Abstract
Myc levels are highly regulated and usually low in vivo. Dimerized with Max, it regulates most expressed genes and so directly and indirectly controls most cellular processes. Intranuclear diffusion of a functional c-Myc-eGFP, expressed from its native locus in murine fibroblasts and 3T3 cells or by transient transfection, was monitored using Two Photon Fluorescence Correlation Spectroscopy, revealing concentration and size (mobility) of complexes. With increased c-Myc-eGFP, a very immobile pool saturates as a ‘mobile' pool increases. Both pools diffuse too slowly to be free Myc-Max dimers. Following serum stimulation, eGFP-c-Myc accumulated in the presence of the proteasome inhbitor MG132. Stimulating without MG132, Myc peaked at 2.5 hrs, and at steady was ~8 ± 1.3 nM. Inhbiting Myc-Max dimerization by Max-knockdown or drug treatment increased the ‘mobile' c-Myc pool size. These results indicate that Myc populates macromolecular complexes of widely heterogenous size and mobility in vivo.
C-MYC is a highly regulated protein controlling the expression of many genes involved in the cell cycle, cell growth, proliferation and apoptosis1,2. c-Myc belongs to a family of transcription factors that contain basic helix-loop-helix motifs (b-HLH) and leucine zipper domains3; c-Myc dimerizes with Max4,5, also a member of the b-HLH family, and potentially tetramerizes (as a dimer of heterodimers) in order to bind to DNA specifically to E-box sequences (CAYGTG). Myc-Max likely carries along transcriptional co-activators or chromatin complexes including their associated histone acetyltransferases or ATPases/helicases5 to the promoters of target genes6. Although historically, c-Myc has been considered to be both an activator and repressor of transcription acting through direct protein-protein interactions with the transcription and chromatin machineries7,8, recent studies suggest that its activation function are dominant- at almost all genes9,10. As expected, c-Myc is tightly regulated by many mechanisms (i.e. the proteasome system11) and its deregulation is an important oncogenic event in cancer1,2.
Disturbed Myc activity or regulation is associated with the pathogenesis of most cancers. Physiologically, c-Myc is among the first genes to be activated by a variety of signals or stresses yielding a cascade of gene regulation12,13,14. Its levels are known to be very stable throughout the cell cycle, despite its short lifetime of 20–30 minutes15, until cell division, when the daughter cells split the parent cell's c-Myc. There is a short lived increase (up to five fold) in the total concentration of c-Myc after a variety of stimuli including restoration of serum to serum-starved cells16. The identifcation of c-Myc's targets as well as its mode and mechanisms of gene regulation remain controversial, despite extensive study. Myc has been reported to associate with a wide range of partners, inviting speculation about multiple modes of action and suggesting that there may be two or more discrete populations of Myc. Although Tworkowsky et al.17 argue that there are two populations that can be considered ‘stable' and ‘unstable' depending on their rate of degradation, few studies have monitored the physiological intracellular/intranuclear trafficking of Myc in vivo. Arabi et al.11 propose that when the c-Myc concentration is high, c-Myc co-localizes with nucleoli. In the same study, FRAP and FLIP experiments demonstrated an 84% recovery with a half-life of 6 minutes, indicative of a very large complex or stable interaction within the nucleolus. They conclude that c-Myc may be sequestered by nucleoli, with subsequent degradation by proteasomes.
Fluorescence correlation spectroscopy (FCS) has been used to study concentrations, interactions and conformational changes of molecules at very low concentrations (~less than μM). It is possible to use FCS to observe the translational motion of fluorescently labeled molecules. The test volumes interrogated, using either one or two photon excitation, are on the order of less than one femtoliter. At nanomolar levels, only a few molecules traverse this active volume, giving rise to a Poisson distribution of particle numbers. The small volumes probed in FCS are ideal to observe molecules that are spatially confined to intracellular compartments (like nuclei). The most basic application of FCS, and the one most commonly used to study biomolecular associations analyzes fluctuations in local concentration (via number (and hence intensity) fluctuations). These intensity fluctuations are recorded over time at a sampling rate faster than the motions under observation. The parameters recovered from subsequent autocorrelation analysis are: 1) Go, the amplitude at delay time zero- simply related to the inverse of the number of particles diffusing in the small volume and 2) the translational diffusion time. To obtain the self diffusion coefficient, the autocorrelation function must be analyzed with a particular physical model such as 3D diffusion, anomalous diffusion or diffusion with binding.
Fluorescence Correlation Spectroscopy (FCS) exploits small fluctuations of concentrations by monitoring very small volumes. In fact, the smaller the concentration that can be observed, the larger the fluctuations that can be recorded and consequently the larger the autocorrelation signal. Hence the maximal correlation (Go) is inversely proportional to the total concentration. For a protein expressed at the few nanomolar level, like c-Myc, FCS is one of the best methods available to monitor dynamics in vivo.
