Abstract
MicroRNAs (miRNAs) post-transcriptionally repress gene expression via the miRNA-induced silencing complex (miRISC), which includes miRNA, Argonaute and a GW182 family member. Here we show that in Caenorhabditis elegans, miRNA-mediated gene silencing is modulated by macroautophagy, a lysosome-mediated degradation process. Loss of autophagy activity suppresses developmental defects caused by partially impaired silencing of miRNA targets including the let-7 family and lsy-6. The C. elegans GW182 homolog AIN-1 is itself selectively degraded by autophagy and colocalizes with the p62 homolog SQST-1 in autophagy mutants. Thus, autophagy activity modulates miRNA-mediated gene silencing and degrades a core miRISC component.
Keywords: Argonaute, autophagy, C. elegans , GW182, miRNA
INTRODUCTION
MicroRNAs (miRNAs), a class of evolutionarily conserved ∼22 nucleotide RNAs, post-transcriptionally repress gene expression through non-perfect base pairing to the 3′ untranslated region (UTR) of target mRNAs, affecting translational efficiency and/or stability 1]. The repressive effect of miRNAs is mediated by the miRNA-induced silencing complex (miRISC), which includes miRNA, Argonaute (AGO) and a GW182 family member [2]. miRNA-mediated gene silencing regulates a variety of developmental and cellular processes. In Caenorhabditis elegans, miRNAs control developmental timing of diverse post-embryonic cell lineages by regulating stage-specific expression of a group of heterochronic genes [3]. For example, the let-7 family miRNAs (mir-48, mir-84 and mir-241) repress the Hunchback homolog hbl-1 to control succession from the L2 to L3 larval stage, while let-7 miRNA downregulates the expression of lin-41 in specifying the L4-adult switch [4], [5], [6]. Loss of function of the miRNA pathway results in retarded heterochronic defects, in which the stage-specific fate is reiterated and terminal differentiation is delayed [3]. The let-7 family miRNAs also negatively regulates let-60/RAS, a key component in vulval cell fate specification [7]. Loss of let-60/RAS activity causes a vulvaless phenotype, while gain of function of let-60 activity results in generation of supernumerary vulva-like structures, a phenotype known as multivulva (Muv) [8]. The lsy-6 miRNA controls left–right asymmetry of the two taste receptor neurons, ASE left (ASEL) and ASE right (ASER) [9]. lsy-6, which is expressed in ASEL, but not in ASER, exerts its effect by repressing the Nkx-type homeobox gene cog-1 [9]. miRNA-mediated silencing of target genes in these developmental processes has been shown to be modulated by various factors, including the TRIM-NHL family protein NHL-2 and Wnt signaling [10], [11].
Autophagy is an evolutionarily conserved intracellular degradation process, involving the formation of a closed double-membrane compartment called the autophagosome, and its subsequent delivery of sequestrated materials to the lysosome for degradation [12]. Several Atg (autophagy-related) and epg (ectopic PGL granules) genes have been identified that act at discrete steps of autophagosome formation in higher eukaryotes [12–14]. For example, the serine/threonine kinase Atg1/Atg13 complex and the class III phosphatidylinositol 3-kinase, Vps34/Atg6 complex are required for initiation and nucleation of the isolation membrane. The two ubiquitin-like systems, consisting of Atg8(ubiquitin-like molecule)/Atg7(E1-like activating enzyme)/Atg3(E2-like conjugating enzyme) and Atg12(ubiquitin-like molecule)/Atg7/Atg10(E2-like enzyme), are essential for the expansion and closure of the isolation membrane to form the autophagosome. The Atg2/Atg18 complex is involved in cycling of the transmembrane protein Atg9, which supplies membranes for the formation of autophagosomes [12]. EPG-5 is involved in the autophagosome maturation [13, 15]. Basal autophagy selectively removes aggregate-prone proteins or damaged organelles, functioning as a quality control system. A family of Atg8/LC3(mammalian Atg8 homolog) interacting proteins act as receptors that mediate delivery of specific cargoes to the autophagic machinery through Atg8 binding [16]. Selective removal of RNA-induced silencing complex (RISC) components by autophagy has recently emerged as a mechanism for modulating small interefering RNA (siRNA) or miRNA activity. In Arabidopsis, the viral suppressor of RNA silencing protein P0, which is a component of an E3 ligase, triggers the degradation of ARGONAUTE1 by autophagy [17]. In mammalian cells, DICER and AGO2 are targeted for selective autophagic degradation by the receptor NDP52 [18].
Here we show that during C. elegans development, autophagy modulates miRNA-mediated cell fate specification and selectively removes AIN-1, a key component of miRISC. Our study reveals a role of autophagy in cell-fate specification by modulating miRNA-mediated gene silencing.
