Abstract
17β-estradiol (E2) plays important roles in functions of many tissues. E2 effects are mediated by estrogen receptor (ER) α and β. ERs regulate transcriptions through estrogen responsive element (ERE)-dependent and ERE-independent modes of action. ER binding to ERE constitutes the basis of the ERE-dependent pathway. Direct/indirect ER interactions with transcription complexes define the ERE-independent signaling. ERs share functional features. Ligand-bound ERs nevertheless induce distinct transcription profiles. Live cell imaging indicates a dynamic nature of gene expressions by highly mobile ERs. However, the relative contribution of ER mobility at the ERE-independent pathway to the overall kinetics of ER mobility remains undefined. We used fluorescent recovery after photo-bleaching (FRAP) approach to assess the ligand-mediated mobilities ERE binding-defective ERs, EREBD. The decrease in ERα mobility with E2 or the selective ER modulator 4-hydroxyl-tamoxifen (4HT) was largely due to the interaction of the receptor with ERE. Thus, ERα bound to E2 or 4HT mediates transcriptions from the ERE-independent pathway with remarkably fast kinetics that contributes fractionally to the overall motility of the receptor. The antagonist ICI 182,780 immobilized ERαs. The mobilities of ERβ and ERβEBD in the presence of ligands were indistinguishable kinetically. Thus, ERβ mobility is independent of the nature of ligands and the mode of interaction with target sites. Chimeric ERs indicated that the carboxyl-termini are critical regions for the subtype-specific mobility. Therefore, while ERs are highly mobile molecules interacting with target sites with fast kinetics, an indication of the hit-and-run model of transcription, they differ mechanistically to modulate transcriptions.
Keywords: Ligand, Nuclear Mobility, FRAP, ERE Binding, Estrogen Receptor α, Estrogen Receptor β
INTRODUCTION
17β-estradiol (E2) plays critical roles in many physiological and pathophysiological processes of a wide range of tissues (Huang, et al. 2005a; Zhao, et al. 2010). E2 effects are primarily mediated by transcription factors, estrogen receptor (ER) α and β that convey E2 signaling through estrogen responsive element (ERE)-dependent and -independent pathways.
Kinetic biochemical assays indicate that the unliganded ERα interacts, albeit inefficiently, with EREs cyclically with a time scale of 20 minutes (Metivier, et al. 2003; Reid, et al. 2003; Shang, et al. 2000). The binding of E2 to ERα leads to a structural reorganization that increases the stability of the ERα dimer (Tamrazi, et al. 2002) and the affinity of ERα to coregulatory proteins (Tamrazi, et al. 2005; Yi, et al. 2002b). The interaction of E2-ERα with ERE extends the duration of promoter engagement to 40–60 minutes (Metivier et al. 2003; Reid et al. 2003; Shang et al. 2000). This is due to the sequential recruitment of preformed co-regulator complexes for initiation and the subsequent dissociation of complexes from promoter for termination of transcription (Metivier et al. 2003; Reid et al. 2003; Shang et al. 2000). This episodic ERα-ERE engagement led to the transcriptional ratchet model that suggests ordered and directional events for ERE-driven gene expressions.
Fluorescent protein technologies together with quantitative live cell imagining also indicate a dynamic transcriptional regulation (Sharp, et al. 2006; Stenoien, et al. 2000a; Stenoien, et al. 2001a; Stenoien, et al. 2001b; Zwart, et al. 2010). These approaches demonstrated that the unliganded ERα exhibit rapid rates of exchange with chromatin, residence time measured in milliseconds. Although the binding of E2 to ERα decreases the mobility of the receptor, the exchange still occurs in seconds, in a clear contrast with longer cycling times determined by kinetic biochemical assays. These fast interactions of ERα with promoters support the alternative hit-and-run model for transcription.
Along with E2, the activity of ERα is modulated by the selective estrogen receptor modulator (SERM) tamoxifen and antagonist ICI (McDonnell 1999). The binding of SERMs or antagonists to ERα alters the nuclear mobility and the ability of ERα to interact with co-regulators and chromatin (McDonnell 1999; Stenoien et al. 2001b; Yi et al. 2002b).
E2-ERs also regulate transcription by interacting with transcription factors (Kushner, et al. 2000; Safe 2001). This nuclear signaling route is called the ERE-independent signaling pathway, which participates in the fine-tuning of cellular responses by regulating the expression of a subset of estrogen responsive genes (Li, et al. 2008; Nott, et al. 2009). However, the underlying mechanisms or the kinetics of events at the ERE-independent signaling pathway remains poorly defined.
Although encoded by distinct gene, ERβ shares structural features with ERα reflected in similar mode of actions through signaling pathways (Huang, et al. 2011). ERβ, nevertheless, regulates transcription with distinct potency and profile in response to ligands at signaling pathways (Huang et al. 2005a; Zhao et al. 2010). Since the nuclear mobility is the sum of ER actions at target sites on chromatin, we addressed how ligands affect the nuclear mobility of an ERE binding defective ER variant (ERαEBD or ERβEBD) that functions exclusively at the ERE-independent pathway to obtain initial insights into mechanisms of ER-mediated gene expressions. To address this issue, we used green fluorescence protein (GFP) fusion-ERs and fluorescent recovery after photo-bleaching (FRAP) approach.
We found that the ligand-mediated nuclear mobility of ERα largely reflects the ability of receptor to interact with ERE, whereas the mobility of ERβ is independent of nature of ligands and the ability of ERβ to bind to ERE. Thus, ERs are highly mobile molecules interacting with target sites with fast kinetics, an indication of the hit-and-run model of transcription, they differ mechanistically to modulate transcriptions.
MATERIALS AND METHODS
Plasmids
The expression vectors bearing human ERα and ERβ cDNAs encoding 595 and 530 amino-acid long receptors, respectively, and the cDNA encoding the designer transcription factor PPVV were described previously (Huang, et al. 2004; Yi, et al. 2002a). The AF2 mutant of ERα contains a three amino acid replacement (D538A, E542A and D545A) that blocks the ligand-dependent activation function (AF2) of ERα (Sathya, et al. 2002; Tzukerman, et al. 1994; Yi et al. 2002a). We initially used an AF2 mutant of ERβ that contains analogous mutations to that of ERα as we described previously (Yi et al. 2002a). However the presence of the GFP at the amino-terminus of this mutant render the receptor toxic to cells as they died prior to experimentation. To circumvent this problem, we used a point mutation that changes only the Glu residue at position 493 to a Lys in ERβ that prevents AF2 (An, et al. 1999). The ERE binding defective ERs were described previously (Li et al. 2008; Nott et al. 2009). The ERE binding defective ERα (ERαEBD) contains Ala, Ala and Glu residues at position 203, 204 and 211, respectively, that replace Glu, Gly and Arg at the corresponding positions in the DNA recognition helix of the first zinc finger critical for ERα-ERE interactions (Nott et al. 2009). The replacement of Glu and Gly at positions 167 and 168, respectively, in the DBD of ERβ with Ala residues generates the ERE binding defective ERβ (ERβEBD) (Li et al. 2008). The chimeric ERαNβC or ERβNαC, generated by genetically exchanging sequences that encode the entire amino terminal region of ERβ or ERα with that of ERα or ERβ, were also described previously (Yi et al. 2002a). cDNAs also contain sequences that encode a Flag epitope at the amino-terminus.
For the engineering of Green Fluorescence Protein (GFP) fusion proteins, a restriction enzyme site was engineered at the 5′ of the start codon of ER cDNAs using an overlapping PCR. The engineered cDNAs were inserted into the 3′ end of the reading frame of the GFP- cDNA in the pAcGFP-C1 expression vector (Clontech, Mountain View, CA) with appropriate restriction enzyme sites. For comparative analysis of GFP-ERs in some biochemical assays, we also generated a GFP cDNA with sequences that encode a Flag epitope at the amino- terminus of the protein. All constructs were sequenced to ensure the fidelity of encoding sequences.
We assessed the effect of ligands on ERE-driven gene expressions using reporter vectors that emulate the ERE-dependent signaling pathway. For the simple TATA box promoter, we used the reporter pGL3 (Promega Corp., Madison, WI) plasmids bearing a TATA box promoter with single (ERE) or two EREs (2XERE) (Sathya et al. 2002; Yi et al. 2002a). We also used the pGL3 reporter vector bearing the promoter of the trefoil factor 1, TFF1, or pS2, gene (TFF1-Luc) (Yi et al. 2002a). To simulate the ERE-independent signaling, we used an MMP1-Luc reporter plasmid that bears a fragment of the proximal promoter of the matrix metallopeptidase 1, MMP1, gene with single AP1 response element (Huang et al. 2004; Li, et al. 2004; Webb, et al. 1995) or an RARA-Luc reporter vector derived from the proximal promoter of the retinoic acid receptor α, RARA, gene that contains two GC-boxes (Huang et al. 2004; Li et al. 2004; Sun, et al. 1998). In all reporter vectors, promoters drive the expression of the firefly luciferase cDNA as the reporter enzyme. A reporter vector driving the expression of the Renilla luciferase cDNA (Promega) was used to assess transfection efficiency (Huang et al. 2004; Yi et al. 2002a). The ratio of the firefly/Renilla luciferase activities of the cell lysate was determined using a dual luciferase assay kit (Promega Corp.) to obtain the relative luciferase activity.
The Flag M2 antibody, ERα-specific H-222 and ERβ-specific antibody D7N were purchased from Sigma-Aldrich, (St. Louis, MO), Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Zymed Laboratories Inc. (San Francisco, CA), respectively.
17β-estradiol (E2) and 4-hyroxyl-tamoxifen (4HT) were purchased from Sigma-Aldrich (St. Louis, MO). Imperial Chemical Industries 182,780 (ICI) was obtained from Tocris Biosciences (Ellisville, MO).
Restriction and DNA modifying enzymes were purchased from New England Bio-Labs (Beverly, MA) and Invitrogen Corp., (Carlsbad, CA).
Transient transfections
Transient transfections for simulated ERE-dependent and ERE-independent pathways were accomplished as described previously (Huang et al. 2004; Li et al. 2004; Yi et al. 2002a). Transfected cells were treated without or with 10−9 M E2 in the absence or presence of 10−7 M 4HT, and/or 10−7 M ICI for 24h to examine the effects of ligands on ER-mediated transcriptional responses from the ERE-dependent and ERE-independent signaling pathways.
