Abstract
We report the first example of antisense RNA regulation in a hyperthermophilic archaeon. In Sulfolobus solfataricus, the transposon-derived paralogous RNAs, RNA-2571–4, show extended complementarity to the 3′ UTR of the 1183 mRNA, encoding a putative phosphate transporter. Phosphate limitation results in decreased RNA-2571 and increased 1183 mRNA levels. Correspondingly, the 1183 mRNA is faster degraded in vitro upon duplex formation with RNA-2571. Insertion of the 1183 3′ UTR downstream of the lacS gene results in strongly reduced lacS mRNA levels in transformed cells, indicating that antisense regulation can function in trans.
Keywords: Sulfolobus solfataricus , non-coding RNA, antisense regulation
INTRODUCTION
In prokaryotes, small non-coding RNAs are involved in various biological processes, including transcriptional and translational regulation, RNA processing, RNA-guided modification of RNA and chromosome replication [1]. In eukaryotes, short interfering RNAs and microRNAs (miRNAs) act as regulators of development, cell death and chromosome silencing. Short interfering RNAs are derived from double-stranded RNA and act by RNA interference (RNAi) resulting in cleavage of the target mRNA [2]. The genome-encoded miRNAs act as components of ribonucleoprotein complexes. Binding of these complexes to the 3′ untranslated region (UTR) of mRNAs leads to translational repression and/or mRNA decay [3]. In contrast to eukaryal miRNAs, in bacteria, small regulatory RNAs (sRNAs) predominantly target the 5′ UTR of mRNAs [1].
In bacteria, two major classes of RNAs involved in gene regulation can be discerned, cis- and trans-acting RNAs. The prototypic trans-acting sRNAs of Enterobacteriaceae have a typical size between 50 and 200 nucleotides, are usually not genetically linked to the loci of their target genes and are often expressed under specific growth or stress conditions [1, 4]. Whereas some sRNAs act to modulate the activity of proteins, the majority appears to modulate gene expression by non-contiguous base-pairing with the 5′ UTR of mRNAs [1]. Regulation is mainly negative and seems to occur largely at the level of translation initiation and mRNA stability control [5]. Cis-acting bacterial RNAs either arise from short convergent transcripts that are complementary to the 5′ UTR and the immediate coding region of their target mRNA or from mRNAs containing a long 5′ or 3′ UTR that overlap with the mRNA encoded by the complementary DNA strand [5, 6]. In bacteria, cis-antisense RNAs are involved in DNA replication control, maintenance of plasmids and in virulence gene regulation [6].
Avenues of research on non-coding RNAs (ncRNAs) in Archaea concerned the identification and characterization of small nucleolar RNAs, RNAs involved in rRNA modification [7] and RNAs involved in CRISPR-based immune systems [8]. In addition, several surveys for small regulatory ncRNAs in Archaea have been conducted [9]. Sulfolobus solfataricus (Sso) is a hyperthermophile that serves as a model organism for the crenarchaeal clade of Archaea. The genome of Sso contains a large number of mobile elements [10]. The genome has apparently undergone, and still undergoes, extensive rearrangements, which can be in part attributed to transposition events [10]. An RNomics approach identified 19 cis-antisense and 11 trans-encoded ncRNA candidates in Sso [11]. The majority of the cis-antisense RNAs were encoded opposite to transposase genes, suggesting that the RNAs could be involved in silencing of transposons. Moreover, high-throughput RNA sequencing (RNAseq) identified 185 cis-antisense and 125 trans-encoded ncRNA candidates in Sso [12]. It has been speculated that some of the trans-acting ncRNAs could regulate mRNAs by interacting with their 3′ UTRs [11], analogously to miRNAs in eukaryotes. However, the function, as well as the mechanism of these putative ncRNAs has remained unclear. With the exception of CRISPR-based immunity systems [13, 14], there is so far only one report on antisense regulation in Archaea. In Halobacterium salinarum, an antisense RNA was shown to interact with the first 151 nucleotides of an early lytic phage transcript. This interaction results in cleavage of the mRNA and leads to removal of the ribosome-binding site, which renders the mRNA non-functional [15].
Here, we show that ncRNAs can regulate gene expression in the hyperthermophile Sso by interacting with complementary sequences present in the 3′ UTR of ORF 1183. As a result, the mRNA is apparently destabilized, which is reminiscent to miRNA-mediated regulation in eukaryotes.