Results
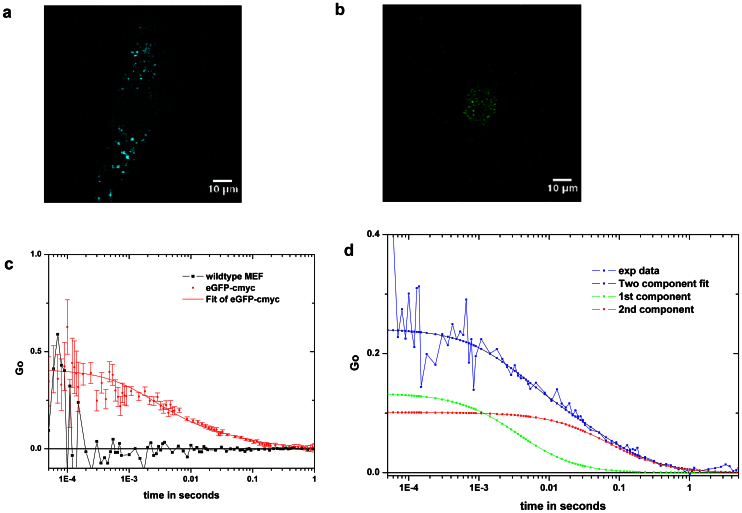
To insure that c-Myc-eGFP was appropriately expressed and physiologically regulated, cells derived from a mouse homozygous for a c-myc allele fused in frame at the end of its coding sequence to DNA encoding a destabilized eGFP; the chimeric Myc-eGFP from these cells is fully functional and expressed at physiological levels, and turns over normally10. Myc-eGFP was readily observed in MEFs by standard fluorescence microscopy only after serum-induction and stabilization using the proteasome inhibitor MG132 (supplementary Figure SN1). To reliably detect the low levels of c-Myc-eGFP within the MEF nuclei, we used a narrow eGFP emission filter coupled with two photon (2p) excitation in the red at 970 nm. We noticed that within the (-GFP) nucleus there was relatively little autofluorescence. As seen in Figure 1, a high resolution scan barely detects the c-Myc-eGFP in a MEF cell stimulated with serum after 30 minutes, even though this level (~10 nM) is higher than the steady state level measured (8 ±1.3 nM). Using FCS, we are able to detect c-Myc-eGFP at very low concentrations without causing visible photodamage to the cell. Figure 1C (in red) compares the autocorrelation traces of the c-Myc-eGFP +ve cells with those of a wild-type MEF lacking GFP. Note the long “tail” in the autocorrelation function(ACF) of the c-Myc-eGFP protein, indicative of a quite immobile pool. At 970 nm excitation, for cells lacking GFP, it is clear there is very little autofluorescence, and better yet, this component has no apparent correlation (Fig. 1C, black).
Figure 1. MYC partitions into less and more mobile populations.
(a) MEF cell with c-Myc-eGFP excited at 800 nm. (b) Same cell excited at 970 nm (high res scan of 1 ms/pixel), this cell contains about 15 nM of c-Myc-eGFP by FCS. (c) Autocorrelation function of a MEF eGFP-cMyc cell excited at 970 nm (red) and a wildtype MEF with no clear autocorrelation (black). (d) An example of the two component fit performed for ACF is shown.
We found that the overall population of c-Myc-eGFP in these MEF cell nuclei was stable, and that it could be divided into two sub-populations with different diffusion coefficients. These two diffusion coefficients were extracted repeatedly from the ACFs of different cell nuclei. Statistically identifying and separating a third translation component is only possible when three very disparate mobilities are evident, and this was not the case in this data. We thus decided to identify only two (arguably mixed) fractions, one denoted “mobile” and one “less mobile”. This simplification was needed despite the fact that the “tail” can be very smooth at times, hinting at lesser contributions from multiple slow diffusion coefficients and/or other dynamic processes like transient binding. Figure 1D shows the ACFs for the two separate components with the appropriate Gi(τ)'s; we separate them into a faster or more mobile fraction (green) and a less mobile or immobile (almost) fraction, blue.
