Abstract
Background
Most patients with non–small cell lung cancer (NSCLC) have responded poorly to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs). We investigated the involvement of insulin-like growth factor 1 receptor (IGF-1R) signaling in primary resistance to EGFR TKIs and the molecular determinants of resistance to IGF-1R TKIs.
Methods
Phosphorylated IGF-1R/insulin receptor (pIGF-1R/IR) was immunohistochemically evaluated in a NSCLC tissue microarray. We analyzed the antitumor effects of an IGF-1R TKI (PQIP or OSI-906), either alone or in combination with a small-molecular inhibitor (PD98059 or U0126) or with siRNA targeting K-Ras or MAPK/extracellular signal-regulated kinase kinase (MEK), in vitro and in vivo in NSCLC cells with variable histologic features and EGFR or K-Ras mutations.
Results
pIGF-1R/IR expression in NSCLC specimens was associated with a history of tobacco smoking, squamous cell carcinoma histology, mutant (mut) K-Ras, and wild-type (wt) EGFR, all of which have been strongly associated with poor response to EGFR TKIs. IGF-1R TKIs exhibited significant antitumor activity in NSCLC cells with wt EGFR and wt K-Ras but not in those with mutations in these genes. Introduction of mut K-Ras attenuated the effects of IGF-1R TKIs on NSCLC cells expressing wt K-Ras. Conversely, inactivation of MEK restored sensitivity to IGF-TKIs in cells carrying mut K-Ras.
Conclusions
The mutation status of both EGFR and K-Ras could be predictive markers of response to IGF-1R TKIs. Also, MEK antagonism can abrogate primary resistance of NSCLC cells to IGF-1R TKIs.
Keywords: EGFR, K-Ras, IGF-1R, lung cancer, TKI
INTRODUCTION
Lung cancer, usually caused by years of tobacco smoking (TS), is the leading cause of cancer deaths in the United States.1. Because conventional chemotherapy has limited efficacy against lung cancer, new targeted therapeutic approaches are being investigated. The epidermal growth factor receptor (EGFR) signaling pathway is an attractive target in the development of lung cancer treatments. However, treatment with erlotinib and gefitinib, the two EGFR tyrosine kinase inhibitors (TKIs) approved by the U.S. Food and Drug Administration, has produced poor response rates in patients with non–small cell lung cancer (NSCLC).2 Although a group of patients with somatic mutations in EGFR respond to these EGFR TKIs,2–4 such mutations have been detected in only 5% of tumors from current or former smokers,2 and a response rate to EGFR TKIs of only 3.9% has been reported in patients with NSCLC and a history of TS compared with 24.7% in NSCLC patients who have never smoked,5 suggesting that EGFR may not be the appropriate target in NSCLC patients with a history of TS.
Signaling through the insulin-like growth factor 1 receptor (IGF-1R) has an essential role in cell mitosis, survival, and transformation6–9 and has been associated with higher risk of multiple neoplasms.10–12 IGF-1 stimulates IGF-1R and the IGF-1R/insulin receptor (IGF-1R/IR) heterodimers. Recently, we demonstrated activation of the IGF-1R signaling axis during the early stages of lung carcinogenesis.13 We found that activation of IGF-1R in the lungs of mice as a result of IGF-1 overexpression led to spontaneous lung tumor development that progressed to adenocarcinoma upon exposure to tobacco carcinogens. This early stage of lung cancer development was suppressed by administration of a selective IGF-1R TKI, cis-3-[3-(4-methyl-piperazin-l-yl)-cyclobutyl]-1-(2-phenyl-quinolin-7-yl)-imidazo[1,5-a]pyrazin-8-ylamine (PQIP).13 Given the importance of IGF-1R signaling in most human cancers and the promising results of clinical trials targeting IGF-1R for cancer therapy,14 we sought to evaluate the potential application of IGF-1R TKIs in a series of NSCLC cells with variable histologic and genetic characteristics to assess potential determinants of response or resistance to these drugs.
Here, we report that the activation of IGF-1R via TS, constitutive activation of EGFR via somatic mutations, and IGF-1R–independent activation of signaling through mutant K-Ras are potential biomarkers of response or resistance of NSCLC cells to small-molecule IGF-1R TKIs, including PQIP and OSI-906. Our findings provide a rationale for the therapeutic use of IGF-1R TKIs, either singly or in combination with MAPK/extracellular signal-regulated kinase (MEK) inhibitors, in TS-related NSCLC, particularly in tumors with K-Ras mutations.
MATERIALS AND METHODS
Cell Lines
NSCLC cell lines were obtained from American Type Culture Collection or provided by Dr. John Minna (University of Texas Southwestern Medical Center), Dallas, TX). The cell lines were authenticated by the Genetic Resources Core Facility at Johns Hopkins University (Baltimore, MD) using DNA (STR) profiling.
Protein Analysis
Total cell or tissue lysates were incubated with anti-IGF-1R antibody (1 µg/mL; Santa Cruz Biotechnology, CA) and protein A agarose for analysis of IGF-1R/IR tyrosine phosphorylation status. The precipitates were analyzed by western blotting with pIGF-1Rβ (Tyr1131)/IRβ (Tyr1146) or pIGF-1Rβ (Tyr1135/1136)/IRβ (Tyr1150/1151) antibody (Cell Signaling Technology, Danvers, MA). Antibodies detecting total IGF-1R (Santa Cruz Biotechnology), pIGF-1Rβ (Tyr1131/1146), pIGF-1Rβ (Tyr1135/1136), pErk1/2, pAkt, pIRS-1, total IRS-1, total Erk1/2, total Akt, actin, tubulin, or cleaved caspase-3 (all from Cell Signaling Technology) were used for western blotting. The culture medium without serum was harvested after 2 days of cell culture and concentrated with a Centricon centrifugal filter unit (Millipore, Billerica, MA), and the free IGF-1 in the medium was measured with an ELISA kit from Diagnostic Systems Laboratories (Webster, TX). PQIP and OSI-906 were provided by OSI Pharmaceuticals (Melville, NY). Reverse-phase protein array was performed as previously described15.
