Abstract
We present active-state structures of the G protein-coupled receptor (GPCRs) rhodopsin carrying the disease-causing mutation G90D. Mutations of G90 cause either retinitis pigmentosa (RP) or congenital stationary night blindness (CSNB), a milder, non-progressive form of RP. Our analysis shows that the CSNB-causing G90D mutation introduces a salt bridge with K296. The mutant thus interferes with the E113Q-K296 activation switch and the covalent binding of the inverse agonist 11-cis-retinal, two interactions that are crucial for the deactivation of rhodopsin. Other mutations, including G90V causing RP, cannot promote similar interactions. We discuss our findings in context of a model in which CSNB is caused by constitutive activation of the visual signalling cascade.
Keywords: G protein-coupled receptors, biased signalling, rhodopsin, congenital stationary night blindness, retinitis pigmentosa
INTRODUCTION
G protein-coupled receptors (GPCRs) are the largest family of signalling proteins in the human genome. Defective signalling of GPCR mutants causes many hereditary human diseases. The dim-light photoreceptor rhodopsin found in retina rod cells is no exception, with about 140 mutations known to cause retinitis pigmentosa (RP), a blinding retinal degeneration affecting 1 in 4,000 persons. Patients characteristically suffer from congenital night blindness and progressive loss of day vision. Virtually all patients lose central vision by the age of 50–80 years. Four mutations in rhodopsin are known to cause congenital stationary night blindness (CSNB) and rod dysfunction similar to the early stages of RP, but without progressive impairment of day vision (supplementary Fig S1 online). Elucidation of the molecular disease mechanisms of these mutations can provide important information about the causes of RP progression and pave the way for novel therapeutic approaches. Of all the rhodopsin mutations causing visual impairments, G90D affects the only position in rhodopsin where mutations can cause either RP [1] or CSNB [2]. The effects of G90D mutation have been carefully characterized in vitro [2–4], as well as in human patients [5] and animal models of the disease [6–8]. In contrast to most RP-causing mutations [9], the G90D rhodopsin does not seem to have folding defects, but causes constant basal activation of the visual system resulting in rod desensitization. Here we report crystal structures of constitutively activated G90D rhodopsin. In combination with a biochemical analysis of its desensitization capabilities, our structural analysis provides new insights into the molecular causes of heritable blindness.
RESULTS AND DISCUSSION
The G90D mutation introduces a stabilizing salt bridge
We attempted to crystallize several rhodopsin forms, including opsin, dark-state and light-activated G90D in the presence or absence of the GαCT2 peptide [10] that specifically binds the active metarhodopsin-II conformation [11]. We obtained crystals of G90D opsin, light-activated G90D and G90D-GαCT2, which diffracted to 3.9, 3.3 and 2.9 Å, respectively (supplementary Table S1 online). No crystals of the G90D dark state could be grown. The presence of retinal and the GαCT2 peptide both increased diffraction quality, consistent with their ability to bind and stabilize an active conformation of the receptor. Overall, the structures are similar to those of metarhodopsin-II obtained by light activation of constitutively active rhodopsin [12] or soaking of opsin with all-trans-retinal [10] (Fig 1; supplementary Table S2 online). While the low resolution obtained for G90D opsin does not allow accurate refinement of an atomic model, no obvious alterations of the protein backbone could be detected. Similarly, presence or absence of the GαCT2 peptide did not alter the overall receptor structure. Although we lack the structure of dark-state G90D rhodopsin, the structural data we have, thus far, imply that the impact of the G90D mutation is limited to the ligand-binding pocket.
Figure 1.
Comparison of the G90D-GαCT2 and G90D structures with metarhodopsin-II [12]. All-trans (yellow) and cis-retinal isomers (orange), the site of retinal attachment K296 (blue slate), the retinal counterion E113 (blue slate), GαCT peptides (cyan) and the constitutively activating mutations G90D and M257Y (green) are shown as spheres. The inset shows a closer view of the side chains that contribute to the retinal-binding pocket (within 4 Å of the retinal+the counterion E113) shown as sticks. Formation of a salt bridge (red dotted lines) between G90D (green) and K296 together with electrostatic interference with the E113 disfavours covalent binding of retinal resulting in a mixed retinal population, illustrated here as 13-cis and 9–13-di-cis-retinal.