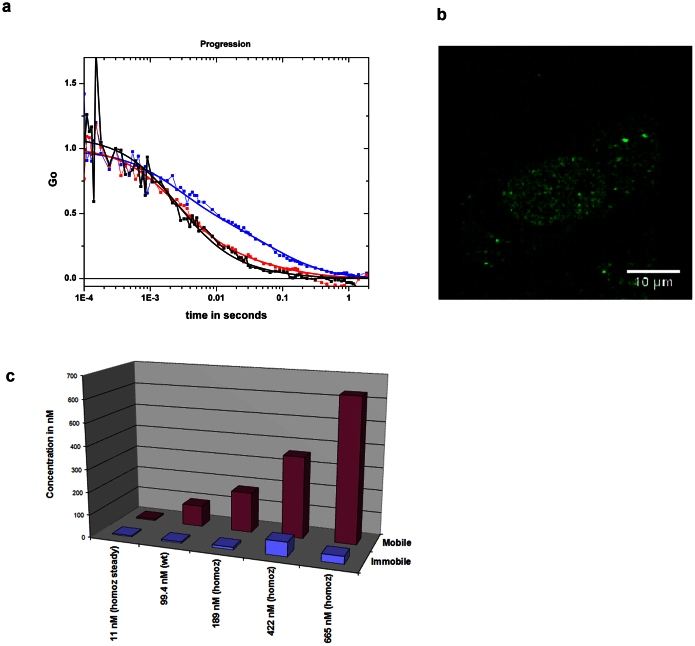
Figure 2a shows the apparent change in populations in three different cells manipulated to produce different concentrations of c-Myc-eGFP. These include transient transfection of eGFP-c-Myc into wild-type or into the homozygous MEFs. (It should be noted that expression of the transfected Myc over the short intervals required for these experiments was not associated with any obvious changes in cellular or subcellular phenotype.) The low levels of expression are exemplified in Figure 2B, where a 2p image of a cell containing 100 nM of c-Myc-eGFP is barely visible. The image was taken over a long exposure (at a 1 ms/pixel duration) and moderate resolution (256 × 256 pixels). As can be seen in Figure 2c, the more “mobile” fraction increases as the total expression of transfected protein increases; concurrently, the “immobile” fraction remains nearly constant. At 100 nM, the more mobile fraction represents >90% of the total c-Myc concentration.
Figure 2.
Increased MYC augments mobile-MYC (a) ACF of three MEF cells with different concentrations of eGFP-cMyc. The concentrations are 10 (blue), 94 (red) and 400 nM (black). All three ACFs have been normalized to G(blue) at τ = 1e − 3. (b) Image of MEF cell at 970 excitation containing about ~50 nM of eGFP-cMyc (high resolution). (c) The fractions of the mobile and immobile populations obtained from transfections of wild type and homozygous cells are shown.
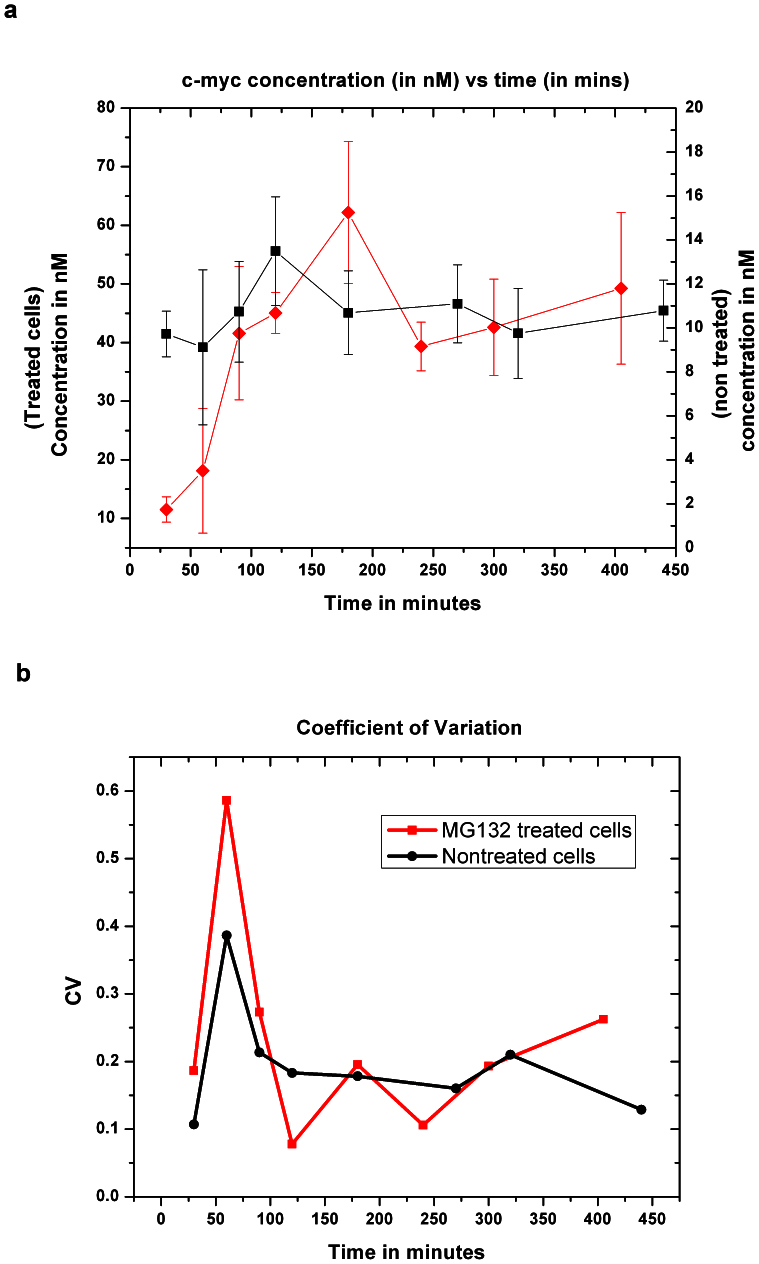
We also studied the physiological accumulation of c-Myc-eGFP over time after serum stimulation of serum-starved MEFs. The addition of the drug MG132 was used in some cells to prevent the degradation of c-Myc-eGFP- allowing us to follow the stimulated accumulation for 6–7 hrs. The results are shown in Figure 3A, and match the expected temporal pattern18 There is a peak at 2–3 hrs for the cells without MG132 (with an increase of about 30% from the final steady state level of about 8 nM). For those with MG132, we see a prolonged and exaggerated increase of cMyc-eGFP concentration. Also, while the concentrations of both fractions increase, the more mobile fraction rapidly becomes dominant (data not shown). We also considered the expected noise for what is essentially an expression counting experiment. Figure 3B displays the coefficient of variation (CV), defined as the ratio of the standard deviation to the mean, for MG132 and nontreated cells. The CV spikes one hour after addition of rich serum medium. The large increase is likely due to lack of synchrony in the cells in response to the impulse and consequent variation in nuclei of different cells. Despite accumulating considerably more Myc protein, the CV of the MG132 treated cells is slightly larger than for nontreated cells. CV for a counting dominated process should decrease by 1/square root of the controlling number; thus the relevant number in these nuclei (perhaps the number of active transcripts, see discussion) did not need to increase to increase c-MYC numbers.