In situ E2 binding assay
To assess the synthesis and function of GFP fusion ERs in transfected cells, we used an in situ E2 binding assay described previously (Huang, et al. 2005b; Li et al. 2008). Briefly, transiently transfected cells in 48-well tissue culture plates were incubated with 10−7 M of [2,4,6,7,16,17-3H] 17β-estradiol (118 Ci/mmol, NEN Life Sciences, Boston, MA) in the absence or presence of 10−6 M 4HT or ICI for 1h. Cells were then washed extensively with PBS, collected and radioactivity remained in cells was measured in a scintillation counter. By the use of in situ ligand binding assay, we estimate that transiently transfected HeLa and MDA-MB-231 cells synthesize about 5.5- and 4-fold, respectively, more ERα compared to MCF-7 cells, a breast adenocarcinoma cell line that endogenously synthesizes ERα (Eckert, et al. 1984).
In situ competition for ERE binding assays
In situ competition for ERE binding assay (Huang et al. 2005b) was used to assess the ability of GFP-ERs to interact with ERE in situ. This assay based on the interference of a constitutively active potent activator (PPVV)-mediated transcription from a single ERE-driven promoter construct by unliganded or ligand bound ERs. The extent of interference is then taken as an indication of ER-ERE interactions (Huang et al. 2005b). In brief, cultured cells in 48-well tissue wells were transfected with 125 ng simple TATA box promoter with one ERE and 300 ng expression vector carrying the PPVV cDNA together with 0, 75, 150, or 300 ng expression vector containing the cDNA for an ER. Appropriate amounts of the parent expression vector were added into a given reaction to equalize the total amount of plasmid DNA. A vector bearing the Renilla luciferase cDNA was used as an internal control in the amount of 0.5 ng to normalize the transfection efficiency. Four hours after transfection, cells were maintained in fresh medium supplemented with 10% CD-FBS in the absence or presence of 10−9 M E2, 10−7 M 4HT or ICI for 24 h.
Western Blot (WB) and Electrophoretic Mobility Shift Assay (EMSA)
Transiently transfected cells with expression vectors in 6-well tissue culture plates were maintained for 24h. Cell extracts (10 μg) were subjected to WB and EMSA as detailed previously (Li et al. 2008; Nott et al. 2009). For WB, proteins were probed with the horseradish peroxidase (HRP) conjugated monoclonal Flag antibody (M2-HRP, Sigma-Aldrich). We also used HC-20 and D7N antibodies (Santa Cruz Inc.) specific to ERα and ERβ respectively to detect receptor proteins, which were visualized with a second antibody conjugated with HRP. The ECL-Plus Western Blotting kit (GE Life Sciences) was used for the detection of receptor proteins. For EMSA, we used the Flag or a receptor-specific antibody to assess the specificity of ER-ERE interactions. Images from WB and EMSA were analyzed by PhosphorImager (Storm 860, GE Life Sciences, Piscataway, NJ) and were quantified with ImageQuant (GE Life Sciences).
We also examined the effects of ER ligands on the detergent extractability and intracellular level of receptor proteins with WB. Cells maintained in 6-well tissue culture plates in 10% CD-FBS containing media for 24h were then transiently transfected for 24h. Cells were subsequently incubated with fresh medium supplemented with or without 10−9 M E2, 10−7 M 4HT or ICI for 1h. At the termination of an experiment, cells were collected, pelleted and subjected to protein extraction using 50 μl of a high salt extraction buffer, HSB, (400 mM KCl, 20% glycerol, 2mM DTT, 1mM PMSF, 1/14 (v/v) protease inhibitor cocktail, Roche Diagnostics, Indianapolis, IN) or RIPA buffer (0.5% sodium deoxycholate, 1% Igepal CA-630, 0.1% SDS, 2mM DTT, 1mM PMSF, 1/14 (v/v) protease inhibitor cocktail). After the HSB or RIPA extraction, the remaining pellet was also subjected to 50 μl of 1x Laemmli Buffer, LB, (60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) to extract insoluble protein aggregates. In addition, we used 100 μl of LB to obtain total cell lysate (TCL) by extracting both soluble and insoluble proteins. Ten μg total protein estimated with Nanodrop (ThermoScientific, Wilmington, DE) was subjected to SDS 10–18% PAGE.
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assay was described previously (Huang et al. 2005b). In brief, cells grown in 6-well tissue culture plates were co-transfected with expression vector and the reporter vector bearing the TATA box promoter with single ERE. Twenty-four hours after transfection, cells were incubated in fresh medium with or without 10−7 M E2, 4HT or ICI for 1h. Cells were then subjected to ChIP using the Flag-M2 antibody conjugated agarose beads (Sigma-Aldrich). The generation of a 366 bp PCR fragment by ChIP indicates the specificity of PCR reactions. Due to the difficulty of assessing the interaction of ER with EREs of endogenous genes by ChIP in transiently transfected cells, we used recombinant adenovirus infected MDA-MB-231 cells with which we previously carried out experiments to assess the interactions of E2-ERs with (Huang et al. 2005b), and transcriptional responses from (Huang et al. 2011; Li et al. 2008; Nott et al. 2009), the ERE of TFF1. MDA-MB-231 cells, 100,000 cells/well in 6 well-tissue culture plates, infected with recombinant adenoviruses in media with 10% CD-FBS for 48h. We used recombinant adenovirus bearing ERα, ERαEBD, ERβ or ERβEBD cDNA with sequences encoding a Flag epitope at the amino-terminus at 100, 150, 600 or 900 multiplicity of infection (MOI), respectively, together with the parent recombinant adenovirus bearing no cDNA at varying MOI to equalize the total amount of adenovirus, 900 MOI, used for infections. At these MOIs, the synthesis of ERs was comparable (Huang et al. 2011; Li et al. 2008; Nott et al. 2009). 48h after infections, cells were incubated in fresh medium supplemented without or with 10−9 M E2, 10−7 M 4HT or ICI for 1h. We also used 10−8 M ERα-selective propyl pyrazole triol (PPT) agonist or 10−8 M ERβ-selective diarylpropionitrile (DPN) agonist for 1h. At these concentrations, ligands maximally affected transcriptional responses from reporter constructs induced by ERs or E2-ERs (data not shown). Cells were collected and subjected to ChIP using M2-Flag antibody conjugated agarose beads (Sigma-Aldrich) as described (Huang et al. 2005b). The production of a 315 bp PCR product indicates specific ER-ERE interactions (Supplemental Data, Fig. 3A).
Live Cell microscopy and Fluoresence Recovery After Photo-bleaching (FRAP)
HeLa or MDA-MB-231 cells grown in 35-mm glass bottom coverslip dishes (MatTek Corp., Ashland, MA) in medium containing 10% CD-FBS without phenol-red for 24h. Cells were then transiently transfected with 1.5 μg of an expression vector bearing the GFP-fusion receptor cDNA. Twenty-four hours after transfections, cells were treated with or without various concentrations (10−10 M to 10−7 M) of E2 for 1h prior to FRAP analysis. We observed in preliminary studies that 10−9 M E2 maximally affected the intracellular mobility of both ERα and ERβ. Based on these findings, we used 10−9 M E2 in subsequent FRAP assays. We also treated cells with 10−7 M 4HT or ICI, a concentration that maximally affected the nuclear mobility of ERs in preliminary experiments. FRAP was performed using an Olympus FV1000 laser scanning confocal microscope containing a full stage incubator equilibrated to 37C housed at the URMC Confocal and Conventional Microscopy Core. Cells were imaged live using a 60x 1.4 NA oil immersion objective.
Cells were initially examined with differential interference contrast (DIC) to assess the cellular health. To prevent experimental artifacts due to over-expression of GFP-fusion ERs (Stenoien et al. 2001a; Stenoien et al. 2001b), cells with low fluorescence intensities (600–1500 arbitrary fluorescence unit) were selected for FRAP analysis. Photo-bleaching was accomplished using a tunnel ROI (region of interest) of the FRAP module. A tunnel ROI as a 9×9 pixel area (4.468 μm2) was used for all photo-bleaching experiments. It should be noted that the FRAP results were independent of the shape of ROI, as we obtained similar results from stripe bleaching in preliminary experiments (data not shown). A single z plane was bleached with the SIM scanner capabilities using the 405 nM laser set to 30% power for 0.2 second and simultaneously imaged in freerun (approximately 0.25 millisecond intervals) mode.
FRAP analysis was carried out with FV1000 Olympus post-processing software. Briefly, frames of the time-lapse data were moved to the point of photo-bleaching and graphs obtained for the bleached ROI. Another ROI at a size and fluorescence intensity that corresponded to those of the experimental ROI prior to photo-bleaching within the same cell was used as the control to assess the background and alterations in total cellular fluorescence due to photo-bleaching (Supplemental Data, Fig. 1). All data were exported to Excel for further analysis. The fluorescence intensity of the control ROI throughout the post-bleach period was used to normalize the recovery of bleached ROI by dividing the fluorescence value of the bleached ROI to that of the control ROI at each time point of imaging. Fluorescence is expressed as relative fluorescence units (RFU) where zero (0) is the fluorescence after photo-bleaching (time 0) and one (1) is the fluorescence of the bleached area reached to pre-bleach levels. Images were exported as Tagged Image File Format (tif) and movie (mov) files. Adobe Photoshop (Adobe Systems Inc., San Jose, CA) was used for image analysis. All experiments were carried out using 5 to 7 individual cells per experiment. Results were repeated for, at least, three independent times.
RESULTS
The synthesis and function of GFP-ERs in HeLa Cells
The interaction of ERα with permutations of a core palindromic DNA sequence 5′-GGTCAnnnTGACC-3′, or ERE as well as ERE half-sites (Ansari, et al. 2012; Kato, et al. 1995), constitutes the ERE-dependent signaling pathway (Huang et al. 2005a; Zhao et al. 2010). The recognition of an ERE by the ERα dimer is mediated by two zinc-binding motifs in each DBD monomer that fold to form a single functional unit (Schwabe, et al. 1993). Distinct residues particularly Glu203 and Gly204 in the DNA recognition helix of the first zinc finger of DBD of the human ERα are critical for DNA sequence discrimination (Schwabe et al. 1993) and also for binding to EREs (DeNardo, et al. 2007). We showed that changing Arg211, a conserved amino acid among nuclear hormone receptors critical for receptor-DNA interactions, to Glu211 together with Ala203 and Ala204 that replace Glu203 and Gly204, respectively, generates ERαEBD that functions only at the ERE-independent signaling pathway (Nott et al. 2009). Similarly, changing Glu167 and Gly168 in the first zinc finger motif of the DBD of the human ERβ to Ala also generated ERβEBD that regulates gene transcriptions exclusively through the ERE-independent signaling pathway (Bjornstrom and Sjoberg 2002; Li et al. 2008).