Tissue Microarray of Primary Tumor Specimens and the Analysis
Primary NSCLC tumor specimens were collected from 354 patients who had been treated at our institution under an Institutional Review Board–approved protocol and had given their informed consent. Demographic information for those patients was described previously.16 Formalin-fixed, paraffin-embedded primary NSCLC sections (5 µm thick) were placed in a tissue microarray (TMA). Immunohistochemical (IHC) evaluation of the NSCLC TMA was performed as previously described.17 Anti-pIGF-1R (Tyr 1135/1136)/IR (Tyr1162/1163) antibody (Biosource, Camarillo, CA; diluted 1:200) or anti-pEGFR (Tyr1068) antibody (Zymed Laboratories, San Francisco, CA; diluted 1:100) was used for staining. Immunostaining for IGF-1R, and pIGF-1R/IR (membrane) was quantified by a lung cancer pathologist who used a four-value intensity score (0, 1+, 2+, and 3+), and the extent of reactivity was expressed as a percentage. A final staining score was calculated by multiplying the intensity score by the extent of reactivity value (range, 0–300). EGFR exons 18–21 and the K-Ras mutational hot spot codons 12, 13, and 61 were amplified as described previously.3–4, 18 Treated polymerase chain reaction (PCR) products were sequenced using a Big Dye Terminator v3.1 sequencing kit (Applied Biosystems, Foster City, CA). Specimens with single or double EGFR and K-Ras mutations were confirmed using repeated PCR and sequencing, and the corresponding normal DNA was sequenced to verify that the mutations were somatic.
In Vitro Drug Sensitivity and Apoptosis Assays
The indicated NSCLC cells were treated with PQIP, either singly or in combination with MEK inhibitors, in 1% FBS. Cell viability was determined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay as described previously.19 At least 6 independent samples were used for the assay. Cell apoptosis was analyzed using immunofluorescence staining with cleaved caspase-3 antibody (BioVision, Mountain View, CA) as described previously.20 Adenovirus expressing dominant-negative MEK1/2 was described previously,21 and siRNA against K-Ras was purchased from Dharmacon (Longmont, CO). Anchorage-independent growth in 0.4% agarose with a 1% agarose underlay was measured as described previously.13
Animal Experiments
All animal procedures were performed in accordance with a protocol approved by the MD Anderson Institutional Animal Care and Use Committee. Athymic nude mice (BALB/c) were obtained from Harlan Laboratories (Madison, WI). Xenograft tumors were generated by subcutaneous injection of H226B-K-Ras. Briefly, nude mice were injected at a single dorsal flank site with 5 × 106 cells in 200 µL of phosphate-buffered saline. When tumors reached a volume of 50–200 mm3, mice were treated orally with vehicle, OSI906 (40 mg/kg per day), U0126 (4 mg/kg, every other day), or both OSI-906 and U0126; the first day of drug treatment was termed day 0. Tumor size was measured every 2 days. Volumes were calculated by 0.5 × a × b2, in which a is the longer and b the shorter diameter. Mean tumor volumes and 95% confidence intervals were determined.
Statistical Analysis
For the TMA data, pIGF-1R expression levels for NSCLC patients with different clinical and demographic characteristics, including sex, history of TS, tumor histologic type, and EGFR and K-Ras mutation status, were compared using Student’s t test, the Mann–Whitney U test, or ANOVA. Correlations among TMA specimens stained for pIGF-1R/IR and pEGFR were identified using the Spearman rank correlation coefficient. For the drug sensitivity analysis, the two-tailed Mann–Whitney U test was used to compare sensitivity between the mut and wt K-Ras groups of cells. All analyses were conducted using SAS (SAS Institute, Cary, NC) or SPSS (SPSS, Chicago, IL). P < 0.05 was considered statistically significant.
RESULTS
Activation of IGF-1R/IR is Associated with Histologic features, History of Tobacco Smoking, and Mutations of EGFR and K-Ras in Human Lung Cancer
We evaluated the expression of pIGF-1R/IR in surgical tumor sections obtained from patients with NSCLC. pIGF-1R/IR staining was detected in the cell membrane (50%), cytoplasm (100%), and nucleus (100%). Given that the nature of IGF-1R as a membrane receptor and the role of nuclear IGF-1R staining are still unclear, we analyzed the membrane staining of pIGF-1R/IR.
Given the frequency of EGFR mutation (exons 18–21) in NSCLC patients who have never smoked, those with adenocarcinoma, and those with wt K-Ras2, 4, 18, 22–24 and the cross-talk between the EGFR and IGF-1R signaling pathways, we assessed the correlation of pIGF-1R/IR staining with the frequency of EGFR and K-Ras mutations in the NSCLC specimens. pIGF-1R/IR expression levels were higher in patients with squamous cell carcinoma than in those with adenocarcinoma (P = 0.019) and were higher in patients with a history of TS (both former and current smokers) than in patients who had never smoked. pIGF-1R/IR level and EGFR mutation were negatively correlated with a marginal significance (Table 1). In addition, pIGF-1R/IR levels were significantly higher in patients with mut K-Ras than in those with wt K-Ras (Table 1). The negative correlation between pIGF-1R/IR expression and mut EGFR and the positive correlation between pIGF-1R/IR expression and mut K-Ras were also observed in patients with adenocarcinoma (data not shown). These findings suggest that activation of the IGF-1R axis is strongly correlated with TS-induced lung carcinogenesis.
Table 1.
Tissue levels of pIGF-1R/IR in case patients with NSCLC
| Variable | Category | pIGF-1R/IR | ||
|---|---|---|---|---|
| n | Mean ± SD | P | ||
| Smoking | Never | 50 | 8.8 ± 29.18 | |
| Former | 134 | 28.28 ± 49.99 | 0.008 | |
| Current | 82 | 30.85 ± 50.08 | 0.027 | |
| Histology | Squamous | 100 | 35.0 ± 53.04 | |
| Adenocarcinoma | 168 | 19.75 ± 47.29 | 0.019 | |
| EGFR | Mutant | 25 | 8.0 ± 28.28 | |
| Wild type | 159 | 24.78 ± 48.63 | 0.066 | |
| K-Ras | Mutant | 22 | 41.82 ± 53.06 | |
| Wild type | 157 | 20.83 ± 45.99 | 0.015 | |
ANOVA test for smoking history, p-values comparing with never smoker.
Independent samples T-test for Histology
Mann-Whitney Test for EGFR mutation and K-Ras mutation.
