Abstract
Mitochondria and peroxisomes can be fragmented by the process of fission. The fission machineries of both organelles share a set of proteins. GDAP1 is a tail-anchored protein of mitochondria and induces mitochondrial fragmentation. Mutations in GDAP1 lead to Charcot-Marie-Tooth disease (CMT), an inherited peripheral neuropathy, and affect mitochondrial dynamics. Here, we show that GDAP1 is also targeted to peroxisomes mediated by the import receptor Pex19. Knockdown of GDAP1 leads to peroxisomal elongation that can be rescued by re-expressing GDAP1 and by missense mutated forms found in CMT patients. GDAP1-induced peroxisomal fission is dependent on the integrity of its hydrophobic domain 1, and on Drp1 and Mff, as is mitochondrial fission. Thus, GDAP1 regulates mitochondrial and peroxisomal fission by a similar mechanism. However, our results reveal also a more critical role of the amino-terminal GDAP1 domains, carrying most CMT-causing mutations, in the regulation of mitochondrial compared to peroxisomal fission.
Keywords: glutathione S-transferase, mitochondrial dynamics, peripheral neuropathy, tail-anchored protein
Introduction
Peroxisomes are dynamic, single membrane-bound organelles. They are responsible for several metabolic processes such as fatty acid β-oxidation, biosynthesis of ether phospholipids, and metabolism of reactive oxygen species (ROS) [1]. Numbers, composition, and morphology of peroxisomes are influenced by extra- and intracellular stimuli [2]. Peroxisomes are generated by two different mechanisms: They can form de novo from the endoplasmic reticulum or by growth and division from pre-existing peroxisomes. The contribution of the two pathways to peroxisome biogenesis is a matter of intensive debate [3–5].
Peroxisomal growth and division occur in morphologically well-defined steps: The spherical peroxisome first elongates in response to exogenous or endogenous stimuli mediated by Pex11 proteins [4]. Next, the membrane of the elongated peroxisome forms constrictions by a not yet clearly defined mechanism [6]. Final fragmentation requires several fission proteins, including the tail-anchored (TA) proteins hFis1 (fission 1), Mff (mitochondrial fission factor) and the cytosolic Drp1/DLP1 (dynamin-related protein 1) [7–12]. These proteins are also essential fission factors at the mitochondrial outer membrane (MOM). Sharing of fission components has probably developed by similar cellular demands, as peroxisomes and mitochondria are metabolically linked [13]. This cooperative interaction likely influences the functionality of both organelles in health and disease [14, 15].
GDAP1 (ganglioside-induced differentiation associated protein 1) is also a TA-protein of the MOM acting as mitochondrial fission factor [16–18]. GDAP1 is the founder of a family of glutathione S-transferases [19] and its expression level influences glutathione (GSH) levels in cultured cells [20]. Over 40 different mutations in GDAP1 lead to Charcot-Marie-Tooth disease (CMT), the most commonly inherited peripheral neuropathy [21, 22]. Recessively inherited disease mutants (rmGDAP1) show reduced mitochondrial fragmentation activity, whereas dominantly inherited disease mutants (dmGDAP1) interfere with mitochondrial fusion. This impaired fusion results in a disturbed mitochondrial membrane potential and increased ROS levels [17].
The aim of this study was to investigate the possible role of the TA-protein GDAP1 as fission factor at peroxisomes and its relevance in disease. We show that GDAP1 is targeted to a subpopulation of peroxisomes in a Pex19-dependent manner. Loss of GDAP1 leads to peroxisomal elongation, whereas overexpression increases peroxisomal fragmentation. Disease-mutated forms of GDAP1 can still induce peroxisomal fragmentation with comparable efficiency unless the mutation interferes with peroxisomal targeting via its TA. GDAP1-induced peroxisomal fragmentation is dependent on Drp1, Mff, and requires the hydrophobic domain 1 (HD1) lying amino-terminally to the transmembrane domain (TMD) of GDAP1 [18]. We conclude that GDAP1 induces fission at the MOM and at the peroxisomes by overlapping mechanisms. However, the finding that N-terminal rmGDAP1-missense mutants retain peroxisomal fission activity, while losing this activity on mitochondria, revealed also mechanistic differences.
