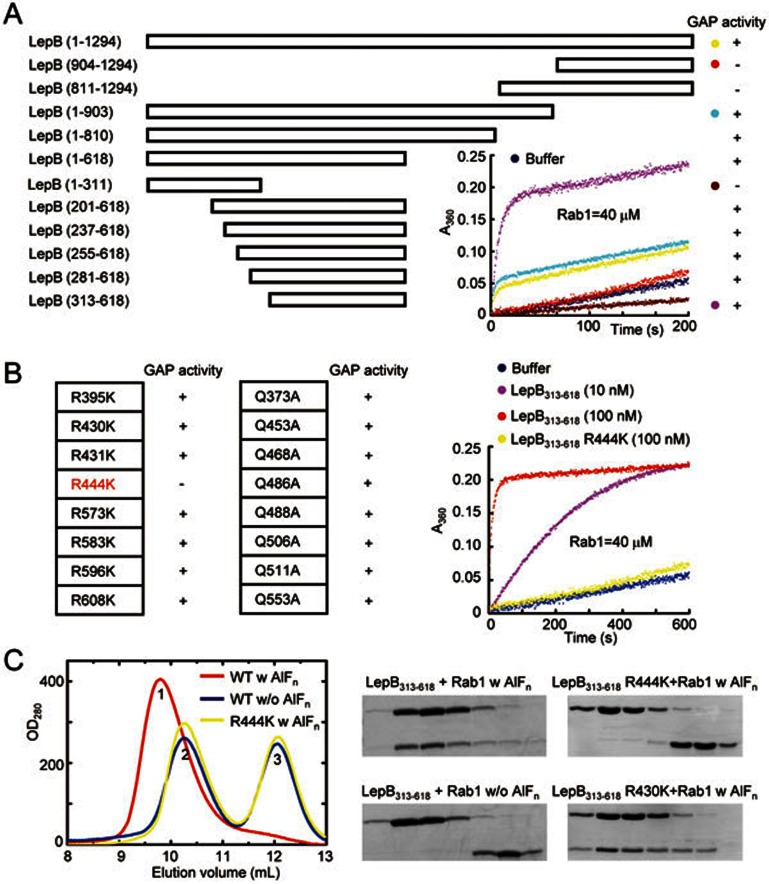
Full-length LepB from L. pneumophila contains 1 294 residues but no recognizable domains/motifs. To locate the domain region responsible for its GAP activity, we started with LepB C-terminal region due to the possible presence of a TBC-like dual-finger motif in primary sequence. In vitro GAP assay showed that bacterially purified LepB (904-1 294) and LepB (811-1 294) fragments harbored no activities of catalyzing GTP hydrolysis in Rab1 (Figure 1A). In contrast, LepB (1-903) displayed a robust GAP activity comparable to that of full-length LepB. Further systematic truncation analyses identified residues 313-618 of LepB (LepB313-618) as the minimal domain capable of catalyzing GTP hydrolysis in Rab1 (Figure 1A). Notably, the activity of LepB313-618 was even higher than that of full-length LepB or LepB (1-903), suggesting a possible autoinhibitory mechanism.
Figure 1.
Identification of the RabGAP domain of LepB and the catalytic arginine finger. (A) Mapping the RabGAP domain of LepB. A series of LepB truncation proteins as indicated were subjected to in vitro GAP assays using Rab1 as the substrate, and the positive and negative results are indicated in the right column by the + and – signs, respectively. Time courses of GTP hydrolysis for Rab1 (40 μM) catalyzed by color-indicated representative LepB truncation mutants are shown in the right corner. (B) Screening for arginine and glutamine residues required for the GAP activity of LepB313-618. Nearly all arginine and glutamine residues in LepB GAP domain (LepB313-618) were individually mutated to lysine and alanine, respectively, and each mutant protein was assayed for GAP activity using Rab1 as the substrate. The results are summarized in the left column. The time course of GTP hydrolysis for the R444K mutant in comparison with that of wild-type LepB313-618 is shown on the right. (C) Gel filtration chromatography assays for AlFn-dependent LepB-Rab1-GDP complex formation. Equal molar amounts of LepB313-618 or the R444K mutant and GDP-loaded Rab11-177 were mixed together at a total concentration of 20 mg/ml, incubated at 4 °C in a buffer supplemented with or without NaF and AlCl3, and then subjected to Superdex-75 gel filtration chromatography. Shown on the left are the chromatograms colored accordingly. Peak 1, 2, and 3 correspond to LepB313-618-Rab1-GDP-AlFn complex, free LepB313-618, and free Rab1, respectively. Elution fractions in the indicated range were analyzed by SDS-PAGE and Coomassie blue-stained gels are shown on the right. LepB313-618 R430K was included as a negative control.
Both eukaryotic TBC-domain RabGAPs20 and the VirA/EspG family of bacterial RabGAPs19 feature an arginine and glutamine dual catalytic finger. To identify the catalytic finger residues, nearly all the arginine (Arg395, 430, 431, 444, 573, 583, 596, and 608) and glutamine residues (Glu373, 453, 468, 486, 488, 506, 511, and 533) in LepB313-618 were individually mutated into lysine and alanine, respectively. Among the 16 mutant proteins, only LepB313-618 R444K was found to be deficient in promoting GTP hydrolysis (Figure 1B). The GAP protein can form a stable complex with the GTPase substrate in the presence of aluminium fluoride (AlFn) and GDP, which approximates the GTP hydrolysis transition state intermediate19,20,21,22. As expected, purified LepB313-618 co-eluted with Rab1-GDP from a gel filtration column in the presence of AlFn, and omission of AlFn prevented the LepB313-618-Rab1-GDP complex formation (Figure 1C). In contrast, LepB313-618 R444K and Rab1-GDP eluted separately from the gel filtration column even in the presence of AlFn, and their elution profile corresponded to that of the two proteins loaded alone. These data suggest that Arg444 is the catalytic arginine finger for LepB GAP activity.