NSCLC Cell Lines Carrying mut EGFR Are Independent of IGF-1R Signaling for Survival and Proliferation
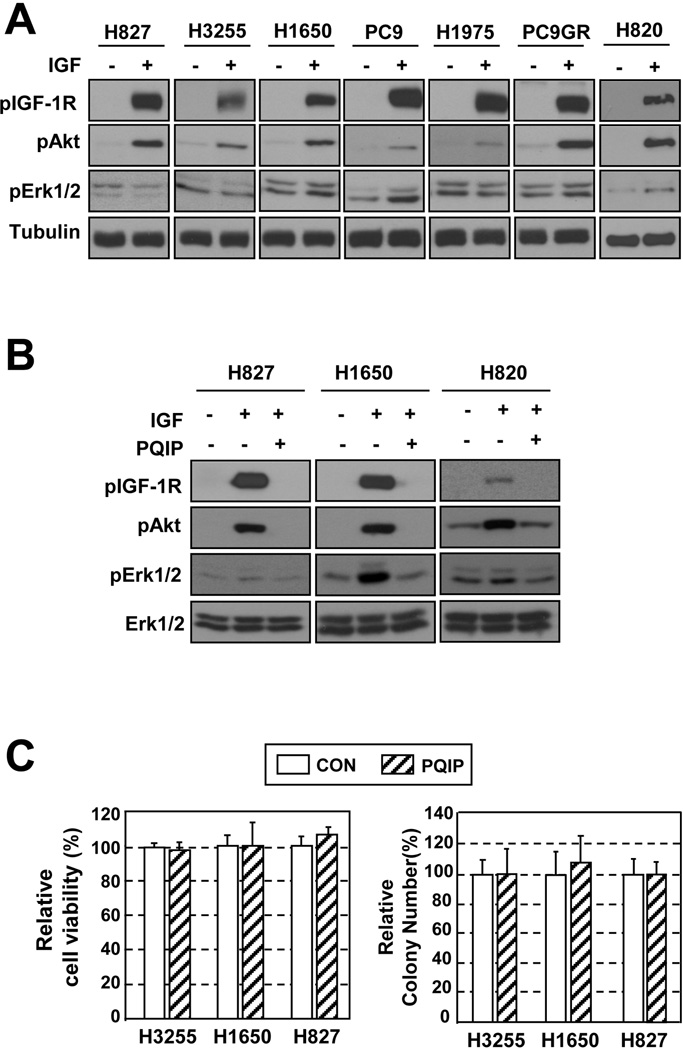
Given the negative association between pIGF-1R/IR level and EGFR mutation, we sought to explore the impact of EGFR mutation on the sensitivity of NSCLC cells to PQIP, an IGF-1R/IR TKI.25 We first examined whether the IGF-1R signaling pathway was functional in six NSCLC cell lines carrying mut EGFR (L858R or Del746–750). IGF-1–induced activation of IGF-1R signaling was well preserved (Fig. 1A) and was effectively inhibited by PQIP (Fig. 1B) in the EGFR mutant cell lines. However, the viability and anchorage-independent colony-forming ability of these cells remained unchanged after PQIP treatment (Fig. 1C). These findings suggest that the NSCLC cells carrying mut EGFR harbor functional IGF-1R signaling but do not rely on the pathway for cell proliferation
Figure 1. Response of NSCLC cell lines carrying mut EGFR to treatment with PQIP.
(A) The IGF-1R signaling pathway is activated by IGF-1 in mut EGFR cells. (B) PQIP inhibits IGF-1R phosphorylation, shown in a subset of mut EGFR cell lines. (C) PQIP does not affect cell viability or anchorage-independent colony formation, shown in 3 mut EGFR cell lines. Columns, means of 3 independent experiments; bars, standard deviation.
K-Ras Mutation Is a Key Determinant of the Response of NSCLC Cell Lines carrying wt EGFR to IGF-1R Inhibitors
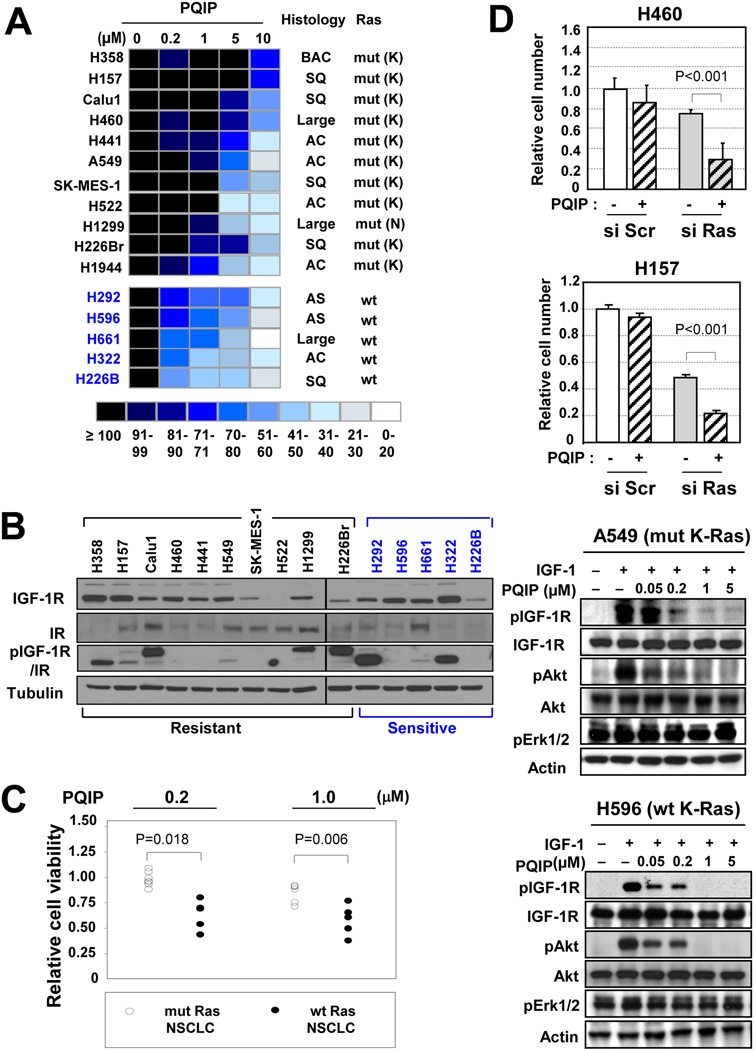
Findings from the NSCLC TMA led us to hypothesize that NSCLC cell lines of which are derived from lung epithelial cells exposed to tobacco smoke,26 could be dependent on IGF-1R signaling for survival and proliferation, thus providing a vulnerable point for pIGF-1R/IR – targeted inhibitors. To test this hypothesis, we examined a panel of 16 NSCLC cell lines carrying wt EGFR with various histologic features (squamous cell carcinoma, bronchioalveolar carcinoma, adenocarcinoma, and large-cell carcinoma) and mutations in K-Ras and p53. We assessed the effects of blockade of IGF-1R signaling by PQIP on the proliferation and viability of these NSCLC cells. When we tested the sensitivity to PQIP at different concentrations (0.2–10 µM), the 16 cell lines displayed differential sensitivity to PQIP treatment (Fig. 2A). We sought to identify predictive biomarkers of PQIP sensitivity in the cells. Although no obvious correlation was seen between PQIP sensitivity and the cells’ histologic features (Fig. 2A) or expression levels of IGF-1R, IR, or pIGF-1R/IR (Fig. 2B), the NSCLC cells with mut K-Ras (listed in black) tended to have poorer sensitivity to PQIP than did those with wt K-Ras (listed in blue) (Fig. 2A). Moreover, cell lines carrying mut K-Ras (n = 7) showed significantly higher viability than those carrying wt K-Ras (n = 5) at doses of 0.2 and 1.0 µM PQIP (P < 0.05, Fig. 2C)
Figure 2. K-Ras mutation is associated with poor response to the IGF-1R TKI PQIP in wt EGFR NSCLC cells.
