Abstract
In renal proximal tubule (PT) cells, sodium-phosphate cotransporter IIa (NaPiIIa) is normally concentrated within the apical membrane where it reabsorbs ∼70% of luminal phosphate (Pi). NaPiIIa activity is acutely regulated by moderating its abundance within the apical membrane. Under low-Pi conditions, NaPiIIa is retained within the apical membrane. Under high-Pi conditions, NaPiIIa is retrieved from the apical membrane and trafficked to the lysosomes for degradation. The present study investigates the role of Shank2 in regulating the distribution of NaPiIIa. In opossum kidney cells, a PT cell model, knockdown of Shank2 in cells maintained in low-Pi media resulted in a marked decrease in NaPiIIa abundance. After being transferred into high-Pi media, live-cell imaging showed that mRFP-Shank2E and GFP-NaPiIIa underwent endocytosis and trafficked together through the subapical domain. Fluorescence cross-correlation spectroscopy demonstrated that GFP-NaPiIIa and mRFP-Shank2 have indistinguishable diffusion coefficients and migrated through the subapical domain in temporal synchrony. Raster image cross-correlation spectroscopy demonstrated these two proteins course through the subapical domain in temporal-spatial synchrony. In the microvilli of cells under low-Pi conditions and in the subapical domain of cells under high-Pi conditions, fluorescence lifetime imaging microscopy-Forster resonance energy transfer analysis of Cer-NaPiIIa and EYFP-Shank2E found these fluors reside within 10 nm of each other. Demonstrating a complexity of functions, in cells maintained under low-Pi conditions, Shank2 plays an essential role in the apical retention of NaPiIIa while under high-Pi conditions Shank2 remains associated with NaPiIIa and escorts NaPiIIa through the cell interior.
Keywords: siRNA, live-cell imaging, fluorescence lifetime imaging microscopy-Forster resonance energy transfer, endocytosis, fluorescence correlation spectroscopy, raster image correlation spectroscopy
phosphorus is an essential mineral that participates in a number of physiologic processes including cellular energetics, posttranslational protein modification, intracellular signaling, extracellular bone formation, and pH buffering of blood (4). Serum phosphate (Pi) concentrations are maintained within a narrow range, and deviations in serum Pi levels have pathophysiologic consequences (2, 21). Hypophosphatemia can lead to rickets and brittle bones while even modest hyperphosphatemia can contribute to increased morbidity and mortality from cardiovascular diseases. In patients with chronic kidney disease, management of hyperphosphatemia is a major therapeutic concern since elevated serum Pi levels correlate directly with the progression of the renal disease, acceleration of atherosclerosis, and increases in vascular stiffness and calcification (30).
The kidney plays a pivotal role in maintaining serum Pi levels by acutely and chronically moderating the fractional excretion of Pi into the urine. The proximal tubule (PT), which has at least three distinct Pi transporters that reside within the apical membrane of PT cells to initiate Pi reabsorption, is the primary site of this regulation (14). Of the three transporters, the sodium-phosphate cotransporter IIa (NaPiIIa) accounts for ∼70% of the bulk reabsorption of Pi from the luminal filtrate and is acutely regulated in response to changes in serum levels of Pi or parathyroid hormone (PTH). Under low serum Pi/PTH conditions, NaPiIIa activity is increased primarily by increasing the abundance of NaPiIIa within the apical membrane (12, 14). Under high serum Pi/PTH conditions, NaPiIIa activity is reduced primarily by decreasing the abundance of NaPiIIa within the apical membrane (12, 14). The recovered NaPiIIa is not recycled but is, instead, trafficked to the lysosomal compartment for degradation.
The COOH-terminal tail of NaPiIIa interacts with at least four specific PDZ domain-containing proteins (18, 20, 28), including EBP50 (a.k.a. NHERF-1), E3KARP (a.k.a. NHERF-2), PDZK1 (a.k.a. NHERF-3), and Shank2 (a.k.a. ProSAP1; CortBP1). These PDZ domain proteins are positioned to moderate the distribution of NaPiIIa within PT cells and impact serum Pi homeostasis. Consistent with a role in retaining NaPiIIa within the apical membrane, EBP50(−/−) mice have diminished NaPiIIa within the microvillar membranes, elevated levels of NaPiIIa within the cell interior, and increased urinary excretion of Pi when compared with wild-type littermates (41). Although less pronounced, PDZK1 is also implicated in retaining NaPiIIa within the apical membrane of PT cells (5). The present study seeks to advance the understanding of the functional significance of Shank2 in regulating the intracellular distribution of NaPiIIa.
The predominant Shank2 spliceoform within the kidney, termed Shank2E (29), is a multiple domain protein that includes an ankyrin repeat domain, a SH3 domain, a single PDZ domain, a proline-rich region, and a sterile-α motif (40). Foreshadowing its importance within epithelial cells, the PDZ domain of Shank2 binds to key transporter and ion channel proteins, including the cystic fibrosis transmembrane conductance regulator (CFTR) Cl− channel, the sodium-proton exchanger 3 (NHE3), and NaPiIIa (19, 23, 28). Rat renal PT cells maintained under low serum Pi conditions have both Shank2 and NaPiIIa concentrated within their apical microvilli. The functional significance of Shank2 on the microvillar retention of NaPiIIa, however, has not been established. NaPiIIa and Shank2 respond to acute increases in serum Pi by undergoing parallel patterns of redistribution through endosomal compartments (12, 28). These observations suggest NaPiIIa and Shank2 might remain associated and comigrate together through the cell interior. The present studies employ opossum kidney (OK) cells, an established PT cell model with regulated expression of NaPiIIa, to explore the relationship between Shank2 and NaPiIIa under both low- and high-Pi conditions. Knocked down expression of Shank2 in cells maintained under low-Pi conditions showed that Shank2 plays a prominent role in the microvillar retention of NaPiIIa. Under high-Pi conditions, live-cell imaging, correlation spectroscopy, and fluorescence lifetime imaging microscopy-Forster resonance energy transfer (FLIM-FRET) techniques extended the capabilities of traditional light microscopy techniques to quantitatively demonstrate on a microsecond time scale that, following endocytosis, Shank2 and NaPiIIa remain closely associated and transit through the cell interior in spatial and temporal synchrony.
MATERIALS AND METHODS
Cell culture.
OK PT cells were maintained in growth media [DMEM/F-12 medium (Sigma, St. Louis, MO), 10% FBS (Hyclone, Logan, UT), 5 mM l-glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin] at 37°C in a humidified atmosphere with 5% CO2. Cells were grown to confluency, cultured in growth media with reduced serum (0.2% FBS) for 24 h and incubated in low-Pi media [phosphate-free DMEM high glucose media (Invitrogen, Carlsbad, CA), 0.2% FBS, 5 mM l-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 0.1 mM Na2HPO4] for 24 h. During the study periods (1, 2, or 3 h), control cells were maintained low-Pi media. Experimental cells were shifted into high-Pi media [phosphate-free DMEM high glucose media (Invitrogen), 0.2% FBS, 5 mM l-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM Na2HPO4].
DNA/small interfering RNA transfections.
Opossum Shank2-specific and scrambled small interfering (si)RNAs were designed and generated by Invitrogen. This included unlabeled siRNAs [Western blotting and quantitative (q)PCR analysis studies] and siRNAs labeled with Alexa-Fluor 546 (immunofluorescence imaging studies). siRNA sequences against two regions of opossum Shank2 included 5′-CCAAGUUCCCAGAGGAAGAGAUGUU-3′ (forward), 5′-AACAUCUCUUCCUCUGGGAACUUGG-3′ (reverse), 5′-Alexa546-GAAGAGAUAUUCGGCGAGGAAA-3′ (forward), and 5′- UUUCCUCGUCGCCGAACAUCACAAC-3′ (reverse). The scrambled sequences 5′-Alexa546-GAAGUAGGCUUAGCGGAGCGAGAAA-3′ (forward) and 5′-UUUCUCGCUCCGCUAAGCCUACUUC-3′ (reverse) were used as a negative control. Transfection of OK cells was performed using Lipofectamine RNAi-MAX as directed by the manufacturer (Invitrogen). The cells were allowed to incubate for 72 h after transfection, and the medium was changed into low-Pi medium for 24 h and processed for Western blotting, qPCR analysis, or immunofluorescence imaging (see below).
To assess the consequences of Shank2 overexpression on NaPiIIa abundance, OK cells were transfected with 1 μg/35 mm well of 3XFlag-Shank2E cDNA using Lipofectamine 2000 reagent as directed by the manufacturer (Invitrogen). Four different fluorescence-tagged plasmid constructs were also used in the present studies. These included mRFP-Shank2E [rat; in pcDNA3.1(−); Invitrogen], EYFP-Shank2E [rat; in pcDNA3.1(−); Invitrogen], GFP-NaPiIIa (mouse; Moshe Levi; University of Colorado, Denver), mCherry-NaPiIIa (mouse; Moshe Levi; University of Colorado, Denver), Cer-NaPiIIa (mouse; Moshe Levi; U Colorado-Denver), and GFP-Rab11a (Rytis Prekeris; University of Colorado, Denver). OK cells were cotransfected with 500 ng of DNA for GFP-NaPiIIa and mRFP-Shank2E; GFP-Rab11a and mRFP-Shank2E; GFP-Rab11a and mCherry-NaPiIIa; and Cer-NaPiIIa and EYFP-Shank2E studies using Lipofectamine 2000 reagent as directed by the manufacturer (Invitrogen).
qPCR analysis.
Total RNA from OK cells was isolated using Qiagen total RNA mini prep kit as directed (Qiagen, Valencia, CA). Two micrograms of total RNA were reverse-transcribed using Super-Script III (Invitrogen). qPCR was performed using Applied Biosystem 7300 with SYBR Green to quantify the production of dsDNA. Shank2 mRNA levels were normalized to GAPDH mRNA levels. Primer sequences for Shank2 were 5′-GCGCGAGAAGGGCATGTACTACCG-3′ (forward) and 5′-GTCGATCTCCATGCTGCTGCCCTGG-3′ (reverse). Primers for GAPDH were 5′-CCCCTTCATTGACCTCAACTAC-3′ (forward) and 5′-CCAAAGTTGTCATGGATGACCC-3′ (reverse).