The rhodopsin ligand is a photoactivateable 11-cis-retinal covalently attached by means of a protonated Schiff base (SB) bond to K296 in the rhodopsin dark state. On absorption of a photon, 11-cis-retinal isomerizes to all-trans-retinal leading to formation of metarhodopsin-II, which activates the visual G protein transducin (Gt). Signalling by the active receptor is quenched via phosphorylation of its C terminus by G protein-coupled receptor kinase 1 (GRK1). Phosphorylation enhances the binding of arrestin-1, resulting in uncoupling of the receptor from Gt. To reset rhodopsin back to its dark state, the SB between K296 and all-trans-retinal is hydrolysed, arrestin-1 dissociates and the apoprotein opsin is regenerated with 11-cis-retinal. The G90D mutation interferes with this cycle by introducing a charged side chain close to the site of retinal attachment. Indeed, during purification of the 11-cis-retinal-regenerated dark state, we observed a shift of the absorption peak to 482 nm resulting from perturbation of SB protonation [2, 3]. The 482 nm/280 nm ratio of the purified protein was 2.0–2.2, indicating a mixture of opsin and rhodopsin. These results are consistent with inhibition of 11-cis-retinal binding by the G90D mutation [13] and a higher hydroxylamine accessibility of the retinal SB in the G90D dark state [3, 4]. This heterogeneity likely prevented us from obtaining crystals of the G90D dark state.
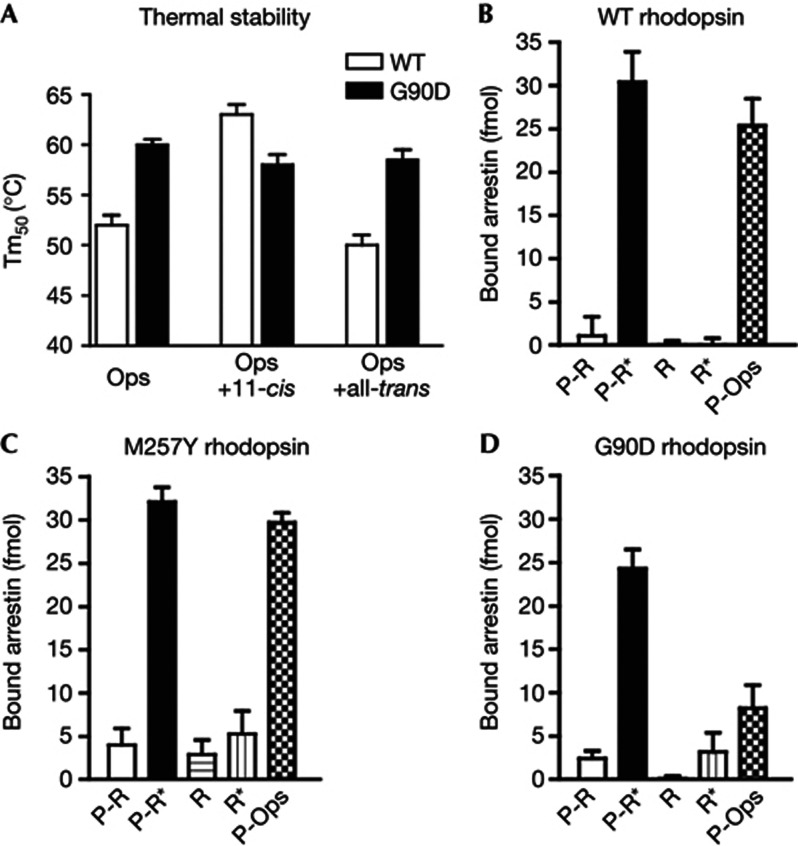
To further characterize the impact of the G90D mutation on rhodopsin integrity, we compared the stability of the mutant with the wild type (Fig 2) in presence or absence of retinal. Incubation with all-trans-retinal does not change the stability of the wild type significantly, as it does not bind opsin efficiently. In contrast, reconstitution of the dark state with 11-cis-retinal, increased the stability of the wild type by 11 °C. The stability at 63 °C for the reconstituted dark state (opsin plus 11-cis-retinal) is in good agreement with values obtained by UV/VIS spectroscopy [14]. Remarkably, the stability of G90D opsin was 8 °C higher compared with that of wild-type opsin. In contrast to the wild type, incubation of G90D opsin with retinal isomers had only an insignificant effect on stability, either because binding of both 11-cis and all-trans-retinal was inhibited or due to lower stability of the mutant dark state.