(A) Role of IGF signaling in NSCLC cells with mut K-Ras (except H1299;N-Ras, listed in black) and wt K-Ras (listed in blue). The effect of PQIP on the survival/proliferation of NSCLC cells was examined (1% FBS in culture medium). Each result is displayed as a colored box; shades of blue represent the cell number relative to the control, as a percentage. The most resistant cells are at the top and the most sensitive at the bottom. AC, adenocarcinoma ;BAC, bronchioalveolar carcinoma; SQ, squamous cell carcinoma; Large, large-cell carcinoma,. (B) Relative expression of IGF-1R, IR, and pIGF-1R/IR in a panel of NSCLC cell lines. (C) Comparison of relative sensitivity to PQIP treatment between the mut and wt K-Ras groups. Relative cell viability decreased by ≥5% in response to treatment with 1 µM PQIP in each group. . (D) (top) H460 and H157 cells, which carry mut K-Ras, were transfected with K-Ras siRNA or Scr (scrambled)siRNA and then seeded in 96-well microplates. The cells were treated with PQIP for 3 days in RPMI containing 1% FBS and then subjected to MTT assay. (bottom) A549 and H596 cells, which carry mut and wt K-Ras, respectively, were treated with the indicated concentrations of PQIP for 6 hours in the absence of FBS and then stimulated with IGF-1 for 30 minutes.
To confirm the role of K-Ras mutation in PQIP resistance, we assessed the effects of PQIP on K-Ras mutant and wild type cells. To investigate the mechanism by which K-Ras mutation rescues NSCLC cells from PQIP treatment, we examined the PQIP-induced antiproliferative activities H460 and H157 cells after mut K-Ras was knocked out by transfection with specific siRNA against K-Ras. Both H460 and H157 cells revealed a significantly enhanced PQIP sensitivity after K-Ras expression was silenced by transfection with specific siRNA (Fig. 2D, top), indicating an essential role of mut K-Ras in mediating PQIP resistance in the NSCLC cell lines. We next assessed the effects of PQIP on IGF-1R signaling in H596 cells, which carry wt K-Ras, and A549 cells, which carry mut K-Ras. We found that PQIP treatment at 1 µM almost completely inhibited IGF-induced IGF-1R and Akt phosphorylation in H596 cells (Fig. 2D, bottom). Similar results were found in A549 cells, indicating that PQIP is effective in blocking IGF-1R signaling in NSCLC cells regardless of K-Ras mutation status. These results indicate that the mechanism by which K-Ras mutation decreases NSCLC cell sensitivity to PQIP is independent of the ligand-induced phosphorylation of IGF-1R.
Mut K-Ras Activates IGF-1R/Akt Signaling but Leads to Resistance to IGF-1R/IR TKI
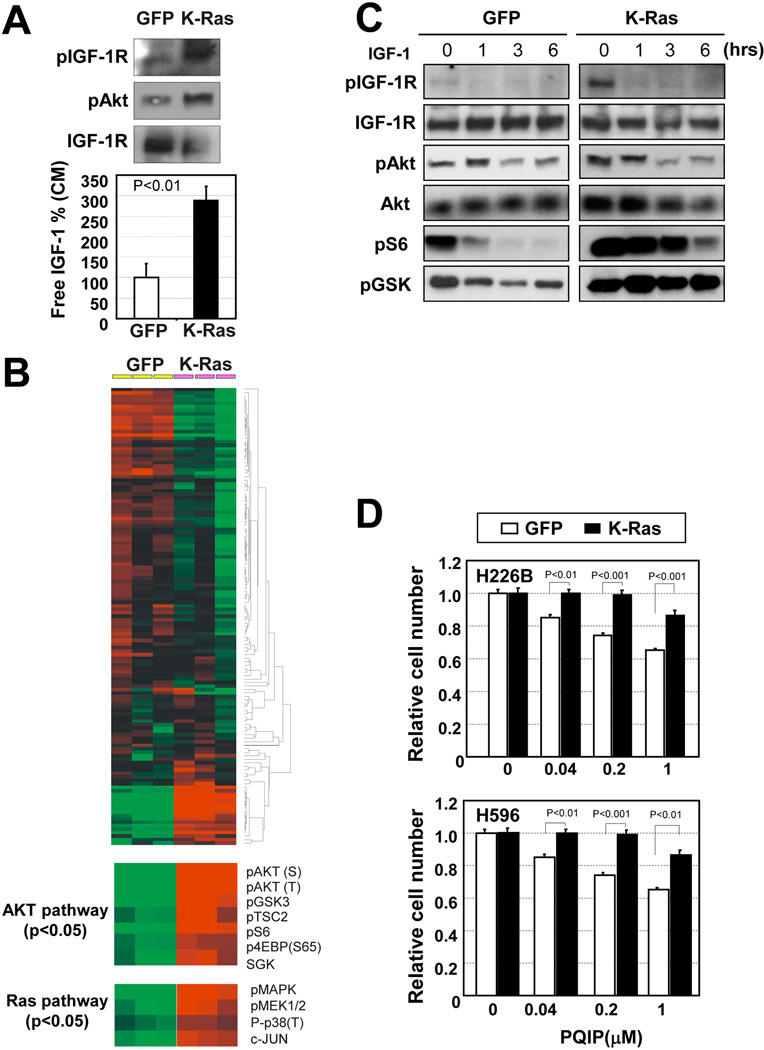
Given the strong positive correlation between IGF-1R activation and K-Ras mutation in the human NSCLC TMA and the inverse correlation between PQIP sensitivity and K-Ras mutation in NSCLC cell lines, we further assessed the role of K-Ras mutation in the IGF-1R pathway and PQIP sensitivity in H226B and H596 cells in which GFP or mut K-Ras had been transduced by retroviral infection. H226B-K-Ras cells showed higher levels of pIGF-1R and pAkt and lower levels of IGF-1R than those in H226B-GFP cells (Fig. 3A). We also observed that H226B-K-Ras cells produced more IGF-1 than H226B-GFP cells did (Fig. 3A). To characterize further molecular sequelae triggered by mut K-Ras, we performed a reverse-phase protein array Unsupervised hierarchical clustering analyses demonstrated that the PI3K/Akt and Ras/MAPK pathways were activated by mut K-Ras (Fig. 3B). Although PQIP treatment decreased pIGF-1R/IR and pAkt levels in both cell lines, phosphorylation of the downstream mediators of Akt, including pS6, and pGSK, was efficiently inhibited by PQIP treatment in H226B-GFP cells but not in H226B-K-Ras cells (Fig. 3C). Furthermore, H226B-K-Ras and H596-K-Ras cells were significantly less sensitive to PQIP treatment than the control cells were (Fig. 3D), suggesting that IGF-1R signaling is enhanced by mut K-Ras; however, K-Ras mutation abrogates NSCLC cell sensitivity to PQIP by activating downstream signaling, including p70S6K