Western blotting.
At the appropriate time points, OK cells were lysed in a 5× PAGE buffer [50 mM Tris-base, 5% sodium dodecyl sulfate, 25% sucrose, and 5 mM EDTA pH 8.0, complete protease inhibitors (Roche, Indianapolis, IN)]. Protein concentrations were measured by bicinchoninic acid assay (Pierce; Rockford, IL), and samples were reduced (10% β-mercaptoethanol). Cell lysates were run on 9% polyacrylamide gels, transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA), and subjected to immunoblotting. Blocking, antibody and wash solutions were diluted in blot buffer (150 mM NaCl, 10 mM Na2HPO4, 5 mM EDTA, and 1% Triton X-100 pH 7.4). Membranes were blocked (5% nonfat dry milk; 60 min); incubated with primary antibody [Shank2 (1:100; Santa Cruz Biotechnology; Santa Cruz, CA), NaPiIIa (1:1,000; Affinity BioReagents; Golden, CO), NaPi-4 (1:500, Moshe Levi; University of Colorado, Denver), NHE3 (1:10; Orson Moe, University of Texas, Southwestern), and actin (1:5,000; Chemicon; Temecula, CA)], washed, incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000 dilution; Jackson ImmunoResearch, West Grove, PA), and washed again. The antibody complexes were detected using enhanced chemiluminescence (Pierce) and imaged using a photodocumentation system (UVP; Upland, CA). Densitometric analysis of Western blot bands was performed using ImageJ [National Institutes of Health (NIH), Bethesda, MD].
Immunofluorescence.
For fixed-cell and live-cell confocal microscopy, cells were seeded onto 35-mm glass-bottom MatTek dishes (MatTek, Ashland, MA). Images were acquired on Zeiss LSM 510 confocal microscope (Zeiss NLO 510 with META; Zeiss Plan-Apochromat 63/1.4NA oil; Thornwood, NY) equipped with a temperature-controlled incubation chamber (Solent Scientific). The imaging settings were initially defined empirically to maximize the signal-to-noise ratio and to avoid saturation. In comparative imaging, all settings were kept constant between samples. The illumination was provided by 30-mW argon (488 nm), 5 mW HeNe (633 nm), and 1 mW HeNe (543 nm) lasers. Image processing was performed using Zeiss ZEN 2009 software. Figures were mounted using Adobe Photoshop CS4 (Adobe System).
For fixed-cell images, cells were fixed in 4% paraformaldehyde in PBS with 0.1% Triton X-100 for 20 min at room temperature. Cells were blocked with 10% normal serum, labeled with primary polyclonal antibodies against Shank2 (1:100; Santa Cruz Biotechnoloy) and NaPiIIa (1:200; Affinity BioReagents), washed, and labeled with the appropriate secondary antibodies that were conjugated to Alexa Fluor 488 or Alexa Fluor 546 (Invitrogen). F-actin was concurrently stained with Alexa-Phalloidin 633 (Invitrogen). For staining of endosomal compartments in cells cotransfected with GFP-NaPiIIa and mRFPShank2E, cells were fixed; blocked; incubated with primary antibodies against EEA1 (1:100; Becton-Dickinson; Franklin Lakes, NJ), LAMP1 (1:100; Enzo Life Sciences, Farmingdale, NY), and Rab11 (1:100; Becton-Dickinson; Franklin Lakes, NJ); washed; and labeled with Alexa Fluor 633-tagged secondary antibodies (Invitrogen). The degree of colocalization of pairs of these proteins within the subapical domain of OK cells under high-Pi conditions was quantified by calculating their intensity correlation quotient (ICQ) values (12, 26). Intensity correlation analysis macros (www.macbiophotonics.ca) for this analysis were applied to ImageJ (NIH). Values for the product of the differences from the mean (PDM) quantify the degree of pixel-by-pixel synchrony of the fluorophore intensities in the two channels [PDM = (Ri − Rave) × (Gi − Gave); Ri = individual pixel intensity-red, Rave = average pixel intensity-red, Gi = individual pixel intensity-green, and Gave = average pixel intensity-green]. The ICQ ratio is equal to the ratio of the number of positive PDM values to the total number of pixel values [ICQ = (N±ve/Ntotal) − 0.5; N±ve = number of pixels with a positive PDM value, Ntotal = total number of pixels with a nonzero value in each channel]. The subapical domain of seven individual OK cells after 60 min of high-Pi incubation were analyzed.
In live-cell studies observing the temporal appearance of GFP-NaPiIIa and mRFP-Shank2E in the subapical domain, images were obtained initially at the apical and then subapical levels with cells in low-Pi media. The media were then exchanged for high-Pi media, cells were incubated for 1 h and then images of subapical domain were collected. In studies observing the short-term comigration of dual-labeled endosomes through the subapical domain of OK cells, cells were shifted from low-Pi media into high-Pi media for 1 h. While focused at the subapical level, 30 images at a 20-s time interval were captured. The series of images were analyzed in ImageJ software (NIH).
Fluorescence cross-correlation spectroscopy measurements.
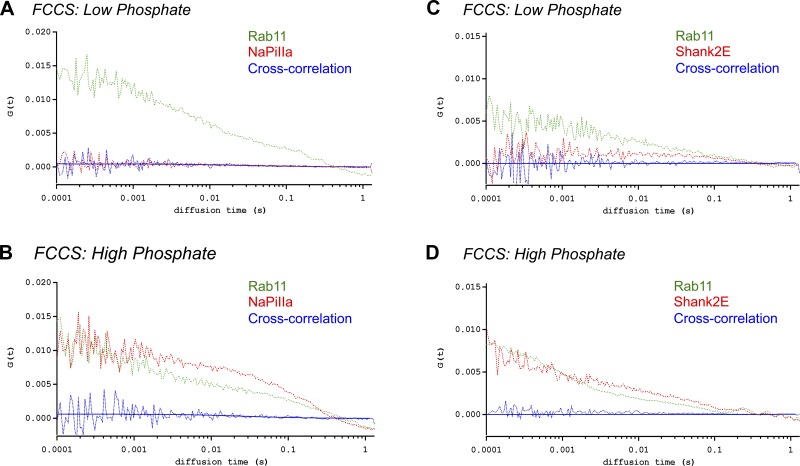
Developed in the early 1970s (27), fluorescence correlation spectroscopy (FCS) and fluorescence cross-correlation spectroscopy (FCCS; Fig. 1A) measure and correlate the temporal intensity fluctuations emitted from fluorescent proteins moving in and out of a small focal volume on a microsecond to millisecond time scale (31). Diffusion coefficients (D) quantify the rates of movement for individual proteins and are derived from the time they dwell within the focal volume and the consequent autocorrelation curves. FCCS measures and compares the degree of temporal synchrony between the fluorescence intensity fluctuations emitted from the two fluorescent proteins within the focal volume. Moving in temporal synchrony in this time frame is a strong indication that the two proteins reside in a common complex. FCS and FCCS studies were performed on an Olympus FV1000 inverted laser scanning confocal microscope with a Plan-Apochromat 60/1.2NA water immersion objective (Olympus, Tokyo, Japan). A 488-nm line (Argon laser) and a 543-nm line (HeNe laser) were used to excite GFP-NaPiIIa and mRFP-Shank2E, respectively. Data were acquired in the photon counting mode using FluoView1000 Software. Calibration measurements were performed using Rhodamine110. The diffusion coefficient (D) for rhodamine110 is 430 μm2/s (15). With the use of the equations D = ω0/4τD (ωo = radius; τD = mobility time), the detection volume was determined to be an ellipsoid with axes of 0.22 and 0.98 μm. The signals were analyzed with SimFCS software (Globals for Images; Enrico Gratton, University of California, Irvine) to determine the autocorrelation of fluctuations within individual channels and the cross-correlation of fluctuations from the two channels. The autocorrelation function used to fit the single channel data was calculated as
where γ, w, and z are geometrical attributes of the focal volume obtained via calibration and N and τD are fitting parameters for determining the mobility coefficients of the fluorescent proteins (36). The cross-correlation function for fitting the data showing temporal synchrony between the fluctuations in the two channels has a similar form with N and τD corresponding to associated proteins moving in synchrony through the focal volume, which two axes are geometrically defined by w and z. The cross-correlation data were obtained for GFP-NaPiIIa:mRFP-Shank2E (n = 10), GFP-rab11:mCherry-NaPiIIa (n = 5), and GFP-rab11:mRFP-Shank2E (n = 5).
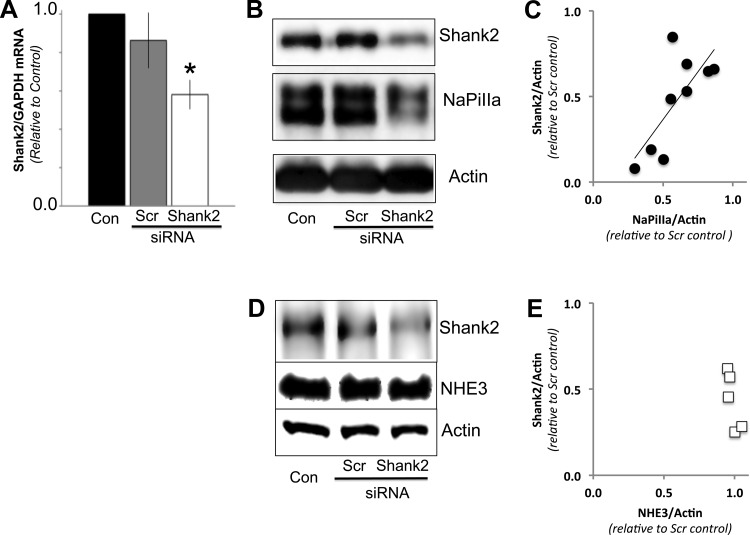
Fig. 1.