RESULTS AND DISCUSSION
Several paralogs of RNA-257 are present in Sso
Tang et al [11] have identified several putative trans-acting Sso ncRNAs, encompassing extended regions of complementarity with distinct mRNAs. In this study, we have focused on RNA-257, termed herein RNA-2571. A Blast search (http://www-archbac.u-psud.fr/projects/sulfolobus/Blast_Search.html) revealed three paralogs of RNA-2571, previously identified as RNA-107, RNA-91 and RNA-20 [11, 12], which are renamed herein as RNA-2572, RNA-2573 and RNA-2574 (supplementary Fig S1A online), respectively. RNA-2571–4 are encoded in intergenic regions of the Sso genome (Fig 1), and all of them are transcribed [12]. The RNA-2571 paralogs differ in length but posses a highly conserved core region (supplementary Fig S1A online). Further bioinformatic analyses disclosed a significant homology of these core regions with the distal coding region of a putative transposase gene of a transposon belonging to the ISC1904 family. This transposon is present in eleven copies in the Sso genome (Fig 1). Most probably, RNA-2571–4 are remnants of transposon rearrangements, whereby subtle nucleotide exchanges (supplementary Fig S1B online) created a promoter sequence, which led to the synthesis of RNA-2571–4. One of the chromosomal copies of the transposons from which RNA-2571–4 are apparently derived, is represented by Sso ORFs 1181 (putative resolvase gene) and 1182 (putative transposase gene; Fig 1). RNA-2571–4, posses consensus-like promoter regions [16] (supplementary Fig S1B online). In contrast, only an imperfect promoter sequence is present upstream of the sequence corresponding to the conserved core sequence of RNA-2571–4 in the distal part of the putative transposase genes of the respective transposons (supplementary Fig S1B online). According to an earlier report [16], the presence of the G at position four of box A (supplementary Fig S1B online) should prevent transcription. In agreement, RNAseq did not show a significant increase in reads corresponding to the 3′ end of ORF1182 [12].
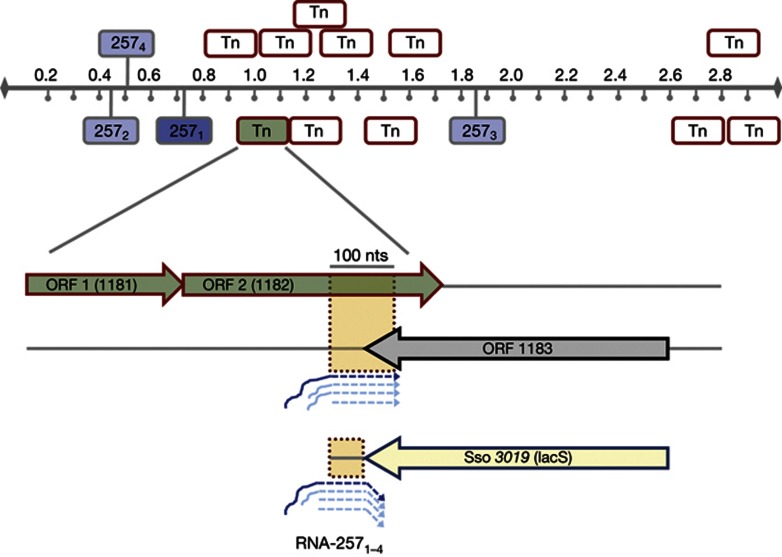
Figure 1.
Several paralogs of the RNA-2571 are present in the Sso genome. Location of RNA-2571–4 (blue) and the 11 transposons (Tn; red boxes) in the Sso genome. The RNA-2571 (dark blue) corresponds to the originally identified RNA-257 [11]. The transposon highlighted in green comprises Sso ORFs 1181 and 1182 (green arrow), which is opposite to Sso ORF 1183 (grey arrow). The RNAs-2571–4 show complementarity to the 3′ end and the 3′ UTR of ORF 1183 (orange box). The lacS-1183 3′ UTR transcript is targeted by RNA-2571–4 in trans.
As the Sso ORFs 1182 and 1183 are convergently transcribed [12], the distal end of the 1182 transcript is complementary to the 3′ end as well as to the 3′ UTR of the 1183 transcript (Fig 1), encoding a putative phosphate transporter. Bioinformatic analyses revealed that ORF 1183 is conserved in other Sulfolobales (∼78% identity). However, the 1183 3′ end and the 3′ UTR with complementarity to ORF 1182 and with RNA-2571–4 is only present in Sso (supplementary Fig S1D online). The high homology of the core regions of RNA-2571–4 to the distal end of 1182 mRNA, and thus the partial complementary to the 3′ end of 1183 mRNA, prompted us to ask whether regulation by antisense RNAs does occur in the hyperthermophile Sso.
Phosphate-dependent abundance of RNA-2571–4
To study the expression pattern of RNA-2571 during different growth/ stress conditions, we first used a RNA-2571-specific probe with complementarity to a less-conserved region (supplementary Fig S1A online). These studies revealed that the steady state levels of RNA-2571 depend on the phosphate availability in the growth medium; the levels of RNA-2571 were decreased under phosphate-limiting conditions (Fig 2A). In addition, northern blot analyses with probes specific for RNA-2572, RNA-2573 and RNA-2574 indicated that their levels were likewise decreased under phosphate-limiting conditions (supplementary Fig S1C online). We can only speculate why expression of all RNA-2571–4 genes is phosphate dependent. Inverted repeats have been described as important elements of phosphate-sensitive promoters in Mycobacterium smegmatis [17]. An inverted repeat preceding boxA is conserved in all four RNA-2571–4 promoters (supplementary Fig S1B online). As the RNA-2571–4 genes are most likely remnants of transposition events, it is conceivable that the inverted repeat, and thus the phosphate sensitivity of all four promoters, were generated during these events. At this junction, we did not further study transcriptional regulation of RNA-2571–4. Instead, we asked whether the increased abundance of RNA-2571–4 in full medium (plus phosphate) might correlate with a downregulation of 1183 mRNA encoding the putative phosphate transporter, that is, whether RNA-2571–4 could be involved in negative antisense regulation of 1183 mRNA. Therefore, the levels of RNA-2571 and 1183 mRNA were quantified in the presence of phosphate and under phosphate-limiting conditions employing qPCR. As shown in Fig 2B, an inverse correlation of the levels of RNA-2571 and 1183 mRNA was observed under both conditions.