Figure 3. Mut K-Ras is a determining factor of IGF-1R TKI sensitivity of NSCLC cells.
(A) Secreted IGF-1 level from NSCLC cells was enhanced by mut K-Ras. H226B-GFP or H226B-K-Ras cells (H226B cells stably transduced with retrovirus expressing GFP or mut K-Ras, respectively) were cultured in 1% FBS for 3 days. The conditioned media (CM) from H226B-GFP and H226B-K-Ras cells were applied to an IGF-1 ELISA. (B) Hierarchical clustering of protein expression data from H226B-GFP and H226B-K-Ras cells (n = 3 each). Reverse-phase protein array data are shown for 131 protein features, The data are presented in a matrix format: rows represent individual protein features, and columns represent individual samples. Each cell in the matrix represents the expression level of a protein feature in an individual cell sample. Red and green reflect relatively high and low expression levels, respectively. A total of 50 proteins showed differences with P < 0.05 by Student’s t test; of these, the molecules related to Akt signaling and Ras/MAPK signaling are shown in the bottom panels. The data were analyzed by using the Cluster and TreeView programs. (C) Molecular changes of the IGF-1R/Akt pathway by mut K-Ras and PQIP. H226B cells transfected with GFP or mut K-Ras were treated with PQIP (1 µM), and the cells were harvested at the indicated time point. (D) Mut K-Ras enhanced the resistance of NSCLC cells to PQIP. Relative cell survival after PQIP treatment for 3 days is shown. Mut K-Ras blunted the sensitivity of 226B and H596 cells to PQIP. Columns, means of 3 independent experiments; bars, standard deviation.
Targeting MEK Overrides the Resistance of mut K-Ras Cells to IGF-1R TKI
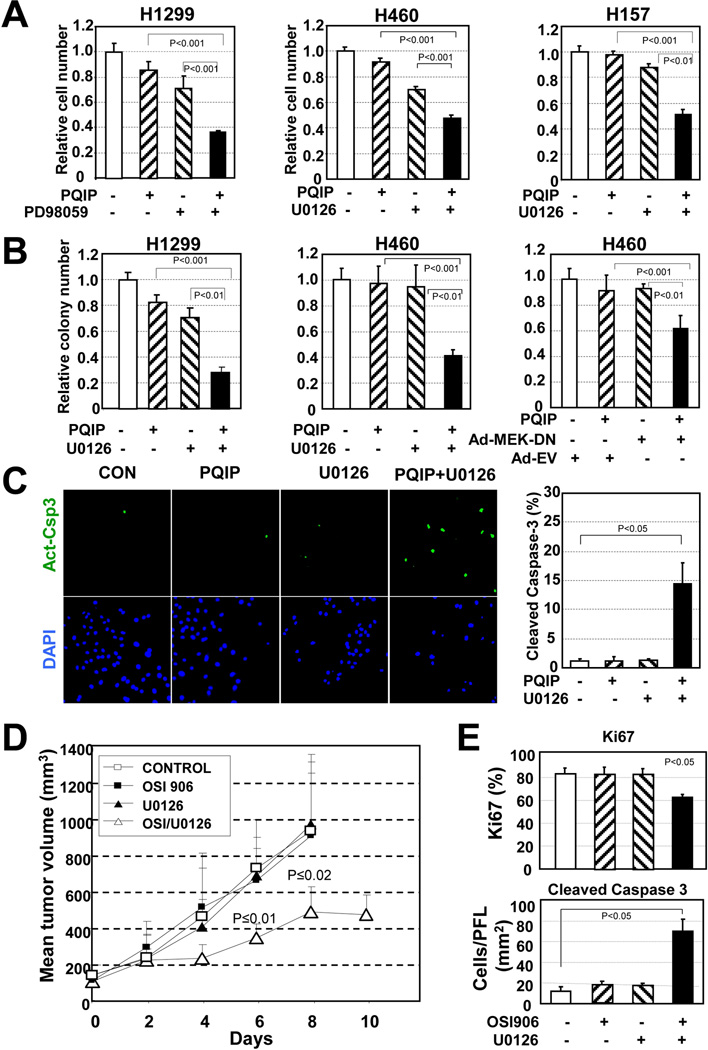
Because p70S6K is known to be activated by the MEK/Erk pathway,27 which can be constitutively activated by K-Ras mutation, we determined whether inactivation of MEK would restore the antitumor effects of PQIP or OSI-906 (a PQIP derivative currently undergoing a phase I clinical trial in our institute in NSCLC cell lines carrying mut K-Ras. We found that co-targeting of MEK, either with a small-molecule MEK inhibitor (PD98059 or U0126) or with adenovirus expressing the dominant-negative form of MEK (Ad-MEK-DN), significantly enhanced the effects of PQIP on cell viability (Fig. 4A) and anchorage-independent colony-forming ability (Fig. 4B) in representative mut K-Ras, resistant cell lines. Furthermore, the percentage of apoptotic cells was significantly increased by the combined treatment (Fig. 4C). These results suggest that inactivation of MEK augments the apoptotic activities PQIP in NSCLC cells carrying mut K-Ras. We finally evaluated the combined effects of OSI-906 and U0126 in vivo. The mice treated with vehicle or OSI-906 alone showed similar H226B K-Ras tumor growth (Fig. 4D). Pharmacologic inhibition of MEK by administration of U0126 dramatically augmented the effects of OSI-906 on the growth of the tumors. On day 8 after the first dose, the mean tumor volume for mice that received combined OSI-906 and U0126 was significantly smaller than the mean tumor volume for mice that received vehicle, OSI-906 alone, or U0126 alone. IHC staining of Ki67 and cleaved caspase-3 in the tumors demonstrated that the combined treatment induced a decrease in cell proliferation in association with an increase in cell apoptosis in vivo (Fig. 4E). Taken together, these findings underscore the pivotal role of activation of the MEK/Erk pathway through K-Ras mutation in the primary resistance of NSCLC cells to IGF-1R TKIs.