The GAP activity of LepB is highly robust and specific to Rab1
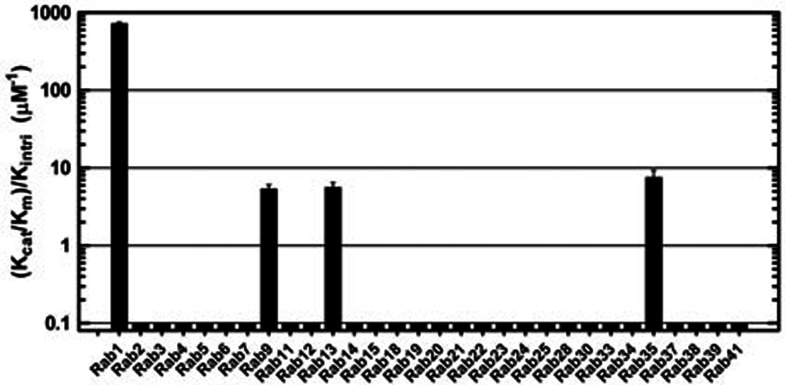
We then profiled the activity of LepB313-618 towards a panel of 30 mammalian Rabs and determined the corresponding kinetic parameters, as we and others have previously performed with Gyp1p and the VirA/EspG family19,20. Compared with Gyp1p and the VirA/EspG family, LepB313-618 showed a much narrower substrate spectrum and acceleration of GTP hydrolysis was only observed with Rab1, Rab9, Rab13 and Rab35 (Figure 2). The activity of LepB313-618 on Rab1 (kcat/Km, 7.24 × 105 M−1S−1) was higher than those on other three Rabs (kcat/Km, 5.40 ∼ 7.60 × 103 M−1S−1) by more than two orders of magnitude (Figure 2). The catalytic efficiency of LepB313-618 on Rab1 was also higher than those of Gyp1p on Rab1 and Rab33 (≤1 × 105 M−1S−1)20 as well as that of VirA/EspG on Rab1 (2 ∼ 3× 105 M−1S−1)19. These enzymatic analyses establish LepB as a highly selective and most potent GAP protein for Rab1, a feature consistent with an essential role of LepB in mediating rapid inactivation and removal of Rab1 from the LCV during infection18.
Figure 2.
The RabGAP activity of LepB is highly robust and specific to Rab1. GTP hydrolysis relative to the intrinsic GTP hydrolysis rate constant was determined for a panel of 30 mammalian Rabs. Data shown are mean values ± standard deviation (error bar) from two independent experiments. The y axis is on the logarithmic scale.
Crystal structure of LepB RabGAP domain
LepB313-618 shows no sequence homology to all known GAP proteins including TBC-domain RabGAPs. To reveal the structural mechanism of LepB RabGAP activity, we crystallized LepB (residues 1-618) that covers the GAP domain (LepB313-618) and solved the 3.2-Å structure by single wavelength anomalous diffraction (SAD) method (Table 1). The structure contains two LepB molecules packing against each other in an asymmetric unit. The interaction between the two molecules likely resulted from crystal packing as purified LepB was exclusively a monomer in solution according to its gel filtration chromatography behavior. No meaningful structural difference was observed between the two molecules in the same asymmetric unit, and chain A was subjected to further analysis. Compared with the N-terminal region, the GAP domain has a higher-quality electron density map and contains residues 326-391, 400-519 and 530-617. Other residues including 317-325, 392-399 and 520-529 are missing and likely in flexible loop regions.
Table 1. Data collection and refinement statistics.
| Crystal statistics | LepB1-618 SeMet | LepBdrancourtii-Rab1 |
|---|---|---|
| Data collection statistics | ||
| Space group | C2221 | P6522 |
| Wavelength (Å) | 0.9792 | 0.9792 |
| a, b, c, (Å) α, β, γ, (°) | 101.53, 159.42, 181.15, 90, 90, 90 | 95.56, 95.56, 197.72, 90, 90, 120 |
| Resolution range (Å)* | 20-3.15 (3.20-3.15) | 20-2.80 (2.85-2.80) |
| No. of unique reflections | 25525 (1163) | 14058 (681) |
| Completeness (%) | 99.5 (94.7) | 100 (99) |
| Redundancy | 13.5 (8.7) | 10.5 (10.7) |
| I/σI | 28.87 (2.00) | 17.56 (3.45) |
| Rmerge (%) | 12.8 (92.6) | 15.8 (77.2) |
| Refinement statistics | ||
| Rwork/Rfree (%)† | 26.40/28.30 | 22.91/26.91 |
| No. of protein atoms | 6894 | 3625 |
| No. of waters | 5 | 4 |
| RMSD bond lengths (Å) | 0.010 | 0.016 |
| RMSD bond angles (°) | 1.367 | 1.419 |
| Ramachandran plot statistics | ||
| Most favored regions (%) | 92.60 | 95.94 |
| Additional allowed regions (%) | 6.70 | 4.06 |
| Outlier region (%) | 0.7 | |
*The data for the highest resolution shell are shown in parentheses.
†Rfree is calculated by omitting 5% of the total number of reflections in model refinement.