Figure 3. Its coefficient of variation indicates that MYC is regulated at several steps.
(a) The c-Myc concentration of MEF cells treated and non-treated with MG132 over a period of 6–7 hrs. Left axis is the concentration of the MG132 treated cells (RED) and right axis is concentration of non-treated cells (BLACK). (b) Coefficient of Variation (CV) for nontreated and MG132 treated cells.
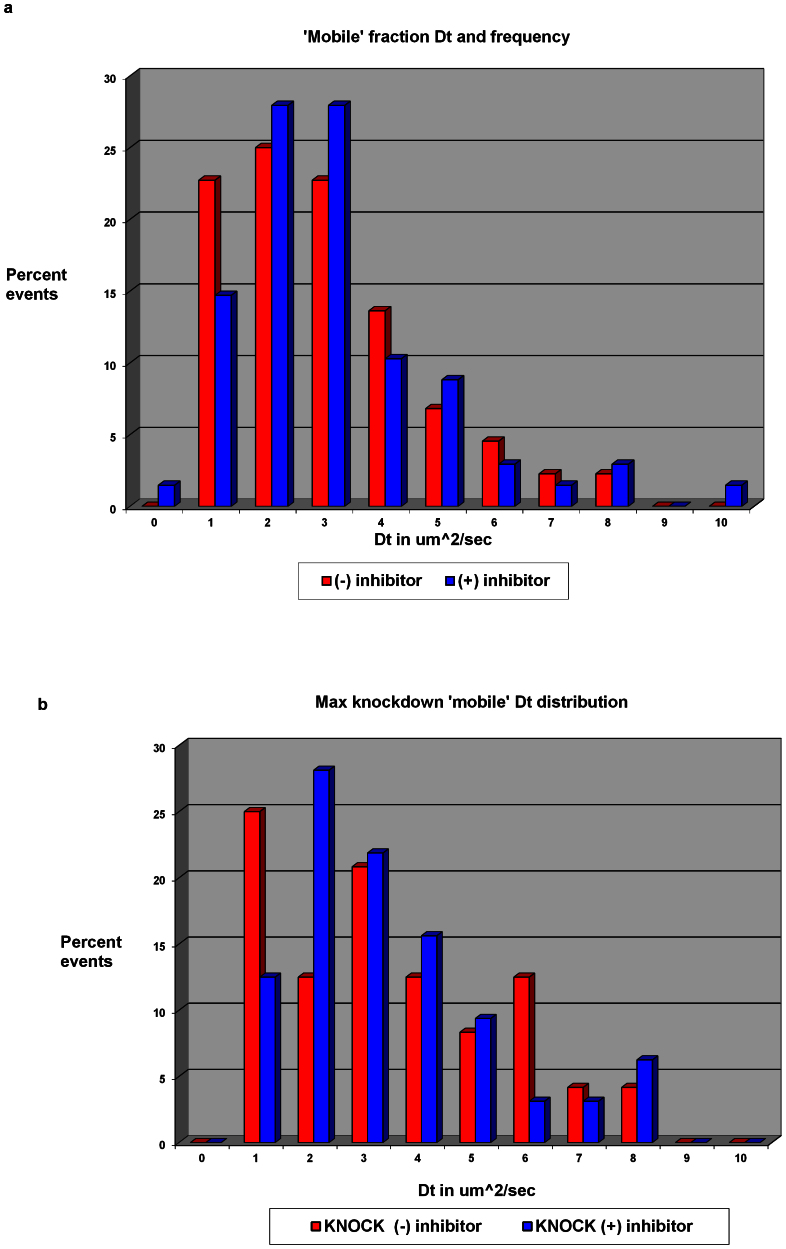
The diffusion of even the most mobile fraction was considerably slower than would be expected for a freely diffusing Myc-eGFP-Max dimer. To test if the mobility of either the immobile or mobile fractions depended on DNA binding, cells at steady state were treated with the inhibitor of Myc-Max dimerization 10058-F419. Because structural studies have revealed that DNA-binding by Myc is stringently dependent on such dimerization20, this inhibitor was expected to distinguish DNA-binding dependent and independent subpopulations of Myc. This same inhibitor has been demonstrated to reduce Myc binding at target promoters genome-wide, in vivo10. Figure 4 shows the distribution of diffusion coefficients of the recovered ‘mobile' fraction. The mean Dt of the mobile fraction for these cells was nearly unchanged: 3.4 ± 1.6 and 3.0 ± 1.8 μm2/sec with and without inhibitor (10058-F4) respectively. Those broad error limits represent biological variation, however, and it is clear upon inspection that the mobile fraction histogram skewed towards a more mobile distribution when the inhibitor was added. Although 10058-F4 shifted c-Myc-eGFP to slightly higher mobilities, it did not provoke the wholesale release of Myc to freely diffusing forms; thus, it seems that even without Max, Myc remains