Figure 2.
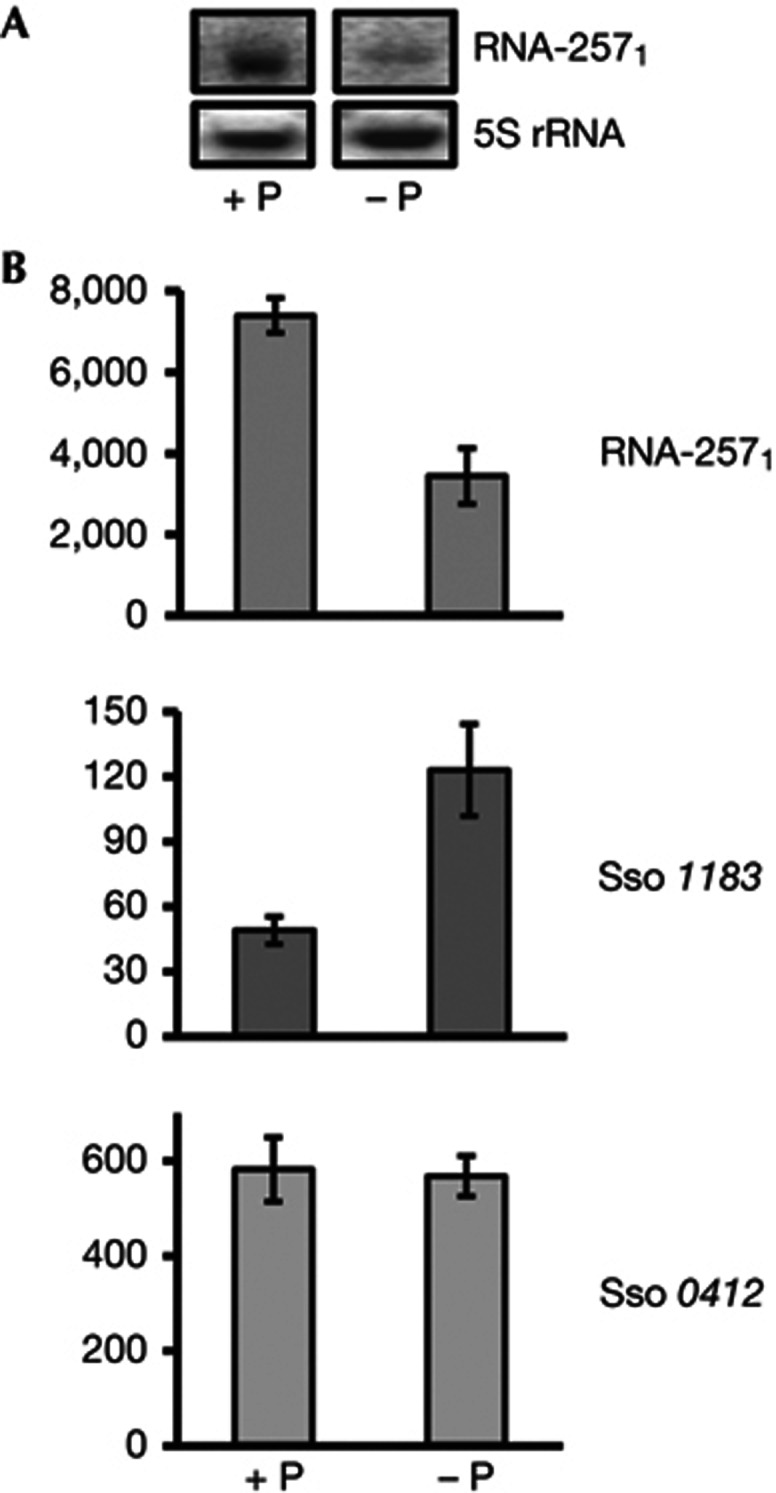
Inverse correlation of RNA-2571 and 1183 mRNA levels in the presence and absence of phosphate. (A) Northern blot analysis of RNA-2571 levels with total RNA from Sso cells grown in either full medium (+P; 280 mg/l KH2PO4) or under phosphate-limiting conditions (−P). 5S rRNA was used as loading control. (B) qPCR analysis of RNA-2571 and 1183 mRNA levels in the presence of phosphate (+P) and under phosphate-limiting conditions (−P). The Sso 0412 mRNA, encoding aIF2-γ, was used as a house-keeping endogenous control. The error bars represent s.d. from triplicate experiments. The numbers represent copies per 10 ng cDNA.