Shank2 small interfering (si)RNAs knockdown levels of Shank2 mRNA and protein. A: quantitative (q)PCR analysis of opossum kidney (OK) cells treated with Shank2 siRNAs had significantly lower levels of Shank2/GAPDH mRNA compared with either untreated controls (Con) or cells transfected with scrambled (Scr) siRNAs (n = 3; *P < 0.05). B: Western blot analysis of OK cells treated with Shank2 siRNAs had significantly lower levels of Shank2/actin protein compared with either untreated controls (Con) or cells transfected with scrambled siRNAs (Scr). The abundance of sodium-phosphate cotransporter IIa (NaPiIIa) was also markedly reduced in the cells treated with Shank2 siRNAs. Actin served as a loading control. C: in OK cells treated with Shank2 vs. scrambled siRNAs, linear regression analysis of the densitometric data from the Western blots showed a linear correlation between level of Shank2 knockdown and NaPiIIa abundance (r2 = 0.59; P = 0.02; n = 9 pairs). D: Western blot analysis of cells treated with Shank2 siRNAs showed a decrease in Shank2 abundance but no significant difference in sodium-proton exchanger 3 (NHE3) abundance. E: graph displays densitometry values and shows that despite a range of Shank2 knockdown levels, the NHE3 levels remain similar to those found in Scr siRNA control cells (n = 5).
Raster image cross-correlation spectroscopy.
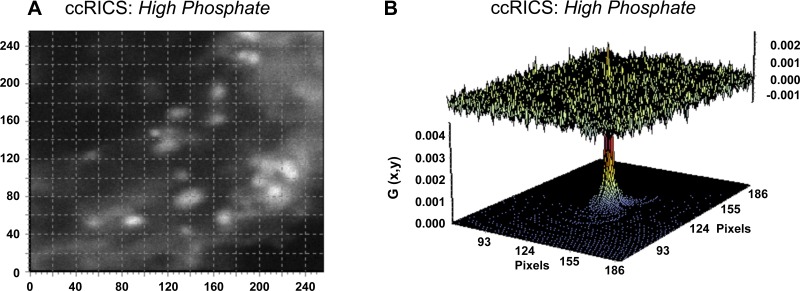
Developed in recent years (8, 10), raster image cross-correlation spectroscopy (ccRICS; Fig. 1B) is a fluorescence correlation technique that repetitively scans a set planar volume within live cells to quantify the spatial and temporal migration patterns of protein pairs (38). ccRICS studies were performed on an Olympus FV1000 inverted laser scanning confocal microscope with a Plan-Apochromat 60/1.2NA water immersion objective (Olympus, Tokyo, Japan). Adjusted to track protein mobility in live cells (i.e., particles moving with mobility coefficients between 1 and 100 μm2/s), the scan speed was set at 20 μs/pixel. Immobile or “slow moving” particles are consequently mathematically removed and do not appear in the acquired images. With the observation plane set in the subapical domain, 100 images were collected for each cell from a scan area of 256 × 256 pixels. The corresponding line time was 6.240 ms, and the frame time was 1.66 s. The pixel size was set to 50 nm, which corresponds to an imaging region of 12.5 × 12.5 μm. A 488-nm line (Argon laser) and a 543-nm line (HeNe laser) were used to excite GFP-NaPiIIa and mRFP-Shank2E, respectively. Data were collected with FluoView1000 Software. Images were analyzed with SimFCS software (Globals for Images; Enrico Gratton, University of California, Irvine). The spatial cross-correlation function for fitting the data was calculated as
where g, w, and z are geometrical attributes of the focal plane being raster scanned with τp as pixel time, τl as line time, ξ as pixel size in the x-direction, and ψ as pixel size in the y-direction (11). D and N correspond to the diffusion and concentration of associated proteins.
FLIM-FRET microscopy.
FLIM measurement of FRET (42) was performed using a Zeiss LSM 510 microscope (Jena, Germany) equipped with a FLIMBox, a digital-frequency-domain setup capable of multiharmonic analysis, as previously detailed (6, 17). Briefly, images of the apical membrane or subapical domain were obtained in the 256 × 256 format with a pixel dwell time of 25.6 μs/pixel and averaging over 20 frames. SimFCS software (Laboratory for Fluorescence Dynamics, University of California, Irvine) was used for the acquisition and analysis of FLIM images following the phasor analysis (16). A digital-frequency-domain setup measured the modulation and phase at each pixel within an image. The modulation and phase determined, respectively, the radial and angular coordinate of the phasor in a polar plot (37). The phasor associated to each cell imaged with FLIM was determined as the average phasor of the pixels corresponding to the apical membrane or subapical domain. In each experiment, the phasor of the unquenched donor (D) was determined as the average phasor of cells transfected only with Cer-NaPiIIa. The phasor of the background (af) was determined as the average phasor of the autofluorescence signal from untransfected cells. The phasor of the donor-acceptor pair (D + A) was determined as the average of cells cotransfected with Cer-NaPiIIa and EYFP-Shank2E.
To quantify FRET, the shift of the (D + A) vs. (D) phasors was determined. The trajectory of variable FRET efficiency is drawn in the plot starting from the (D) position (E = 0) to the (af) position (E = 1; see Fig. 10A). The quenched donor (Dq) position yields the value of efficiency (E) associated with the FRET interaction and represents the maximum FRET detectable if all donors are ideally paired with acceptors. Any experimental phasor along this trajectory corresponds to a pure species of (Dq) with FRET efficiency E. If the donors are not all paired with an acceptor, then a pixel will contain a mixture of quenched and unquenched donors with relative fractions (fq) and (funq). In this case, the (D + A) phasor is a normalized linear combination of (Dq) and (D) phasors and will lie along the line connecting these two absolute phasors. The fraction of interacting donors, (fq), in a given mixture was calculated from the distance of the phasor (D + A) from (D) divided by the distance between (Dq) and (D) (see Fig. 10A).
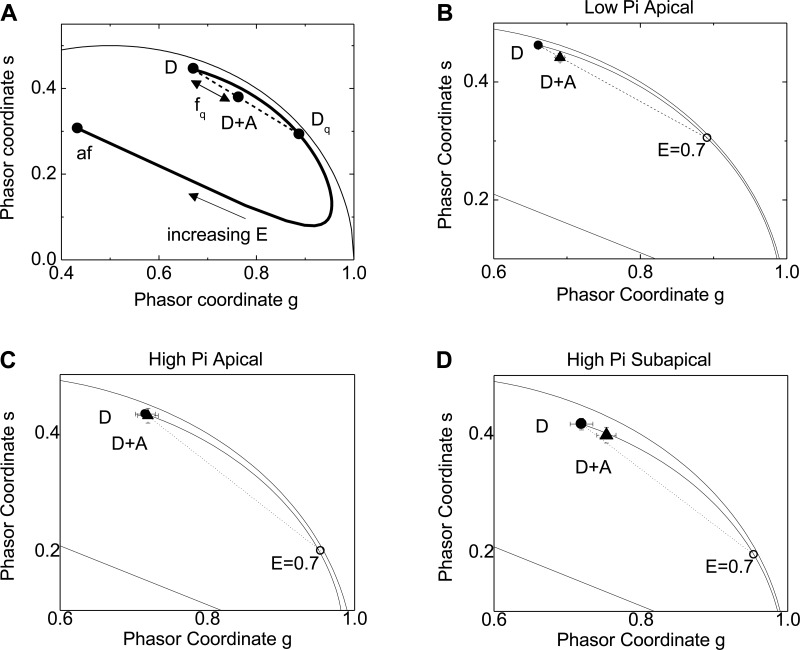
Fig. 10.
fluorescence lifetime imaging microscopy-Forster resonance energy transfer (FRET-FLIM) analysis of Cer-NaPiIIa and EYFP-Shank2E association. A: schematic representation of a phasor plot from a FLIM-FRET measurement showing how FRET is quantified in terms of an efficiency (E) and fraction of quenched donors (fq). Phasor position of the fluorescence lifetime values from cells cotransfected with donor and acceptor proteins (D + A) determines if FRET occurred. The distance between (D + A) and (D) normalized to the distance between (D) and (Dq) equals the fraction of donor fluorophors whose fluorescence lifetimes were decreased by FRET to the acceptor fluorophors (fq). B: within microvilli under low-Pi conditions, there was a significant separation in the phasor coordinates of (D) vs. (D + A). Analysis found there was FRET between 11 ± 2% of donor-acceptor pairs. C: within microvilli under high-Pi conditions, there was no discernible FRET observed. D: within the subapical domain under high-Pi conditions, FRET occurred between 10 ± 5% of donor-acceptor pairs.
Statistical analysis.
All data are presented as means ± SE. Statistical analysis was performed using t-tests for pairs of data and one-way ANOVA for data groups of three or more using Prism-4 GraphPad software. Tukey's post hoc test was applied to the ANOVA data. Values were considered statistically significant when P < 0.05.
RESULTS
Shank2 knockdown impacts NaPiIIa abundance and distribution.
In intact rat renal PT cells and cultured OK cells, low-Pi conditions in the serum or culture medium induce NaPiIIa and Shank2 to concentrate within the apical microvilli with little of either protein found in the cell interior (12, 28). To determine if Shank2 contributed to the microvillar retention of NaPiIIa, the expression levels of Shank2 were either knocked down by transfecting Shank2 siRNAs or were elevated by transfecting Flag-Shank2 cDNA into OK cells. qPCR analyses showed that compared with cells that were untransfected or transfected with scrambled siRNAs, OK cells receiving Shank2 siRNAs had significantly lower Shank2 mRNA levels (Fig. 1A; n = 3). Western blot analysis of OK cells treated with scr siRNA showed Shank2 and NaPiIIa protein levels were not different from control cells (94 ± 9 and 98 ± 9% of controls, respectively; n = 9). In Shank2 siRNA-treated cells, however, Shank2 levels were decreased to 58 ± 4% of levels in control cells (Fig. 1B; n = 9). Analysis of NaPiIIa protein levels in these same cell lysates showed NaPiIIa levels were also significantly diminished (67 ± 8% vs. controls; Fig. 1B; n = 9). Plotting the relative Shank2/actin levels against NaPiIIa/actin levels from pairs of scrambled vs. Shank2 siRNA cells shows a linear correlation between the degree of Shank2 knockdown and the resultant decrease in NaPiIIa levels (Fig. 1C; r2 = 0.59; P = 0.02). To evaluate the specificity of the Shank2 knockdown effect on NaPiIIa abundance, the effect of Shank2E knockdown on NHE3 abundance was measured (Fig. 1D). Like NaPiIIa, NHE3 is concentrated within the microvillar membrane of PT cells and is bound by Shank2 (19). In contrast to NaPiIIa, densitometric analysis showed that in OK cells with knocked down levels of Shank2E, NHE3 levels remained at control levels (Fig. 1E; n = 5).