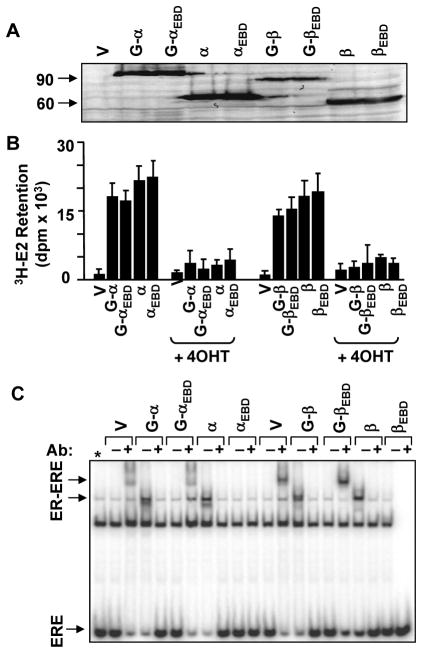
To examine the effects of ligands on the kinetics of nuclear movement of EREBD, we initially assessed the synthesis and biochemical features of GFP fusion receptors in comparison with the wild-type counterparts in transiently transfected HeLa cells derived from an ER-negative cervical carcinoma. Cellular extracts were subjected to WB using an antibody specific to the Flag epitope present at the amino-terminus of each receptor (Fig. 1A). Results revealed that HeLa cells synthesize GFP-ERs with expected molecular mass. The treatment of cells with a saturating concentration of 3H-E2 (10−7 M) showed that the radiolabeled E2 is retained in cells synthesizing a GFP-ER as observed in cells synthesizing an ER (Fig. 1B). The treatment of cells with 10−6 M 4HT or ICI (data not shown) effectively prevented the retention of 3H-E2. Thus, the presence of GFP at the amino-termini of ERs has little effect on the synthesis and the ligand binding abilities of the fusion receptors.
Fig. 1.
The synthesis of functional GFP-ERs in HeLa cells. A. The synthesis of ER species. HeLa cells were transiently transfected with an expression vector bearing none (V) or an ER cDNA with or without sequences encoding GFP (G) genetically conjugated to the 5′ of ER coding sequences. Constructs also contain sequences that encode for a Flag epitope present at the amino-terminus of the resulting protein. Cell extracts (10 μg) were subjected to WB using a horseradish peroxidase conjugated monoclonal Flag antibody. Molecular mass in KDa is indicated. B. In situ E2 binding assay. Twenty-four hour after transient transfections with an expression vector bearing none (V) or an ER cDNA with (G) or without GFP, HeLa cells were incubated in medium containing 10−7 M of 3H-E2 in the absence or presence of 10−6 M 4HT (+4HT) for 1h. The medium containing the radioactive 3H-E2 was removed and cells were extensively washed with PBS prior to dislodging. Radioactivity retained in cells was then quantified by scintillation counting. The graph represents the mean ± SEM of three independent experiments performed in duplicate. C. The interaction of ERs with ERE in vitro. Cell extracts (10 μg) of transfected cells were also subjected to electrophoretic mobility shift assay (EMSA) without (−) or with (+) a receptor-specific antibody (Ab). ERE specifies unbound and ER-ERE denotes ER-bound radiolabeled ERE. Asterisk denotes the free ERE lane. A representative result from a minimum of two independent experiments of WB or EMSA is shown.
To assess the interaction of GFP-ERs with ERE, we employed EMSA with cellular extracts from transiently transfected HeLa cells (Fig. 1C). Displaying similar mobilities to those of corresponding parent ERs, the GFP-ERα and GFP-ERβ retarded the migration of the radiolabeled ERE. Whereas GFP-ERαEBD, GFP-ERβEBD or the ERE binding defective ERs showed no binding.
To further ensure that GFP-ERs in response to ligands mimic the effects of the parent ERs on the transcription and ERE binding defective GFP-ERs are functional only at ERE-independent signaling pathways, we used reporter vectors with promoters emulating ERE-dependent and ERE-independent signaling routes (Supplemental Data, Fig. 2 & 3). For the simulated ERE-dependent signaling pathway, we used reporter plasmid bearing two EREs in tandem located at the upstream of the simple TATA box promoter (2XERE-Luc) or the proximal promoter region derived from the TFF1 gene (TFF1-Luc) bearing an ERE. ERα increased the activity of the reporter enzyme in response to a physiological concentration (10−9 M) of E2 from both promoters in transfected cells (Supplemental Data, Fig. 2A). Although the extent of activations was lower than those induced by ERα, GFP-ERα also increased the enzyme levels in response to E2. The treatment of cells with 10−7 M 4HT or 10−7 M ICI alone had little effect on transcriptional responses mediated by ERs or GFP-ERs. However, 4HT or ICI effectively countered the effect of E2 on the reporter enzyme when cells were co-treated. Whereas, ERαEBD or GFP-ERαEBD did not affect the enzyme activity whether or not cells were exposed to ligands alone or in combination. Similarly ERβ or GFP-ERβ, but not the ERE binding defective counterparts, augmented the activity of the reporter enzyme only in the presence of E2, which was blocked by the co-treatment of cells with 4HT or ICI (Supplemental Data, Fig. 3A). Thus, GFP-ERs, but not the ERE binding defective ERs with or without GFP, in response to ligands mimic the effects of the parent ERs on the transcription of the reporter enzyme mediated through the ERE-dependent signaling pathway.
We previously showed that the MMP1 and RARA genes are targets for E2-ER signaling as they are regulated by ERs in response to 10−9 M E2 (Li et al. 2008; Nott et al. 2009). Simulated systems suggest that the functional interaction of ERs with AP1 bound to an AP1 element provides the basis for the regulation of MMP1 gene promoter in an ER subtype, nature and concentration of ligand, promoter and cell type dependent manner (Kushner et al. 2000; Webb et al. 1995). Similarly, the interaction of ER with SP1 bound to GC boxes is critical for the ligand-mediated regulation of the RARA gene promoter in reporter assays (Safe 2001; Sun et al. 1998). To ensure that GFP-ERs also mimic the effects of ER counterparts on transcription, an expression vector bearing none or an ER cDNA was transfected into HeLa cells together with MMP1-Luc or the RARA-Luc reporter vector (Supplemental Data, Fig. 2B & 3B). Although we did not observe a significant effect of 10−9 M E2 mediated by ERs with or without GFP on the activity of the reporter enzyme from both promoters, 10−7 M 4HT mediated transcriptional responses to ERs similarly. ICI at 10−7 M also affected the luciferase activity mediated by ERα proteins, but not by ERβ with or without GFP. Importantly, ERE binding defective ERs mimicked the effects of the parent ERs on transcriptions in response to ligands from reporter vectors emulating only ERE-independent signaling pathways. Thus, the presence of GFP at the amino-termini of ERs does not affect the transregulatory functions of the receptors at simulated ERE-independent signaling pathways as well.
Ligand-Mediated Nuclear mobility of GFP-ERs
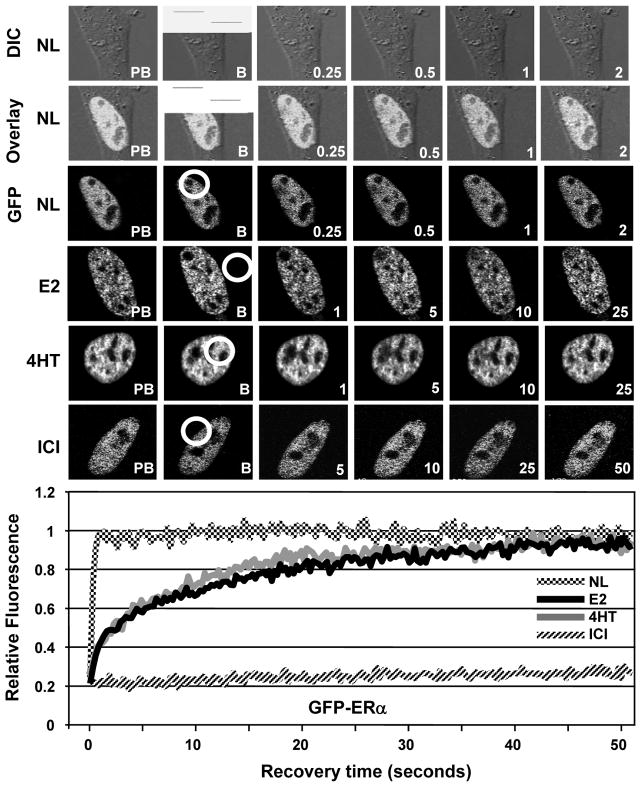
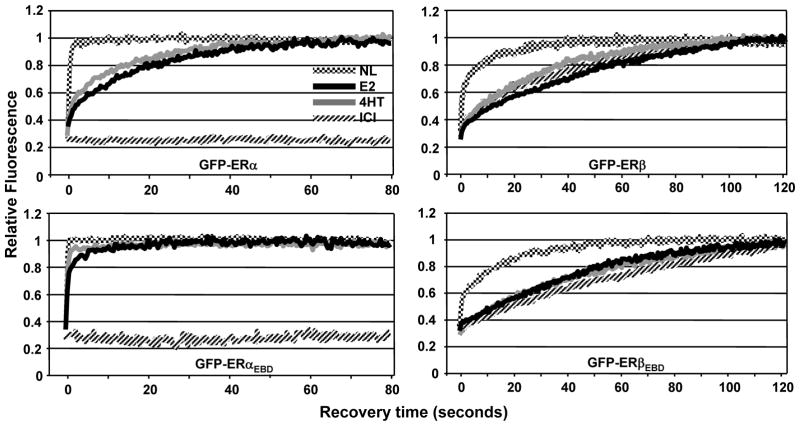
To examine the effects of ligands on nuclear mobility of ERs with or without ERE binding function, transfected HeLa cells were treated with a vehicle (EtOH, 0.01%) for 1h and then subjected to FRAP analysis (Fig. 2). GFP-ERα showed a diffuse distribution throughout the nucleus but it is excluded from nucleoli. After a 0.2 sec photo-bleaching, the bleached area equilibrated to pre-bleach levels within 1 sec with a half-maximum recovery rate (½mRR) of less than 0.2 sec. On the other hand, the treatment of cells with 10−9 M E2 or 10−7 M 4HT for 1h, reduced the mobility of the receptor. Fluorescence after photo-bleaching was fully recovered within 40 sec of post-bleaching with a ½mRR of about 5 sec. In contrast, the treatment of cells with 10−7 M ICI immobilized GFP-ERα as no fluorescence recovery was observed (up to 15 min, data not shown) in post-bleaching. Consistent with previous studies (Sharp et al. 2006; Stenoien et al. 2000a; Stenoien et al. 2001a; Stenoien et al. 2001b; Zwart et al. 2010), our results also demonstrate that GFP-ERα in the unliganded state is a highly mobile molecule and shows different kinetics of mobility in response to ligands.