Antisense regulation by trans-acting RNAs in Sso
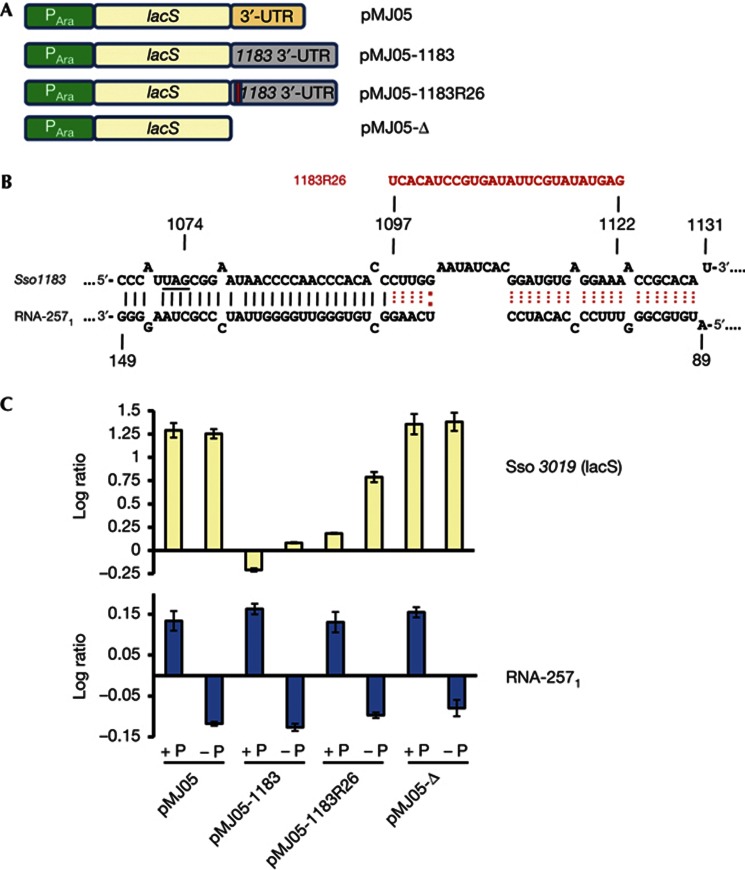
As the levels of 1183 mRNA were decreased at elevated levels of RNA-2571 (Fig 2B), these pilot studies provided a first hint for antisense regulation in a hyperthermophile. However, as ORF 1182 is convergently transcribed with 1183 mRNA, the above experiment did not distinguish whether 1183 mRNA is downregulated in trans by RNA-2571–4 or by a cis-antisense mechanism mediated by the full-length 1182 transcript. To address whether antisense regulation by RNA-2571–4 can function in trans, the 3′ UTR of ORF 1183 was inserted downstream of the lacS gene (Fig 3A). The Sso strain PH1-16 was transformed with plasmid pMJ05 [18] (lacS with the authentic 3′ UTR) and plasmid pMJ05-1183 (lacS with the 3′ UTR of 1183), respectively. In contrast to strain PH1-16(pMJ05), very low levels of ß-galactosidase activity were observed with strain PH1-16(pMJ05-1183) (supplementary Fig S2A online). As the transcripts are in both cases under the control of an arabinose-inducible promoter it was likely that the lacS-3′UTR-1183 mRNA is targeted by RNA-2571–4 in trans, and that the mRNA is subsequently degraded. Therefore, we next compared the lacS mRNA levels of strain PH1-16(pMJ05) and strain PH1-16(pMJ05-1183) grown in the presence of phosphate, using RT–PCR (supplementary Fig S2B online) and qPCR (Fig 3C). In contrast to strain PH1-16(pMJ05), no (supplementary Fig S2B online) or very low levels (Fig 3C) of lacS-3′UTR-1183 mRNA were detected in strain PH1-16(pMJ05-1183), suggesting that lacS-3′UTR-1183 mRNA is rapidly degraded in vivo.
Figure 3.
Replacement of the lacS 3′ UTR with the 1183 3′ UTR results in destabilization of lacS mRNA. (A) Schematic depiction of gene constructs in plasmids pMJ05, pMJ05-1183, pMJ05-1183R26 and pMJ05-Δ. The lacS gene is preceded by an arabinose-inducible promoter (PAra). The lacS 3′ UTR is replaced by the 3′ UTR of Sso 1183 in the pMJ05-1183 construct. In pMJ05-1183R26, 26 nt are replaced by an unrelated sequence (see B). In pMJ05-Δ the lacS gene is devoid of a 3′ UTR. (B) A part of the complementary region between RNA-2571 and the 3′ UTR of ORF 1183 is depicted. Twenty-six nucleotides of the 1183 3′ UTR were replaced by an unrelated sequence (1183R26 shown in red). (C) qPCR analysis of lacS and RNA-2571 expression levels in the presence of phosphate (+P) and under phosphate-limiting conditions (−P). The transcript levels were normalized against that of Sso 0412, encoding aIF2-γ. The numbers represent the log ratio normalized to expression values of Sso 0412. The ratio of lacS mRNA: RNA-2571 was calculated for strain PH1-16(pMJ05-1183) with 0.4 (+P) and 1.6 (−P) and for strain PH1-16(pMJ05-1183R26) with 1.3 (+P) and 7 (−P), respectively. The error bars represent s.d. from triplicate experiments.