associated with its other macromolecular partners. At very high concentrations of transfected c-Myc-eGFP, a diffusion coefficient of 5.5 ± 0.3 μm2/sec was recovered for the most mobile fraction, suggesting that MYC may eventually saturate those lower mobility partners and “spill over” into a somewhat higher mobility niche. We considered using a ‘global' approach for the analysis of the diffusion coefficients, but at this stage we were more interested in obtaining data on Dt variations in many cells and the qualitative patterns that could be discerned. Eventually, when we return to the mosaic nature of expression, global analysis will become a key element in quantifying populations and rates.
Figure 4. Inhbition of MYC-Max dimerization does not liberate freely diffusing MYC.
(a) Diffusion coefficient shifted slightly with the addition of inhibitor. (n = 42 for no inhibitor and 57 for inhibitor). (b) Max knockdown cells have a slightly larger diffusion coefficient to start and the inhibitor further increases this fraction (n = 24 for no inhibitor and 32 for inhibitor).
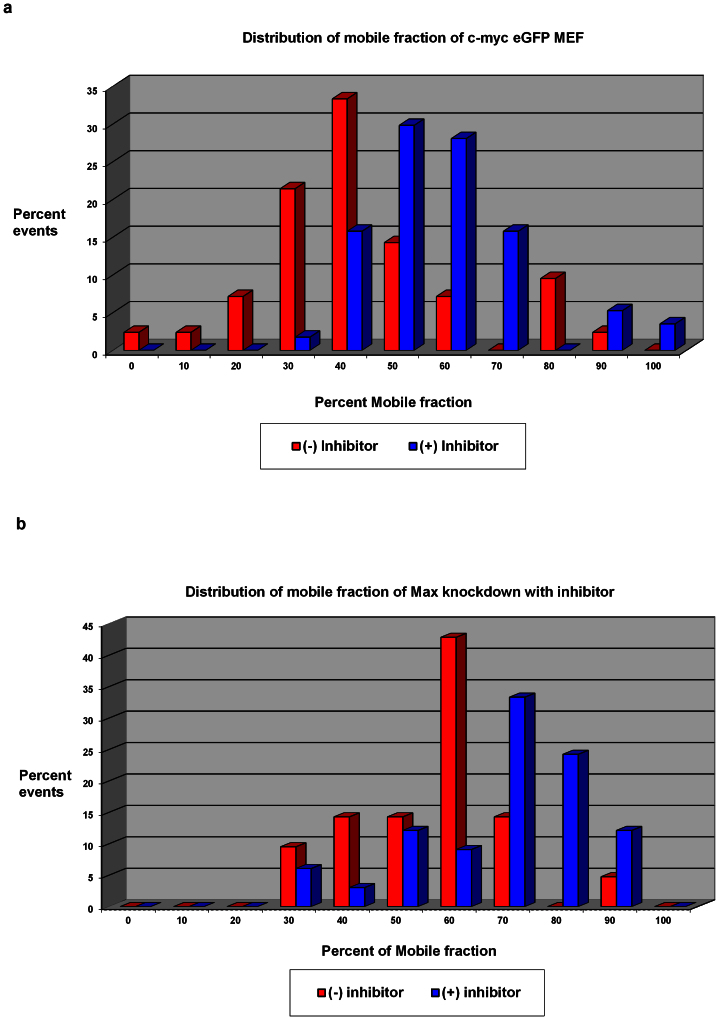
RNAi or shRNA that targeted Max efficiently (supplementary Fig. SN2) were also used in combination (or not) with 10058-F4 to liberate Myc from Myc-Max dimers in cells at steady state. While these manipulations failed to expose a new highly mobile population of “free” Myc-eGFP, the diffusion times recovered from these measurements indicated noticeable increases in the mobile fraction. The Dt of the immobile fraction remained near 0.20 ± 0.06 μm2/sec with all treatments. Figure 5 shows the visibly altered distribution of the mobile fraction Dt values.
Figure 5. Without Max, MYC complexes are more mobile.