RESULTS AND DISCUSSION
Role of autophagy in the heterochronic pathway
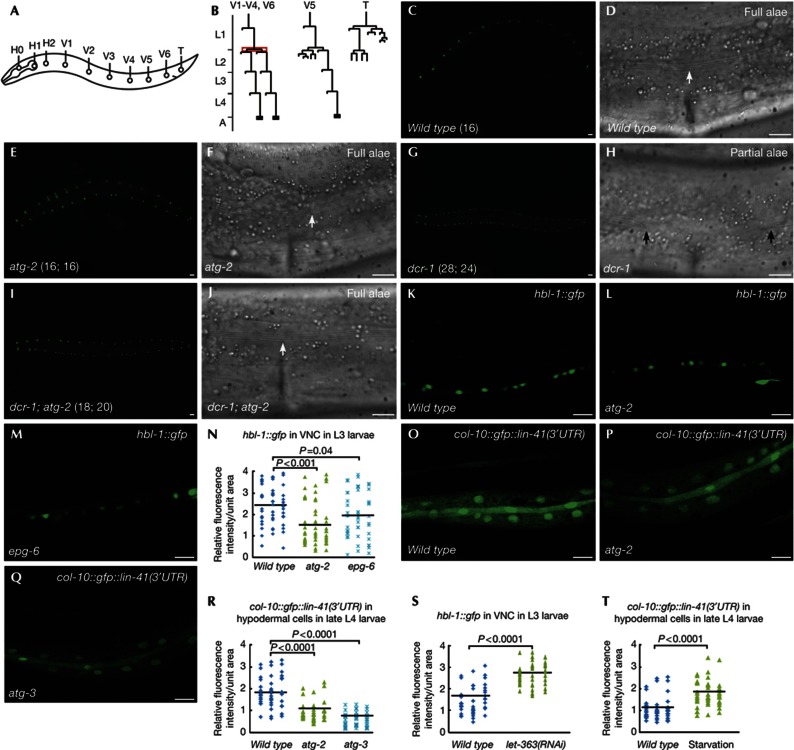
miRNAs play an important role in controlling the developmental timing of a row of lateral hypodermal seam cells, which undergo stage-specific developmental programs and increase in number from 10 at hatching to 16 from the L2 larval stage onwards (Fig 1A–C). At the late L4 larval stage, seam cells terminally differentiate, resulting in synthesis of adult-specific cuticular structures called alae (Fig 1D). Mutants of autophagy genes acting at distinct steps of autophagosome formation, including lgg-1/Atg8, atg-3, epg-8/Atg6, epg-6 and atg-2, exhibited normal seam cell development (Fig 1E,F; Table I).
Figure 1.
Loss of autophagy activity suppresses the retarded heterochronic defects in miRISC mutants. (A) Schematic illustration of seam cells in a wild-type L1 larva. (B) Division pattern of seam cells at the postembryonic stage in hermaphrodites. Seam cells V1–V4 and V6 divide symmetrically at the L2 stage (highlighted in red), thus doubling in number. (C–F) At the young adult stage, wild-type and atg-2 mutant animals have 16 seam cells on each side, visualized by the seam cell-specific marker scm::gfp. Cuticular alae (arrow) run continuously from anterior to posterior. The number of seam cells is shown in brackets. (G,H) dcr-1 young adults have an increased number of seam cells (shown in brackets) and gaps in the alae (between arrows). (I,J) atg-2 mutations suppress the retarded heterochronic defect in dcr-1 mutants. Complete alae (arrow) are formed in dcr-1; atg-2 double mutants. (K–M) Confocal images showing expression of hbl-1::gfp::hbl-1 reporter in VNC in wild type (K), atg-2 (L) and epg-6 (M) L3 larvae. (N) Distribution of relative fluorescence intensity of HBL-1::GFP in 50 unit areas in VNC in the indicated strains. (O–Q) Confocal images showing the expression of the col-10::gfp::lin-41(3′UTR) reporter in wild type (O), atg-2 (P) and atg-3 (Q) late L4 larvae. (R) Distribution of relative fluorescence intensity of col-10::gfp::lin-41(3′UTR) in 50 unit areas of hypodermal cells in the indicated strains. (S,T) Distribution of relative fluorescence intensity of hbl-1::gfp in VNC and col-10::gfp::lin-41(3′UTR) in hypodermal cells in 50 unit areas in wild type, let-363(RNAi) or starvation-treated animals. Black lines in (N,R–T) represent the average fluorescence intensity of the reporter in each strain. P values are determined by Student’s two-tailed unpaired t-test in (N,R–T). (C,E,G,I) Scale bar, 20 μm; (D,F,H,J–M,O–Q) scale bar, 10 μm. GFP, green fluorescent protein; miRISC, miRNA-induced silencing complex; RNAi, RNA-mediated interference; UTR, untranslated region; VNC, ventral nerve cord.