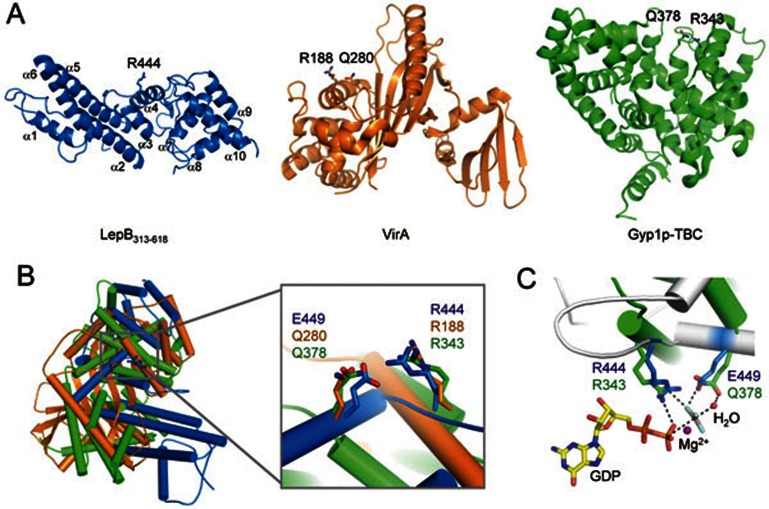
LepB GAP domain adopts a wide “V”-shape architecture, consisting of 10 α-helices and no β-strands (Figure 3A). The more elongated left arm of the “V” shape is mainly formed by a four-helix bundle (α2, α3, α5 and α6) packed by α1 and α4 on opposite sides. α1 at the top end of the left arm connects the GAP domain to the N-terminal domain (not shown in the picture) through a linking loop; α4 is situated in the valley bottom. The right arm is made up of four helices (α7-α10) organized into a more globular shape. α7 and α8, projecting perpendicular to the plane of the V shape, is located closer to the valley bottom; α9 and α10, parallel to each other, cross α7 and pack against α7/α8 from outside. Notably, the overall shape and structural architecture of LepB GAP domain shows no similarity to that of eukaryotic TBC domain (illustrated using Gyp1p TBC domain)20 (Figure 3A and 3B). LepB GAP domain is also structurally distinct from the VirA/EspG family of bacterial RabGAPs that adopt an α/β fold despite that they are also of prokaryotic origin and highly specific for Rab1. The structural divergence of LepB and the VirA/EspG family from eukaryotic TBC GAPs shall bring insights into the evolution of bacterial virulence activity.
Figure 3.
Crystal structure of LepB313-618 and comparison with the VirA/EspG-family bacterial RabGAP and the TBC domain of Gyp1p. (A) Overall structure of LepB313-618 in comparison with those of VirA (PDB ID: 4FMB) and Gyp1p (PDB ID: 2G77). Secondary structure elements are drawn with α-helices as coils, β-strands as flat arrows and loops as lines. LepB313-618, VirA and Gyp1p are colored blue, orange and green, respectively. (B) Superimposition of LepB313-618 structure onto those of VirA and Gyp1p using Arg444/Glu449 in LepB313-618, Arg188/Gln280 in VirA and Arg343/Gln378 in Gyp1p as the references. Shown on the right is a close-up view with marked reference residues in stick models. α-helices are shown as cylinders colored as that in A. (C) Structural comparison of Arg444/Glu449 in LepB313-618 with Arg343/Gln378 in Gyp1p. GDP and AlF3 in the Gyp1p-Rab1 complex structure are shown as yellow sticks. Mg2+ and the nucleophilic water are depicted as magenta and red sphere, respectively. Polar interactions between GDP-AlF3 and Arg343/Gln378 in Gyp1p are denoted by black dashed lines.
The candidate arginine finger residue Arg444 is located in the loop between α3 and α4 on the concave surface of the V-shaped valley; its side chain is solvent exposed and projects from the valley bottom upwards to the open space (Figure 3A). Structural superimposition of LepB GAP domain with VirA-Rab1 or Gyp1p-Rab33 complex (see below), which places Arg444 in a position equivalent to that of the catalytic arginine in VirA/Gyp1p and amenable to interacting with GDP and AlFn, revealed no structural clashes but instead a well accommodation of Rab1/33 in the open space of the V-shaped valley. These structural analyses lend further support to Arg444 as the canonical arginine finger for LepB GAP activity. It is also worth noting that LepB Arg444 is situated in a loop region rather than supplied by a helix as observed with those in Gyp1p and VirA/EspG (Figure 3A and 3B), highlighting an additional structural difference between LepB and known RabGAPs.
Glu449, occupying the structural position of TBC GAP glutamine finger in apo-LepB, is critical for LepB GAP activity
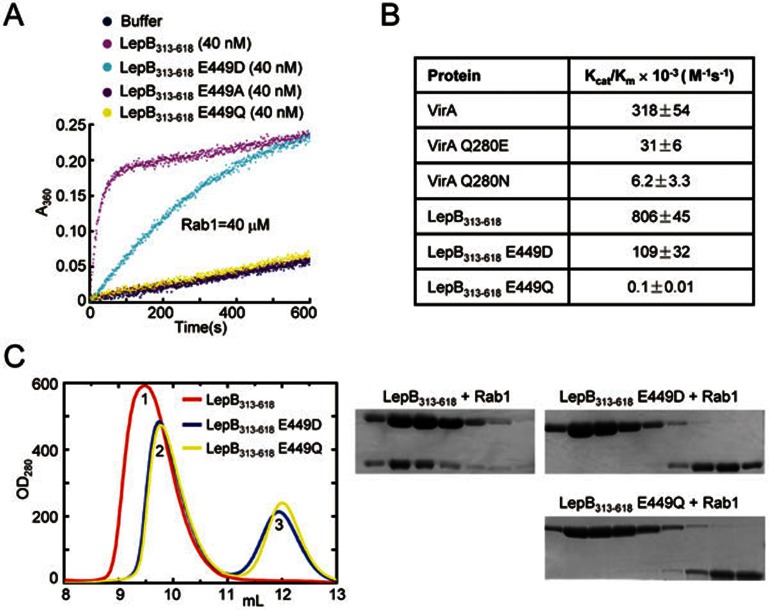
Different from GAPs for Rho and Ras families, both eukaryotic TBC GAPs and the VirA/EspG family of bacterial GAPs feature a catalytic glutamine finger in addition to the canonical arginine finger19,20. The glutamine side chain is at a distance of ∼ 6 Å from the guanidine group of the arginine finger; its carbonyl amide mediates bipartite hydrogen-bonding interactions with an equatorial fluoride ion and the axial water in the AlFn complex. In catalysis, the glutamine carbonyl acts as a general base to deprotonate and orientate the water molecule for nulceophilically attacking on the γ-phosphate in GTP. Consistent with the fact that none of the glutamines in LepB313-618 are essential for its GAP activity (Figure 1B), no glutamine residues could be found within the 8-Å distance from Arg444, confirming that LepB does not employ a TBC-like catalytic glutamine finger. Notably, we found a glutamate residue (Glu449) whose side-chain carboxyl carbon is ∼ 4 Å away from the carbon atom of the guanidine group of Arg444 (Figure 3B). When LepB GAP domain was structurally superimposed onto Gyp1p TBC domain and VirA using Arg444/Glu449 in LepB and the catalytic arginine/glutamine fingers in Gyp1p/VirA as the reference, presentation and stereochemistry of Glu449 side chain appeared to be analogous to those of the catalytic glutamine in TBC and TBC-like GAPs despite that the carboxyl group of Glu449 is slightly closer to AlFn (Figure 3C). In fact, mutation of Glu449 to alanine, glutamine or aspartate either completely abolished or largely attenuated the activity of LepB313-618 in promoting GTP hydrolysis in Rab1 (Figure 4A and 4B). Consistently, both LepB313-618 E449Q and E449D mutants were deficient in forming an AlFn-dependent complex with Rab1-GDP (Figure 4C). Prompted by this observation, we also examined the TBC-like glutamine finger in VirA and found that substitution of Q280 by either a glutamate or an asparagine could also largely abolish VirA GAP activity by more than 90% (Figure 4B). These analyses establish a critical role of Glu449 for the GAP activity of LepB effector.