Results and Discussion
GDAP1 is targeted to peroxisomes
TA-proteins with positively charged amino acids surrounding the carboxy-terminal TMD are inserted into the MOM and can be inserted into the peroxisomal membrane of mammalian cells [23]. As GDAP1 is a TA-protein with such features [18], we tested its co-localization with peroxisomal markers. GDAP1 was transiently expressed in COS7 cells stably expressing GFP–SKL, a GFP with a C-terminal peroxisomal targeting sequence [24]. As expected, GDAP1 staining predominantly co-localized with the mitochondrial marker cytochrome c [16]. Additionally, some peroxisomes co-localized with the GDAP1 signal (Fig 1A). We also analysed endogenous GDAP1 in mouse primary hippocampal cell cultures infected with lentivirus encoding GFP–SKL. The signal of endogenous GDAP1 protein predominantly co-localized with cytochrome c and partially with GFP–SKL-positive peroxisomes (Fig 1B). Taken together, GDAP1 shows dual targeting to mitochondria and peroxisomes.
Figure 1.
GDAP1 is targeted to peroxisomes. (A) COS7 cells stably expressing GFP–SKL were transfected with GDAP1 expression constructs and stained for GDAP1 and cytochrome c. GDAP1 is mainly localized to mitochondria. Higher magnification (zoom box) shows GFP–SKL-positive structures that are positive for GDAP1 but negative for cytochrome c (arrows). A neighbouring, untransfected cell (light yellow broken line). (B) Endogenously GDAP1-expressing primary hippocampal cell cultures were infected with lentivirus encoding GFP–SKL, and stained for GDAP1 and cytochrome c. Higher magnification (zoom box) shows that endogenous GDAP1 predominantly co-localizes with the cytochrome c, and also with GFP–SKL-positive structures, which are negative for cytochrome c (arrows). (C) Measured GDAP1 fluorescence intensity in the peroxisomal population was decreased in Pex19 knockdown cells but unaffected in the mitochondrial population. Values represent mean and s.e.m. of three independent experiments. Fluorescence intensity of the peroxisomal/mitochondrial population was analysed for 6–9 cells per condition per experiment: *P<0.05, two-tailed unpaired t-test. Scale bar, 10 μm. ctrl shRNA, non-targeting shRNA control; GDAP1, ganglioside-induced differentiation associated protein 1; GFP–SKL, green fluorescent protein with peroxisomal targeting sequence; shPex19, shRNA against Pex19.
TA-proteins are targeted to peroxisomes via the Pex19 shuttle receptor [25–28]. In an interaction assay, in vitro-translated GFP-GDAP1 interacted with recombinant Pex19 and formed a trimeric complex with Pex19 and Pex3 (supplementary Fig S1A online). To confirm the functional relevance of this interaction, we infected COS7 cells stably expressing GFP–SKL with lentiviruses encoding two different shRNAs targeting Pex19 or a non-silencing control shRNA. When reduced expression of Pex19 was significant (supplementary Fig S2A online), cells were transiently co-transfected with GDAP1 and mitochondrially-targeted DsRed2 (mtDsRed) expression constructs. After 16 h, cells were fixed, immunostained for GDAP1, and the signal intensity of GDAP1 was determined at peroxisomes (GFP–SKL-positive structures) and mitochondria (mtDsRed-positive structures) on single plane confocal images (supplementary Fig S1B online). The GDAP1-signal intensity at peroxisomes was significantly lower in Pex19 knockdown cells compared to controls. Under the same conditions, targeting of GDAP1 to mitochondria was not affected (Fig 1C). We conclude that GDAP1 is targeted to peroxisomes via the peroxisomal import receptor Pex19.