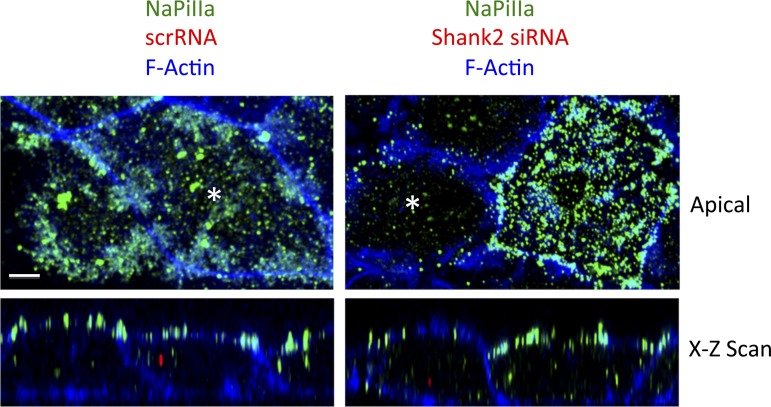
To further investigate this functional relationship between Shank2 and NaPiIIa, the comparative abundance of NaPiIIa was evaluated by confocal immunofluorescence in OK cells that were transfected with Shank2 siRNA. To identify the cells that were successfully transfected with the scrambled or Shank2 siRNA molecules, the siRNA molecules were constructed with an Alexa-546 (red) fluor covalently attached to the siRNA molecules. Positively transfected cells were denoted by the presence of red puncta within the cell interior. In the analysis, neighboring cells without any observed siRNA in the cell interior served as an internal control. In OK cells transfected with scr siRNA molecules, the total fluorescence (i.e., abundance) of NaPiIIa (green) was similar to that observed in neighboring nontransfected cells (Fig. 2, left). NaPiIIa was concentrated within the apical domain with lesser levels of NaPiIIa distributed within the cell interior in both untransfected and scr siRNA-transfected cells. In OK cells transfected with Shank2 siRNA molecules, however, the total fluorescence (i.e., abundance) of NaPiIIa was markedly less than the fluorescence observed in adjacent untransfected cells. In untransfected cells, NaPiIIa was abundantly expressed and remained concentrated within the apical domain while Shank2 siRNA-transfected cells had markedly less GFP-NaPiIIa signal (Fig. 2, right). The remaining signal in the Shank2 siRNA-transfected cells was distributed both within the apical membrane and within the cell interior.
Fig. 2.
Shank2 knockdown alters the abundance and distribution of NaPiIIa. Left: in OK cells transfected with scrambled siRNAs (red puncta seen in the x-z scan; denoted by * in the apical x-y scan), imaging in both the x-y and x-z axes shows NaPiIIa (green) concentrated within the apical domain. This pattern and the signal abundance is similar to that seen in adjacent cells with no observed uptake of scrambled siRNAs (no red observed in cell interior). Right: OK cells transfected with Shank2 siRNAs (red puncta seen in the x-z scan; denoted with an asterisk in the apical x-y scan) had markedly less NaPiIIa (green) than adjacent, untransfected cells (no red observed in cell interior). NaPiIIa remaining in the Shank2 siRNA-transfected cells was observed in both the apical membrane and the cell interior (n = 4 preps; 10 cells/prep). Bar = 5 μm. Apical sections shown are from four 0.5-μm stacked sections.
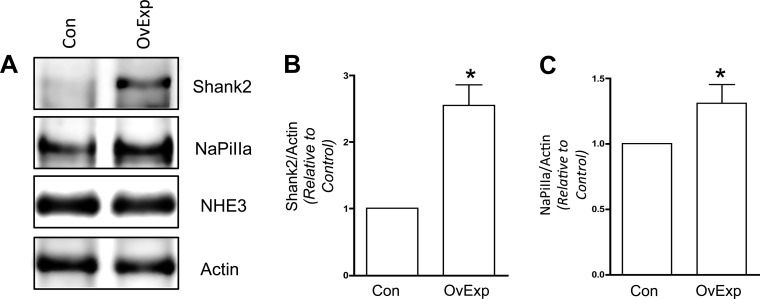
Although more modest, this dependent relationship between Shank2 and NaPiIIa protein abundance was also observed in OK cells that overexpress Shank2 in OK cells (Fig. 3). Densitometric analysis of Shank2 showed OK cells transfected with Flag-Shank2 cDNA had 2.55 ± 0.31 times the amount of Shank2 compared with mock transfected cells (n = 4). NaPiIIa levels in these Shank2 overexpressing cells were modestly but significantly increased (1.31 ± 0.14 times control) compared with mock transfected cells. No changes in NHE3 abundance were observed. The observations made in the Shank2 knockdown and overexpression studies provide the first direct evidence that Shank2 impacts NaPiIIa function by promoting the retention of NaPiIIa within the apical membrane of OK cells maintained under low-Pi conditions.
Fig. 3.
Shank2 overexpression (OvExp) increases NaPiIIa abundance. A: in OK cells transfected with Flag-Shank2 cDNA, densitometric analysis found Shank2 abundance increased by 2.55 ± 0.31 times compared with control cells (Con; n = 4; *P < 0.05). No change in NHE3 levels were observed. B: Flag-Shank2-transfected cells had a modest but significant increase in the abundance of endogenous NaPiIIa (1.31 ± 0.14 times vs. control) measured in Shank2-transfected cells (n = 4; *P < 0.05). C: NaPiIIa/actin.
High-Pi media induces internalization of GFP-NaPiIIa and mRFP-Shank2.
When PT cells experience an acute increase in serum Pi levels, NaPiIIa is retrieved from the microvillar membrane, endocytosed, and trafficked to lysosomes for degradation. Shank2 redistributes into the intracellular compartments in a pattern that directly parallels that of NaPiIIa (12). OK cells were further considered as a model to explore the relationship between Shank2 and NaPiIIa during the regulated endocytosis of NaPiIIa in response to high extracellular Pi. The marked decrease in NaPiIIa abundance following Shank2 knockdown in OK cells maintained under low-Pi conditions precluded the use of the knockdown approach in cells shifted into high-Pi media to study the role of Shank2 on NaPiIIa endocytosis. The OK cells do allow for live-cell studies and the dynamic nature of migrating proteins to be exploited to test if Shank2 and NaPiIIa dissociate or remain closely associated following the high-Pi-induced endocytic recovery from the apical membrane. This includes both correlation spectroscopy techniques that are capable of quantitatively resolving the comigration characteristics of two distinct proteins and FLIM-FRET studies to document the close (<10 nm) apposition of NaPiIIa and Shank2.