As multiple gene knockouts are not feasible in Sso, we could not delete the RNA-2571–4 genes to unequivocally test whether these RNAs mediate antisense regulation of 1183 mRNA or of the lacS-3′UTR-1183 transcript. Therefore, the putative RNA-2571–4 base-pairing sequence within the 1183 3′ UTR of the lacS-3′UTR-1183 transcript was altered to reduce binding of RNA-2571–4. First, 26 nucleotides of the base-pairing site (Fig 3B) within the 3′ UTR of the 1183 ORF were replaced in plasmid pMJ05-1183R26 by an unrelated sequence with the aim to weaken the interaction between RNA-2571–4, and the 1183 3′ UTR. Second, the entire 1183 3′ UTR of the lacS-3′UTR-1183 transcript was deleted in plasmid pMJ05-Δ to provide a mock control. When the cells were grown in the presence of phosphate, the replacement restored the lacS mRNA levels to some extent, whereas the entire deletion of the 1183 3′ UTR within the lacS-3′UTR-1183 transcript resulted in lacS mRNA levels comparable to that observed with plasmid pMJ05 (supplementary Fig S2B online; Fig 3C). In agreement, the strains PH1-16(pMJ05) and PH1-16(pMJ05-Δ) displayed equivalent β-galactosidase activities, whereas the strains PH1-16 (pMJ05-1183) and PH1-16(pMJ05-1183R26) displayed very low β-galactosidase activities (supplementary Fig S2A online).
The experiments shown in Fig 2 did not exclude the possibility that Sso 1183 levels could be as well regulated in a phosphate-dependent manner, that is, independently of RNA-2571–4. To obtain additional support for the phosphate-dependent regulation of 1183 mRNA by RNA-2571–4 (Fig 2), we tested whether the abundance of the lacS transcript in strains PH1-16(pMJ05), PH1-16(pMJ05-1183), PH1-16(pMJ05-1183R26) and PH1-16(pMJ05-Δ) is likewise phosphate dependent. Using qPCR, the lacS transcript abundance was compared in cells grown in full medium (+P) and under phosphate-limiting conditions (−P). As anticipated, the lacS (Fig 3C) and ß-galactosidase (supplementary Fig S2A online) levels were comparable under both conditions in strains PH1-16(pMJ05) and PH1-16(pMJ05-Δ). In contrast, in the absence of phosphate, that is, at reduced levels of RNA-2571, an increased abundance of both, the lacS-3′UTR-1183 and the lacS-3′UTR-1183R26 transcript was observed (Fig 3C). The abundance of the lacS-3′UTR-1183R26 transcript (lacS: RNA-2571 ratio=7) was higher than that of the lacS-3′UTR-1183 transcript (lacS: RNA-2571 ratio=1.6), which was anticipated as the alteration within the 1183 3′ UTR already attenuated the apparent regulation by RNA-2571–4 (supplementary Fig S2B online; Fig 3C), which was obviously augmented by a simultaneous reduction of the RNA-2571–4 levels. However, the ß-galactosidase activities obtained with plasmids pMJ05-1183 and pMJ05-1183R26 (supplementary Fig S2A online) were very low and not distinguishable when the cells were cultivated in the presence or absence of phosphate. Apparently, there is enough RNA-2571–4 under both conditions to drastically reduce the transcript levels of the corresponding lacS transcript (Fig 3D; supplementary Fig S2B online), which results in rather low ß-galactosidase activities (supplementary Fig S2A online), and in turn precludes a reasonable read out dependent on the levels of RNA-2571–4. In summary, as a mutation of the base-pairing site as well as the phosphate-dependent modulation of the RNA-2571–4 levels lead to increased or decreased levels of fusion constructs containing the 1183 3′ UTR, these experiments supported the hypothesis that the 1183 3′ UTR is targeted by the antisense RNAs in trans.
To verify the studies performed with the lacS-3′UTR-1183 construct(s), the same sequence of the 1183 3′part was fused to the coding region of the Sso Sm1 protein (ORF 6454) in plasmid pMJ05 (supplementary Fig S3A online). In brief, with this fusion construct the same results as with the lacS-3′ UTR-1183 construct were obtained. The same inverse correlation between the abundance of the 6454-3′UTR-1183 transcript and that of RNA-2571 were observed in the presence of phosphate and under phosphate-limiting conditions (supplementary Fig S3B online). In addition, under phosphate-limiting conditions the Sm1 protein levels of Sso strain PH1-16(pMJ05-6454-1183) were increased when compared with the same strain grown in the presence of phosphate (supplementary Fig S3C online).