Figure 2.
Impact of the G90D mutation on thermal stability and binding of arrestin. Thermal stability (A) of wild type (WT) and G90D opsin (all in the stabilizing N2C/D282C background) in presence and absence of retinal. Graphs indicate the mean melting temperature (Tm50)±s.d. of the melting temperatures obtained from four fluorescent thermal shift experiments. (B–D) Direct binding assay of radiolabeled arrestin-1 to different forms of rhodopsin (phosphorylated rhodopsin (R-P), phosphorylated opsin (P-Ops) and unphosphorylated rhodopsin (R)) in nanodiscs. Binding experiments of wild type (B), constitutively active M257Y (C) and G90D (D) rhodopsin were performed either in the dark or under room light (*). Means and s.d. were obtained from four experiments.
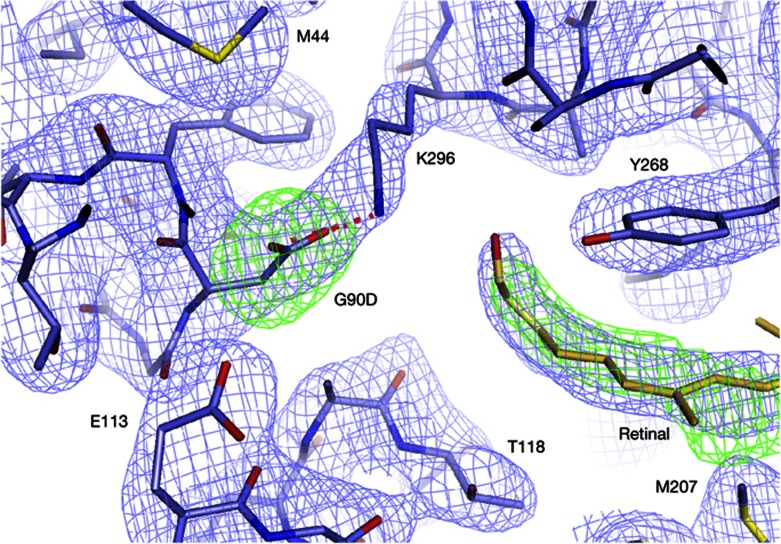
Even though binding of retinal to G90D rhodopsin was impaired, electron density in the ligand-binding pocket of the mutant incubated with retinal showed that some retinal was retained during crystallization (Fig 3). Crystallographic refinement with different retinal isomers indicated a heterogeneous, non-covalently bound population that we attribute to a mixture of cis isomers. One reason for this heterogeneity is the close proximity of the introduced G90D side chain to E113, with 3.6 Å separating the two carboxyl groups. Residue E113 is critical for retinal binding, because it provides the counterion that is involved in maintenance and hydrolysis of the retinal SB, depending on the conformational state of the protein [15]. E113 is also critical for maintaining the inactive conformation through a stabilizing salt bridge with the protonated SB. Disruption of the interhelical salt bridge by light-induced isomerization of retinal and proton transfer from the protonated SB to its counterion E113 is a critical protonation switch that controls the conformational transition to the active receptor-state metarhodopsin-II [16]. In the absence of retinal, a similar salt bridge forms directly between E113 and K296 to stabilize the inactive conformation of the apoprotein opsin [17]. Interference of G90D with the E113-K296 activation switch is thus one explanation for the observed constitutive activity of the mutant, as had been suggested even before any structural data on rhodopsin were available [2]. Our structure of the G90D mutant shows that the impact of the mutation goes further, as the introduced charged carboxyl group forms a salt bridge with the J;ε-amino group of K296, thereby providing an alternative for the deactivating E113-K296 interaction and preventing effective formation of a SB with retinal (Fig 1, inset). Formation of this alternative salt bridge provides an elegant explanation for the 80 times slower 11-cis-retinal uptake [13], and increased thermal stability of G90D opsin observed in the thermal shift assays (Fig 2). Interestingly, neither the introduced salt bridge nor the non-covalently bound retinal affects the overall conformation of the ligand-binding pocket, which remains virtually identical to those of opsin [18] and metarhodopsin-II [10, 12]. The active conformation of opsin thus seems to preform a binding pocket into which several retinal isomers can bind. Once the right retinal isomer is in place, a SB is formed and snaps into the strong salt bridge with its counterion E113 to force the seven-helix bundle into the inactive rhodopsin dark state [2, 19]. The G90D mutation prevents an efficient snapping by forming a salt bridge with K296, the site of SB formation. Even when the SB has been formed, the structurally close side chain of G90D destabilizes the inactive conformation [2–4, 6] by electrostatic interaction with K296, replacing E113 as SB counterion.