(a) Mobile fraction shifted slightly with the addition of inhibitor (n = 42 for no inhibitor and 57 for inhibitor). (b) Knockdown cells have a slightly larger population of mobile fraction to start and the inhibitor further increases this fraction (n = 24 for no inhibitor and 32 for inhibitor).
In short, the mobile fraction for cMyc-eGFP in untreated cells was 45 ±19%; addition of inhibitor shifted this to 58 ± 15% (see Fig. 5A). Knockdown cells had a mobile fraction of 56 ± 14%; synergy of 10058-F4 and RNAi against Max increased the mobile fraction to 68 ± 16% (see Fig. 5B).
Discussion
We demonstrate the ability of FCS to measure the very low physiological in vivo (nM) levels of GFP-tagged c-MYC expressed from the enodgenous c-MYC locus in the nuclei of normal MEFs derived from a knock-in mouse. We find a very immobile fraction is present at the level of a few nanomolar. The most likely explanation for the immobility of this population of Myc is that it is chromatin bound; if so, this localization is likely to play an important role in the regulation of the c-Myc protein and its targets; alternatively, this Myc would need to be associated with some stationary structure, compartment or network within the nucleus. Upon flooding the cell with large quantities of exogenously expressed c-Myc-eGFP, the excess protein partitions mainly into a mobile fraction suggesting saturation of the immobile compartment. Because cancers often over-express c-Myc, these results suggest that Myc may also overpopulate the immobile sites available in tumor cells. It may be of considerable importance to accurately define Myc expression levels in transfection experiments in order to achieve an accurate reflection of the physiology or pathology of normal versus malignant cells. Certainly, we show by transfection that at c-MYC levels beyond physiological, distinctly non-native mobility patterns are manifest.
Serum stimulation yielded the expected peak of Myc-eGFP 2–3 hours post-stimulation; treatment with the proteasome inhibitor MG132 converted such a pulse of Myc into a monotonic accumulation of c-Myc-eGFP rising to 400% of the steady state levels. On statistical grounds, the coefficient of variation (CV) reporting the cell-to-cell variation in c-Myc levels in MG132-treated cells would thus have been expected to be halved relative to the untreated cells; the fact that MG132 instead slightly increased MYC's CV suggested that proteosomal degradation may help to buffer fluctuations in MYC levels within single cells. From another viewpoint, the counting error expected for a protein with copy number near 1000 is only about 3%, while our increased CV lies within the range 10–20% over the length of the experiment. The difference could be thought of as a direct measure of the transcript number and/or translational burst size. A 10% CV would correspond to a transcript number of 100, each generating (on average) about 10 c-MYC. Precise determination of exactly why CV is elevated above protein copy number –predicted levels will, of course, require additional experiments that manipulate translation, but those are beyond the scope of this manuscript.
The mobile fraction increases upon either the addition of the inhibitor 10058-F4 and/or knockdown of the Max protein, while the diffusion coefficients are not changed in serum starved cells. In serum stimulated cells, however, there was a shift to a higher diffusion coefficient. Mehmet et al.21, in measurements performed by ELISA, demonstrated that there are ~450 molecules of c-Myc per cell at steady state levels. To put the measured values of ~10 nM in perspective, the volume of a typical MEF nucleus is ~300 fL (Daniel Larson, NCI, Bethesda, personal communication). This combination yields about 1800 fluorescent particles per cell.
Max is expressed at higher levels than Myc protein in the cell. Therefore, to observe a Max-Myc complex using two color fluorescence cross correlation spectroscopy (FCCS), Max-eGFP must be expressed at levels high enough to overwhelm endogeneous Max; these levels are suboptimal for FCCS. The alternative would be to fuse a FP with Max at its native chromosomal site (as has been done with Myc); such a Max-FP knock-in is not currently available.
The ability of FCS to nondestructively report on both mobile and chromatin-associated populations of c-Myc in nuclei, despite low copy numbers, points toward a future where this technique is employed to follow the dynamics of transcriptional activation processes throughout the cell cycle and to assess the possibly functional heterogeneity within a population of cells. Few methods can directly monitor concentration of a key protein at nM levels; fewer still can do so nondestructively (allowing sequential observation throughout cell cycle or experiment condition changes) and even fewer can explore the mosaic nature of expression via histograms. FCS is thus a desirable technique for studies of regulators.