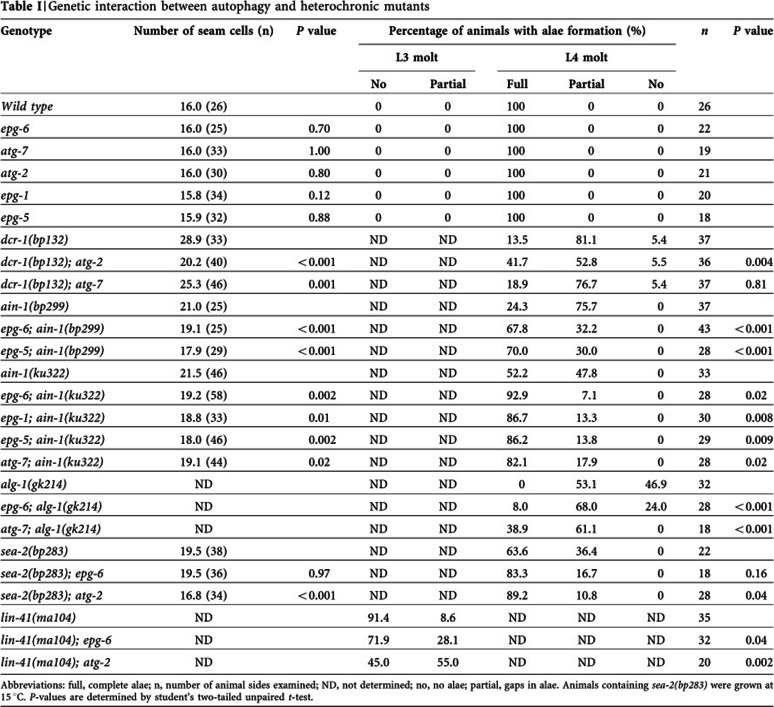
Table 1. Genetic interaction between autophagy and heterochronic mutants.
Abbreviations: full, complete alae; n, number of animal sides examined; ND, not determined; no, no alae; partial, gaps in alae.
Animals containing sea-2(bp283) were grown at 15 °C.
P-values are determined by student’s two-tailed unpaired t-test.
We tested whether loss of autophagy activity modulates phenotypes associated with loss-of-function mutants of miRISC components. Reduced activity of dcr-1 (the single C. elegans Dicer homolog), alg-1 (one of two Ago homologs) and ain-1 (ALG-1 interacting protein 1, one of two GW182 homologs) impairs miRNA-mediated downregulation of two heterochronic genes, hbl-1 and lin-41, and causes reiteration of the L2-specific seam cell proliferation programs at the L3 larval stage and delayed terminal differentiation [19–21]. dcr-1(bp132) contains an aspartate-to-asparagine mutation in the RNase III domain and causes reduced levels of mature miRNAs [11]. dcr-1(bp132) young adults have an average of 28.9 seam cells compared to 16.0 in wild type and 81.1% of mutants exhibited incomplete terminal differentiation of seam cells with gaps in the alae (Fig 1G,H). Loss of function of atg-2 suppressed the retarded heterochronic defect in dcr-1(bp132) mutants. In dcr-1; atg-2 double mutants, the average number of seam cells was 20.2, and 41.7% of dcr-1; atg-2 double mutants showed completely fused alae structures (Figs 1I,J; Table I). Loss of activity of autophagy genes, including epg-1, atg-7, epg-6 and epg-5, also partially suppressed the retarded heterochronic defect in ain-1 or alg-1 loss-of-function mutants (Table I). Suppression of the heterochronic defect in ain-1 and alg-1 mutants by loss of autophagy activity suggests that additional targets might function to modulate miRNA activity.
We further examined whether autophagy interacts with other heterochronic mutants. The zinc-finger gene sea-2 regulates expression of the heterochronic gene lin-28 at the post-transcriptional level [22]. Mutations in sea-2 cause retarded heterochronic defects. We found that atg-2 mutants suppressed the retarded terminal differentiation defect in sea-2 mutants (Table I). In contrast, loss of activity of autophagy genes, including epg-6 and atg-2, enhanced the precocious heterochronic defect associated with lin-41 hypomorphic mutants (Table I).
To determine whether autophagy regulates miRNA-mediated silencing of downstream targets, we examined the expression of reporters for hbl-1 and lin-41, two genetically verified targets of let-7 miRNAs. Expression of a hbl-1::gfp::hbl-1 reporter is strong in the ventral nerve cord (VNC) at larval stages before early L4, then greatly diminishes in late L4 and adult animals [23, 24]. Downregulation of HBL-1::GFP in VNC is mediated by let-7 and also lin-4 miRNAs [23, 24]. The fluorescence intensity of HBL-1::GFP in VNC was significantly reduced in atg-2 and epg-6 autophagy mutants (Fig 1K–N). We further examined the expression of a hypodermally expressed reporter, col-10::gfp::lin-41 (3′UTR), which contains the lin-41 3′UTR. The let-7 binding sites present in the lin-41 3′UTR sufficiently mediate the downregulation of lin-41 by let-7 [4]. Expression of this reporter is strong at larval stages, but decreases from the late L4 stage onwards [25]. In autophagy mutants, the same transgene was much more weakly expressed in late L4 animals (Fig 1O–R). Activating autophagy by inactivation of Tor signaling or starvation increased the expression of hbl-1::gfp::hbl-1 in VNC and col-10::gfp::lin-41 (3′UTR) in hypodermal cells (Fig 1S,T). The elevated expression of these reporters in let-7 mutants was not affected by loss of autophagy activity (supplementary Figs S1A–H online). These results indicate that loss of autophagy activity enhances let-7-mediated downregulation of target genes.