Degradation of 1183 mRNA by RNA-2571
Duplex formation between a ncRNA and mRNA can create a cleavage site for dedicated riboendonucleases recognizing double-stranded RNA [18, 19]. We therefore asked whether on addition of Sso S30 extracts a preformed duplex between RNA-2571 and 1183 mRNA is subjected to accelerated degradation when compared with 1183 mRNA alone. We first tested whether duplex formation between RNA-2571 and the 3′ UTR of 1183 mRNA occurs in vitro at 75 °C. An electrophoretic mobility shift assay (supplementary Fig S4A online) revealed that RNA-2571 forms a duplex with the 3′ UTR of ORF 1183 mRNA, showing that base pairing between these RNAs is feasible. To further assess the stability of the duplex at high temperature, a melting curve analysis was performed. This analysis showed that the duplex between RNA-2571 and the 3′ UTR of 1183 mRNA is stable below 75 °C, begins to melt at temperatures above 75 °C, and that full denaturation occurs at temperatures above 90 °C (supplementary Fig S4B online).
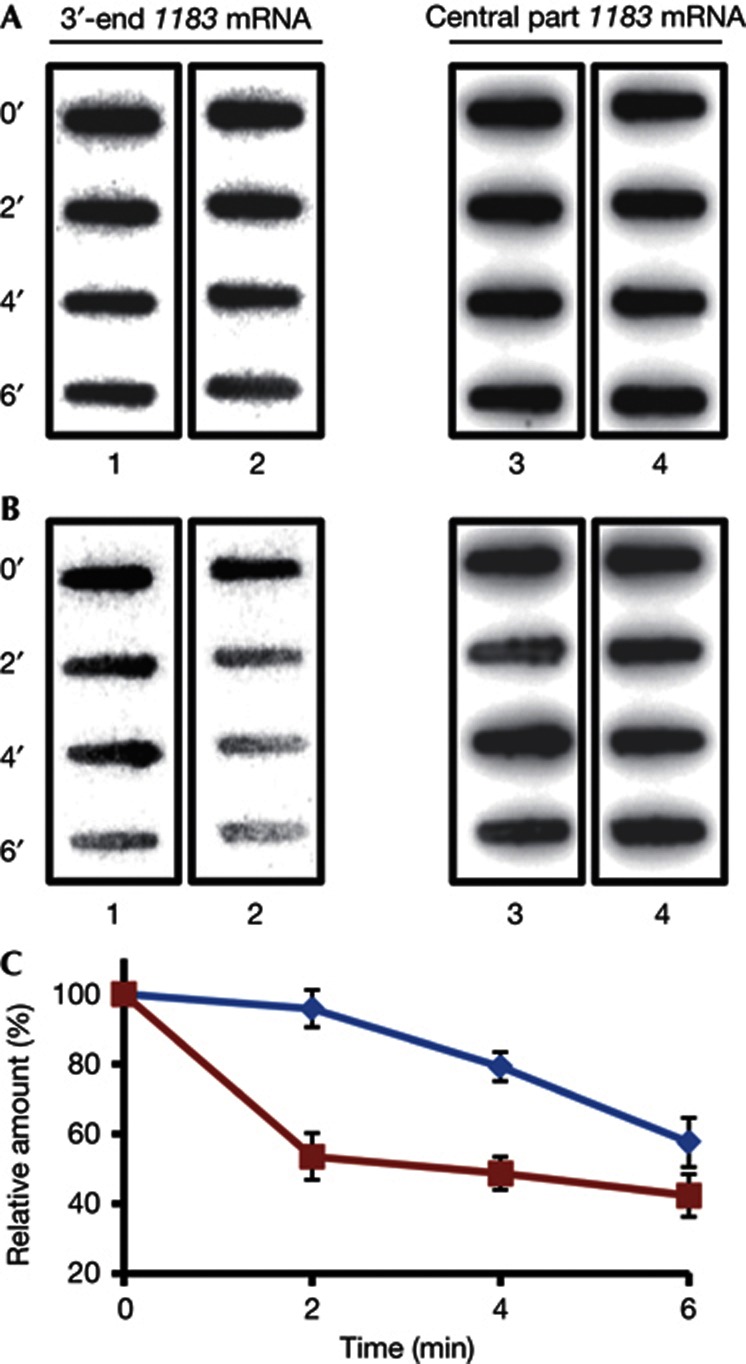
The 1183 mRNA and the preformed 1183 mRNA-RNA2571 duplex, respectively, were added to Sso extracts. Samples were withdrawn after 0, 2, 4 and 6 min, transferred to a nylon membrane and degradation of 1183 mRNA was monitored using a probe specific for the 3′ end of the 1183 coding region (Fig 4B, lanes 1 and 2) as well as for the central part of 1183 mRNA (Fig 4B, lanes 3 and 4). The experiments were done in triplicate and the remaining 1183 mRNA was quantified. The degradation of the 3′ end of the 1183 coding region started after 4 min on addition of Sso extracts, whereas after 2 min 50% of the 3′ end of 1183 mRNA was degraded when the 1183 mRNA-RNA2571 duplex was incubated with Sso extracts (Fig 4B, C). In contrast, during the 6-min time course no differences were observed in the stability for 1183 mRNA alone and of the 1183 mRNA-RNA2571 duplex on addition of Sso extracts when the remaining 1183 mRNA was monitored with a probe directed against the central part of the mRNA (Fig 4B). The same set of experiments was performed with heat-inactivated S30 extracts to ascertain that degradation is dependent on components present in the Sso extracts. As shown in Fig 4A, no significant degradation of 1183 mRNA was observed on addition of heat-inactivated Sso extracts. Hence, we could recapitulate the apparent in vivo antisense regulation of 1183 mRNA and of the lacS-3′UTR-1183 transcript in vitro using RNA-2571.