Figure 4. Co-targeting of IGF-1R and Ras signaling overrides the resistance to IGF-1R TKI driven by the K-Ras mutation in vitro and in vivo.
(A) Relative cell survival of mut Ras, resistant NSCLC cells after treatment with an IGF-1R TKI (PQIP, 5 µM), a MEK inhibitor (PD98059 [10 µM] or U0126 [2 µM or 1 µM]), or both. (B) Effect of combined IGF-1R and MEK inhibition on anchorage-independent colony-forming ability of NSCLC cells with mut Ras. The indicated NSCLC cells seeded in soft agar were treated with PQIP (1 µM), U0126 (2 µM), or both. Ad-MEK-DN, dominant negative form MEK expressing adenovirus ; Ad-EV, empty adenovirus. The relative colony numbers are shown. (C) Synergistic apoptotic effect of combined treatment with PQIP (5 µM) and U0126 (1 µM) on apoptosis of mut K-Ras-H157 NSCLC cells. The active form of caspase-3 was stained, and the percentage of apoptotic cells is plotted. (D) Mice bearing H226B-K-Ras xenograft tumors (2 tumors per mouse, 4 or 5 mice per group) were treated with vehicle (control), OSI-906 (40 mg/kg, once a day), U0126 (4 mg/kg, every other day), or both OSI-906 and U0126 as indicated. Day 0 represents the first day of drug treatment. Data are means and 95% confidence intervals. (E) IHC staining of the xenograft tumors in (D) with Ki67 and cleaved caspase-3 antibody was performed, and results are shown for at least 4 tumors in each group.
DISCUSSION
In the present study, we elucidate potential predictive markers of response of NSCLC cells to IGF-1R TKIs. We show that: 1) the expression of IGF-1R/IR in NSCLC specimens are positively associated with a history of TS, squamous cell carcinoma, wt EGFR, and mut K-Ras; 2) somatic mutation of EGFR, which confers addiction to the EGFR signaling pathway, induces a lack of primary response to IGF-1R TKIs in NSCLC cells; and 3) K-Ras mutation causes increased production of IGF-1 and activation of the IGF-1R pathway but induces resistance to IGF-1R TKIs. Moreover, our findings provide a proof of principle that targeted inactivation of IGF-1R by a TKI, in combination with MEK inhibition, can achieve a favorable outcome in the treatment of NSCLC patients with a history of TS and mut K-Ras.
Several preclinical and clinical studies have shown encouraging therapeutic efficacy of EGFR TKI in NSCLC with mut EGFR;2–3 however, the limited response rates to EGFR TKIs underscore the need to develop effective treatment strategies for patients with wt EGFR. Targeting the IGF-1R pathway is one emerging strategy. The two major approaches are small-molecule IGF-1R TKIs and anti-IGF-1R monoclonal antibodies. However, limited data are available about predictors of sensitivity to the anti-IGF-1R approaches. In this study, we identified predictors that could be used in clinical trials of IGF-1R TKIs in NSCLC patients. Previous studies have shown high levels of IGF-1R expression in squamous cell carcinoma histology28. By analyzing a TMA of specimens from 354 patients with NSCLC, we extended this observation by showing that high levels of pIGF-1R/IR in patients with squamous cell carcinoma. These data suggest that squamous cell carcinoma may be more sensitive to IGF-1R TKIs than lung adenocarcinoma is. However, previous reports and our current results show that tumor histology is not a predictive marker of response to IGF-1R-targeted strategies. We also observed significantly elevated pIGF-1R/IR levels in patients with a history of TS, those with mut K-Ras, and those with wt EGFR, all of which have been strongly associated with poor response to EGFR TKIs.