Figure 3.
Electron density (2Fo-Fc contoured at 1.5 sigma, blue and Fo-Fc contoured at 3.5 sigma, green) of the G90D-GαCT retinal-binding pocket calculated after simulated annealing refinement with the G90D side chain omitted. The obtained map shows a clear difference peak for the introduced G90D mutation and the salt bridge (red dashes) with K296. Clear positive density close to the position of all-trans-retinal in metarhodopsin-II indicates that some retinal was retained in the binding pocket during crystallization. Formation of the G90D-K296 salt bridge interferes with formation of a covalent bond and results in density that is most compatible with a mixture of retinals. For comparison, the 9,13-di-cis retinal isomer is shown as orange sticks.
Structural comparison of CSNB-causing mutations
To investigate what characterizes the active state of CSNB-causing mutations, we have modelled the three other known amino-acid substitutions causing CSNB (T94I, A292E and A295V) into our G90D structure (Fig 4). In this model, the charged side chain of A292E forms a salt bridge with K296, analogous to that between G90D and K296 but from transmembrane helix (TM) 7 instead of TM2. T94I is one helical turn away from G90D and forms a hydrophobic van der Waals contact with K296. The fourth CSNB mutant, A295V, is located one residue away from K296. It might thus induce a similar stabilizing effect on the active-state position of TM7, possibly by bridging TM6 and TM7 through van der Waals interactions with W265, another activation switch in close contact with retinal [20]. The common theme of all four CSNB-causing mutations is therefore not only constitutive activity and interference with retinal binding in the rhodopsin dark state [21] but also the formation of specific additional interactions involving K296 in the active state. Notably, replacing T94 with eight other amino acids caused constitutive activity [22], yet only T94I has been reported to cause CSNB. Similarly, both G90D and G90V are constitutively active [4] but only G90D leads to CSNB while G90V causes RP [1]. Placement of other nearby mutants causing progressive loss of vision by RP into our structure shows that none of them favours similar interactions with K296 as the four CSNB mutations. It is well known that most RP-causing mutations reduce stability and/or lead to misfolding of the protein [9], whereas CSNB mutants generally fold well [21]. Although similar in their early symptoms, both diseases are likely caused by different molecular mechanisms. A better structural characterization of these differences might open the way for medical treatment of RP, possibly by small molecules stabilizing interactions with K296 similar to the ones preventing disease progression in CSNB mutants. Although this approach would not cure night blindness, it could prevent progression of RP towards severe day vision impairment.