Methods
2P FCS
Two photon imaging and FCS measurements were carried out using a system built in the Ultrafast Laser Microscopy laboratory. The excitation source was a tunable Ti-Sapphire wideband Mai Tai laser (Spectra-Physics-Newport) set at 970 nm. The excitation power was set at a level where no bleaching or visible damage to the cell occurred (~10 mW at the microscope entrance). The microscope was a Zeiss Axiovert 135 M using an E680 SP 2P dichroic filter (Chroma Technology Corporation) to eliminate the IR exciting light. The objective was a 100× Plan-Neofluar oil objective (Zeiss) with NA 1.3. The microscope was equipped with a piezo-electric stage for x-y control (Mad City Labs.) and the objective was also equipped with a piezo-electric device for z-control (Mad City Labs). Detection used an Alba 3 channel fluorescence correlation system (ISS, Inc). The detected light was split the into two channels with a 495 nm dichroic mirror (Chroma). A 515 +/− 30 nm bandpass emission filter was placed before channel 1 for eGFP detection, and a 450 +/− 40 nm bandpass filter was used with channel 2 to detect autofluorescence. Excitation volumes were calibrated by raster scanning sub-difraction limit (e.g. 40 nm) fluorescent beads. The1/e2 beam waist in plane, wo, was found to be 0.34 μm, while the axial extent zo was 1.8 μm. The diffusion coefficient of eGFP in a cell has been measured before to be 26 ± 7 μm2/s. Also, a mutant of the transcription factor VBP that does not bind DNA and has only a “leucine zipper” domain was measured to have a diffusion coefficient of 13 ± 4 μm2/s 22. These values are in agreement with the diffusion coefficients of NLS-eGFP proteins23.
Cell culture and transfections
In some experiments, a plasmid vector transiently directing the expression of the same MYC-eGFP fusion was transfected into these same homozygous or non-targeted (wild-type) fibroblasts. Cells were incubated with the inhibitor 10058-F4 for at least six hours. Knockdown treated cells were incubated for 48 hours before measurements.
Preparation of MEF and 3T3 cells
MEF cells were prepared following standard protocols (http://www.fhcrc.org/science/labs/fero/protocols/MEF.html). In brief, E13.5 embryos were harvested from wt or homozygous c-Myc-eGFP knock-in mice and washed with sterile PBS to remove any remaining maternal blood cells. After removing the umbilical cord, liver, spleen and tubular intestine, the bulk of the CNS tissue was trimmed by dissecting the head above the level of the oral cavity. The remaining embryo was treated with trypsin (5 ml/embryo) and minced with sterile forceps and scissors for 2–3 minutes and incubated in 37°C for 15 minutes. The embryonic tissue of embryo was further dissociated by triturating with a10 ml pipet. Isolated primary MEF cells were pelleted in 5 ml of DMEM with 10% FBS by centrifugation at 1000 RPM for 5 min. After aspirating the supernatant, the P0 primary MEF cells were re-suspended in fresh medium and incubated at 37°C in 5% CO2. The 3T3 cells were generated by passage of 1.2 × 106 primary MEF cells every 3 days on P100 dishes (http://labs.fhcrc.org/fero/Protocols/MEFs. html; Nilausen k. Green H. Exp Cell res. 1965). After a rapid growth period (passage 1–5), a slower growth period (passage 5–10) and senescence (little or no growth, passage 10–25) the immortalized 3T3 grow out.
Serum starvation and stimulation
The WT, c-Myc-eGFP primary and/or NIH 3T3 cells were cultured in DMEM containing 0.03% FBS for 48 hrs, then tripsinized and re-cultured in the DMEM medium containing 10% FBS and 1× MEM Non-Essential Amino Acids (MEM NEAA, GiBCO 11140) in the presence and/or absence of proteasome inhibitor MG132 or Myc-MAX inhibitor, 10058-F4. The cells were seeded in the Lab-tek Chambered Coverglass w/cvr #1German borosilicate sterile two chambers slides (NUNC cat. # 155380) and incubated at 37°C in 5% CO2. Pheno-less DMEM (GIBCO, 21063) and charcoal/dextran treated FBS (HyClone, SH30068.01) were used during analysis.
Knock-down MAX gene
Transient knock down of MAX
Custom designed stealth siRNA for MAX accession No. BC138671 position 313, 5′-CCAUACGCACCAGCAAGACAUUGAU-3′ and control, 5′-AUCAAUGUCUUGCUGGUGCGUAUGG-3′ (Invitrogen) were electroporated to either primary or NIH 3T3 cells with Amaxa MEF2 Nucleofector Kit (LONZA, VPD-1005) using program MEF2/A023 followed manufactory's protocols. About 2.5 nmole of each stealth siRNA were used for one well of a 6 well plate and incubated at 37°C, in 5% of CO2 for 40–48 hrs for assay.
MAX knock-down stable cell line
PLKO.1 lentiviral vector carrying MAX shRNA/RNAi sequences 5′-AAAGCTGTCTTTGATGTGGTC-3′ (Thermo-Open Biosystem, cat. no. RHS3979-9607256, clone ID: TRCN0000039867) was transfected into HEK 293T cells with arrest-in reagent according to manufactorer's protocol. Viral particles were packaged, titered and transduced into the c-Myc-eGFP NIH 3T3 cells to create the MAX stable knock-down cell line. The PLKO.1 vector plasmid was the negative controls for these assays.
Q-real-time PCR analysis
Total RNAs were purified from WT, c-Myc-eGFP and c-Myc-eGFP-Max knock-down NIH 3T3 cells at various time points as described (Nie et al., 2012);, 0.25 mg of total RNA from each time point were used for first-strand c-DNA synthesis (Enhanced Avian HS RT-PCR-100 kit, SIGMA, Cat. No. HSRT100-1kt). The primers and probes for each gene were designed using Roche Universal Probe library Assay Design Center Web (Figure S7F). q-PCR was performed with the Roche LightCycler 480 system (LightCycler 480 Probe Master, Ref. No. 04 707 494 001; Universal Probe library set, Human, Ref. No. 04 683 633 001). The expression levels of c-Myc, Max and MAD were normalized with GAPDH.