Numerous studies have suggested that human cancer cells can be highly dependent on single or multiple pathways that are overly activated, conferring tumorigenic potential,29–31 and successful anticancer therapeutic strategies would rely on the selection of patients harboring tumors that rely on those pathways for cell growth and survival. Our previous and current findings show that transformed lung epithelial cell lines induced by TS components had an elevated expression of pIGF-1R/IR and were sensitive to the molecularly targeted strategies against the IGF-1R system.32–33 TS components such as NNK have been shown to induce genetic changes in p53 and PTEN, which regulate IGF-2 and IGF-1R expression.34–35 NNK can also induce phosphorylation and degradation of p53 and inactivation of PTEN via activation of Akt.40 Although we did not have mechanistic evidence for TS-induced activation of IGF-1R/IR signaling in lung carcinogenesis, impact of the IGF-1R pathway in cell proliferation and survival suggested that targeting IGF-1R could be an effective therapeutic strategy for NSCLC patients with TS history. This notion and our subsequent findings, including (1) the characteristics of patients with NSCLC harboring elevated pIGF-1R/IR levels were negatively correlated with those of patients harboring EGFR mutation, and (2) PQIP treatment effectively inhibited stimulation of the IGF-1R pathway but had little antitumor activity in mut EGFR–expressing NSCLC cells, led us to hypothesize that a history of TS and EGFR mutation are predictive biomarkers for no responsiveness to IGF-1R TKIs. However, we found that only a subset of human NSCLC cell lines with high pIGF-1R/IR levels and wt EGFR were sensitive to PQIP treatment. These observations suggest that EGFR mutation is not a predictive marker to response to IGF-1R TKI-based therapies.
Considering the potential mechanisms of cross-talk between EGFR and IGF-1R signaling,19, 36–38 inhibition of IGF-1R signaling could have been compensated for by enhanced activation through EGFR. However, NSCLC cells expressing mut Ras did not exhibit significantly enhanced sensitivity in response to co-targeting of IGF-1R and EGFR by treatment with PQIP and the EGFR TKI erlotinib, whereas the same regimen significantly reduced cell viability in a subset of head and neck squamous cell carcinoma cell lines carrying wt Ras (data not shown). It has been suggested that sensitivity of NSCLC cells to TKIs of IGF-1R and EGFR, either alone or their combination, is determined by the epithelial-to-mesenchymal transition (EMT)36, 39. However, EMT status was not a consistent predictive marker for insensitivity to antagonism against IGF-1R or to co-targeting IGF-1R and EGFR36. These findings indicate the involvement of additional biomolecules that differentiate the NSCLC cell response to IGF-1R TKIs.
Our current findings from several in vitro and in vivo experiments indicate that mut K-Ras differentiates the response to IGF-1R inhibitors. In the current study, we found evidence that activation of the IGF-1R pathway is correlated with K-Ras mutation, which may increase IGF-1 production, as shown by significantly higher levels of IGF-1 in the conditioned media from H226B cells harboring mut K-Ras compared with those harboring wt K-Ras (Fig. 3A). Therefore, K-Ras mutation could be a driving force for activation of the IGF-1R pathway and may thus be a predictive marker of sensitivity to IGF-1R blocking. However, our subsequent results clearly show that mut K-Ras is a poor predictive marker of the therapeutic efficacy of the drugs: (1) mut K-Ras lead increased resistance to PQIP in many assay systems, and (2) the inactivation of K-Ras or MEK by genomic approaches (siRNA targeting K-Ras, Ad-MEK-DN) or pharmacologic approaches (PD98059, U0126) induced antitumor activity of IGF-1R TKIs (PQIP, OSI-906) in vitro and in vivo in mut K-Ras cell lines. These findings highlight the need for stratification of patients on the basis of K-Ras mutation, in addition to history of TS and EGFR mutation, when an IGF-1R–targeted therapeutic regimen is considered in clinical trials.
In summary, this study characterizes potential predictive markers of actions of IGF-1R TKIs. Our findings show that activation of IGF-1R/IR is mutually exclusive with activation of EGFR and is associated with TS in NSCLC, suggesting that transformed lung epithelial cells and NSCLC cells are dependent on IGF-1R/IR signaling for survival and sustained proliferation. However, we also provide evidence for the first time that mutation in K-Ras is associated with activation of IGF-1R and the development of physiologically redundant signaling in patients with NSCLC, implicating mut K-Ras as an important predictive marker to optimize the clinical efficacy of the IGF-1R–targeting strategy. Further investigation is warranted into the discovery of the predictive biomarkers for IGF-1R-targeted therapy and the exact mechanism of synergy between IGF-1R TKIs and MEK inhibitors
Acknowledgments
We thank OSI Pharmaceuticals for providing PQIP, OSI-906, and erlotinib. This work was supported in part by the National Institutes of Health, through grants R01 CA-109520-01 and CA-100816 (to HYL) and through MD Anderson’s Cancer Center Support Grant (CA-016672), and in part by the Department of Defense, through grants W81XWH-04-1-0142 VITAL and W8XWH-06-1-0303 BATTLE (to WKH)
Footnotes
The authors declare that they have no competing financial interests.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 6.Kozma LM, Weber MJ. Constitutive phosphorylation of the receptor for insulinlike growth factor I in cells transformed by the src oncogene. Mol Cell Biol. 1990;10(7):3626–3634. doi: 10.1128/mcb.10.7.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppola D, Ferber A, Miura M, et al. A functional insulin-like growth factor I receptor is required for the mitogenic and transforming activities of the epidermal growth factor receptor. Mol Cell Biol. 1994;14(7):4588–4595. doi: 10.1128/mcb.14.7.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodt P, Samani A, Navab R. Inhibition of the type I insulin-like growth factor receptor expression and signaling: novel strategies for antimetastatic therapy. Biochem Pharmacol. 2000;60(8):1101–1107. doi: 10.1016/s0006-2952(00)00422-6. [DOI] [PubMed] [Google Scholar]
- 9.Baserga R. The insulin-like growth factor-I receptor as a target for cancer therapy. Expert Opin Ther Targets. 2005;9(4):753–768. doi: 10.1517/14728222.9.4.753. [DOI] [PubMed] [Google Scholar]
- 10.Renehan AG, Atkin WS, O'Dwyer ST, Shalet SM. The effect of cigarette smoking use and cessation on serum insulin-like growth factors. Br J Cancer. 2004;91(8):1525–1531. doi: 10.1038/sj.bjc.6602150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Spitz MR, Mistry J, Gu J, Hong WK, Wu X. Plasma levels of insulin-like growth factor-I and lung cancer risk: a case-control analysis. J Natl Cancer Inst. 1999;91(2):151–156. doi: 10.1093/jnci/91.2.151. [DOI] [PubMed] [Google Scholar]