Fig. 2.
The assessment of nuclear mobility of GFP-ERα by FRAP. HeLa cells transiently transfected with GFP-ERα for 24h were treated without (NL) or with 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells were then subjected to FRAP analysis. Images were obtained before bleaching (pre-bleach, PB), at bleaching for 0.2 second (bleach, B) and at the indicated times in seconds after bleaching. The overlay image (Overlay) was generated with the superimposition of images from differential interference contrast (DIC) and fluorescence (GFP). The time-dependent equilibration of the bleached area (within the white circle) was used to estimate the recovery rate of ER in response to ligands. The recovery rate was based on a control region of interest with the size and fluorescence intensity that corresponded to those of the region of interest (ROI; bleached area) prior to photo-bleaching within the same cell to normalize the background and alterations in total cellular fluorescence after bleaching (Supplemental Data, Fig. 1). The control ROI values obtained throughout the post-bleach period were then used for data normalization. Fluorescence intensity is expressed as the relative fluorescence (RF) where zero (0) is the RF at the photo-bleaching (time 0) and one (1) is the fluorescence of the bleached area equilibrated to pre-bleach levels. Graph represents the normalized mean fluorescence recovery of GFP-ERα with or without a ligand in three independent experiments with a minimum of five individual cells per experiment. SEM, which was less than 15% of the mean, is not shown for simplicity.
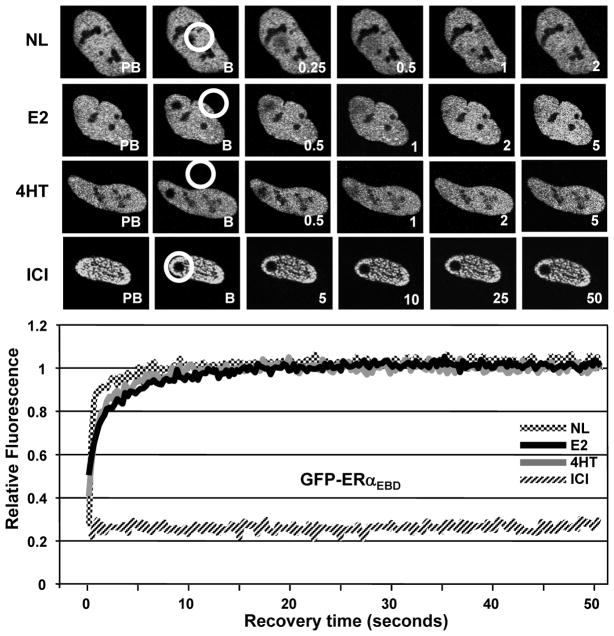
Similar to the unliganded GFP-ERα, GFP-ERαEBD in the absence of a ligand showed a rapid mobility with ½mRR of less than 0.2 sec (Fig. 3). Although the treatment of cells with 10−9 M E2 or 10−7 M 4HT for 1h slowed the nuclear mobility of GFP-ERαEBD with a ½mRR of about 1 sec, the full fluorescence recovery occurred within 10 sec of post-bleaching, a much faster kinetics than that observed with E2 or 4HT liganded ERα. These results indicate that the E2- or 4HT-mediated decrease in the nuclear mobility of ERα is primarily due to the interaction of the receptor with ERE. ICI, on the other hand, prevented the mobility of GFP-ERαEBD in the majority of cells (more than 80%). However, in the remaining cell population, GFP-ERαEBD in response to ICI showed mobility with varying ½mRRs (Supplemental data, Fig. 4). These results suggest that the DBD of ERα contributes to but it is not sufficient for the ICI-mediated immobilization of ERα.
Fig. 3.
The kinetics of nuclear mobility of GFP-ERαEBD. Transiently transfected HeLa cells were treated without or with ligands for 1h and subjected to FRAP analysis as described in legend of Fig. 2. Graph represents the normalized mean fluorescence recovery of GFP-ERαEBD in three independent experiments with a minimum of five individual cells per experiment. SEM, which was less than 15% of the mean, is not shown for simplicity.
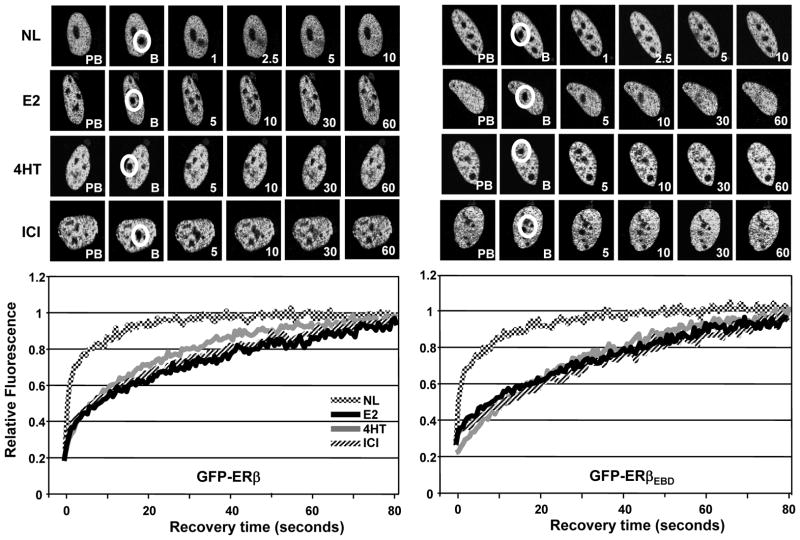
GFP-ERβ (Fig. 4A) and GFP-ERβEBD (Fig. 4B) displayed similar patterns of intra-nuclear distribution and kinetics of mobility. In cells synthesizing ERβ or ERβEBD in response to the vehicle control, the fluorescence recovery of the region after a 0.2 sec photo-bleaching occurred with a ½mRR of about 1 sec that reached to pre-bleach fluorescence intensities within 40 sec of post-bleaching. These results, as shown for ERβ (Damdimopoulos, et al. 2008), indicate that the mobilities of the unliganded ERβ variants are kinetically slower than the corresponding ERα species. This was also the case for the E2 or 4HT liganded ERβ proteins. Treatment of cells with 10−9 M E2 or 10−7 M 4HT decreased the mobility of the receptors that was reflected in ½mRRs of about 15 sec with full recoveries occurring within 90 sec after bleaching. Remarkably, both GFP-ERβ and GFP-ERβEBD in the presence of 10−7 M ICI displayed mobilities that were kinetically indistinguishable from those of the E2- or 4HT-bound receptors, in contrast to ERα species that were stationary in the presence of ICI. Thus, the ability of ERβ to bind to ERE is uncoupled from the nuclear mobility of the receptor independent of the nature of ligand.
Fig. 4.
The assessment of the ligand-mediated mobility of GFP-ERβ (A) and GFP-ERβEBD (B) by FRAP. Transient transfection and processing of HeLa cells for FRAP were carried out as described in legend of Fig. 2. The normalized mean fluorescence recovery of GFP-ERβ species without (NL) or with a ligand from three independent experiments with a minimum of five individual cells per experiment was graphed without SEM, which was less than 15% of the mean.
In addition to alterations in the stability, turnover and intracellular location of ERα (Dauvois, et al. 1992), ICI rapidly sequesters the receptor to a sub-compartment that also involves the nuclear matrix resistant to detergent and salt extractions (Long and Nephew 2006; Lupien, et al. 2007; Stenoien et al. 2000a; Stenoien et al. 2001a; Stenoien et al. 2001b). This sequestration appears to be responsible for the immobilization (Reid et al. 2003; Stenoien et al. 2000a; Stenoien et al. 2001a; Stenoien et al. 2001b) and the absence of interaction with ERE (Reid et al. 2003) of the receptor. In contrast, ICI does not affect the turnover of ERβ (Long, et al. 2010; Peekhaus, et al. 2004).
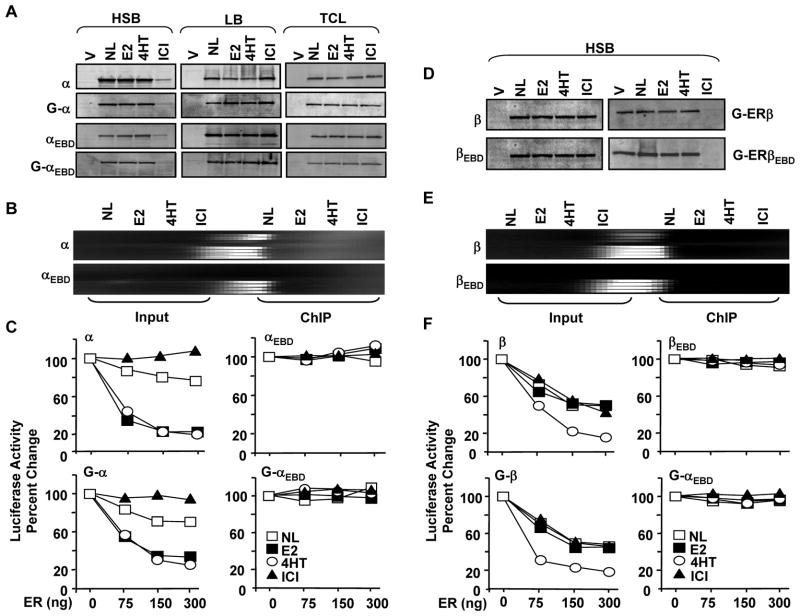
Indeed, ERα (Fig. 5A) and ERβ (Fig. 5D) with or without GFP showed different intracellular levels in the presence of ICI. Transiently transfected HeLa cells synthesizing ERα or GFP-ERα for 24h were treated with or without 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells were then subjected to protein extractions using a buffer containing high salt (HSB) or detergent (RIPA, data not shown). Extracts (10 μg) were then subjected to WB. While E2 or 4HT had minimal effect on ERα levels at 1h post-treatment, ICI substantially reduced the receptor level in HSB extracts. This was inversely correlated with the detection of a higher receptor amount in ICI, but not E2 or 4HT, treated cell extracts obtained with 1x Laemmli buffer (LB) to solubilize the insoluble aggregates following HSB extractions. In contrast, WB of total cell lysates (TCLs) generated with LB to extract both soluble and insoluble protein aggregates revealed that ligands had minimal effects on total receptor levels. This suggests that ICI-mediated rapid immobilization of ERα variants is primarily independent of the receptor degradation.