Figure 4.
Comparison of congenital stationary night blindness (CSNB)- (green, lower left panel) and retinitis pigmentosa (RP)- (blue, lower right panel) causing mutations near the retinal-binding pocket of light-activated rhodopsin. All four CSNB mutations could be placed into the structure of active G90D rhodopsin in a favourable rotamer conformation and without introducing major clashes with the rest of the protein. Three CSNB mutations are able to form either van der Waals (blue dashes), or salt bridge interactions (red dashes) with K296, whereas A295V might alter the position of K296 more indirectly and interacts with the W265 activation switch. Nearby RP mutations do not show a similar pattern and point away from the retinal Schiff base (SB). The only exceptions are RP-causing mutations of K296, which cannot covalently bind retinal, and the uncharged G90V mutation that cannot form the same salt bridge towards K296 as the G90D mutation.
The G90D-K296 salt bridge reduces binding of arrestin
In vivo rhodopsin signalling is rapidly quenched by phosphorylation, followed by the binding of arrestin-1, which blocks Gt binding. Disruption of this desensitization mechanism through mutations in arrestin [23] or rhodopsin kinase [24] leads to Oguchi disease, a recessively inherited form of CSNB. Arrestin does not efficiently compete with Gt for G90D opsin [25]. To quantify this effect, we incorporated G90D, wild type and constitutively active M257Y rhodopsin [26] into nanodiscs [27] and quantified their ability to undergo phosphorylation and bind arrestin-1. Phosphorylation in all samples reached levels of 2–3 phosphates per rhodopsin (supplementary Fig S2 online), comparable to the levels observed for rhodopsin in native disc membranes. Whereas the phosphorylation level of G90D was slightly higher compared with wild-type rhodopsin, several functional forms of this mutant demonstrated reduced ability to bind arrestin-1 (Fig 2B–D). A particularly striking difference between wild-type rhodopsin and the G90D mutant was observed in case of phosphorylated opsin, where arrestin-1 binding was reduced by 70%. Normal binding of arrestin-1 to M257Y rhodopsin shows that this is not a general feature of constitutively active rhodopsin mutants. Structurally, the G90D mutation is located in a strong helix distortion in TM2 introduced by two adjacent glycines at positions 89 and 90. These two glycines are highly conserved among visual pigments, whereas in other GPCRs proline residues at the equivalent position 2.57 produce similar kinks near ligand-binding pocket. In β-adrenergic receptors, ligands that bias the receptor towards arrestin binding form additional contacts to TM2, TM3, and TM7 [28] and predominantly affect the conformational state of TM7 [29]. In rhodopsin, pump-probe experiments suggested a vital role of TM7/TM8 dynamics in arrestin binding [30]. Likely the reduced arrestin binding observed for G90D opsin is thus due to the unnatural salt bridge between TM2 and TM7 and the resulting reduced conformational flexibility of the receptor. Most importantly, these data indicate that the conformation of Gt-bound opsin is different from arrestin-bound opsin.