Autophagy mutants suppress the let-60(gf) Muv defect
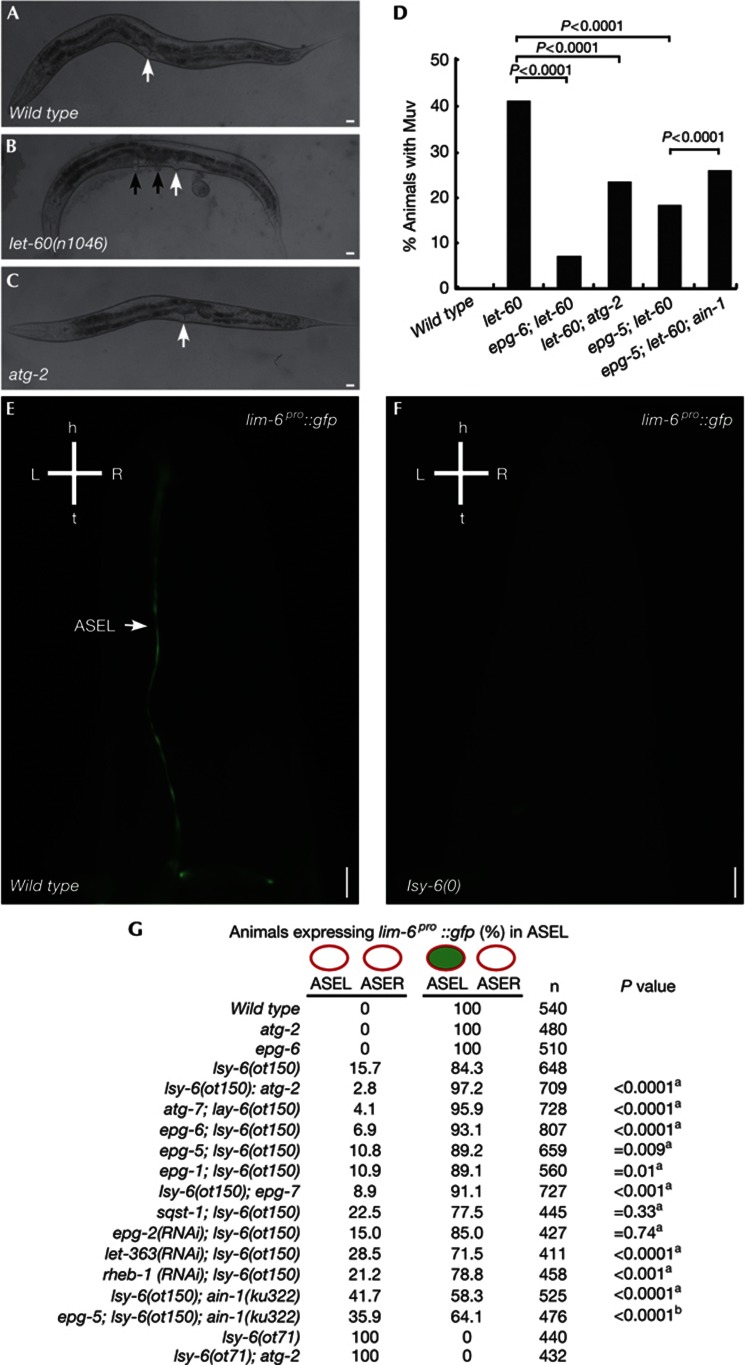
We determined if autophagy activity regulates expression of another let-7 family miRNA target, let-60/RAS [7], which is essential for vulval cell fate specification. The vulval precursor cell fate is sensitive to the dosage of let-60 activity [8]. let-60(n1046gf), a gain-of-function mutation, results in elevated LET-60 activity and a Muv phenotype (Figs 2A,B). Autophagy mutants did not show defects in vulval development, but simultaneous loss of autophagy gene activity in let-60(n1046gf) mutants, including epg-6, atg-2 and epg-5, dramatically reduced the Muv phenotype (Figs 2C,D). The suppression effect was partially reduced by loss of ain-1 activity (Fig 2D). mpk-1, encoding a mitogen-activated protein kinase, acts downstream of let-60 in controlling vulval development [8]. Hyperactive mpk-1 mutants, gaIs37, exhibited a Muv phenotype in 98.8% of animals (n=322) at 24 °C. The Muv phenotype in gaIs37 mutants was not affected by loss of epg-5 activity (98.3% Muv, n=294).