Fig. 5.
Effects of ligands on protein levels and ERE interactions of ERα and ERαEBD with or without GFP. A. Transiently transfected HeLa cells for 24h were incubated with fresh media supplemented with or without 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells were then collected, washed, re-suspended in 1 ml PBS and divided into two equal portions. One portion of collected cells was pelleted and subjected to protein extraction using a high salt extraction buffer (HSB). The remaining pellet was subjected to 1x Laemmli Buffer (LB) to extract insoluble receptor aggregates. The other portion of the suspended cells was pelleted and the pellet was suspended with LB to extract both soluble and insoluble proteins for total cell lysate (TCL). Ten μg total protein was subjected to SDS 10–18%PAGE. Proteins with (G) or without GFP were probed with a receptor specific antibody. All experiments were replicated at least two independent times. B. Chromatin Immunoprecipitation (ChIP) of transiently transfected HeLa cells. Cells co-transfected with expression vector expressing an ERα cDNA and the reporter vector bearing the TATA box promoter with single ERE for 24h were treated without (NL) or with 10−9 M E2, 10−7 4HT or ICI for 1h. Cells were then subjected to ChIP using Flag-M2 antibody conjugated agarose beads. A 366 bp PCR fragment indicates the ER-ERE interactions. Experiments were replicated at least three independent times. C. The in situ ERE binding competition assay. HeLa cells were co-transfected with 125 ng the TATA box promoter with one ERE that drives the expression of the firefly luciferase cDNA as the reporter enzyme and 300 ng expression plasmid bearing the designer transcription factor, PPVV, without (0 ng ER) or with 75, 150 or 300 ng expression vector bearing an ER cDNA with or without GFP. Cells were then grown in the medium supplemented without (NL) or with 10−9 M E2, 10−7 M 4HT or ICI for 24h. Normalized luciferase activity is presented as percent change compared to the control (PPVV, 0 ng ER) without ligand, which was set to 100. Graph represents the mean of three independent experiments performed in duplicate; SEM, which was less than 15% of the mean, is not shown for simplicity. D. Transfected HeLa cells with an expression vector bearing none (V) or an ERβ cDNA were treated without (NL) or with 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells were collected, pelleted and subjected to protein extraction using HSB. Ten μg total protein was subjected to SDS 10–18%PAGE. Proteins were probed with a receptor specific antibody. The image is from an experiment that was repeated at least three independent times. E. ChIP assays for in situ interactions of ERβ and ERβEBD with ERE in HeLa cells were carried out using the M2-Flag antibody conjugated agarose beads as described for ERα proteins. A representative image from an experiment repeated three independent times is shown. F. Transient transfections of HeLa cells for the assessment of the binding of ERβ proteins with (G) or without to ERE using the in situ ERE binding competition assay are accomplished as described for ERα. The mean of three independent experiments performed in duplicate without the SEM, which was less than 15% of the mean, is shown.
A rapid sequestration of ERα with or without GFP by ICI to a nuclear sub-compartment resistant to HSB extraction also predicts that ICI prevents the interaction of GFP-ERα with ERE, as shown previously for ERα (Reid et al. 2003). To address this point, we employed ChIP assay (Fig. 5B). The expression vector bearing none or an ER cDNA was co-transfected with the reporter TATA box promoter vector bearing one ERE into HeLa cells. Cells were treated with or without 10−9 E2, 10−7 4HT or ICI for 1h, and processed for ChIP using a Flag antibody. Results revealed that the binding of apoERα to ERE is augmented when cells were treated with E2 or 4HT. ICI effectively prevented ERα-ERE interaction, as there was no PCR product. In clear contrast, E2 or ICI had minimal effects on the binding of ERβ to ERE (Fig. 5E). On the other hand, 4HT enhanced the binding of ERβ to ERE. As expected, ERαEBD or ERβEBD did not interact with ERE whether or not cells were treated with a ligand.
To correlate the intracellular mobility of GFP-ERs to ERE binding using ChIP with various antibodies directed to different structural domains of fusion receptor with or without Flag epitope was proved to be difficult. To circumvent this problem, we used an in situ ERE binding competition assay (Huang et al. 2005b). This assay is based on the ability of ER to compete for ERE binding with a designer activator, PPVV, that constitutively and potently induces transcription from the TATA box promoter construct bearing single ERE (ERE-TATA) at which ERs have minimal effect on transcription (Huang et al. 2005b). Thereby, interference of activator-mediated transcription by unliganded or liganded ERs is taken as an indication of ER-ERE interaction.
The reporter ERE-TATA plasmid was co-transfected with an expression vector encoding the PPVV cDNA into HeLa cells in the absence (0) or presence of varying amounts (75, 150 and 300 ng/well) of an expression vector bearing an ER cDNA (Fig. 5C). Cells were then treated with or without 10−9 M E2, 10−7 M 4HT or ICI for 24h. Since PPVV does not bind to a ligand and consequently ligands do not affect the transregulatory potential of PPVV (Huang et al. 2005b), the normalized luciferase activity mediated by PPVV alone in the absence of a ligand was set to 100%. Alterations in the reporter enzyme activity as a result of a co-transfected ER in the absence or presence of a ligand are depicted as percentage change compared to the activity induced by PPVV alone (0 ng ER). Similar to results obtained with ChIP assay, E2 or 4HT increased the ability of ERα or GFP-ERα to interact with ERE reflected in a further repression of the PPVV-induced luciferase activity by the unliganded ERα with or without GFP. The treatment of cells with ICI, on the other hand, had no effect on enzyme levels induced by PPVV. This suggests that ICI prevents the binding of ERα or GFP-ERα to ERE. The effect of ERα in the absence or presence of ligand on PPVV-mediated enzyme activity requires ERE interactions as ERαEBD or GFP-ERαEBD did not alter enzyme levels whether or not cells were exposed to a ligand. Thus, the decrease in the nuclear mobility of ERα mediated by E2 or 4HT is dependent upon the ability of the receptor to interact with ERE, whereas ICI sequesters the majority of the receptor to and immobilizes at a nuclear sub-compartment, thereby preventing ERα-ERE interactions.
In clear contrast to ERα, the short-term treatment (1h) of cells with ICI as E2 or 4HT did not affect intracellular levels of ERβ or ERβEBD with or without GFP (Fig. 5D). The absence of an effect of ICI, as E2 or 4HT, on levels and mobilities of ERβ proteins also predicts that ICI does not alter ERβ-ERE interactions in situ. Indeed, ChIP (Fig. 5E) or the in situ ERE binding competition assay (Fig. 5F) revealed that E2 or ICI did not affected the binding of ERβ or GFP-ERβ to ERE, whereas 4HT increased ERβ-ERE interactions. As expected, the ERE binding defective ERβ with or without GFP did not bind to ERE. These findings imply that the ligand-mediated nuclear mobility of ERβ is independent of nature of ligands and the ability of ERβ to bind to ERE.
Effects of Ligands on Transregulatory Function and Nuclear Mobility of GFP-ERs in MDA-MB-231 cells
To examine whether or not the effects of ligands on the nuclear mobilities of ERs are cell type-specific, we also used ER-negative MDA-MB-231 cells derived from a breast adenocarcinoma. Exogenously introduced ERα or ERβ in MDA-MB-231 cells modulates genomic and cellular responses in the presence of E2 (Garcia, et al. 1992; Huang et al. 2011; Lazennec, et al. 1999; Li et al. 2008; Nott et al. 2009; Zajchowski, et al. 1993). In this cell line, 4HT acts as an ERα subtype-specific agonist by mimicking the effects of E2 on cellular responses when mediated by the ERE-dependent signaling pathway, whereas ICI is an antagonist for both ER subtypes (Bentrem, et al. 2001; Tonetti, et al. 2003). The ERE binding defective ERs do not interact with the ERE sequence of the estrogen responsive TFF1 gene with or without ligands, while ligands differentially alter the parent ER-ERE interactions (Supplemental Data, Fig. 5A). Moreover, providing evidence for a functional ERE-independent signaling pathway, we recently showed that the DNA binding defective ERs participate in the fine-tuning of phenotypic features of MDA-MB-231 cells by regulating the expression of a subset of estrogen responsive genes (Li et al. 2008; Nott et al. 2009).
Transient transfections of MDA-MB-231 cells with heterologous reporter vectors emulating ERE-dependent and ERE-independent signaling pathways revealed that the GFP fusion-ERs mimic the abilities of the parent receptors to regulate transcription in response to ligands (Supplemental Data, Fig. 5 & 6).
The nuclear motilities of GFP-ERα with or without ligands in MDA-MB-231 cells showed patterns indistinguishable from those observed in HeLa cells (Fig. 6). However, the rate and the time of the total recovery of ERα variants in response to E2 or 4HT were about two-fold slower than those of the receptor synthesized in HeLa cells. The ½mRR of the unliganded GFP-ERα or GFP-ERαEBD was less than 0.2 sec with a total recovery within 5 sec after photo-bleaching. The treatment of cells with 10−9 M E2 or 10−7 M 4HT in cells synthesizing GFP-ERα increased the ½mRR of the bleached region to about 11 and 9 sec, respectively, with a full fluorescence recovery occurring within 60 sec of post-bleaching. ICI at 10−7 M effectively halted the fluorescence recovery of GFP-ERα. As observed in HeLa cells, ICI also prevented the recovery of the bleached region in the majority of cells (more than 80%) synthesizing GFP-ERαEBD, while the rate of fluorescence recovery vastly varied in individual cells in the remaining population (data not shown). These finding support our conclusion that the DBD contributes to ICI-mediated immobilization of ERα. On the other hand, the unliganded GFP-ERαEBD showed a very rapid recovery with a ½mRR of less than 0.2 sec with a full recovery occurring within one second. This was similar to the rate of recovery of GFP-ERαEBD in cells exposed to E2 or 4HT with the fluorescence equilibration occurring within 10 sec of post-bleaching. Thus, the E2- or 4HT-mediated decrease in the nuclear mobility of ERα is dependent upon the ability of the receptor to interact with ERE and is independent of cell-type.
Fig. 6.