Figure 4.
Glu449 is required for the RabGAP activity of LepB as well as AlFn-mediated LepB-Rab1-GDP complex formation. (A) Effects of Glu449 mutations on the RabGAP activity of LepB. Time courses of GTP hydrolysis for Rab1 (40 μM) catalyzed by LepB313-618 or its E449A, E449D and E449Q mutants are shown as color indicates. (B) Summary of the kinetic parameters of LepB Glu449 and VirA Gln280 mutants. The Michaelis-Menten kinetic parameters (kcat/Km) determined by using the Lineweaver-Burk plot for LepB313-618 and VirA or their indicated mutants are listed. Data shown are mean values ± standard deviation (error bar) from two independent experiments. (C) Effects of Glu449 mutations on AlFn-mediated LepB-Rab1-GDP complex formation. The assay was performed similarly as that in Figure 1C. Shown on the left are the gel filtration chromatograms colored accordingly. Peek 1, 2, and 3 correspond to LepB313-618-Rab1-GDP-AlFn complex, free LepB313-618, and free Rab1, respectively. Elution fractions in the indicated range were analyzed by SDS-PAGE and Coomassie blue-stained gels are shown on the right.
Crystal structure of LepBdrancourtii GAP domain-Rab1-GDP-AlF3 complex
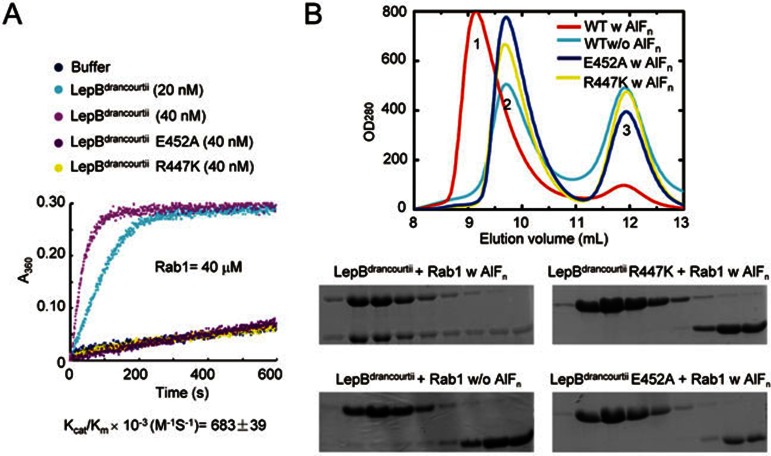
To understand the exact mechanism of LepB recognizing Rab1 and catalyzing GTP hydrolysis, we efforted to determine the structure of LepB GAP domain in complex with Rab1, but unfortunately we were unable to obtain diffractable crystals. We then turned to LepB homologues from other Legionella species. LDG_7216 from L. drancourtii strain LLAP12, a strictly intracellular amoebal pathogen23, contains 1 189 amino acids and shares about 55% overall sequence similarity to LepB from L. pneumophila. Residues 316-620 at the N-terminal region of LDG_7216 harbored 54% sequence homology to the GAP domain of L. pneumophila LepB. For convenience, a recombinant fragment containing residues 316-620 of LDG_7216 was referred as LepBdrancourtii GAP domain hereafter. Biochemical assays confirmed that LepBdrancourtii GAP domain (residues 316-620) harbored a robust GAP activity using Rab1 as the substrate, comparable to that of L. pneumophila LepB (Figure 5A). LepBdrancourtii GAP domain could also form an AlFn-dependent complex with Rab1-GDP (Figure 5B). Also similarly to that observed with L. pneumophila LepB, the RabGAP activity and formation of the transition state-approximating ternary complex required Arg447 and Glu452 in LepBdrancourtii GAP domain (equivalent to Arg444 and Glu449 in L. pneumophila LepB) (Figure 5A and 5B).
Figure 5.
The RabGAP activity of LepB from L. drancourtii. (A) In vitro GAP assay of LepB from L. drancourtii using Rab1 as the substrate. LepBdrancourtii is the purified GAP domain of LepB from L. drancourtii (residues 316-620). Arg447 and Glu452, corresponding to Arg444 and Glu449 in L. pneumophila LepB, were subjected to mutation analysis in this assay. (B) Effects of Arg447 and Glu452 mutations on AlFn-mediated LepBdrancourtii-Rab1-GDP complex formation. The assay was performed similarly as that in Figure 1C. Shown at the top are the Superdex-75 gel filtration chromatograms colored accordingly. Peek 1, 2, and 3 correspond to LepBdrancourtii-Rab1-GDP-AlFn complex, free LepBdrancourtii, and free Rab1, respectively. Elution fractions in the indicated range were analyzed by SDS-PAGE and Coomassie blue-stained gels are shown at the bottom.