Figure 4.
Duplex formation between RNA-2571 and 1183 mRNA augments degradation of 1183 mRNA in vitro. (A) Heat-inactivated Sso extracts and (B) ‘active’ S30 extracts were added to 1183 mRNA (lanes 1 and 3) or to the 1183 mRNA-RNA2571 duplex (lane 2 and 4). After the indicated times in minutes, samples were withdrawn and the 1183 mRNA levels were determined with a probe specific for the 3′ end of 1183 mRNA (lane 1 and 2) and with a probe specific for the central part of 1183 mRNA (lane 3 and 4). The experiment was performed in triplicate. One representative experiment is shown. (C) Quantification of the results. The graph shows the relative amounts of the 3′ part of 1183 mRNA remaining in the absence of RNA-2571 (blue) and when in duplex with RNA-2571 (red) plotted as a function of time. The error bars represent s.d. from triplicate experiments.
Concluding remarks
As noted above, we cannot completely dismiss a cis-acting mechanism or a direct, phosphate-dependent regulation of Sso 1183 in the natural setting. However, as the phosphate-dependent modulation of the RNA-2571–4 levels led to increased or decreased levels of lacS/6454 fusion transcripts containing the 1183 3′ UTR, we conclude that antisense regulation can function in trans in the hyperthermophile Sso. This raises the question as regarding RNases/factors involved in degradation of the target mRNA. In eukaryotes, two families of proteins have been shown to be required for RNAi pathways, the Dicer and Argonaute protein families [3]. Dicers belong to the RNase III family of riboendonucleases containing a PAZ domain, that bind and cleave double-stranded RNAs [3]. Argonaute proteins also possess endonuclease activity and require guide RNAs to cleave target mRNAs [3]. Besides the PAZ domain, Argonaute proteins contain an RNase H-like Piwi domain responsible for target RNA cleavage [3]. Proteins containing a Piwi domain on the basis of the Argonaute homologue of Pyrococcus furiosus [20] were only found in five strains belonging to the clade of Euryarchaeota [21]. In Sulfolobus tokadaii, a homologue of RNase HI [22], one of the three prokaryotic RNase H classes, was reported to cleave dsRNA [23]. In Sso, a gene encoding a homologue of RNase HII was identified [22]. It remains to be elucidated whether these proteins or proteins such as CRISPR crRNA-related Cas proteins [13, 24] or as yet unidentified endoribonucleases are involved in degradation of Sso mRNAs targeted by antisense RNAs.
The most prominent group of Sso antisense RNAs identified by an RNomics approach is transcribed in the opposite orientation to transposase genes. Therefore, it has been speculated that these antisense RNAs regulate transposition events by inhibiting expression of transposase mRNA [11]. Similarly, the majority of eukaryal PIWI-interacting RNAs are antisense to transposon sequences, suggesting that they are involved in silencing of transposons [25]. In contrast, RNA-2571–4 are sense to the transposase gene (ORF 1182). The 3′ end of ORF 1183 and the 3′ UTR are not present in three other Sulfolobales (supplementary Fig S1D online). Thus, apparently only in Sso the transposon (ORF1182) inserted within ORF 1183, thereby altering the 3′ end of this reading frame. This event apparently rendered 1183 mRNA vulnerable to antisense regulation by RNA-2571–4 being descendants of the transposon ORF 1182. Thus, RNA-2571–4 could exemplify a means for the origin of trans-encoded regulatory RNAs.
METHODS
Strains and growth conditions. S. solfataricus P2 was grown at 75 °C in Brock’s medium containing either KH2PO4 to a final concentration of 280 mg/l (+P) or shifted to Brocks medium without KH2PO4 (−P; phosphate-limiting conditions). Electroporation of S. solfataricus pyrEF lacS mutant PH1-16 with plasmids pMJ05, pMJ05-1183, pMJ05-1183R26, pMJ05-Δ and pMJ05-6454-1183, and the isolation of transformants was performed as described [26]. See supplementary information online for further details.
Northern blot analysis. Total RNA from S. solfataricus strain P2 (OD600=0.6) grown in the presence and absence of KH2PO4, was isolated and separated on 8% polyacrylamide/urea gels and transferred to nylon membranes. After crosslinking, the membrane was incubated with either the [32P]-5′-end labelled oligonucleotide 5′-GGCAGACCCGTTCATAC-3′ specific for RNA-2571, the oligonucleotide 5′-GGTGGTGCGTCATCAGATTAT-3′ specific for RNA-2573 or the oligonucleotide 5′-GATTGTCTTACCAACCTTTC-3′ specific for RNA-2572 and RNA-2574. The 5S rRNA was probed with oligonucleotide 5′-CACTAACGTGAGCGGCTTAAC-3′ and served as loading control.