The effects of ligands on the nuclear mobility of GFP fusion ER proteins in MDA-MB-231 cells. Transiently transfected cells for 24h were incubated in the absence (NL) or presence of 10−9 M E2, 10−7 M 4HT or ICI for 1h and subjected to FRAP analysis as described in legend of Fig. 2. Graph represents the normalized mean fluorescence recovery of GFP-ER with or without a ligand in three independent experiments with a minimum of five individual cells per experiment. The SEM, which was less than 15% of the mean, is not shown.
The fluorescence recovery of GFP-ERβ in the absence or presence of a ligand was kinetically similar to that of GFP-ERβEBD in MDA-MB-231 cells and mirrored those observed in HeLa cells wherein the overall rate of recoveries was faster for both receptor species. In the absence of ligand, the bleached region synthesizing ERβ or ERβEBD recovered within 40 sec of post-bleaching with a ½mRR of about 1 sec. Treatment of cells with E2, 4HT or ICI slowed the rate of fluorescence recovery to about 25 sec with a full recovery within 120 sec post-bleaching. Thus, the ability of ERβ to bind to ERE is not reflected in the nuclear mobility of the receptor, which is also independent of the nature of ligand and cell-context.
Structural domains responsible for ER subtype-specific nuclear mobility
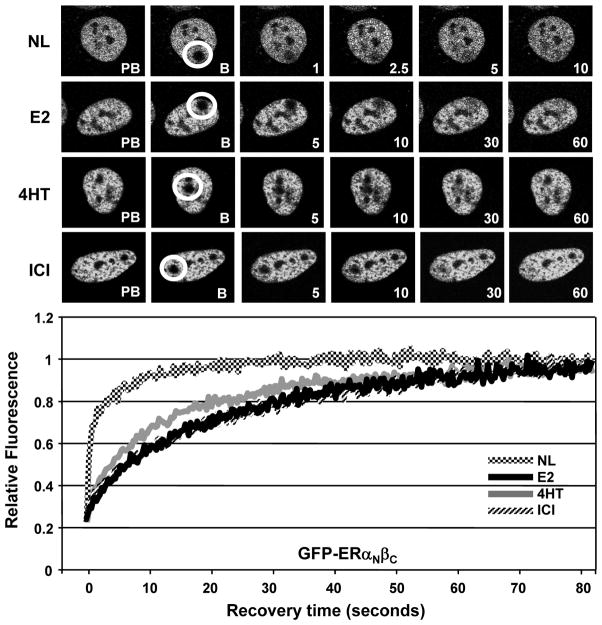
The amino- and carboxyl-termini of ERs functionally differ (Cowley and Parker 1999; Hall and McDonnell 1999; Huang et al. 2005b; Yi et al. 2002a). To examine the roles of structural termini on the nuclear mobility of ERs, we used GFP fusion ER chimera proteins. In ERαNβC and ERβNαC the entire amino-termini of the receptors are genetically interchanged (Yi et al. 2002a). We found in transiently transfected HeLa cells that the fluorescence recoveries of ERαNβC with or without a functional ERE binding (data not shown) were kinetically similar to those observed with ERβs in the absence or presence of ligands (Fig. 7). Conversely, ERβNαC or the ERE binding defective ERβNαC mimicked the nuclear mobilities of ERα proteins with or without a ligand (data not shown). These results indicate that the carboxyl-termini are the structural basis for the difference in the nuclear mobility of ER subtypes in the absence or presence of a ligand.
Fig. 7.
The nuclear mobility of the chimeric ERαNβC in HeLa cells. Transiently transfected cells for 24h were incubated in the absence (NL) or presence of 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells were subjected to FRAP analysis as described in legend of Fig. 2. Graph represents the normalized mean fluorescence recovery of GFP- ERαNβC from three independent experiments with a minimum of five individual cells per experiment. SEM, which was less than 15% of the mean, is not shown for simplicity.
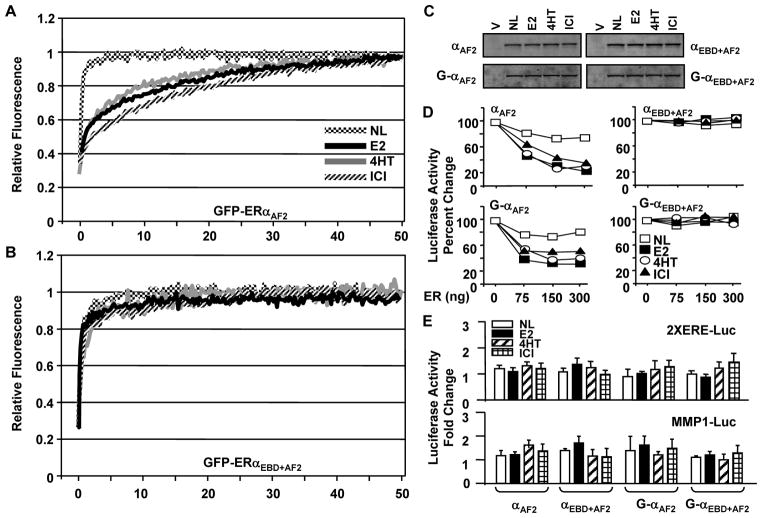
Distinct conformational features of ER carboxyl-termini induced by a ligand determine the formation of a functional co-regulator interacting surface responsible for AF2 of the receptors. We also addressed whether the changing of the critical residues that prevent AF2 affects the subtype-specific nuclear mobility of ERs in response to ligands. To examine this issue, we used the GFP-fusion ERs with abrogated AF2 (ERAF2). In transiently transfected HeLa cells, the nuclear mobility of the unliganded, E2- or 4HT-bound GFP-ERαAF2 showed kinetics of mobility (Fig. 8A) similar to that of ERα (Fig. 2). GFP-ERαAF2 was also mobile when ICI was present. This suggests that the absence of AF2 renders ERα mobile when ICI is present. ICI, as E2 or 4HT, also had minimal effects on the fluorescence recovery after photo-bleaching of the ERE binding defective receptor with abrogated AF2, GFP-ERαEBD+AF2, (Fig. 8B). ERαAF2 or ERαEBD+AF2 without or with GFP was HSB extractable (Fig. 8C). Moreover, ICI-ERαAF2 gained the ability to interact with ERE in situ (Fig. 8D) in stark contrast to ICI-ERα, which was immobile due to the sequestration to a nuclear sub-compartment resistant to HSB extraction. However, ERαAF2 with or without GFP was transcriptionally inactive when cells were treated with ICI, E2 or 4HT (Fig. 8E). Thus, it appears that the ligand-mediated nuclear mobility and the ability to interact with and to induce transcription from target sites of ERα are discernable. A much faster kinetics of mobility of ERαEBD compared to that of ERα in response to E2 or 4HT also indicate that the mobility of ERα at the ERE-independent signaling pathway contributes fractionally to the overall nuclear mobility of the receptor.
Fig. 8.
The nuclear mobility of the GFP-ERαAF2 with GFP-ERαEBD+AF2 in HeLa cells. Transiently transfected cells for 24h were incubated in the absence (NL) or presence of 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells synthesizing GFP-ERαAF2 (A) or GFP-ERαEBD+AF2 (B) were subjected to FRAP analysis as described in legend of Fig. 2. Graphs represent the normalized mean fluorescence recovery of ERα proteins from three independent experiments with a minimum of five individual cells per experiment. The SEM, which was less than 15% of the mean, is not shown for simplicity. C. Intracellular levels of ERαAF2 and ERαEBD+AF2 with (G) or without in transiently transfected in HeLa cells for 24h. Cells were then treated in the absence or presence of 10−9 M E2, 10−7 M 4HT or ICI for 1h. Cells were collected, pelleted and subjected to protein extraction using HSB. Ten μg total protein was subjected to SDS 10–18%PAGE. Proteins were probed with a receptor specific antibody. Experiments were repeated two independent times. D. Assessing the effects of ligands on in situ ERE binding abilities of ERαAF2 and ERαEBD+AF2 with (G) or without GFP. Transiently transfected HeLa cells were treated in the absence or presence of a ligand for 24h. Graphs depict the mean of three independent experiments performed in duplicate. The SEM, which was less than 15% of the mean, is not shown for simplicity. E. The effects of ligands on transcription by ERαAF2 and ERαEBD+AF2 with (G) or without GFP. Cells were transfected with the 2XERE or the MMP1-Luc promoter reporter vector emulating the ERE-dependent or ERE-independent signaling pathway. Cells were also co-transfected with a vector expressing ERαAF2 and ERαEBD+AF2 with (G) or without GFP. Cells were then treated in the absence or presence of a ligand for 24h for the luciferase activity for 24h. Graphs represent the mean of three independent experiments performed in duplicate. The SEM, which was less than 15% of the mean, is not shown.
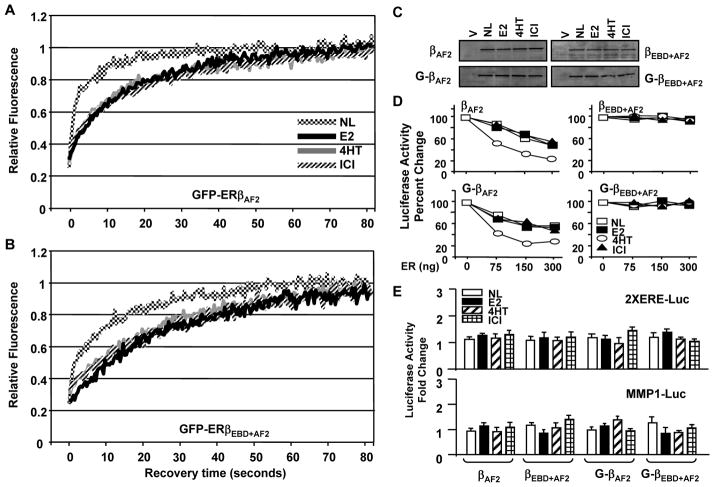
The prevention of AF2 did not alter the pattern or the kinetic of mobility of the GFP-ERβAF2 mutant compared to that of GFP-ERβ in the absence or presence of ligands (Fig. 9A). The pattern of fluorescence recovery of GFP-ERβEBD+AF2 was also similar to that of GFP-ERβAF2 (Fig. 9B). Interestingly, however, the nuclear movement of GFP-ERβEBD+AF2 occurred at slower kinetics than GFP-ERβEBD or GFP-ERβAF2 in the absence or presence of ligands. The fluorescence level of the bleached region in cells synthesizing ERβEBD+AF2 in the absence of ligand reached to pre-bleach levels with a ½mRR of about 6 sec, whereas the fluorescence recovery in cells synthesizing GFP-ERβEBD or GFP-ERβAF2 was about 1 sec. The treatment of cells with E2, 4HT or ICI decreased the mobility of ERβEBD+AF2 similarly, reflected in a ½mRR of about 20 sec in comparison with liganded GFP-ERβEBD that showed recovery rates of about 15 sec. This suggests that integrated effects of the DBD and the LBD of ERβ are important for the mobility characteristics of ERβ.