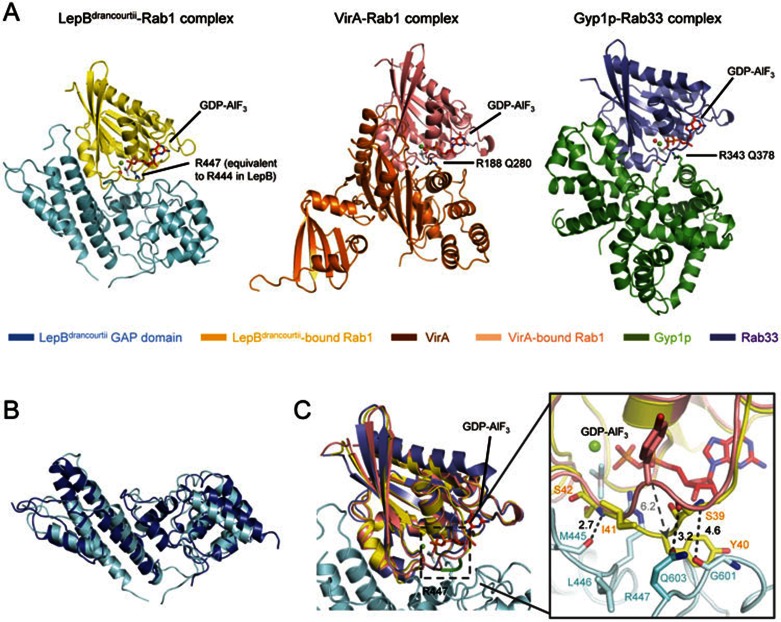
We succeeded in obtaining diffraction-quality crystals of LepBdrancourtii GAP domain in complex with Rab1-GDP and AlF3 and determined 2.8-Å structure of the ternary complex (Figure 6A and Table 1). The structure contains one LepBdrancourtii and one Rab1 in an asymmetric unit. Rab1 is cradled by the concave surface of LepBdrancourtii GAP domain along the V-shaped valley. Consistent with the fact that LepB GAP domain has a completely different fold from both the eukaryotic TBC domain and the VirA/EspG family of bacterial RabGAPs, the Rab1-binding mode of LepBdrancourtii is unique and shows no similarity to those of the two known classes of RabGAPs (Figure 6A). Rab1-bound LepBdrancourtii GAP domain is structurally similar to apo-LepB GAP domain with RMSD value of 2.642 for 233 selected Cα atoms (Figure 6B). The overall structure of Rab1 is also nearly identical to those of VirA-bound Rab1 and Gyp1p-bound Rab33 (Figure 6C, right). The interaction of LepBdrancourtii with Rab1 engages residues from helices α3, α4 and more importantly the connecting loop in LepBdrancourtii GAP domain. The interaction involves a network of extensive hydrogen bonds and hydrophobic interactions (Figure 6C). Specifically, the backbone carbonyl oxygen of Met445 in LepBdrancourtii forms a hydrogen bond with the backbone amide of Rab1 Ser42. Rab1 Ser39 has two hydrogen bonds with LepBdrancourtii GAP domain; its hydroxyl group donates one hydrogen to the side-chain carbonyl of LepBdrancourtii Gln603 and its backbone carbonyl oxygen accepts one hydrogen from the backbone amide of LepBdrancourtii Gly601. Rab1 Ile41 contributes to hydrophobic interactions with LepBdrancourtii Leu446. Accompanying these interactions, the Cα atom of Rab1 Tyr40 undergoes a drastic movement of 6.2 Å towards LepBdrancourtii GAP domain and its side chain is flipped by ∼ 120° around the peptide bond, which results in a more opened switch I with altered surface features (Figure 6C). The conformational change of Tyr40 and switch I in Rab1 has not been seen with Rabs in Gyp1p and VirA/EspG, highlighting the distinct mode of Rab recognition by LepBdrancourtii GAP domain.
Figure 6.
Crystal structure of LepBdrancourtii GAP domain-Rab1-GDP-AlF3 complex. (A) Overall structure of LepBdrancourtii GAP domain-Rab1-GDP-AlF3 complex in comparison with those of VirA-Rab1-GDP-AlF3 (PDB ID: 4FMB) and Gyp1p-Rab33-GDP-AlF3 complexes (PDB ID: 2G77). The overall structures are shown in cartoons with color scheme indicated underneath of the pictures. GDP and AlF3 in the complexes are shown in red stick models. Mg2+ and water are shown as green and red sphere, respectively. (B) Structural comparison of Rab1-bound LepBdrancourtii GAP domain (light cyan) with apo-LepB GAP domain (blue). The alignment was performed by using the two sets of Cα chains as the reference and the least-square algorithm. (C) Interactions of Rab1 with LepBdrancourtii GAP domain and structural comparison of Rab1 with Rabs in TBC and TBC-like RabGAPs. The superimposition was performed using Cα chains in Rabs as the references and the least-square algorithm. Left panel is the cartoon diagram of the superimposition. The color scheme follows that in A. The structurally rearranged loop in Rab1 is highlighted in green and boxed with black dashed line. Right panel is a close-up view of structural changes in Rab1. Residues involved in the interaction between Rab1 and LepBdrancourtii are shown in sticks colored as their residing chains. Polar interactions are depicted by black dashed lines with a number to denote the distance in angstrom. The drastic positional movement of Tyr40 in Rab1 is indicated by a dashed arrow.