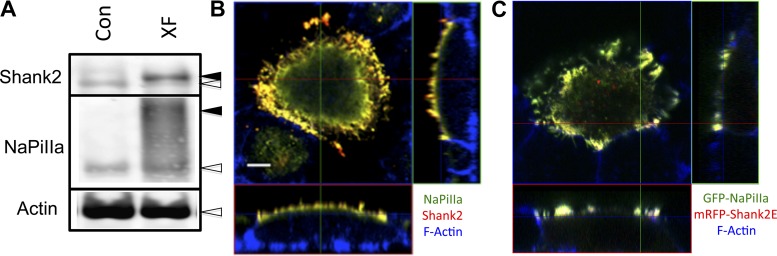
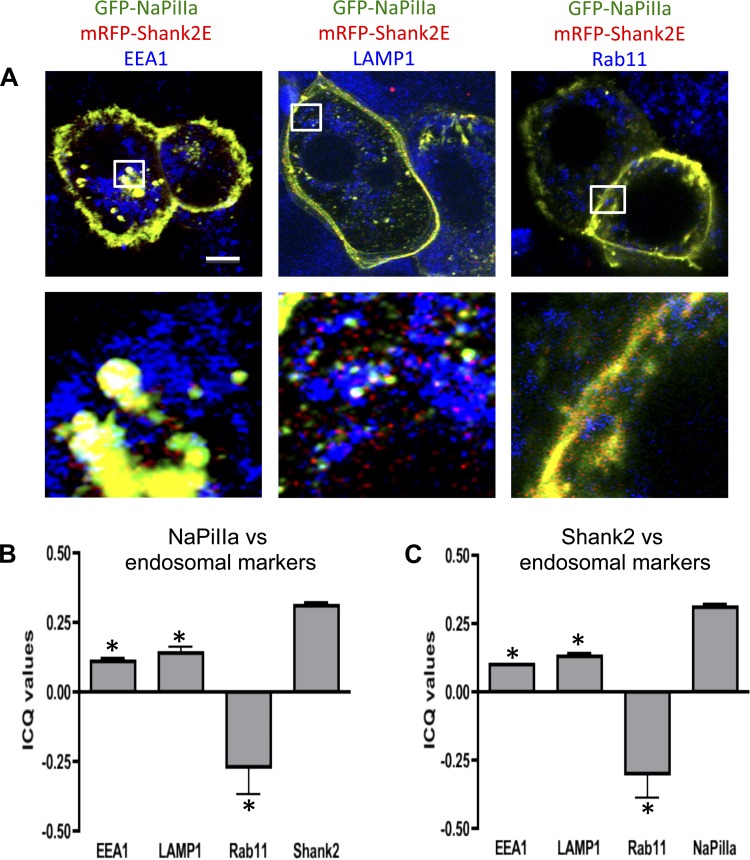
These live-cell studies utilize OK cells that are transfected with fluorescence-tagged forms of NaPiIIa [e.g., GFP−, mCherry−, and Cerulean (Cer) −] and Shank2 (e.g., mRFP− and EYFP−). The expression levels and cellular distribution of these recombinant proteins were initially evaluated as part of an effort to establish that the recombinant proteins behaved similar to their native counterparts. Western blot analysis demonstrates that the expression levels of these recombinant proteins were approximately two to three times the levels of the native forms of these two proteins (Fig. 4A). Under low-Pi conditions, native NaPiIIa and Shank2 are both concentrated within the apical microvilli of in vivo rat renal PT cells (12, 28) and cultured OK cells (Fig. 4B) (12, 28). Mirroring the native proteins, GFP-NaPiIIa and mRFP-Shank2 are both concentrated in the microvillar region of OK cells when cultured in low-Pi media (Fig. 4C). In rat renal PT cells exposed to high serum Pi levels, quantitative analyses show that NaPiIIa and Shank2 are diminished in the microvillar domain and redistribute into clathrin-associated compartments, early endosomes and lysosomes (12). Neither protein, however, was found within rab11a-containing recycling endosomes. GFP-NaPiIIa and mRFP-Shank2 expressed in cultured OK cells show a similar pattern (Fig. 5A). In cells fixed after 60 min in high-Pi media, GFP-NaPiIIa showed a high degree of colocalization with mRFP-Shank2. GFP-NaPiIIa and mRFP-Shank2E colocalization was greatest in the upper reaches of the cell. Within the mid-cell region, colocalization was also readily observed but distinct GFP-NaPiIIa and mRFP-Shank2E signals were also present. Both NaPiIIa and Shank2E also showed partial colocalization with EEA1, a marker of early endosomes, and with LAMP1, a lysosome marker. In contrast, while rab11 also distributed within the subapical domain, little rab11 was found to colocalize with either GFP-NaPiIIa or mRFP-Shank2. The degree of colocalization within the subapical domain was quantified by determining the ICQ for each pair of proteins (Fig. 5B; ICQmax = +0.5; ICQmin = −0.5). The distributions of the subapical pixel intensities for GFP-NaPiIIa and mRFP-Shank2 were highly correlated (GFP-NaPiIIa vs. mRFP-Shank2: 0.31 ± 0.01; n = 7). The subapical pixel intensity distributions of EEA1 and LAMP1 with either GFP-NaPiIIa or mRFP-Shank2 showed a modest degree of correlation (GFP-NaPiIIa vs. EEA1: 0.11 ± 0.01; mRFP-Shank2 vs. EEA1: 0.10 ± 0.02; GFP-NaPiIIa vs. LAMP1: 0.14 ± 0.02; mRFP-Shank2 vs. LAMP1: 0.13 ± 0.01; n = 7). The subapical pixel intensity distributions of rab11 with either GFP-NaPiIIa or mRFP-Shank2 were markedly distinct (GFP-NaPiIIa vs. Rab11: −0.26 ± 0.09; mRFP-Shank2 vs. Rab11: −0.29 ± 0.09; n = 7). The findings above establish that the regulated redistribution of GFP-NaPiIIa and mRFP-Shank2 in OK cells directly parallels the regulated redistribution of native NaPiIIa and Shank2 previously documented in in vivo rat renal PT cells.
Fig. 4.
GFP-NaPiIIa and mRFP-Shank2E expression characteristics in OK cells. A: Western blotting shows recombinant GFP-NaPiIIa and mRFP-Shank2E (closed arrowheads) are expressed at modestly higher levels than native NaPiIIa and Shank2 (open arrowheads). Actin served as a loading control (XF, transfected). B: confocal imaging in the X-Z (bottom rectangle) and Y-Z planes (right rectangle) shows that native NaPiIIa (green) and Shank2 (red) are concentrated at the apical domain. F-actin (blue) delineates the cell periphery. X-Y scans (top left box) are at the level of the subapical domain. C: confocal imaging shows that recombinant GFP-NaPiIIa (green) and mRFP-Shank2E (red) are similarly concentrated at the apical domain. Bar = 5 μm.
Fig. 5.
GFP-NaPiIIa and mRFP-Shank2E colocalize in the cell interior. A: after a 1-h incubation in high phosphate media, GFP-NaPiIIa (green) and mRFP-Shank2E (red) are present within the cell interior and colocalize with each other (i.e., yellow). Partial colocalization of GFP-NaPiIIa and mRFP-Shank2E with EEA1 (left; blue) and LAMP1 (middle; blue) but not with rab11 (right; blue) is observed (i.e., white). bar = 5 μm. White boxes (5 × 5 μm) in upper panels are shown at higher magnification in lower panels. B: ICQ values for NaPiIIa showed a modest positive correlation with EEA1 (0.11 ± 0.01; n = 7) and LAMP1 (0.14 ± 0.02; n = 7), a negative correlation with rab11 (−0.26 ± 0.09; n = 7) and a strong positive correlation with Shank2E (0.31 ± 0.01; n = 7). C: likewise, ICQ values for Shank2E showed a modest positive correlation with EEA1 (0.10 ± 0.02; n = 7) and LAMP1 (0.13 ± 0.01; n = 7), a negative correlation with rab11 (−0.29 ± 0.09; n = 7) and a strong positive correlation with NaPiIIa (0.31 ± 0.01; n = 7). *P < 0.05 vs. NaPiIIa/or Shank2.
Live-cell imaging shows dynamic comigration of GFP-NaPiIIa and mRFP-Shank2.
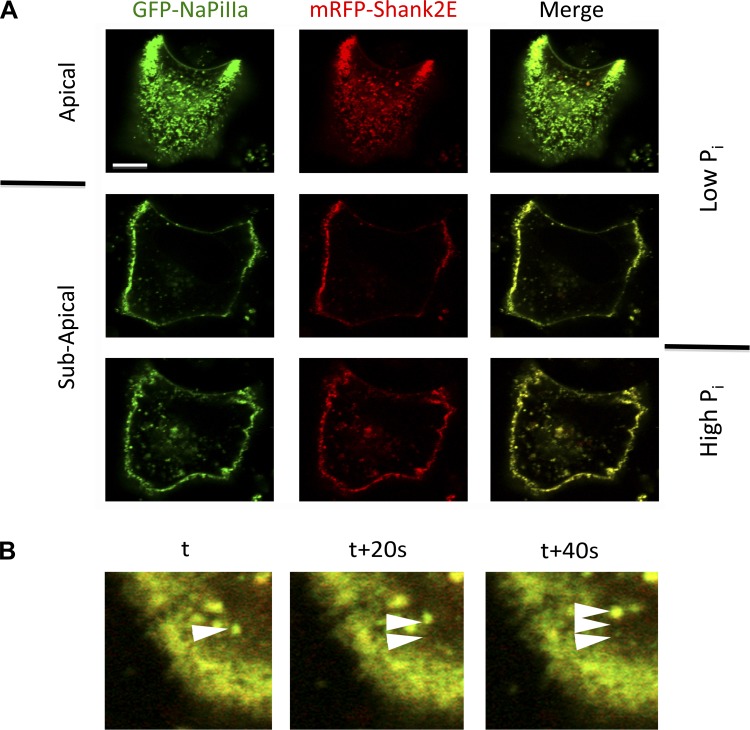
Having validated cotransfected OK cells as a faithful model to study the regulated NaPiIIa-Shank2 relationship, efforts were made to determine if Shank2 dissociates from or comigrates with NaPiIIa during the regulated recovery and internalization of NaPiIIa. First, live-cell confocal microscopy was employed to follow the fates of GFP-NaPiIIa and mRFP-Shank2 in OK cells subjected to high extracellular Pi. Under low-Pi conditions, both GFP-NaPiIIa and mRFP-Shank2 are concentrated within the apical microvilli with very little abundance within the subapical domain (Fig. 6A). The apical aspect of OK cells is typically dome shaped. This allows for orientation in confocal sections to be seen with the apical membrane ringing the cell periphery and the subapical domain residing within the membrane ring. Maintaining the confocal image plane in the subapical domain, the cells were shifted into high-Pi medium. Over time, both GFP-NaPiIIa and mRFP-Shank2 accumulated within the subapical domain. Merged images of the subapical domain demonstrate these two proteins colocalize. When followed at 20-s intervals, colocalized GFP-NaPiIIa and mRFP-Shank2 proteins are observed emerging together from the apical membrane and tracking together through the cell interior (Fig. 6B). These live-cell images indicate the large majority of putative GFP-NaPiIIa-laden endosomes have mRFP-Shank2 associated with them.
Fig. 6.
Live-cell imaging of GFP-NaPiIIa and mRFP-Shank2-containing endosomes. A: live-cell confocal imaging of GFP-NaPiIIa (green) and mRFP-Shank2 (red) in OK cells shows that in low-Pi media both proteins are concentrated within the apical domain with very little of either protein in the subapical domain. Upon shifting cells into high-Pi media, both proteins migrate into the subapical domain. High-Pi images were taken after 60 min in high-Pi media. Merged images of both channels show a high degree of colocalization of both proteins within the apical and subapical domains. Bar = 5 μm. B: merged images of GFP-NaPiIIa (green) and mRFP-Shank2E (red) captured at sequential 20-s time intervals. Arrowheads highlight the sequential migration of a dual-labeled endosome after 0, 20, and 40 s. Bar = 5 μm.
Correlation spectroscopy quantifies the comigration patterns of NaPiIIa and Shank2.
For mobile proteins, fluorescence correlation techniques can greatly extend the ability of light microscopy to demonstrate an association between two proteins by determining if the two proteins move in temporal or spatial synchrony on a microsecond time scale. In the present study, FCCS and ccRICS were employed to determine in a quantitative manner if GFP-NaPiIIa and mRFP-Shank2E traversed through the subapical domain in synchrony. Rab11 also resides within the subapical domain but does not distribute with either NaPiIIa or Shank2 (Fig. 5). Consequently, the synchrony of movement between GFP-rab11 and either mCherry-NaPiIIa or mRFP-Shank2E was evaluated as negative controls.