Fig. 9.
The mobilities of GFP-ERβAF2 and GFP-ERβEBD+AF2 in HeLa cells. The transfection and processing of cells synthesizing (A) GFP-ERβAF2 or (B) GFP-ERβEBD+AF2 for FRAP were carried out as described in legend of Fig. 2. C. Intracellular level of receptor proteins were analyzed with high salt extracts of transfected HeLa cells which were treated and process as described in legend of Fig. 8C. D. The effects of ligands on in situ ERE binding abilities of ERβAF2 and ERβEBD+AF2 with (G) or without GFP were assessed as described in legend of Fig. 8D. The SEM, which was less than 15% of the mean, is not shown for simplicity. E. The effects of ligands on transcription by ERβAF2 and ERβEBD+AF2 with (G) or without GFP were assessed as described in legend of Fig. 8E. Graphs show the mean of three independent experiments performed in duplicate. The SEM, which was less than 15% of the mean, is not shown.
Ligands did not affect the intracellular levels of the receptor species (Fig. 9C). The treatment of cells with or without a ligand did not alter the ability of ERβAF2 or GFP-ERβAF2 to interact with ERE in situ (Fig. 9D), despite the fact that the receptors were transcriptionally silent at simulated ERE-dependent and ERE-independent signaling pathways (Fig. 9E). Showing similar intracellular levels in the absence or presence of a ligand (Fig. 9C), ERβEBD+AF2 with or without GFP did not bind to ERE (Fig. 9D) nor modulated the reporter enzyme levels whether or not cells were treated with a ligand (Fig. 9E).
Thus, the nuclear mobility of ERβ is independent from the nature of ligand and from the ability of receptor to interact with target sites. These results imply that ERβ mediates gene transcriptions through the ERE-dependent and ERE-independent signaling pathways with similar kinetics.
DISCUSSION
ERs are highly mobile proteins partitioned dynamically between the nucleoplasm and target sites on the chromatin that constitute the ERE-dependent and ERE-independent signaling pathways. We here assessed the relative contribution of ER mobility at the ERE-independent signaling pathway to the overall mobility of receptors to gain insights into mechanisms of action.
Our observations revealed several distinct features of ERβ mobility compared to ERα. These are: 1) ERβ mobility with or without ligands is slower than ERα mobility. 2) The interaction of ERβ with ERE is augmented with 4HT but not with E2 or ICI, whereas E2 and 4HT enhances and ICI prevents ERα-ERE interactions. 3) ICI does not sequester ERβ with or without ERE binding and/or AF2 functions to a nuclear sub-compartment, whereas the sequestration of ERα is dependent upon AF2. 4) The ability of ERβ to interact with and to induce transcription from target sites is largely uncoupled from the receptor mobility. 5) Cooperation between DBD and LBD contributes to ERβ motility.
Based on these observations, we conclude that while ICI immobilizes ERα to a sub-nuclear compartment, E2 or 4HT decreases ERα mobility by increasing ERα-ERE interactions. We therefore suggest that ERα in response to E2 and 4HT mediates transcriptions from the ERE-independent pathway with fast kinetics that contributes fractionally to the overall motility of the receptor. On the other hand, the ligand-mediated mobility of ERβ is independent of the nature of ligands or the mode of interaction with target sites. It therefore appears that although ERs interact with target sites with fast kinetics, they use distinct mechanisms to regulate transcriptions at ERE-dependent and signaling pathways.
We show here, as previous studies (Sharp et al. 2006; Zwart et al. 2010), that 4HT, as E2, decreases the mobility of ERα by enhancing ERα-ERE interactions (Huang et al. 2005b; Shang et al. 2000). However the underlying mechanism(s) remains unclear. The binding of tamoxifen to ERα alters conformation (Paige, et al. 1999) that affects co-activator recruitment (Yi, et al. 2002b). Tamoxifen-ERα can also recruit co-repressors for transcription repression (Delage-Mourroux, et al. 2000; Lavinsky, et al. 1998; Shang et al. 2000). While 4HT is an antagonist for ERα in HeLa cells, it acts as an agonist in MDA-MB-231 cells (Dardes et al. 2002; Tonetti et al. 2003). 4HT also augmented ERα-ERE interactions. 4HT-ERα showed mobility similar to E2-ERα. On the other hand, ERαEBD with or without AF2 was kinetically much faster when bound to 4HT or E2 compared to ERα. Our findings therefore imply that E2- and 4HT-mediated decreases in ERα mobility are due to the residency time of the receptor on ERE independent of transcription. In contrast, ICI immobilized ERα. ICI binding prevents ER-coregulator interactions (Yi, et al. 2002b) but drives ERα to interact with cytokeratins through LBD (Long and Nephew 2006). Leading to the association of ICI-ERα with nuclear matrix (Long and Nephew 2006; Lupien et al. 2007), this could result in the immobilization and complete prevention of ERE interactions, as shown here and previously (Reid et al. 2003). However, it was also shown that a fraction of ICI-ERα remains associated with the prolactin promoter array, which is composed of 52 prolactin gene promoters containing multiple EREs (Sharp et al. 2006). It is possible that while the majority of ERα bound to ICI is immobilized to a sub-nuclear region, a fraction of ERα bound to EREs of the promoter array cooperatively and hence stably (Yi, et al. 2002b) cannot be readily sequestered away from the array in contrast to the single ERE of the reporter system and the endogenous gene we used here. Another puzzling observation is that while ICI immobilized ERα, ICI-ERα still modulated the reporter gene transcription from ERE-independent pathways. Immobilization of ERα by ICI could prevent the interaction of ER with co-regulators/transfactors thereby countering the ERα-mediated repressed or activated state of transcriptions.
The changing (Fig. 8) or deletion (Sharp et al. 2006), of critical residues to block AF2 rendered ICI-ERα mobile in cells. This was reflected in the increased extractability of ERα with HSB or detergent likely due to the inability of the receptor to interact with cytokeratins (Long and Nephew 2006). Nevertheless, ICI-ERαAF2 was transcriptionally silent despite the fact that the receptor interacted with ERE. Moreover, the increased mobility of ERαEBD+AF2 regardless of the nature of ligand strengthens the conclusion that the duration of ERE occupancy reflects the ligand-mediated mobility of ERα independent of transcription status. This lends further credence to the hit-and-run model of transcription for ERα regardless of signaling pathway.
In contrast to ERα, ERβ and ERβEBD showed indistinguishable mobility rates independent of the nature of ligands, the ability of receptor to interact with target sites and cellular context. Consequently, it appears that ERβ mediates gene transcriptions through the ERE-dependent and ERE-independent signaling pathways with similar kinetics. ERs share structural features reflected in similar functional properties. ERs nevertheless exhibit distinct transregulatory potentials at signaling pathways. The amino-termini are critical regions that contribute to subtype-specific transcriptional responses. In contrast to ERα, ERβ amino-terminus impairs ER-ERE interactions (Huang et al. 2005b), lacks an activation function (Cowley and Parker 1999; Yi et al. 2002a) and does not interact with the carboxyl-terminus (Yi et al. 2002a). However, the amino-termini do not appear to contribute to distinct receptor mobility. We observed that the mobilities of the ERαNβC chimeras were kinetically similar to those observed with ERβ variants. ERβNαC, on the other hand, mimicked ERα mobility in response to ligands. These imply that the carboxyl-termini are critical regions in defining mobility differences of ERs. Studies also showed that the carboxyl-termini contribute to transcriptional potencies of ERs (Yi et al. 2002a) by differentially interacting with coregulators (Kressler, et al. 2002; Seol, et al. 1998). Moreover, some coregulator interactions with ERs are specific to the nature of ligand. The unliganded ERα interacts with co-repressors SMRT/NCoR (Lavinsky et al. 1998; Webb, et al. 2003). The binding of E2 releases co-repressors from ERα (Lavinsky et al. 1998; Webb et al. 2003). The unliganded ERβ also interacts with SMRT/NCoR through the carboxyl-terminus (Webb et al. 2003). However, the binding of E2 does not promote co-repressor dissociations (Webb et al. 2003). In contrast, the binding of 4HT or ICI releases SMRT/NCoR from ERβ but not from ERα (Lavinsky et al. 1998; Webb et al. 2003). Since ERβ requires E2 to regulate transcription from the ERE-dependent signaling pathway, the E2 binding could act as a switch to convert ERβ to an active state by concurrently recruiting co-activators likely through a distinct surface.
We also found that 4HT enhanced the ERβ-ERE interaction in contrast to E2 or ICI. Although is unclear, distinct trans-conformational changes in ERβ-DBD mediated by the binding of 4HT to LBD could underlie the effect of 4HT on ERβ-ERE interactions. We observed that 4HT- or ICI-bound ERβ, as ERβAF2, showed mobility similar to E2-ERβ despite the fact that the receptor was transcriptionally inactive at the ERE-dependent pathway. Furthermore, ERβEBD mobility was indistinguishable from that of ERβ independently of ligands. However, ERβEBD+AF2 showed slower mobility than ERβEBD. This suggests that the cooperation of AF2 with the ability of the receptor to interact with target sites is a critical feature for the nuclear mobility of ERβ.
In summary, our results indicate that while ERs use hit-and-run mode of action, they differ mechanistically to modulate transcriptions. The use of integrated promoter arrays mimicking various signaling pathways would yield further insights into mechanisms of ER actions. This in turn could aid in the development of better strategies to combat estrogen target tissue malignancies.
Supplementary Material
Acknowledgments
FUNDING
The work was supported by a grant from NIH CA113682 (MM).
We express our gratitude to Melinda Huang and Aja Kalkanoglu for their help in western blot analyses. We thank Drs. James McGrath and Thomas R. Gaborski for the establishment of FRAP assays and Brian Fluharty for preliminary studies.
Footnotes
DECLERATION OF INTEREST
The authors declare that they have no conflict of interest.