LepB employs a RasGAP-like catalytic mechanism
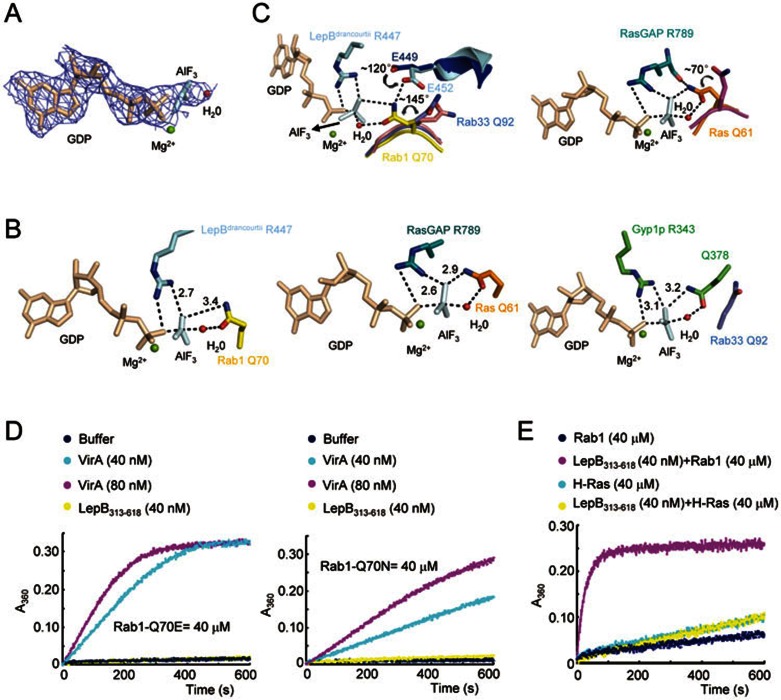
In the refined map of the LepBdrancourtii GAP domain-Rab1 complex, clear electron density is observed for GDP, Mg2+, a putative nucleophilic water and AlF3 (Figure 7A). AlF3 is coordinated by an axial oxygen from GDP and occupies the site of γ-phosphate in the GTP-bound state24. The conformational arrangement and chemical geometry of GDP, Mg2+ and AlF3 are similar to those observed in other GAP-GTPase complex including the Gyp1p-Rab33 and VirA-Rab1 complexes (Figure 7B), confirming a GTP-hydrolysis transition state-approximating intermediate. As expected, Arg447 in LepBdrancourtii mediates bidentate polar interactions with an oxygen from β-phosphate of GDP and an equatorial fluoride ion. This structural feature also resembles those described for other GAP-GTPase complexes19,20,25, reassuring Arg447 being the arginine finger in catalyzing GTP hydrolysis.
Figure 7.
Structural basis for a RasGAP-like catalytic mechanism of LepB. (A) Sigma-A weighted 2mFo-DFc map contoured at 1.5 σ for the modeled GDP, AlF3, Mg2+ and putative nucleophilic water in the LepBdrancourtii GAP domain-Rab1 complex. (B) Structural comparison of GDP, AlF3, and the catalytic arginine and glutamine fingers among LepBdrancourtii GAP domain-Rab1 complex (left), RasGAP-Ras complex (middle, PDB ID: 1WQ1) and Gyp1p-Rab1 complex (right, PDB ID: 2G77). The catalytic arginine and glutamine and other key residues are shown in sticks and colored the same as their residing chains. Mg2+ and the nucleophilic water are shown as green and red spheres, respectively. Polar interactions are indicated by black dashed lines. (C) Conformational changes of Gln70 in Rab1 and Glu449/452 in LepB and comparison with related structural elements in the RasGAP-Ras complex. Shown on the left is structural alignment of key residues in the catalytic sites of VirA-bound Rab1 and Gyp1p-bound Rab33 with those in the LepBdrancourtii-Rab1 complex. The right panel shows the alignment of free Ras with RasGAP-Ras complex. Alignments were performed using Cα chains of Rabs or Ras as the reference. In the left panel, apo-LepB GAP domain was superimposed onto the LepBdrancourtii-Rab1 complex using Cα atoms of LepB Arg444 (Arg447 in LepBdrancourtii) and E449 (Glu452 in LepBdrancourtii) as the reference. Free Ras, RasGAP-bound Ras and RasGAP are colored in magenta, orange and teal blue, respectively. Coloring of LepBdrancourtii-Rab1 complex and LepB GAP domain alone follows that in Figure 6A and Figure 3A, respectively. Rotational movement of the cis-glutamine in Rab1 and Ras and the critical Glu452 in LepBdrancourtii are denoted by black curved arrows. (D) Effects of mutation of Gln70 in Rab1 on LepB- and VirA-catalyzed GTP hydrolysis. The indicated concentrations of purified VirA and LepB313-618 were subjected to in vitro GAP assays using Rab1 Q70E (Left) and Q70N (Right) as the substrate. (E) In vitro GAP assay of LepB313-618 using H-Ras as the substrate.
To our surprise, the side chain of Glu452 in LepBdrancourtii rotates away from its original position in the apo-LepB structure by ∼ 120 °C (Figure 7C), which no longer occupies the structural position equivalent to the glutamine finger in TBC and TBC-like RabGAPs. Most notably, Glu452 in the Rab1-GDP-AlF3 complex now bears a strong hydrogen-bond or ionic interaction with Gln70 in Rab1, and this interaction renders a drastic conformational change of the side chain of Gln70 (Figure 7C). Such structural rearrangement of the glutamine residue, though more dramatic, echoes what has been observed in the RasGAP-Ras complex, which is induced by interaction of Gln61 in Ras with the backbone carbonyl oxygen of RasGAP arginine finger (Arg789)26 (Figure 7C). In fact, Gln61 in Ras serves as the catalytic cis-glutamine to facilitate GTP hydrolysis. Similar to Gln61 in the RasGAP-Ras complex, Gln70 in the LepBdrancourtii GAP domain-Rab1 complex also mediates bipartite polar interactions with equatorial fluoride ion and the putative nucleophilic water (Figure 7B and 7C). The importance of Gln70 was supported by results of the in vitro GAP assay, in which Q70E or Q70N mutants of Rab1 did not significantly affect the activity of VirA but were completely resistant to LepB-catalyzed GTP hydrolysis (Figure 7D). These observations clearly distinguish LepB RabGAP from eukaryotic TBC GAPs and the VirA/EspG family of TBC-like RabGAPs that both supply the catalytic glutamine finger in trans (Figure 7B). Thus, the structural and chemical similarities between LepBdrancourtii Arg447/Rab1 Gln70 and RasGAP Arg789/Ras Gln61 establish that Legionella LepB effector does not harbor the TBC RabGAP-like dual catalytic fingers but instead employs a RasGAP-like catalytic mechanism in promoting GTP hydrolysis in Rab1. RasGAP has extensive specific interactions with Ras for productive substrate binding26. Consistently, LepB did not catalyze GTP hydrolysis when Ras was used a substrate (Figure 7E).