RT–PCR and qPCR. Total RNA from S. solfataricus strains P2, PH1-16(pMJ05), PH1-16(pMJ05-1183), PH1-16(pMJ05-1183R26), PH1-16(pMJ05-Δ), PH1-16(pMJ05-6454-1183), grown in either full medium (phosphate present) or under phosphate-limiting conditions was isolated and complementary DNA was synthesized using random hexamer oligos (Fermentas) and SuperScriptIII Reverse Transcriptase (Invitrogen). See supplementary information online for further details.
In vitro RNA degradation assay. S30 extracts of Sulfolobus solfataricus P2 were either pre-incubated for 20 min at 75 °C or heat-inactivated for 20 min at 100 °C. To achieve duplex formation, full-length 1183 mRNA (0.16 μM) was incubated with RNA-2571 (0.48 μM) followed by addition of either the ‘active’ or the heat-inactivated extracts (0.3 μg protein/μl) and incubated at 75 °C. Samples were withdrawn at 0, 2, 4 and 6 min, the reaction was terminated by addition of 1 mM EDTA and an equal amount of 2 × RNA loading dye, and then transferred to a nitrocellulose membrane. After crosslinking either the [32P] 5′-end labelled oligonucleotide 5′-GAAATCCCATGAAGCCCAAAC-3′ (complementary to nucleotides 979–999 of 1183 mRNA) or the [32P] 5′-end labelled oligonucleotide 5′-CTGTGTTCAGCCATTATCG-3′ (complementary to nucleotides 410–428 (central part) of 1183 mRNA) was added to detect 1183 mRNA. See supplementary information online for further details.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by grant 22888 from the Austrian Science Fund (FWF) to U.B. We thank R. Garrett and V. Sedlyarov for helpful discussions and P. Blum for materials.
Author contributions: B.M., D.H. and U.B. designed experiments; B.M., S.M. and A.M. performed experiments; B.M. and U.B. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J (2010) RNAs: regulators of bacterial virulence. Nat Rev Microbiol 8: 857–866 [DOI] [PubMed] [Google Scholar]
- Kaberdin VR, Bläsi U (2006) Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev 30: 967–979 [DOI] [PubMed] [Google Scholar]
- Georg J, Hess WR cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Omer A (2005) Small non-coding RNAs in Archaea. Curr Opin Microbiol 8: 685–694 [DOI] [PubMed] [Google Scholar]
- Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, Erdmann S, She Q (2011) CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans 39: 51–57 [DOI] [PubMed] [Google Scholar]
- Marchfelder A, Fischer S, Brendel J, Stoll B, Maier LK, Jager D, Prasse D, Plagens A, Schmitz RA, Randau L (2012) Small RNAs for defence and regulation in Archaea. Extremophiles 16: 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redder P, Garrett RA (2006) Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J Bacteriol 188: 4198–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Hüttenhofer A (2005) Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol 55: 469–481 [DOI] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R (2009) A single-base resolution map of an archaeal transcriptome. Genome Res 20: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A, Zebec Z, Teichmann D, Schleper C (2011) In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80: 481–491 [DOI] [PubMed] [Google Scholar]
- Stolt P, Zillig W (1993) Antisense RNA mediates transcriptional processing in an archaebacterium, indicating a novel kind of RNase activity. Mol Microbiol 7: 875–882 [DOI] [PubMed] [Google Scholar]
- Hain J, Reiter WD, Hudepohl U, Zillig W (1992) Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res 20: 5423–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR (2007) The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol 189: 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A, Afonyushkin T, Lombo TB, McDowall KJ, Bläsi U, Kaberdin VR (2008) Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 14: 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 The same inverse correlation between the abundance of the 6454-3′UTR-1183 transcript and that of RNA-2571 were observed in the presence of phosphate and under phosphate limiting conditions nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Fujishima K, Sato A, Tsuchiya D, Tomita M, Kanai A (2010) Characterization of RNase HII substrate recognition using RNase HII-argonaute chimaeric enzymes from Pyrococcus furiosus. Biochem J 426: 337–344 [DOI] [PubMed] [Google Scholar]
- Wei KF, Wu LJ, Chen J, Chen YF, Xie DX (2012) Structural evolution and functional diversification analyses of argonaute protein. J Cell Biochem 113: 2576–2585 [DOI] [PubMed] [Google Scholar]
- Kochiwa H, Tomita M, Kanai A (2007) Evolution of ribonuclease H genes in prokaryotes to avoid inheritance of redundant genes. BMC Evol Biol 7: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Yanagawa H, Tomita M, Itaya M (2004) Cleavage of double-stranded RNA by RNase HI from a thermoacidophilic archaeon, Sulfolobus tokodaii 7. Nucleic Acids Res 32: 5809–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Brouns SJ (2009) RNAi: prokaryotes get in on the act. Cell 139: 863–865 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ (2009) Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonuscheit M, Martusewitsch E, Stedman KM, Schleper C (2003) A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol 48: 1241–1252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.