References
- An J, Ribeiro RC, Webb P, Gustafsson JA, Kushner PJ, Baxter JD, Leitman DC. Estradiol repression of tumor necrosis factor-alpha transcription requires estrogen receptor activation function-2 and is enhanced by coactivators. Proc Natl Acad Sci U S A. 1999;96:15161–15166. doi: 10.1073/pnas.96.26.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- Bentrem D, Dardes R, Liu H, MacGregor-Schafer J, Zapf J, Jordan V. Molecular mechanism of action at estrogen receptor alpha of a new clinically relevant antiestrogen (GW7604) related to tamoxifen. Endocrinology. 2001;142:838–846. doi: 10.1210/endo.142.2.7932. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mutations in the estrogen receptor DNA-binding domain discriminate between the classical mechanism of action and cross-talk with Stat5b and activating protein 1 (AP-1) J Biol Chem. 2002;277:48479–48483. doi: 10.1074/jbc.C200570200. [DOI] [PubMed] [Google Scholar]
- Cowley SM, Parker MG. A comparison of transcriptional activation by ER alpha and ER beta. J Steroid Biochem Mol Biol. 1999;69:165–175. doi: 10.1016/s0960-0760(99)00055-2. [DOI] [PubMed] [Google Scholar]
- Damdimopoulos AE, Spyrou G, Gustafsson JA. Ligands differentially modify the nuclear mobility of estrogen receptors alpha and beta. Endocrinology. 2008;149:339–345. doi: 10.1210/en.2007-0198. [DOI] [PubMed] [Google Scholar]
- Dauvois S, Danielian PS, White R, Parker MG. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc Natl Acad Sci U S A. 1992;89:4037–4041. doi: 10.1073/pnas.89.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage-Mourroux R, Martini PG, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS. Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J Biol Chem. 2000;275:35848–35856. doi: 10.1074/jbc.M001327200. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Cuba VL, Kim H, Wu K, Lee AV, Brown PH. Estrogen receptor DNA binding is not required for estrogen-induced breast cell growth. Mol Cell Endocrinol. 2007;277:13–25. doi: 10.1016/j.mce.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Mullick A, Rorke EA, Katzenellenbogen BS. Estrogen receptor synthesis and turnover in MCF-7 breast cancer cells measured by a density shift technique. Endocrinology. 1984;114:629–637. doi: 10.1210/endo-114-2-629. [DOI] [PubMed] [Google Scholar]
- Garcia M, Derocq D, Freiss G, Rochefort H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci U S A. 1992;89:11538–11542. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. The estrogen receptor beta-isoform (ER beta) of the human estrogen receptor modulates ER alpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Hilf R, Bambara RA, Muyan M. Molecular basis of therapeutic strategies for breast cancer. Curr Drug Targets Immune Endocr Metabol Disord. 2005a;5:379–396. doi: 10.2174/156800805774912944. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Maguire CA, Hilf R, Bambara RA, Muyan M. Binding of estrogen receptor beta to estrogen response element in situ is independent of estradiol and impaired by its amino terminus. Mol Endocrinol. 2005b;19:2696–2712. doi: 10.1210/me.2005-0120. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Yi P, Hilf R, Bambara RA, Muyan M. Targeting estrogen responsive elements (EREs): design of potent transactivators for ERE-containing genes. Mol Cell Endocrinol. 2004;218:65–78. doi: 10.1016/j.mce.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li X, Muyan M. Estrogen receptors similarly mediate the effects of 17beta-estradiol on cellular responses but differ in their potencies. Endocrine. 2011;39:48–61. doi: 10.1007/s12020-010-9411-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Sasaki H, Suzawa M, Masushige S, Tora L, Chambon P, Gronemeyer H. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol Cell Biol. 1995;15:5858–5867. doi: 10.1128/mcb.15.11.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci U S A. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. ER beta inhibits proliferation and invasion of breast cancer cells. Endocrinology. 2001;142:4120–4130. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Huang J, Yi P, Bambara RA, Hilf R, Muyan M. Single-chain estrogen receptors (ERs) reveal that the ERalpha/beta heterodimer emulates functions of the ERalpha dimer in genomic estrogen signaling pathways. Mol Cell Biol. 2004;24:7681–7694. doi: 10.1128/MCB.24.17.7681-7694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Nott SL, Huang Y, Hilf R, Bambara RA, Qiu X, Yakovlev A, Welle S, Muyan M. Gene expression profiling reveals that the regulation of estrogen-responsive element-independent genes by 17beta-estradiol-estrogen receptor beta is uncoupled from the induction of phenotypic changes in cell models. J Mol Endocrinol. 2008;40:211–229. doi: 10.1677/JME-07-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Fan M, Nephew KP. Estrogen receptor-alpha-interacting cytokeratins potentiate the antiestrogenic activity of fulvestrant. Cancer Biol Ther. 2010;9:389–396. doi: 10.4161/cbt.9.5.10926. [DOI] [PubMed] [Google Scholar]
- Long X, Nephew KP. Fulvestrant (ICI 182,780)-dependent interacting proteins mediate immobilization and degradation of estrogen receptor-alpha. J Biol Chem. 2006;281:9607–9615. doi: 10.1074/jbc.M510809200. [DOI] [PubMed] [Google Scholar]
- Lupien M, Jeyakumar M, Hebert E, Hilmi K, Cotnoir-White D, Loch C, Auger A, Dayan G, Pinard GA, Wurtz JM, et al. Raloxifene and ICI182,780 increase estrogen receptor-alpha association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol Endocrinol. 2007;21:797–816. doi: 10.1210/me.2006-0074. [DOI] [PubMed] [Google Scholar]
- McDonnell DP. The Molecular Pharmacology of SERMs. Trends Endocrinol Metab. 1999;10:301–311. doi: 10.1016/s1043-2760(99)00177-0. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Nott SL, Huang Y, Li X, Fluharty BR, Qiu X, Welshons WV, Yeh S, Muyan M. Genomic responses from the estrogen responsive element-dependent signaling pathway mediated by estrogen receptor alpha are required to elicit cellular alterations. J Biol Chem. 2009;284:15277–15288. doi: 10.1074/jbc.M900365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige LA, Christensen DJ, Gron H, Norris JD, Gottlin EB, Padilla KM, Chang CY, Ballas LM, Hamilton PT, McDonnell DP, et al. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc Natl Acad Sci U S A. 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peekhaus NT, Chang T, Hayes EC, Wilkinson HA, Mitra SW, Schaeffer JM, Rohrer SP. Distinct effects of the antiestrogen Faslodex on the stability of estrogen receptors-alpha and -beta in the breast cancer cell line MCF-7. J Mol Endocrinol. 2004;32:987–995. doi: 10.1677/jme.0.0320987. [DOI] [PubMed] [Google Scholar]
- Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Safe S. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- Sathya G, Yi P, Bhagat S, Bambara RA, Hilf R, Muyan M. Structural regions of ERalpha critical for synergistic transcriptional responses contain co-factor interacting surfaces. Mol Cell Endocrinol. 2002;192:171–185. doi: 10.1016/s0303-7207(01)00673-6. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Seol W, Hanstein B, Brown M, Moore DD. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner) Mol Endocrinol. 1998;12:1551–1557. doi: 10.1210/mend.12.10.0184. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TP, Ingber DE, Mancini MA. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci. 2006;119:4101–4116. doi: 10.1242/jcs.03161. [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol Endocrinol. 2000a;14:518–534. doi: 10.1210/mend.14.4.0436. [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, O’Malley BW, Smith CL, Belmont AS, Mancini MA. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol Cell Biol. 2001a;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, Mancini MA. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat Cell Biol. 2001b;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Sun G, Porter W, Safe S. Estrogen-induced retinoic acid receptor alpha 1 gene expression: role of estrogen receptor-Sp1 complex. Mol Endocrinol. 1998;12:882–890. doi: 10.1210/mend.12.6.0125. [DOI] [PubMed] [Google Scholar]
- Tamrazi A, Carlson KE, Daniels JR, Hurth KM, Katzenellenbogen JA. Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol Endocrinol. 2002;16:2706–2719. doi: 10.1210/me.2002-0250. [DOI] [PubMed] [Google Scholar]
- Tamrazi A, Carlson KE, Rodriguez AL, Katzenellenbogen JA. Coactivator proteins as determinants of estrogen receptor structure and function: spectroscopic evidence for a novel coactivator-stabilized receptor conformation. Mol Endocrinol. 2005;19:1516–1528. doi: 10.1210/me.2004-0458. [DOI] [PubMed] [Google Scholar]
- Tonetti DA, Rubenstein R, DeLeon M, Zhao H, Pappas SG, Bentrem DJ, Chen B, Constantinou A, Craig Jordan V. Stable transfection of an estrogen receptor beta cDNA isoform into MDA-MB-231 breast cancer cells. J Steroid Biochem Mol Biol. 2003;87:47–55. doi: 10.1016/j.jsbmb.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- Webb P, Lopez GN, Uht RM, Kushner PJ. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- Webb P, Valentine C, Nguyen P, Price RH, Jr, Marimuthu A, West BL, Baxter JD, Kushner PJ. ERbeta Binds N-CoR in the Presence of Estrogens via an LXXLL-like Motif in the N-CoR C-terminus. Nucl Recept. 2003;1:4. doi: 10.1186/1478-1336-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Bhagat S, Hilf R, Bambara RA, Muyan M. Differences in the abilities of estrogen receptors to integrate activation functions are critical for subtype-specific transcriptional responses. Mol Endocrinol. 2002a;16:1810–1827. doi: 10.1210/me.2001-0323. [DOI] [PubMed] [Google Scholar]
- Yi P, Driscoll MD, Huang J, Bhagat S, Hilf R, Bambara RA, Muyan M. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ER alpha and ER beta. Mol Endocrinol. 2002b;16:674–693. doi: 10.1210/mend.16.4.0810. [DOI] [PubMed] [Google Scholar]
- Zajchowski DA, Sager R, Webster L. Estrogen inhibits the growth of estrogen receptor-negative, but not estrogen receptor-positive, human mammary epithelial cells expressing a recombinant estrogen receptor. Cancer Res. 1993;53:5004–5011. [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen signaling via estrogen receptor beta. J Biol Chem. 2010;285:39575–39579. doi: 10.1074/jbc.R110.180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, de Leeuw R, Rondaij M, Neefjes J, Mancini MA, Michalides R. The hinge region of the human estrogen receptor determines functional synergy between AF-1 and AF-2 in the quantitative response to estradiol and tamoxifen. J Cell Sci. 2010;123:1253–1261. doi: 10.1242/jcs.061135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.