- 12.Papadimitrakopoulou VA, Brown EN, Liu DD, et al. The prognostic role of loss of insulin-like growth factor-binding protein-3 expression in head and neck carcinogenesis. Cancer Lett. 2006;239(1):136–143. doi: 10.1016/j.canlet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Kim WY, Jin Q, Oh SH, et al. Elevated epithelial insulin-like growth factor expression is a risk factor for lung cancer development. Cancer Res. 2009;69(18):7439–7448. doi: 10.1158/0008-5472.CAN-08-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallis AG, Serfass L, Dziadziusko R, et al. Targeted therapies in the treatment of advanced/metastatic NSCLC. Eur J Cancer. 2009;45(14):2473–2487. doi: 10.1016/j.ejca.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Kim WY, Chang DJ, Hennessy B, et al. A novel derivative of the natural agent deguelin for cancer chemoprevention and therapy. Cancer Prev Res (Phila) 2008;1(7):577–587. doi: 10.1158/1940-6207.CAPR-08-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim WY, Kim MJ, Moon H, et al. Differential Impacts of Insulin-Like Growth Factor-Binding Protein-3 (IGFBP-3) in Epithelial IGF-Induced Lung Cancer Development. Endocrinology. 2011 doi: 10.1210/en.2010-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim W-YJQ, Oh S-H, Kim ES, Yang YJ, Lee DH, Feng L, Behrens C, Prudkin L, Miller YE, Lee JJ, Lippman SM, Hong WK, Wistuba II, Lee H-Y. Elevated epithelial insulin-like growth factor expression is a risk factor for lung cancer development. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-08-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12(5):1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 19.Morgillo F, Woo JK, Kim ES, Hong WK, Lee HY. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. 2006;66(20):10100–10111. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- 20.Woo JK, Choi DS, Tran HT, Gilbert BE, Hong WK, Lee HY. Liposomal encapsulation of deguelin: evidence for enhanced antitumor activity in tobacco carcinogen-induced and oncogenic K-ras-induced lung tumorigenesis. Cancer Prev Res (Phila Pa) 2009;2(4):361–369. doi: 10.1158/1940-6207.CAPR-08-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HY, Moon H, Chun KH, et al. Effects of insulin-like growth factor binding protein-3 and farnesyltransferase inhibitor SCH66336 on Akt expression and apoptosis in non-small-cell lung cancer cells. J Natl Cancer Inst. 2004;96(20):1536–1548. doi: 10.1093/jnci/djh286. [DOI] [PubMed] [Google Scholar]
- 22.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugio K, Uramoto H, Ono K, et al. Mutations within the tyrosine kinase domain of EGFR gene specifically occur in lung adenocarcinoma patients with a low exposure of tobacco smoking. Br J Cancer. 2006;94(6):896–903. doi: 10.1038/sj.bjc.6603040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyooka S, Tokumo M, Shigematsu H, et al. Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res. 2006;66(3):1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]
- 25.Ji QS, Mulvihill MJ, Rosenfeld-Franklin M, et al. A novel, potent, and selective insulin-like growth factor-I receptor kinase inhibitor blocks insulin-like growth factor-I receptor signaling in vitro and inhibits insulin-like growth factor-I receptor dependent tumor growth in vivo. Mol Cancer Ther. 2007;6(8):2158–2167. doi: 10.1158/1535-7163.MCT-07-0070. [DOI] [PubMed] [Google Scholar]
- 26.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91(14):1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 27.Deguil J, Perault-Pochat MC, Chavant F, Lafay-Chebassier C, Fauconneau B, Pain S. Activation of the protein p7OS6K via ERK phosphorylation by cholinergic muscarinic receptors stimulation in human neuroblastoma cells and in mice brain. Toxicol Lett. 2008;182(1–3):91–96. doi: 10.1016/j.toxlet.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y, Yao E, Shen R, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS ONE. 2009;4(10):e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haber DA, Bell DW, Sordella R, et al. Molecular targeted therapy of lung cancer: EGFR mutations and response to EGFR inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:419–426. doi: 10.1101/sqb.2005.70.043. [DOI] [PubMed] [Google Scholar]
- 30.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68(9):3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 80. [DOI] [PubMed] [Google Scholar]
- 31.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3(8):448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 32.Klein-Szanto AJ, Iizasa T, Momiki S, et al. A tobacco-specific N-nitrosamine or cigarette smoke condensate causes neoplastic transformation of xenotransplanted human bronchial epithelial cells. Proc Natl Acad Sci U S A. 1992;89(15):6693–6697. doi: 10.1073/pnas.89.15.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun KH, Kosmeder JW, 2nd, Sun S, et al. Effects of deguelin on the phosphatidylinositol 3-kinase/Akt pathway and apoptosis in premalignant human bronchial epithelial cells. J Natl Cancer Inst. 2003;95(4):291–302. doi: 10.1093/jnci/95.4.291. [DOI] [PubMed] [Google Scholar]
- 34.Ohlsson C, Kley N, Werner H, LeRoith D. p53 regulates insulin-like growth factor-I (IGF-I) receptor expression and IGF-I-induced tyrosine phosphorylation in an osteosarcoma cell line: interaction between p53 and Sp1. Endocrinology. 1998;139(3):1101–1107. doi: 10.1210/endo.139.3.5832. [DOI] [PubMed] [Google Scholar]
- 35.Yi HK, Kim SY, Hwang PH, et al. Impact of PTEN on the expression of insulin-like growth factors (IGFs) and IGF-binding proteins in human gastric adenocarcinoma cells. Biochem Biophys Res Commun. 2005;330(3):760–767. doi: 10.1016/j.bbrc.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 36.Buck E, Eyzaguirre A, Rosenfeld-Franklin M, et al. Feedback mechanisms promote cooperativity for small molecule inhibitors of epidermal and insulin-like growth factor receptors. Cancer Res. 2008;68(20):8322–8332. doi: 10.1158/0008-5472.CAN-07-6720. [DOI] [PubMed] [Google Scholar]
- 37.Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13(9):2795–2803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 38.Barnes CJ, Ohshiro K, Rayala SK, El-Naggar AK, Kumar R. Insulin-like Growth Factor Receptor as a Therapeutic Target in Head and Neck Cancer. Clin Cancer Res. 2007;13(14):4291–4299. doi: 10.1158/1078-0432.CCR-06-2040. [DOI] [PubMed] [Google Scholar]
- 39.Yauch RL, Januario T, Eberhard DA, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11(24 Pt 1):8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]