Relevance to CSNB
Rod cells of human CSNB patients [5, 21] and of animal models carrying the G90D mutation [6–8] are functionally desensitized, as if under constant low basal stimulation that confounds dim-light vision. Three explanations (Fig 5), which are not mutually exclusive, have been proposed for the increased basal activity characteristic for CSNB: (1) constitutive Gt activation by G90D opsin [2, 7]; (2) spontaneous activation by thermal isomerization of retinal [5]; (3) constant basal activation by a preactivated dark state [8]. Our structures of light-activated and unliganded G90D provide information concerning all three proposed mechanisms. First, the structures show that D90 displaces the retinal counterion E113 by forming a salt bridge with K296, stabilizing an active opsin conformation. In addition, this salt bridge reduces binding of opsin to the desensitizing protein arrestin-1. Second, it seems likely that interference of G90D with the site of retinal attachment also increases the rate of thermal isomerization. Several retinal isomers, including all-trans and 11-cis-retinal, can transiently activate opsin until SB formation with K296 deactivates the protein [31, 32]. Interference of CSNB mutations with SB formation and stability can lead to increased background activity in the presence of non-covalently bound cis-retinal, as evidenced in our structure (Fig 3). The third mechanism, that of a dark state with increased basal activity, is supported by studies using FTIR spectroscopy [3], spin-label experiments [17] and increased hydroxylamine reactivity indicating a perturbation of the G90D dark state [6]. It is valid to ask what the structure of preactivated rhodopsin, that is, an active rhodopsin species with bound cis-retinal, would look like. It is commonly accepted that G protein activation by rhodopsin and other GPCRs is characterized by a conformational change in the seven-helix bundle, most prominently in TM6. So far, all active-state rhodopsin structures, be it in absence [18], or presence of non-covalently [33] or covalently bound retinal [10, 12] show these structural changes. Spin labelling of the cytoplasmic ends of TM3, TM5, TM6 and H8 has shown that G90D rhodopsin can adopt an active-like dark state, that is strikingly similar to the state produced by light activation of the wild-type receptor [17]. FTIR spectroscopy indicated that the preactivated dark state of G90D rhodopsin has a protonated E113 and increased hydrogen-bond strength resulting from rearrangement of the hydrogen-bonding network involving D83, both hallmarks of the light-activated metarhodopsin-II state [3]. Although the dark state of G90D is most likely structurally heterogeneous (supplementary information online), we speculate that the activated fraction of it will be similar to the G90D opsin with the bound retinal we have crystallized.
Figure 5.
The molecular causes of congenital stationary night blindness. In healthy dim-light vision, (A) basal G protein transducin (Gt) activation by opsin and dark-state rhodopsin (red circles) is prevented by a salt bridge (dashed red line) between K296, the site of retinal attachment by means of a protonated Schiff base (SB) (black line) and the retinal counterion E113. Absorption of a photon leads to isomerization of retinal (to trans), proton transfer to E113, opening of the E113-K296 activation switch and a series of conformational changes in the seven-helix bundle (yellow circles). Subsequent phosphorylation by GRK1 increases affinity for arrestin-1, which quenches signalling by blocking the Gt-binding site. (B) The G90D mutation causes congenital stationary night blindness (CSNB) through constant background activation that desensitizes the visual system. Three not mutually exclusive mechanisms might cause this phenotype: (1) constant simulation by constitutively active opsin, (2) spontaneous activation by thermal isomerization of retinal or (3) a preactivated rhodopsin dark state. The G90D mutation introduces an extra charge that interferes (|_|) with the deactivating E113-K296 salt bridge [2, 17] and instead forms a salt bridge with K296 in the active opsin state. This salt bridge interferes with SB formation and prevents efficient opsin deactivation through covalent binding of 11-cis-retinal. Interference of G90D with stability of the retinal SB increases the rate of thermal isomerization and the basal activity of the receptor dark state.
The question remains why only four mutations in rhodopsin cause CSNB, whereas many other known mutations increase constitutive activity of opsin [26], interfere with retinal binding [19], increase thermal isomerization [34] or preactivate dark-state rhodopsin [17, 35]. Our structural data indicate that substitutions that specifically perturb the E113-K296 activation switch result in CSNB (Fig 4), likely by destabilizing the inactive conformation and selectively favouring a short-lived preactivated conformation at the same time. In addition, reduced binding of arrestin could contribute to the disease just as loss of arrestin causes night blindness in patients with Oguchi disease. Whereas the contribution in each of the four CSNB mutations might vary, we suggest that together these mechanisms increase the basal activity in rods and thus cause night blindness in CSNB patients.
METHODS
Mutant rhodopsin was expressed in stably transfected HEK293S cells. Different forms of the receptor were purified using 1D4 immuno-affinity resin and crystallized as described [12, 33]. Diffraction data were collected at the Swiss Light Source and structures were solved by molecular replacement.