Figure 2.
Autophagy regulates developmental processes specified by the let-7 miRNA family and lsy-6 miRNA. (A–C) Wild-type and atg-2 hermaphrodites have a single vulva (arrow). (B) let-60(n1046gf) mutants show a Muv phenotype. Black arrows indicate pseudovulvae. (D) Loss of autophagy activity suppresses the Muv phenotype in let-60(n1046) mutants. The number of adult animals examined was: wild type (n=450), let-60 (n=406), epg-6; let-60 (n=444), let-60; atg-2 (n=803), epg-5; let-60 (n=672), epg-5; let-60; ain-1(ku322) (n=635). P values are determined by a two-tailed χ2 test. (E,F) lsy-6 mutants show defects in ASEL fate specification and do not express the ASEL-specific reporter lim-6pro::GFP. (G) Expression of lim-6pro::GFP in various strains. aCompared with lsy-6(ot150).bCompared with lsy-6(ot150); ain-1(ku322). P values are determined by a two-tailed χ2 test. (A–C) Scale bar, 20 μm; (E,F) scale bar, 10 μm. ASEL, ASE left; ASER, ASE right; GFP, green fluorescent protein; miRNA, microRNA; Muv, multivulva.
Loss of autophagy activity suppresses lsy-6(hypo)
The non-let-7-family miRNA lsy-6 specifies the fates of two neurons, ASEL and ASER, by downregulating cog-1 in ASEL [9]. Loss of lsy-6 activity causes the ASEL neuron to adopt the ASER fate (Figs 2E,F) [9]. Transformation of ASEL to ASER was complete in lsy-6(ot71) null animals but incomplete in animals bearing a hypomorphic allele of lsy-6(ot150) (Fig 2G). Loss of function of autophagy genes did not cause defects in ASEL fate specification, visualized by the ASEL-specific reporter lim-6pro::GFP (Fig 2G). Autophagy mutants, including epg-1, atg-2, atg-7, epg-6 and epg-5, partially rescued the ASEL fate specification phenotype in lsy-6(ot150) mutants (Fig 2G). Elevated autophagy activity, caused by TOR signaling mutations including let-363/Tor and rheb-1, exacerbated the ASEL specification defect in lsy-6(ot150) mutants (Fig 2G). Loss of autophagy activity did not suppress the defect in lsy-6(ot71) null animals (Fig 2G), indicating that autophagy mutants could not bypass the requirement of lsy-6 activity for specification of ASEL fate. In summary, loss of autophagy activity suppresses developmental defects associated with impaired gene silencing mediated by diverse miRNAs.
AIN-1/GW182 is selectively removed by autophagy
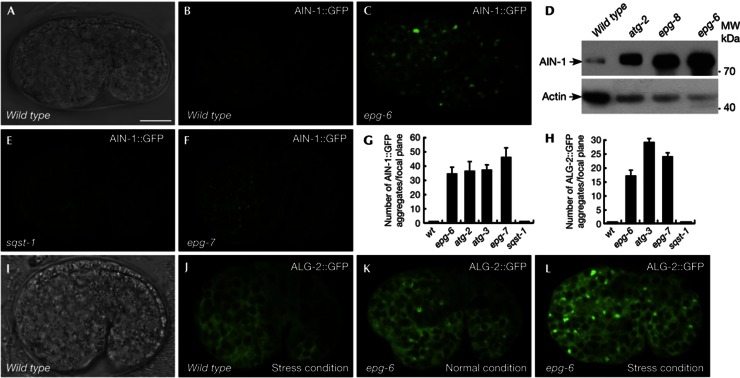
We next examined the expression of C. elegans miRISC components, including the GW182 homologs AIN-1 and AIN-2, and the Ago homologs ALG-1 and ALG-2.ain-1 and ain-2 or alg-1 and alg-2 function redundantly. ain-1 and alg-1 single mutants show retarded heterochronic defects, while ain-2 and alg-2 single mutants have no evident defects. ain-1/-2 or alg-1/-2 double mutants display much stronger defects than either single mutant [19–21]. In wild-type embryos, AIN-1::GFP was diffusely expressed in the cytoplasm during embryogenesis (Figs 3A,B). In autophagy mutants, including epg-6, atg-3, epg-8 and atg-2, AIN-1::GFP was strongly expressed and accumulated into a large number of aggregates (Figs 3C,G; supplementary Figs S2A,B online). Levels of endogenous AIN-1 protein, but not ain-1 mRNA, were increased in autophagy mutants (Fig 3D; supplementary Figs S2C,D online). These results indicate that AIN-1 is degraded by autophagy. Expression of AIN-2::GFP, which is diffusely localized in wild-type embryos, was unaffected in autophagy mutant embryos (supplementary Figs S2E,F online).
Figure 3.