Immunoblotting
Equal amounts of c-Myc-eGFP-MEF and MAX knock-down cells at different time points were harvested in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% NaDOC, 0.05% SDS, 50 mM Tris, pH 7.5 and 1× protease inhibitor). Proteins were separated on 4–12% SDS-PAGE and blotted with anti-c-Myc (Epitomics, Cal. No. Y69), anti-Max (C-17) (Santa Cruz Biotech, Cal. No. SC-197), anti-Mad 1 (FL-221) (Santa Cruz Biotech, Cal. No. SC-766) and anti actin (1-19) (Santa Cruz Biotech, Cal. No. SC-1616).
Author Contributions
T.R., Z.N., J.K. and D.L. planned experiments, T.R., Z.N., V.K. and R.C. performed experiments, T.R., Z.N., J.K. and D.L. analyzed data, T.R., J.K. and D.L. wrote main text, Z.N. and D.L. wrote the supplement, T.R., Z.N., V.K. and R.C. made figures in text and supplement. All authors reviewed the manuscript.
Supplementary Material
supplementary information
Acknowledgments
This research was supported by the Intramural Research Programs of the NIH at National Cancer Institute, Center for Cancer Research and NHLBI.
References
- Dang C. V. Enigmatic MYC Conducts an Unfolding Systems Biology Symphony. Genes Cancer 1, 526–531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylishen A. R. & Penn L. Z. Myc: the beauty and the beast. Genes Cancer 1, 532–541 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell Tk Kretzner L. B. E. E. R. W. H. Sequence-specific DNA-Binding by the c-myc protein. Science 250, 1149–1151 (1990). [DOI] [PubMed] [Google Scholar]
- Blackwood Em Eisenman R. N. Max- a Helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251, 1211–1217 (1991). [DOI] [PubMed] [Google Scholar]
- Nilsson J. A. & Cleveland J. L. Myc pathways provoking cell suicide and cancer. Oncogene 22, 9007–9021 (2003). [DOI] [PubMed] [Google Scholar]
- Cole M. D. & Nikiforov M. A. Transcriptional activation by the Myc oncoprotein. CurrTopMicrobiolImmunol 302, 33–50 (2006). [DOI] [PubMed] [Google Scholar]
- Amati B., Littlewood T., Evan G. & Land H. The C-MYC protein induces cell-cycle progression and apoptosis through dimerization with Max. Embo J 12, 5083–5087 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inghirami G. et al. Down-regulation of LFA-1 adhesion receptors by C-MYC oncogene in human B-lymphoblastooid cells. Science 250, 682–686 (1990). [DOI] [PubMed] [Google Scholar]
- Lin C. Y. et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A., Rustum C., Hallberg E. & Wright A. P. H. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J Cell Sci 116, 1707–1717 (2003). [DOI] [PubMed] [Google Scholar]
- Levens D. You Don't Muck with MYC. Genes Cancer 1, 547–554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. & Levens D. Making myc. Curr Top Microbiol Immunol 302, 1–32 (2006). [DOI] [PubMed] [Google Scholar]
- Wierstra I. & Alves J. The c-myc promoter: still MysterY and challenge. Adv Cancer Res 99, 113–333 (2008). [DOI] [PubMed] [Google Scholar]
- Hann Sr Eisenman R. N. Proteins encoded by the human C-MYC oncogene- Differential expression in neoplastic cells. Mol Cell Biol 4, 2486–2497 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H., Gray H. E. & Godeau F. Growth-dependent synthesis of C-MYC encoded proteins- Early stimulation by serum factors in synchronized mouse 3T3 cells. Mol Cell Biol 5, 2903–2912 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkowski K. A., Salghetti S. E. & Tansey W. P. Stable and unstable pools of Myc protein exist in human cells. Oncogene 21, 8515–8520 (2002). [DOI] [PubMed] [Google Scholar]
- Dean M. et al. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem 261, 9161–9166 (1986). [PubMed] [Google Scholar]
- Wang H. et al. Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther 6, 2399–2408 (2007). [DOI] [PubMed] [Google Scholar]
- Nair S. K. & Burley S. K. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205 (2003). [DOI] [PubMed] [Google Scholar]
- Mehmet H. et al. Large induction of c-Myc is not essential for the mitogenic response of Swiss 3T3 fibroblasts. Cell Growth Differ 8, 187–193 (1997). [PubMed] [Google Scholar]
- Michelman-Ribeiro A. et al. Direct Measurement of Association and Dissociation Rates of DNA Binding in Live Cells by Fluorescence Correlation Spectroscopy. Biophys J 97, 337–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Corbett A. H. & Berland K. M. The Intracellular Mobility of Nuclear Import Receptors and NLS Cargoes. Biophys J 96, 3840–3849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information