The drastic side-chain movement of Rab1 Gln70 towards GDP-AlF3 and therefore serving as the catalytic cis-glutamine is unexpected. Given that Glu449/Glu452 in LepB plays an absolutely essential role for LepB to form an AlFn-dependent ternary complex with Rab1 and catalyze GTP hydrolysis (Figures 4 and 5), the strong interaction observed between Gln70 in Rab1 and Glu452 in LepBdrancourtii is likely a determining structural factor for Gln70 being able to play the catalytic role. Gln70 in Rab1, compared with Gln61 in Ras, is more towards the GTPase inside and relatively difficult to move out. These analyses together help to explain why TBC or TBC-like RabGAPs have to supply the glutamine finger as they do not harbor a glutamate equivalent to Glu449 in LepB.
Discussion
In this study, we map the RabGAP domain of the Legionella Dot/Icm type IV effector LepB and determine crystal structures of the GAP domain alone and in complex with Rab1-GDP-AlF3. LepB represents a novel RabGAP fold and employs a distinct GTPase-recognition mode. Combining extensive biochemical analyses together with the structural characterization, we identify the canonical arginine finger residue responsible for LepB to catalyze GTP hydrolysis. As a RabGAP, the VirA/EspG family of bacterial effectors shares the arginine and glutamine dual catalytic fingers, but no primary sequence and three-dimensional structural similarities with eukaryotic TBC-domain RabGAPs19. Notably, the overall structure of LepB GAP domain is also clearly distinct from those of TBC domain and the VirA/EspG family. The three different classes of RabGAPs all employ a common catalytic arginine finger, representing a striking example of convergent evolution of bacterial virulence activity. The sequence and structural divergence of the three classes of RabGAPs also suggests a possible existence of other structural classes of RabGAPs in eukaryotes that have escaped from identification by conventional sequence and domain analyses. S. flexneri is free living after gaining access to host cytosol and VirA inactivation of Rab1 contributes to suppression of autophagy-mediated host defense; GTP hydrolysis and inactivation of Rab1 by EspG and the consequent disruption of the general secretory pathway is important for inhibition of cytokine secretion by EPEC that resides extracellularly; intracellular L. pneumophila relies on LepB-mediated inactivation of Rab1 to achieve sophisticated manipulation of Rab1 cycle and decorate LCV into an ER-like structure. The three pathogens have completely different pathogenesis and life cycle, but all have evolved a virulence activity of catalyzing GTP hydrolysis in Rab1 for their own purposes. These observations are insightful in deepening and broadening our understanding of hijacking Rab function by bacterial pathogens as well as the associated diverse bacterial virulence mechanism. Therefore, future studies will likely identify more bacterial virulence factors that are RabGAP proteins either similar to or distinct from the above three classes.
Our analyses also identify a glutamate in the catalytic site of LepB that is indispensable for its RabGAP activity and formation of the AlF3-depenendent complex with Rab1-GDP. In apo-LepB, this glutamate occupies the structural position of the catalytic glutamine finger in eukaryotic TBC GAPs and the VirA/EspG family of TBC-like bacterial RabGAPs. Interestingly, this residue, upon LepB binding to GTPase substrate, moves away from contacting AlF3 and bears a strong ionic interaction with Gln70 in Rab1. The interaction is accompanied by a drastic side-chain flipping of Rab1 Gln70 towards GDP-AlF3. Such conformational changes provide the structural basis for Gln70 acting as the catalytic cis-glutamine in promoting GTP hydrolysis. The catalytic mechanism of LepB indeed mirrors that of RasGAP26 and also represents the first example that Rabs can supply a catalytic glutamine finger in GAP-mediated GTP hydrolysis20. This unexpected finding breaks the boundary of GAP classification and highlights an extraordinary catalytic diversity of GAP proteins for small GTPases.
While this manuscript was being prepared for publication, Goody and their colleagues also reported the structure of L. pneumophila LepB in complex with Rab127. The Goody structure is overall highly similar to our structure of LepBdrancourtii-Rab1 complex. However, beryllium fluoride (rather than aluminum fluoride) is used as the γ-phosphate surrogate in their study, and therefore the critical nucleophilic water is missing in the catalytic center and direct evidences for Gln70 acting as the cis-glutamine are not provided. In fact, the beryllium fluoride complex structure more mimics the ground state of GAP-catalyzed GTP hydrolysis while our structure, obtained by using aluminum fluoride, approximates the subsequent transition-state intermediate.
Materials and Methods
Plasmids and reagents
DNA for LepB was amplified from L. pneumophila genomic DNA and cloned into the pQE-80L vector (QIAGEN) for recombinant expression in E. coli. DNA encoding the GAP domain of LepBdrancourtii (residues 316-620 of ZP_09620807 from Legionella drancourtii LLAP12 strain) was artificially synthesized. Human Rab1a (residues 1-177) were PCR cloned into pET28a vector with an N-terminal SUMO fusion. Truncation and deletion mutants were constructed by standard PCR cloning strategy. All point mutations were generated by QuikChange™ Site-Directed Mutagenesis Kit (Stratagene) and the plasmids were verified by DNA sequencing. Bacterial expression constructs for 30 Rabs were kindly provided by Dr David G Lambright (University of Massachusetts Medical School, USA). All chemicals were Sigma-Aldrich products.