AIN-1 is selectively degraded by autophagy. (A,B) AIN-1::GFP is weakly and diffusely expressed in the cytoplasm during embryogenesis in wild-type animals. (A) Nomarski image of the embryo shown in (B). Scale bar, 10 μm for whole embryo. C. elegans embryos remain the same size during embryogenesis and loss of autophagy activity has no effect on the embryo size. Thus, scale bars for embryos are only shown once in each figure. (C) AIN-1::GFP forms a large number of aggregates in epg-6 mutant embryos. (A–C) are confocal images. (D) Western blotting shows that endogenous AIN-1 protein levels are dramatically elevated in autophagy mutant embryos compared with wild type. (E) No AIN-1::GFP aggregates are formed in sqst-1 mutant embryos. (F) AIN-1::GFP accumulates into numerous aggregates in epg-7 mutant embryos. (G,H) Number of AIN-1::GFP (G) and ALG-2::GFP (H) aggregates per focal plane in the indicated strains. Error bars indicate the s.d. of four examined embryos. (I,J) ALG-2::GFP is diffusely localized in the cytoplasm in wild-type embryos. (I) Nomarski image of the embryo shown in (J). (K) ALG-2::GFP forms a few aggregates in epg-6 mutant embryos. (L) ALG-2::GFP forms a large number of aggregates in epg-6 mutant embryos under stress conditions. GFP, green fluorescent protein; s.d., standard deviation.
Translational fusion reporters for gfp::alg-1 and alg-2::gfp were both diffusely expressed in the cytoplasm of most embryonic cells (Figs 3I,J; supplementary Figs S2I,J online). A few ALG-1 and ALG-2 aggregates were formed in autophagy mutants but the number of aggregates dramatically increased under certain stress conditions (Figs 3H,K; supplementary Figs S2G,H,K,L online). ALG-1 levels also increased in autophagy mutants (supplementary Fig S2M online). alg-1 and alg-2 mRNA levels appeared normal in autophagy mutants (supplementary Figs S2N,O online). These results indicate that ALG-1 and ALG-2 accumulate into aggregates in autophagy mutants under certain stress conditions.
AIN-1 associates with SQST-1 aggregates
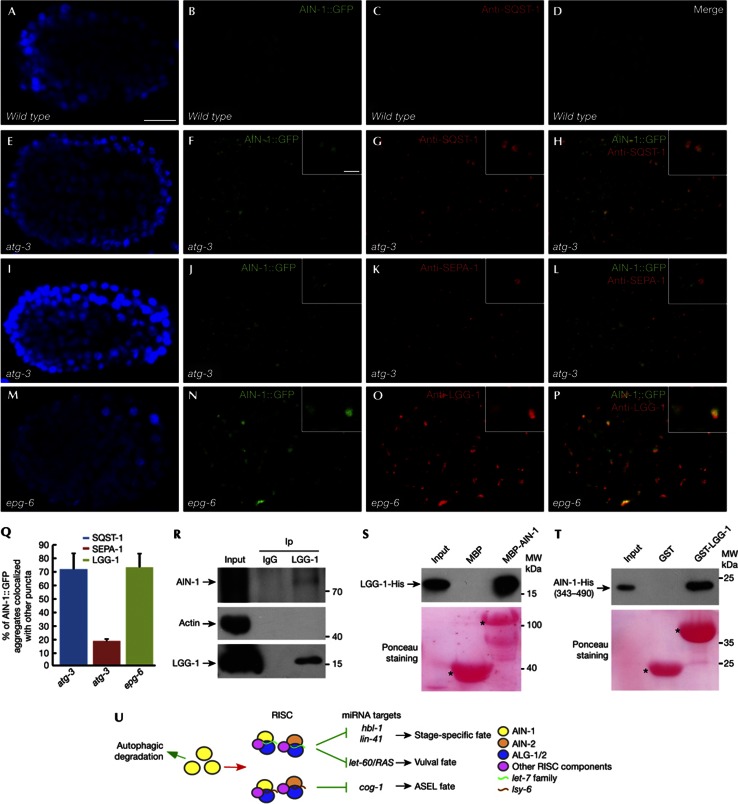
We next determined whether AIN-1 colocalizes with known protein aggregates in autophagy mutants. Maternally loaded germline P granule components PGL-1 and PGL-3 are degraded in somatic cells by autophagy during embryogenesis [26]. This process requires the self-oligomerizing receptor protein SEPA-1. In autophagy mutants, PGL-1/-3 and SEPA-1 accumulate into numerous aggregates, called PGL granules [26]. The C. elegans p62 homolog SQST-1 is also removed by autophagy and forms numerous aggregates in autophagy mutants [13]. PGL granules and SQST-1 aggregates are separable in various autophagy mutants, including mutants of the LGG-1/Atg8 conjugation system [13]. We found that in atg-3 mutants, AIN-1::GFP aggregates largely colocalized with SQST-1 aggregates, but were separable from PGL granules (Figs 4A–L). The EPG-6/ATG-2 complex is involved in progression of PI(3)P-enriched subdomains of the ER, called omegasomes, to autophagosomes [14]. epg-6 and atg-2 mutants show accumulation of LGG-1, the C. elegans Atg8 homolog, which associates with autophagosomal structures. In epg-6 and atg-2 mutants, LGG-1 puncta are enlarged and colocalize with SQST-1 aggregates [14]. Coimmunostaining showed that AIN-1 aggregates colocalized with LGG-1 puncta in epg-6 mutants (Figs 4M–Q).