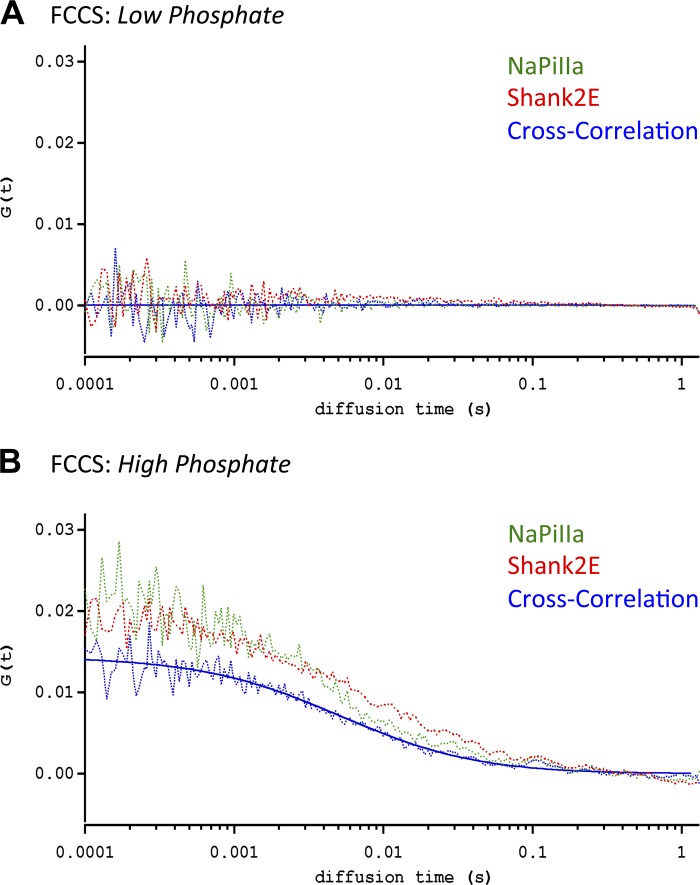
In FCS/FCCS studies of OK cells maintained in low-Pi media, essentially no GFP-NaPiIIa or mRFP-Shank2E was observed trafficking through the focal volume within the subapical domain (Fig. 7A). In contrast, GFP-rab11 did migrate within the subapical domain under the low-Pi conditions (Fig. 8, A and C; Table 1; n = 5 each). One hour after transitioning into high-Pi media, FCS measurements were repeated within the subapical domain of the same cells. GFP-NaPiIIa and mRFP-Shank2E were now observed migrating within the subapical domain (Fig. 7B). The diffusion coefficients (i.e., rates of movement) for GFP-NaPiIIa (DNaPiIIa = 3.9 ± 0.3 μm2/s; n = 10; Table 1) and mRFP-Shank2E (DShank2E = 3.8 ± 0.3 μm2/s; n = 10; Table 1) were remarkably similar to each other. Demonstrating that these two proteins were migrating through the focal volume in synchrony, FCCS analysis showed a high degree of cross-correlation between GFP-NaPiIIa and mRFP-Shank2E (Fig. 7B; Table 1). These observations are in contrast to the analyses of OK cells cotransfected with GFP-rab11 and mCherry-NaPiIIa or GFP-rab11 and mRFP-Shank2E under high-Pi conditions. While each protein migrates within the subapical domain, the diffusion coefficients are significantly different for GFP-rab11 vs. either mCherry-NaPiIIa or mRFP-Shank2E (Table 1) and there are no observed cross-correlations in their temporal patterns of entering and exiting the focal volume (Fig. 8, B and D). In short, while there is no observable synchrony in the movements of rab11 with either NaPiIIa or Shank2, there is direct synchrony in the movements of NaPiIIa and Shank2.
Fig. 7.
Fluorescence correlation spectroscopy (FCS) and fluorescence cross-correlation spectroscopy (FCCS) analysis of GFP-NaPiIIa and mRFP-Shank2 comigration. A: FCS analysis of OK cells under low-Pi conditions found little evidence of GFP-NaPiIIa (green line) and mRFP-Shank2 (red line) migration through a focal volume within the subapical domain. B: after 60 min under high-Pi conditions, FCS analysis readily detected the migration of GFP-NaPiIIa (green line) and mRFP-Shank2 (red line) within the subapical domain. FCCS analysis showed a strong degree of cross-correlation (blue line; both raw data and fitted line are shown) in the temporal patterns of GFP-NaPiIIa and mRFP-Shank2 transit through the focal volume.
Fig. 8.
FCS/FCCS analysis of rab11 vs. NaPiIIa and Shank2 comigration. A: FCS analysis of OK cells under Low-Pi conditions measured the temporal migration of GFP-rab11 (green line) through a focal volume within the subapical domain but found little evidence of mCherry-NaPiIIa (red line) migration through the same volume. B: after 60 min under High-Pi conditions, FCS analysis readily detected the migration of GFP-rab11 (green line) and mCherry-NaPiIIa (red line) within the subapical domain. FCCS analysis (blue line; both raw data and fitted line are shown), however, measured no appreciable cross-correlation in the temporal patterns of GFP-rab11 and mCherry-NaPiIIa migration. C and D: similar to rab11 vs. NaPiIIa above, FCCS analysis found no cross-correlation in the temporal patterns of GFP-rab11 and mRFP-Shank2 migration under high-Pi conditions.
Table 1.
Migration coefficients for FCS, FCCS, and ccRICS studies
| Proteins |
Low Phosphate |
High Phosphate |
||||||
|---|---|---|---|---|---|---|---|---|
| Protein1 | Protein2 | Dprotein1, μm2/s | Dprotein2, μm2/s | DFCCS, μm2/s | Dprotein1, μm2/s | Dprotein2, μm2/s | DFCCS, μm2/s | DccRICS, μm2/s |
| GFP | mCherry | 9.5 ± 1.0 (n = 5) | 34.5 ± 6.0 (n = 5) | 28.9 ± 3.1 (n = 5) | 99.3 ± 5.5 (n = 5) | |||
| NaPiIIa* | — | — | — | 3.5 ± 0.6 (n = 5) | — | |||
| — | Shank2† | — | — | — | 3.1 ± 0.5 (n = 5) | |||
| NaPiIIa* | Shank2† | — | — | — | 3.9 ± 0.3 (n = 10) | 3.8 ± 0.3 (n = 10) | 3.7 ± 0.3 (n = 10) | 2.5 ± 0.7 (n = 4) |
| Rab11* | NaPiIIa‡ | 5.8 ± 1.2 (n = 5) | — | — | 7.6 ± 0.6 (n = 5) | 3.3 ± 0.3 (n = 5) | — | |
| Rab11* | Shank2† | 6.0 ± 0.6 (n = 5) | — | — | 6.8 ± 0.2 (n = 5) | 3.0 ± 0.5 (n = 5) | — | |
Values are means ± SE. NaPiIIa, sodium-phosphate cotransporter IIa; D, migration coefficient; FCS, fluorescence correlation spectroscopy; FCCS, fluorescence cross-correlation spectroscopy ccRICS, raster image cross-correlation spectroscopy.
Proteins were GFP tagged;
proteins were mRFP-tagged;
proteins were mCherry-tagged.
FCS analysis of OK cells incubated under low-Pi vs. high-Pi also yielded an unanticipated observation. Same-cell diffusion coefficient measurements of GFP-only (low-Pi: DGFP = 9.5 ± 1.0 μm2/s; high-Pi: DGFP = 28.9 ± 3.1 μm2/s; n = 5) and mCherry-only proteins (low-Pi: DmCherry = 34.5 ± 6.0 μm2/s; high-Pi: DmCherry = 99.3 ± 5.5 μm2/s; n = 5) documented a roughly three-fold increase in their mobility rates after one h of incubation in high-Pi media (Table 1). Cells maintained under low-Pi conditions did not have a change in their GFP-only and mCherry-only diffusion coefficients. This observation requires more direct investigation but suggests that increasing the extracellular Pi concentration altered the intracellular environment that resulted in a decreased resistance to movement within the subapical domain of OK cells.
OK cells expressing GFP-NaPiIIa:mRFP-Shank2, GFP-rab11:mCherry-NaPiIIa and GFP-rab11:mRFP-Shank2 protein pairs were also evaluated by ccRICS (Fig. 9). By repetitively scanning in register over a set planar volume, ccRICS is capable of evaluating both the temporal and spatial synchrony of movements between two proteins. The fitting function for the distribution of the individual spatial cross-correlation values is presented graphically as a two-dimensional function (Fig. 9B, bottom plane). The tall, narrow conical shape is consistent with the mobility coefficient being near the lower end of the range of detection (slower diffusion) and indicates that the distinct cross-correlated protein pairs are predominantly moving together. Figure 9B, top plane, presents graphically the fitting residuals, which is the difference between the measured data and the fitting function and highlights the tightness of the fitted data. The cross-correlated signal can be visualized in images of cross-correlated signals obtained from individual raster scans (Fig. 9A). When the correlation of the spatial redistribution of moving particles between both channels was evaluated, a positive cross-correlation diffusion coefficient (DccRICS = 2.7 ± 0.7 μm2/s; n = 4; Table 1) was observed. The strong degree of cross-correlation measured for GFP-NaPiIIa:mRFP-Shank2E was contrasted by the lack of cross-correlation between GFP-rab11:mCherry-NaPiIIa and GFP-rab11:mRFP-Shank2E protein pairs (data not shown). The ccRICS studies confirm the FCCS observations and extend the observations by demonstrating that NaPiIIa and Shank2 migrate through the subapical domain of OK cells in both temporal and spatial synchrony.
Fig. 9.
Raster image cross-correlation spectroscopy (ccRICS) analysis of GFP-NaPiIIa and mRFP-Shank2 comigration. A: single 256 × 256 pixel frame from a 100-frame ccRICS raster scan in the subapical domain taken after one h in High-Pi media shows an abundance of cross-correlated GFP-NaPiIIa and mRFP-Shank2E signal. Immobile and slow-moving signals were mathematically removed and are not present in the image. B: three-dimensional presentation of the two-dimensional fitting function of the ccRICS data (top) shows a distinct conical peak, representing a high degree of synchrony in the temporal-spatial migration patterns of GFP-NaPiIIa and mRFP-Shank2. The residuals, the difference between the measured data and fitting function (bottom), demonstrate the tightness of the fit.