Recombinant protein expression and purification
E. coli BL21 (DE3) strain (Novagen) was used as the host for expression of recombinant proteins. Protein expression was induced for 16 h at 16 °C or 22 °C with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) after OD600 reached 0.6-0.8. N-terminal 6× His-tagged LepB and its mutants in the pQE-80L vector were purified by using Ni-NTA agarose beads (QIAGEN) followed by Hitrap Q HP ion exchange chromatography (GE Healthcare). Recombinant proteins were kept in a buffer containing 25 mM HEPES (pH 7.4) and 150 mM NaCl. LepBdrancourtii GAP domain and Rab1 proteins were also overexpressed in BL21 (DE3) followed by Ni-affinity purification. The SUMO-fusion parts were removed by Ulp1 protease digestion and the proteins were further purified by Hitrap Q ion exchange column. Protein concentrations were determined spectrophotometrically using the theoretical molar extinction coefficients at 280 nm and protein purity was examined by Coomassie blue staining of SDS-PAGE gels. For crystallization, LepB (residues 1-618) were also cloned into the pQE-80L vector for bacterial expression, and proteins were further purified by Mono Q ion exchange and Sephadex 75 gel filtration chromatography.
GAP assay
The GAP assay using the EnzChek Phosphate Assay Kit (Invitrogen) and determination of kinetics were carried out by strictly following the previously described procedure19. The absorbance at 360 nm was monitored on a UV-2450 spectrometer (Shimadzu).
Crystallization and data collection
The LepB1-618 crystals were grown by the hanging-drop vapor diffusion method at 22 °C by mixing equal volumes (1 μl each) of the protein and the reservoir solution. Diffraction-quality crystals appeared at the condition of 15 mg/ml of LepB (residues 1-618) with a reservoir solution containing 0.1 M citrate (pH 6.5) and 20% MPEG 2000. For cryo-protection, crystals were transferred into the cryoprotectant solution containing 0.1 M citrate (pH 6.5), 25% MPEG 2000 and 5% glycerol, soaked for several seconds, and then flash-cooled in liquid nitrogen. X-ray diffraction data were collected at SPring-8 beamline BL41XU at the wavelength of 0.9792 Å. To crystallize the LepBdrancourtii GAP domain-Rab1 complex, LepBdrancourtii GAP domain was mixed with GDP-loaded Rab1 at the molar ratio of 1:1.1 in a buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20 mM NaF, 2 mM AlCl3, 5 mM MgCl2 and 2 mM DTT, and the mixture was incubated on ice overnight to obtain the LepBdrancourtii GAP domain-Rab1-GDP-AlF3 complex. Crystallization of the complex was performed by hanging-drop vapor diffusion method using the concentrated mixture at 24 mg/ml. The crystals were grown at 20 °C for one week in 0.2 M ammonium acetate, 0.1 M Tris·HCl pH 8.5, and 25% w/v polyethylene glycol 3350. Crystals were coated by cryoprotectant oil containing 50% v/v Parabar 10312 (Hampton Research) and 50% v/v paraffin oil, and then subjected to flash freezing in liquid nitrogen. Data were collected at Shanghai Synchrotron Radiation Facility Beamline 17U.
Structural determination and refinement
Raw diffraction data of LepB1-618 were processed with the HKL package28. The phase was solved using selenium single wavelength anomalous dispersion method29 in the program SHARP (http://www.globalphasing.com/sharp/). Initial experimental map was density-modified in DM30. The model was automatically built in Phenix.autobuild31 and Buccaneer32, and manually adjusted in COOT33. The structure was refined by using the program phenix.refine in PHENIX. Diffraction data of the LepBdrancourtii GAP domain-Rab1 complex were processed with HKL2000 and scaled with Scalepack28. The complex structure was solved by molecular placement using VirA-bound Rab1 and sculptured LepB-GAP domain34 as searching models in the program Phaser35. The complete model was manually built in COOT33 and refined in refmac36. All the structural figures were generated using PyMol (www.pymol.org). The atomic coordinates of L. pneumophila LepB (residues 1-618) and LepBdrancourtii GAP domain-Rab1 complex have been deposited in the Protein Data Bank with the accession codes 4JVS and 4JW1, respectively.
Acknowledgments
We thank Dr David G Lambright for kindly providing bacterial expression constructs for 30 Rabs. We thank Dr Nieng Yan's group (Tsinghua University, China) for helping to collect crystal diffraction data at the Spring-8 synchrotron (Hyogo, Japan) as well as staff at Shanghai Synchrony Radiation Facility (Shanghai, China). We also thank Yuqing Zheng and other members of the Shao lab for technical assistance and helpful discussions. The research was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute to FS. YZ was supported by new faculty start-up funds from Zhejiang University. This work was also supported by the National Basic Research Program of China (973 Programs; 2010CB835400 and 2012CB518700) to FS and (2011CB910304 and 2011CB911103) to DCW.
References
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci USA. 2009;106:14185–14186. doi: 10.1073/pnas.0907725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- Ge J, Shao F. Manipulation of host vesicular trafficking and innate immune defense by Legionella Dot/Icm effectors. Cell Microbiol. 2011;13:1870–1880. doi: 10.1111/j.1462-5822.2011.01710.x. [DOI] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36:1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Suh HY, Lee DW, Lee KH, et al. Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J. 2010;29:496–504. doi: 10.1038/emboj.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu L, Zhou Y, Yao Q, Liu L, Shao F. Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA. Proc Natl Acad Sci USA. 2010;107:4699–4704. doi: 10.1073/pnas.0914231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr, Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Arnold RJ, Luo ZQ. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci USA. 2011;108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, Shao F. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- La Scola B, Birtles RJ, Greub G, Harrison TJ, Ratcliff RM, Raoult D. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int J Syst Evol Microbiol. 2004;54:699–703. doi: 10.1099/ijs.0.02455-0. [DOI] [PubMed] [Google Scholar]
- Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Mihai Gazdag E, Streller A, Haneburger I, et al. Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep. 2013;14:199–205. doi: 10.1038/embor.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W.Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology Academic Press 1997276part A, 307–326. [DOI] [PubMed] [Google Scholar]
- Pahler A, Smith JL, Hendrickson WA. A probability representation for phase information from multiwavelength anomalous dispersion. Acta Crystallogr A. 1990;46(Pt 7):537–540. doi: 10.1107/s0108767390002379. [DOI] [PubMed] [Google Scholar]
- Cowtan K. An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]