Figure 4.
AIN-1 colocalizes with SQST-1 aggregates in autophagy mutants and directly interacts with LGG-1. (A–D) Both AIN-1::GFP and SQST-1, detected by anti-SQST-1 antibody, are very weakly expressed and diffusely localized in wild-type embryos. (E–H) AIN-1::GFP aggregates colocalize with SQST-1 aggregates in atg-3 mutant embryos. Scale bars, 10 μm for insets that show a magnified view. (I–L) AIN-1::GFP aggregates are separable from PGL granules, detected by anti-SEPA-1, in atg-3 mutant embryos. (M–P) Colocalization of AIN-1::GFP aggregates with LGG-1 puncta in epg-6 mutant embryos. (Q) % of AIN-1::GFP aggregates colocalized with SQST-1 aggregates, SEPA-1 aggregates and LGG-1 puncta in autophagy mutants. Error bars indicate the s.d. of four examined embryos. (R) A co-IP assay shows that AIN-1 associates with LGG-1 in vivo. (S) Direct interaction between AIN-1 and LGG-1 in an in vitro pull-down assay. 5% of the His fusion proteins used for pull-down are shown as input. Corresponding Ponceau-stained blots show the amount of proteins used for pull-down. (T) A fragment of AIN-1 directly binds to LGG-1 in an in vitro pull-down assay. (U) Model for the role of autophagy in modulating the miRNA pathway. co-IP, coimmunoprecipitation; GFP, green fluorescent protein; MBP, maltose-binding protein; miRNA, microRNA; IP, immunoprecipitant; RISC, RNA-induced silencing complex; s.d., standard deviation.
Colocalization of AIN-1 and SQST-1 aggregates in autophagy mutants prompted us to investigate the role of SQST-1/p62 in autophagic removal of AIN-1. Mammalian p62 acts as a receptor mediating the formation and autophagic degradation of ubiquitin-positive protein aggregates [16]. C. elegans SQST-1, as p62, self-interacts and directly associates with LGG-1/Atg8. In sqst-1 mutants, AIN-1::GFP expression was unchanged and no aggregates were formed (Figs 3E,G). AIN-1::GFP still formed aggregates in lgg-1(RNAi); sqst-1 animals (supplementary Fig 2P online). Thus, sqst-1 is not required for the removal of AIN-1 and its formation of aggregates in autophagy mutants. SQST-1 aggregates contain several other unrelated self-oligomerized proteins that are removed by autophagy in a sqst-1-independent manner [27], suggesting that AIN-1 can be recruited into SQST-1 aggregates by other components. Selective degradation of autophagy substrates under physiological conditions requires a scaffold protein, which bridges the cargo/receptor complex to the autophagic machinery [27]. EPG-7 acts as a scaffold protein in mediating autophagic degradation of SQST-1 aggregates, but not PGL granule components [27]. EPG-7 itself is weakly expressed and diffusely localized in wild-type embryos and accumulates into numerous aggregates in autophagy mutants [27]. We found that loss of function of epg-7 caused accumulation of AIN-1 aggregates (Figs 3F,G), indicating that EPG-7 mediates the autophagic degradation of AIN-1. GFP::ALG-1 and ALG-2::GFP aggregates also colocalized with SQST-1 aggregates but were separable from PGL granules in atg-3 mutants (supplementary Figs S3A–Z online). GFP::ALG-1 and ALG-2::GFP accumulated in epg-7 but not in sqst-1 mutants (Fig 3H; supplementary Figs S3A2,B2 online). Consistent with the role of epg-7 in degradation of AIN-1, the ASEL specification defect in lsy-6(ot150) mutants was suppressed by loss of activity of epg-7, but not sqst-1 (Fig 2G). Loss of function of epg-2, which functions as a scaffold protein in mediating autophagic degradation of PGL granules but not SQST-1 aggregates [13], had no effect on the ASEL specification defect in lsy-6(ot150) mutants (Fig 2G).