FLIM-FRET analysis of NaPiIIa and Shank2 interactions in OK cells.
To determine if NaPiIIa and Shank2 remained closely associated during endocytosis and trafficking through the subapical domain, OK cells were subjected to FLIM-FRET analysis. FRET can only occur when donor and acceptor fluorophors are positioned within 10 nm of each other. The occurrence of FRET between Cer-NaPiIIa (donor fluorophore) and EYFP-Shank2E (acceptor fluorophor) was determined using fluorescence lifetime measurements of Cer-NaPiIIa. The fluorescence lifetime of donor fluorophors is diminished when FRET occurs. To facilitate interpretation, the FLIM data were subjected to phasor analysis (9). As shown in an “idealized” phasor plot in Fig. 10A, FRET between a donor and acceptor (D + A) is seen as a shift along the line established between the unquenched donor (D) and the quenched donor (Dq). The degree of shift depends on the efficiency of the interaction and the fraction (fq) of molecules undergoing FRET. In OK cells, FLIM measurements were taken within the microvilli of cells under low-Pi conditions (Fig. 10B), within the microvilli of cells under high-Pi conditions (Fig. 10C) and within the subapical domain of cells under high-Pi conditions (Fig. 10D). In each study, the average phasor position of (D) (i.e., Cer-NaPiIIa) in the phasor plot was determined and compared with the phasor position of (D + A) (i.e., Cer-NaPiIIa and EYFP-Shank2E) species. Within the microvilli of cells under low-Pi conditions, the phasors from (D + A) cotransfected cells were significantly shifted in the direction of lower lifetimes to demonstrate the occurrence of FRET (Fig. 10B). The fraction of donors undergoing FRET was 0.11 ± 0.02 (n = 18) with a FRET efficiency of 0.7. Within the microvilli of cells under high-Pi conditions, the phasors of (D + A) cotransfected cells did not show a significant shift in position compared with (D) transfected cells (Fig. 10C). This absence of FRET indicates the remaining Cer-NaPiIIa and EYFP-Shank2E within the microvilli of OK cells under high-Pi conditions do not reside within 10 nm of each other. This is in contrast to the same fluorophors with the subapical domain of OK cells under high-Pi conditions. When compared with (D) transfected cells, the phasors of (D + A) cotransfected cells are again shifted toward the direction of lower lifetime (Fig. 10D). The fraction of donors undergoing FRET was 0.10 ± 0.05 (n = 14) with a FRET efficiency of 0.7. Thus the FLIM-FRET studies confirm the close association between NaPiIIa and Shank2 within the microvilli of PT cells in low-Pi conditions and demonstrate this close association is maintained as both proteins are endocytosed into the subapical domain of PT cells in high-Pi conditions.
Together, the ICQ analysis, the qualitative comigration observations, the quantitative FCCS/ccRICS measurements, and the FLIM-FRET analysis of protein-protein apposition provide robust evidence that Shank2 remains closely associated with NaPiIIa during the regulated recovery and intracellular trafficking of NaPiIIa.
DISCUSSION
The Shank family of proteins (Shank1, Shank2, and Shank3) continues to emerge as critical regulators of protein complexes and cellular functions. Initially discovered in neurons, Shank proteins direct the formation of nascent dendrites, coordinate the organization of proteins within the postsynaptic density regions, and moderate synaptic activities (13, 39). This role in neuronal plasticity and function translates into Shank proteins being important in learning, memory and development. For example, Shank1(−/−) mice have smaller dendrites, altered protein compositions within their postsynaptic densities, and impaired patterns of learning and memory (22). Furthermore, genetic studies in humans have established a direct linkage between mutations in either Shank2 or Shank3 and autism (3, 35). While Shank1 expression appears constrained to neurons, Shank2 and Shank3 are more broadly expressed and both Shank2 and Shank3 have important physiologic roles in these other tissues.
In epithelial cells, Shank2 is concentrated within the apical domain and binds to specific ion transporter and channel proteins (19, 23, 28, 29). In cultured colonic epithelial cells, Shank2 binds to and moderates the activity of both CFTR and NHE3, in part, by serving as a platform to sequester regulatory proteins within the same microdomain as CFTR and NHE3 (19, 23–25). These Shank2-bound regulatory proteins moderate the open probability of CFTR and cycling rate of NHE3 while they reside within the apical membrane. Unlike CFTR and NHE3, the primary means of moderating NaPiIIa activity is not by posttranslational modification (e.g., phosphorylation) while remaining in the apical membrane but is, instead, by controlling its relative abundance within the microvillar membrane. Consequently, it was unclear if and how Shank2 might influence NaPiIIa activity. Studies employing EBP50(−/−) mice and PDZK1(−/−) mice showed that these two PDZ domain-expressing proteins promote the retention of NaPiIIa in the apical membrane (5, 41). The present studies show that EBP50 and PDZK1 are not acting alone in this role and provide the first direct observation that Shank2 impacts the microvillar retention of NaPiIIa. In OK cells maintained under low-Pi conditions, decreases in Shank2 expression resulted in a decrease in the cellular abundance of NaPiIIa (Figs. 1 and 2). Regression analysis demonstrated a linear correlation between the percent of Shank2 knockdown vs. the decrease in NaPiIIa abundance (Fig. 1). Extrapolation of the regression line suggests only ∼20% of NaPiIIa would remain in the cells independent of any Shank2 interaction. Future studies will be important to determine how EBP50, PDZK1, and Shank2 coordinate their efforts to deliver, retain, and release NaPiIIa in a regulated fashion.
When PT cells are acutely shifted from a low-Pi to a high-Pi environment, NaPiIIa is retrieved from the apical microvilli, endocytosed, and trafficked to the lysosomes for degradation (12, 14). Very little is known regarding the molecular mechanisms that drive these steps. Simiilar to the response to high-Pi, PT cells respond to PTH treatment by recovering NaPiIIa from the microvillar membrane and traffic NaPiIIa to lysosomes for degradation. In response to PTH, the EBP50-NaPiIIa binding interaction is reduced and EBP50 remains within the microvillar domain (7, 43, 44). In contrast, in vivo rat studies show that following an acute increase in serum Pi levels, Shank2 redistributes into the intracellular compartments in a pattern that directly parallels that of NaPiIIa (12). This could maintain the Shank2 in position to direct the regulated endocytic recovery and intracellular trafficking of NaPiIIa. Unfortunately, the profound effect of Shank2 knockdown on NaPiIIa levels under low-Pi (i.e., pretreatment) conditions precludes the use of the knockdown approach to directly assess the functional roles of Shank2 in the regulated endocytic recovery of NaPiIIa. A number of lines of evidence do, however, strengthen the support for the hypothesis that Shank2 moderates NaPiIIa endocytosis and trafficking.
Many of the molecular mechanisms underlying the formation, detachment, and initial internalization of endosomes from the plasma membrane are well documented (34). Among the contributing proteins, dynamin-2 is a “pinchase” that facilitates the detachment of endosomes and cortactin binds Arp2/3 and facilitates actin polymerization that drives endosomes into the cell interior. Both dynamin-2 and cortactin are Shank2 binding partners (32, 33) and coimmunoprecipitate with Shank2 from renal cortex lysates (data not shown). Consequently, Shank2 has the physical characteristics that would allow it to coordinate the formation and initial internalization of nascent NaPiIIa-laden endosomes.
Once liberated from the apical membrane, both NaPiIIa and Shank2 behave like they are associated with a common endosome. Although the resolution of light microscopy (∼200 nm) limits the ability to discern the true colocalization of two independent proteins (often <25 nm in diameter), live-cell confocal imaging of GFP-NaPiIIa and mRFP-Shank2E shows both proteins migrating through the cell interior in close apposition (Fig. 6). The FCS/FCCS diffusion coefficients for both GFP-NaPiIIa and mRFP-Shank2 under a variety of conditions are both consistently ∼3.5 μm2/s (Table 1), indicating both proteins traverse through the cell at the same rate. This value is similar to the FCS diffusion coefficients determined for cholera toxin-labeled endosomes in live Vero cells (1). Cross-correlation spectroscopy analyses of mobile proteins minimize the limitations of light microscopy resolution by tracking the comparative positional fluorescence over microsecond time scales. The FCCS and ccRICS performed clearly demonstrated a persistent temporal and spatial synchrony in the intracellular movements of NaPiIIa and Shank2 (Figs. 7, 8, and 9). Finally, the FLIM-FRET studies directly demonstrated that the Cer-NaPiIIa and EYFP-Shank2E proteins in the subapical domain remain closely associated and reside within 10 nm of each other (Fig. 10). Together, this study provides definitive evidence that Shank2 remains in close association with NaPiIIa during the regulated endocytosis and trafficking of NaPiIIa.
In summary, Shank2 has emerged as a pivotal regulator of key epithelial receptors, transporters, and ion channels. Previous modes of regulation by Shank2 centered around its capability to cluster interactive sets of regulatory and effector proteins within the confines of the microvillar membrane. The present study brings to light novel mechanisms by which Shank2 might impact its binding partners. Under low-Pi conditions, Shank2 plays an essential role in retaining NaPiIIa within the apical membrane. Under high-Pi conditions, the association is retained and Shank2 escorts NaPiIIa through its endocytic recovery and intracellular trafficking.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-080769 (to R. B. Doctor).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: E.D., R.M., T.L., E.G., and R.B.D. conception and design of research; E.D., L.L., J.R., and D.C. performed experiments; E.D., L.L., D.C., R.M., and R.B.D. analyzed data; E.D., L.L., J.R., D.C., R.M., T.L., E.G., and R.B.D. interpreted results of experiments; E.D. and L.L. prepared figures; E.D. and R.B.D. drafted manuscript; E.D., L.L., R.M., T.L., E.G., and R.B.D. edited and revised manuscript; E.D., L.L., J.R., D.C., R.M., T.L., E.G., and R.B.D. approved final version of manuscript.