Thermal denaturation was followed using binding of the thiol-sensitive dye CPM to cysteins exposed during unfolding. Phosphorylation and arrestin-1 binding to rhodopsin incorporated into nanodiscs were measured using radioactively labelled ATP and arrestin [27]. Full methods are included in the supplementary information online.
Coordinates and structure factors have been deposited under pdb code 4bey for G90D-GaCT2, 4bez for G90D.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank the staff at the Swiss Light Source for excellent support during data collection. We are grateful for financial support from the Swiss National Science Foundation grant 31003A_132815 (to J.S., X.D. and G.F.X.S.) and 31003A_141235 (to J.S.). The work was financially supported by the National Center for Competence in Research in Structural Biology Program (to G.F.X.S.) and National Institutes of Health grants EY011500 (V.V.G.) and HL071818 (J.J.G.T.).
Author contributions: A.S. performed cloning, purification, crystallization and thermal stability measurements. A.S., M.K.O. and J.S. collected data and refined the structures. M.K.O., V.P. and D.V. prepared nanodiscs containing rhodopsin that was phosphorylated by S.A.V and V.V.G. using GRK1 from K.T.H. and J.J.G.T. Binding of radiolabeled arrestin to rhodopsin in nanodiscs were performed by S.S.V. and V.V.G. Data were analysed by A.S., G.F.X.S., V.V.G, X.D. and J.S. The manuscript was written by V.V.G. and J.S. The overall project management was done by J.S.
Footnotes
The authors declare that they have no conflict of interest.
References
- Neidhardt J, Barthelmes D, Farahmand F, Fleischhauer JC, Berger W (2006) Different amino acid substitutions at the same position in rhodopsin lead to distinct phenotypes. Invest Ophthalmol Vis Sci 47: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Rao VR, Cohen GB, Oprian DD (1994) Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature 367: 639–642 [DOI] [PubMed] [Google Scholar]
- Zvyaga TA, Fahmy K, Siebert F, Sakmar TP (1996) Characterization of the mutant visual pigment responsible for congenital night blindness: a biochemical and Fourier-transform infrared spectroscopy study. Biochemistry 35: 7536–7545 [DOI] [PubMed] [Google Scholar]
- Toledo D, Ramon E, Aguilà M, Cordomí A, Pérez JJ, Mendes HF, Cheetham ME, Garriga P (2011) Molecular mechanisms of disease for mutations at Gly-90 in rhodopsin. J Biol Chem 286: 39993–40001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieving PA, Richards JE, Naarendorp F, Bingham EL, Scott K, Alpern M (1995) Dark-light: model for nightblindness from the human rhodopsin Gly-90-->Asp mutation. Proc Natl Acad Sci USA 92: 880–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieving PA, Fowler ML, Bush RA, Machida S, Calvert PD, Green DG, Makino CL, McHenry CL (2001) Constitutive ‘light’ adaptation in rods from G90D rhodopsin: a mechanism for human congenital nightblindness without rod cell loss. J Neurosci 21: 5449–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Cornwall MC, Oprian DD (2003) Opsin activation as a cause of congenital night blindness. Nat. Neurosci 6: 731–735 [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Woodruff ML, Olshevskaya EV, Cilluffo MC, Cornwall MC, Sieving PA, Fain GL (2008) Night blindness and the mechanism of constitutive signaling of mutant G90D rhodopsin. J Neurosci 28: 11662–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MP, Holden DC, Joshi P, Clark CL, Lee AH, Kaushal S (2010) Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. J Mol Biol 395: 1063–1078 [DOI] [PubMed] [Google Scholar]
- Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP (2011) Crystal structure of metarhodopsin II. Nature 471: 651–655 [DOI] [PubMed] [Google Scholar]
- Hamm HE, Deretic D, Arendt A, Hargrave PA, Koenig B, Hofmann KP (1988) Site of G protein binding to rhodopsin mapped with synthetic peptides from the α subunit. Science 241: 832–835 [DOI] [PubMed] [Google Scholar]
- Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J (2012) Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA 109: 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross AKA, Xie GG, Oprian DDD (2003) Slow binding of retinal to rhodopsin mutants G90D and T94D. Biochemistry 42: 2002–2008 [DOI] [PubMed] [Google Scholar]
- Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GFX (2007) Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol 372: 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovsky EA, Oprian DD (1989) Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science 246: 928–930 [DOI] [PubMed] [Google Scholar]
- Mahalingam M, Martínez-Mayorga K, Brown MF, Vogel R (2008) Two protonation switches control rhodopsin activation in membranes. Proc Natl Acad Sci USA 105: 17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-M, Altenbach C, Kono M, Oprian DD, Hubbell WL, Khorana HG (2004) Structural origins of constitutive activation in rhodopsin: Role of the K296/E113 salt bridge. Proc Natl Acad Sci USA 101: 12508–12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454: 183–187 [DOI] [PubMed] [Google Scholar]
- Piechnick R, Ritter E, Hildebrand PW, Ernst OP, Scheerer P, Hofmann KP, Heck M (2012) Effect of channel mutations on the uptake and release of the retinal ligand in opsin. Proc Natl Acad Sci USA 109: 5247–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX (2004) Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol 343: 1409–1438 [DOI] [PubMed] [Google Scholar]
- McAlear SD, Kraft TW, Gross AK (2010) 1 rhodopsin mutations in congenital night blindness. Adv Exp Med Biol 664: 263–272 [DOI] [PubMed] [Google Scholar]
- Gross AK, Rao VR, Oprian DD (2003) Characterization of rhodopsin congenital night blindness mutant T94I. Biochemistry 42: 2009–2015 [DOI] [PubMed] [Google Scholar]
- Fuchs SS, Nakazawa MM, Maw MM, Tamai MM, Oguchi YY, Gal AA (1995) A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet 10: 360–362 [DOI] [PubMed] [Google Scholar]
- Yamamoto SS, Sippel KCK, Berson ELE, Dryja TPT (1997) Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet 15: 175–178 [DOI] [PubMed] [Google Scholar]
- Rim JJ, Oprian DDD (1995) Constitutive activation of opsin: interaction of mutants with rhodopsin kinase and arrestin. Biochemistry 34: 11938–11945 [DOI] [PubMed] [Google Scholar]
- Han M, Smith S, Sakmar T (1998) Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry 37: 8253–8261 [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV (2010) Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem 286: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne T, Edwards PC, Leslie AGW, Tate CG (2012) Crystal structures of a stabilized β(1)-adrenoceptor bound to the biased agonists Bucindolol and Carvedilol. Structure 20: 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K (2012) Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335: 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberg K, Kim T-Y, Möller M, Skegro D, Dasara Raju G, Granzin J, Büldt G, Schlesinger R, Alexiev U (2011) Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proc Natl Acad Sci USA 108: 18690–18695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono MM, Goletz PWP, Crouch RKR (2008) 11-cis- and all-trans-retinols can activate rod opsin: rational design of the visual cycle. Biochemistry 47: 7567–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefalov VJ, Crouch RK, Cornwall MC (2001) Role of noncovalent binding of 11-cis-retinal to opsin in dark adaptation of rod and cone photoreceptors. Neuron 29: 749–755 [DOI] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D'Antona A, Fransen M, Xie G, Oprian DD, Schertler GF (2011) The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu MY, Nguyen JB, Bhagat A, Mooney V, Yan ECY (2011) Thermal properties of rhodopsin: insight into the molecular mechanism of dim-light vision. J Biol Chem 286: 27622–27629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JMJ, Altenbach CC, Thurmond RLR, Khorana HGH, Hubbell WLW (1997) Structure and function in rhodopsin: rhodopsin mutants with a neutral amino acid at E134 have a partially activated conformation in the dark state. Proc Natl Acad Sci USA 94: 14273–14278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.