AIN-1 directly interacts with LGG-1/Atg8
Direct interaction of substrates with Atg8/LC3 contributes to cargo specificity and degradation efficiency. Thus, we examined whether AIN-1 interacts with LGG-1. Using in vivo coimmunoprecipitation assays, we found that LGG-1 directly interacted with AIN-1, but not ALG-1 or ALG-2 (Fig 4R; supplementary Figs S4A–C online). His-fused LGG-1 interacted with maltose-binding protein-tagged AIN-1 in vitro (Fig 4S). His-fused AIN-1 also associated with GST-tagged LGG-1 and the LGG-1 binding region in AIN-1 was delineated to amino acids 343–490 (Fig 4T; supplementary Figs S4D–F online). This region contains two LIR motifs (Atg8/LC3-interacting region), 437WGEL440 and 462WNDL465. Mutating either one or two LIR motifs did not alter LGG-1 binding, indicating that the LIR motif is not required for binding of AIN-1 to LGG-1 (supplementary Figs S4G–I online).
In conclusion, autophagy modulates diverse developmental processes specified by the miRNA pathway during C. elegans development. In mammalian cells, DICER and AGO2, but not GW182, are targeted for selective autophagic degradation [18]. Degradation of DICER and AGO2 is mediated by the selective autophagy receptor NDP52, which associates with the cargoes and also with Atg8/LC3 [18]. No obvious NDP52 homologs are found in C. elegans. The presence of distinct autophagy receptor and scaffold proteins might help determine whether different components of the RISC complex are degraded by autophagy in C. elegans and mammals. In contrast to C. elegans, the efficiency of miRNA-mediated silencing is reduced in autophagy-deficient mammalian cells [18]. The effect of autophagy on miRNA activity might depend on whether the degraded component acts before or after miRISC assembly. Our study reveals that autophagy modulates miRNA activity under physiological conditions.
METHODS
See supplementary information online for a list of strains, sequence for reporter construction and RNAi, antibody preparation and stress assay.
Indirect immunofluorescence. Permeabilization of embryos was performed by freeze-cracking methods. Freeze-cracked slides were fixed, blocked and incubated with diluted antibody at 4 °C overnight. Animals were then washed three times with phosphate-buffered saline with 2% Tween-20 (PBST) and incubated with fluorescently labeled secondary antibody at room temperature for 1 h. Slides were viewed using a confocal microscope (Zeiss LSM 510 Meta plus Zeiss Axiovert zoom, LSM Image Browser software). To compare the relative fluorescence intensity of gfp reporters, images were taken using the same exposure time and were measured using ImageJ software.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Dr Isabel Hanson for editing the manuscript. Some strains were provided by the Caenorhabditis Genetics Center (CGC). This work was supported by the National Basic Research Program of China (2013CB910100, 2011CB910100) and also a grant from the National Natural Science Foundation of China (31225018) to H.Z. The research of H.Z. was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute.
Author contributions: P.P.Z. and H.Z. designed the experiments, analysed the data and wrote the manuscript. P.P.Z. performed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Han M (2007) GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol 17: 411–416 [DOI] [PubMed] [Google Scholar]
- Ambros V (2011) MicroRNAs and developmental timing. Curr Opin Genet Dev 21: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5: 659–669 [DOI] [PubMed] [Google Scholar]
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V (2005) The let-7 MicroRNA family members mir-48, mir-84 and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 3: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE (2005) Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell 9: 415–422 [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Han M (1998) Genetics of RAS signaling in C. elegans. Trends Genet 14: 466–472 [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O (2003) A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849 [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren HY, Zhang H (2010) Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev Biol 345: 144–155 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 931–937 [DOI] [PubMed] [Google Scholar]
- Tian Y et al. (2010) C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141: 1042–1055 [DOI] [PubMed] [Google Scholar]
- Lu Q, Yang PG, Huang XX, Hu WQ, Guo B, Wu F, Lin L, Kovács AL, Yu L, Zhang H (2011) The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 21: 343–357 [DOI] [PubMed] [Google Scholar]
- Zhao HY, Zhao GY, Wang XW, Xu LJ, Miao L, Feng D, Chen Q, Kovács AL, Fan DS, Zhang H (2013) Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J Cell Biol 200: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA 109: 15942–15946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O (2012) Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol 14: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal rnas that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M (2005) The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell 19: 437–447 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR III, Han M (2007) Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell 28: 598–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XX, Zhang H, Zhang H (2011) The zinc-finger protein SEA-2 regulates larval developmental timing and adult life span in C. elegans. Development 138: 2059–2068 [DOI] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ (2003) The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell 4: 639–650 [DOI] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE (2003) The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell 4: 625–637 [DOI] [PubMed] [Google Scholar]
- Cai QC, Sun YY, Huang XX, Guo C, Zhang YX, Zhu ZY, Zhang H (2008) The Caenorhabditis elegans PcG-like gene sop-2 regulates the temporal and sexual specificities of cell fates. Genetics 178: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX et al. (2009) SEPA-1 mediates the specific recognition and degradation of p granule components by autophagy in C. elegans. Cell 136: 308–321 [DOI] [PubMed] [Google Scholar]
- Lin L, Yang PG, Huang XX, Zhang H, Lu Q, Zhang H (2013) The scaffold protein EPG-7 links cargo/receptor complexes with the autophagic assembly machinery. J Cell Biol 201: 113–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.