ACKNOWLEDGMENTS
Technical guidance on qPCR studies and analysis was graciously provided by Drs. Eric Campbell and Doug Kominsky. The NaPiIIa plasmid constructs were graciously provided by Dr. Moshe Levi (University of Colorado, Denver). The rab11a plasmid construct was graciously provided by Dr. Rytis Prekeris (University of Colorado, Denver). The NHE3 antibody was gracioiusly provided by Dr. Orson Moe (University of Texas-Southwestern). Technical guidance in the correlation spectroscopy methods was graciously provided by Dr. Michelle Digman (University of California, Irvine). The microscopy studies were performed in the Advanced Light Microscopy Core at the University of Colorado-Anschutz Medical Campus.
REFERENCES
- 1. Bacia K, Majoul IV, Schwille P. Probing the endocytic pathway in live cells using dual-color fluorescence cross-correlation analysis. Biophys J 83: 1184–1193, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bastepe M, Juppner H. Inherited hypophosphatemic disorders in children and the evolving mechanisms of phosphate regulation. Rev Endocr Metab Disord 9: 171–180, 2008. [DOI] [PubMed] [Google Scholar]
- 3. Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, Bonin M, Reiss A, Engels H, Sprengel R, Scherer SW, Rappold GA. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet 42: 489–491, 2010. [DOI] [PubMed] [Google Scholar]
- 4. Berndt T, Kumar R. Novel mechanisms in the regulatioin of phosphorus homeostasis. Physiology 24: 17–25, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capuano P, Bacic D, Stange G, Hernando N, Kaissling B, Pal R, Kocher O, Biber J, Wagner CA, Murer H. Expression and regulation of the renal Na/Pi cotransporter NaPi-IIa in a mouse model deficient for the PDZ protein, PDZK1. Pflügers Arch 449: 392–402, 2005. [DOI] [PubMed] [Google Scholar]
- 6. Colyer RA, Lee C, Gratton E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microsc Res Tech 71: 201–213, 2008. [DOI] [PubMed] [Google Scholar]
- 7. Deliot N, Hernando N, Horst-Liu Z, Gisler SM, Capuano P, Wagner CA, Bacic D, O'Brien S, Biber J, Murer H. Parathyroid hormone treatment induces dissociation of type IIa Na-Pi cotransporter-NHERF1 complexes. Am J Physiol Cell Physiol 289: C159–C167, 2005. [DOI] [PubMed] [Google Scholar]
- 8. Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions in cells with a laser scanning technique. Biophys J 89: 1317–1327, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DIgman MA, Caiolfa VR, Zamai M, Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys J 94: L14–16, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Digman MA, Sengupta P, Wiseman PW, Brown CM, Horwitz AR, Gratton E. Fluctuation correlation spectroscopy with a laser-scanning microscope: exploiting the hidden time structure. Biophys J 88: L33–36, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Digman MA, Wiseman PW, Horwitz AR, Gratton E. Detecting protein complexes in living cells from laser scanning confocal image sequences by the cross correlation raster image spectroscopy method. Biophys J 96: 707–716, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dobrinskikh E, Giral H, Caldas Y, Levi M, Doctor RB. Shank2 redistributes with NaPiIIa during regulated endocytosis. Am J Physiol Cell Physiol 299: C1324–C1334, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci 10: 87–99, 2009. [DOI] [PubMed] [Google Scholar]
- 14. Forster IC, Hernando N, Biber J, Murer H. Proximal tubular handling of phosphate: a molecular perspective. Kidney Int 70: 1548–1559, 2006. [DOI] [PubMed] [Google Scholar]
- 15. Gendron PO, Avaltroni F, Wilkinson KJ. Diffusion coefficients of several rhodamine derivatives as determined by pulse field gradient-NMR and fluorescence correlation spectroscopy. J Fluoresc 18: 1093–1101, 2008. [DOI] [PubMed] [Google Scholar]
- 16. Giral H, Cranston D, Lanzano L, Caldas Y, Sutherland E, Rachelson J, Dobrinskikh E, Weinman EJ, Doctor RB, Gatton E, Levi M. NHE3 regulatory factor 1 (NHERF1) modulates intestinal sodium-dependent phosphate transporter (NaPi-2b) expression in apical microvilli. J Biol Chem 287: 35047–35056, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giral H, Lanzano L, Caldas Y, Blaine J, Verlander JW, Lei T, Gratton E, Levi M. Role of PDZK1protein in apical membrane expression of renal sodium-coupled phosphate transporters. J Biol Chem 286: 15032–15042, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gisler SM, Stagljar I, Traebert M, Bacic D, Biber J, Murer H. Interaction of the type IIa Na/Pi cotransporter with PDZ proteins. J Biol Chem 276: 9206–9213, 2001. [DOI] [PubMed] [Google Scholar]
- 19. Han W, Kim KH, Jo MJ, Lee JH, Yang J, Doctor RB, Moe OW, Lee J, Kim E, Lee MG. Shank2 associates with and regulates NHE3. J Biol Chem 281: 1461–1469, 2006. [DOI] [PubMed] [Google Scholar]
- 20. Hernando N, Deliot M, Gisler SM, Lederer E, Weinman EJ, Biber J, Murer H. PDZ-domain interactions and apical expression of type IIa Na/Pi cotransporters. Proc Natl Acad Sci USA 99: 11957–11962, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hruska KA, Methew S, Lund R, Qui P, Pratt R. Hyperphosphatemia of chronic diseases. Kidney Int 74: 148–157, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci 28: 1697–1708, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim JY, Han W, Namkung W, Lee JH, Kim KH, Shin H, Kim E, Lee MG. Inhibitory regulation of CFTR anion-transporting activities by Shank2. J Biol Chem 279: 10389–10396, 2004. [DOI] [PubMed] [Google Scholar]
- 24. Lee JH, Lee YM, Kim JY, Park HW, Grinstein S, Orlowski J, Kim E, Kim KH, Lee MG. βPix upregulates NHE3 through a Shank2-mediated protein-protein interaction. J Biol Chem 285: 8104–8113, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee JH, Richter W, Namkung W, Kim KH, Kim E, Conti M, Lee MG. Dynamic regulation of cystic fibrosis transmembrane conductance regulator by competitive interactions of molecular adaptors. J Biol Chem 282: 10414–10122, 2007. [DOI] [PubMed] [Google Scholar]
- 26. Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin1, Ga and N-type Ca2+ channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J Neurosci 24: 4070–4081, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Magde D, Elson EL, Webb WW. Thermodynamic fluctuations in a reacting system: measurements by fluorescence correlation spectroscopy. Phys Rev Lett 29: 705–708, 1972. [Google Scholar]
- 28. McWilliams RR, Breusegem SY, Brodsky KF, Kim E, Levi M, Doctor RB. Shank2E binds Na/Pi cotransporter at the apical membrane of proximal tubule cells. Am J Physiol Cell Physiol 289: C1042–C1051, 2005. [DOI] [PubMed] [Google Scholar]
- 29. McWilliams RR, Gidey E, Fouassier L, Weed SA, Doctor RB. Characterization of an ankyrin repeat-containing Shank2 isoform (Shank2E) in liver epithelial cells. Biochem J 380: 181–191, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Molony DA, Stephens BW. Derangements in phosphate metabolism in CKD/ESRD: therapeutic considerations. Adv Chr Kid Dis 18: 120–131, 2011. [DOI] [PubMed] [Google Scholar]
- 31. Mutze J, Ohrt T, Schwille P. Fluorescence correlation spectroscopy in vivo. Laser Photo Rev 5: 52–67, 2011. [Google Scholar]
- 32. Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley RF, Sheng M. Shank, a novel family of PSD proteins that binds to the NMDA receptor/PSD95/GKAP complex and cortactin. Neuron 23: 569–582, 1999. [DOI] [PubMed] [Google Scholar]
- 33. Okamoto PM, Gamby C, Wells D, Fallon J, Vallee RB. Dynamin isoform-specific interaction with shank/ProSAP scaffolding proteins of the postsynaptic density and actin cytoskeleton. J Biol Chem 276: 48458–48465, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Orth JD, McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol 15: 31–39, 2003. [DOI] [PubMed] [Google Scholar]
- 35. Phelan MC, Rogers RC, Saul RA, Stapleton GA, Sweet K, McDermid H, Shaw SR, Claytor J, Willis J, Kelly DP. 22q13 deletion syndrome. Am J Med Genet 101: 91–99, 2001. [DOI] [PubMed] [Google Scholar]
- 36. Rarbach M, Kettling U, Koltermann A, Eigen M. Dual-color fluorescence cross-correlation spectroscopy for monitoring the kinetics of enzyme-catalyzed reactions. Methods 24: 104–116, 2001. [DOI] [PubMed] [Google Scholar]
- 37. Redford GI, Clegg RM. Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J Fluoresc 15: 805–815, 2005. [DOI] [PubMed] [Google Scholar]
- 38. Rossow MJ, Sasaki JM, DIgman MA, Gratton E. Raster image correlation spectroscopy in live cells. Nat Prot 5: 1761–1774, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115–130, 2001. [DOI] [PubMed] [Google Scholar]
- 40. Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci 113: 1851–1856, 2000. [DOI] [PubMed] [Google Scholar]
- 41. Shenolikar S, Voltz J, Minkoff CM, Wade J, Weinman EJ. Targeted disruption of the mouse gene encoding a PDZ domain-containing protein adaptor, NHERF-1, promotes Npt2 internalization and renal phosphate wasting. Proc Natl Acad Sci USA 99: 11470–11475, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tietz P, Levine S, Holman R, Fretham C, LaRusso N. Characterization of apical and basolateral plasma membrane domains derived from cultured rat cholangiocytes. Anal Biochem 254: 192–199, 1997. [DOI] [PubMed] [Google Scholar]
- 43. Weinman EJ, Steplock D, Cha B, Kovbasnjuk O, Frost NA, Cunningham R, Shenolikar S, Blanpied TA, Donowitz M. PTH transiently increases the percent mobile fraction of Npt2a in OK cells as determined by FRAP. Am J Physiol Renal Physiol 297: F1560–F1565, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weinman EJ, Steplock D, Shenolikar S, Blanpied TA. Dynamics of PTH-induced disassembly of Npt2a/NHERF-1 complexes in living OK cells. Am J Physiol Renal Physiol 300: F231–F235, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]