Loss of GDAP1 reduces peroxisomal fragmentation
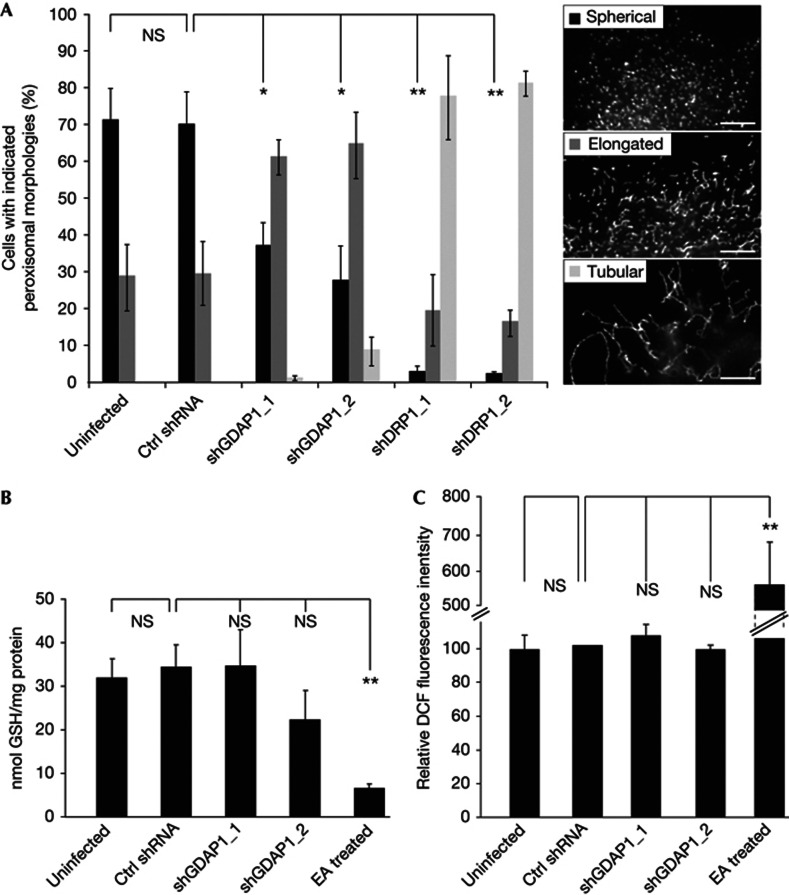
To assess the functional relevance of GDAP1 at peroxisomes, we infected mouse neuroblastoma N1E-115 cells with lentiviruses encoding two different shRNAs against GDAP1. Knockdown of Drp1 by two different shRNAs served as positive controls, while a non-targeting shRNA and uninfected cells were used as negative controls. Five days after infection, cells were fixed and stained for Pex14 as peroxisomal marker. Sister plates were analysed by immunoblotting to confirm the knockdowns (supplementary Fig S2B,C online). To quantify the morphologies, we distinguished three different categories: spherical peroxisomes, elongated peroxisomes and tubular peroxisomes (Fig 2A). Uninfected and control-infected cells generally showed a spherical peroxisomal morphology. Reduced levels of GDAP1 led to elongated peroxisomes. This effect was even more pronounced after knockdown of Drp1, where most peroxisomes were tubular as described (Fig 2A) [6, 8]. This experiment demonstrated that loss of GDAP1 alters peroxisomal morphology. Peroxisomal proliferation can be mediated by elongation of existing peroxisomes. Elongated peroxisomes get constricted and subsequently fragmented by the fission machinery [14]. Therefore, both reduced fission activity and enhanced elongation might shift the overall peroxisomal morphology towards a more tubular, less spherical phenotype. GDAP1 expression levels influence GSH levels in cultured cells [20], thereby potentially influencing peroxisomal biogenesis by increasing elongation due to cellular stress. Thus, we measured GSH and ROS levels in acute GDAP1 knockdown cells by the enzymatic recycling method and 2,7-dichlorofluorescin (DCF) fluorescence intensity measurements, respectively [17, 29]. Cellular GSH levels after GDAP1 knockdown by two individual shRNAs showed no significant difference to control cells (Fig 2B). As positive control, uninfected cells were treated with ethacrynic acid (EA), which led to significant GSH depletion. DCF fluorescence intensities were also identical between GDAP1 knockdown cells and controls, while EA treatment increased DCF fluorescence intensity (Fig 2C). In summary, short-term knockdown of GDAP1 did not affect intracellular GSH levels or cause ROS stress in cultured cells and cannot account for the peroxisomal elongation due to cellular stress. Thus, our results indicate that the observed morphological changes of peroxisomes in GDAP1 knockdown cells are caused by reduction in fission activity.
Figure 2.
Loss of GDAP1 leads to peroxisomal elongation. (A) N1E-115 cells were infected with lentiviruses encoding shRNA against GDAP1, Drp1 or a non-targeting control (ctrl) shRNA, or were left uninfected. Five days after infection, peroxisomal morphologies were assessed after immunostaining for Pex14 according to the three categories: spherical, elongated or tubular and were quantified in blinded countings. (B) Measurements of cellular GSH levels and (C) ROS levels show no significant alterations at day five of the GDAP1 knockdown compared to control cells. EA treated cells served as positive control (50 μg/ml for 2 h). Values represent mean and s.e.m. of three independent experiments. For morphology analysis, 100 cells were counted per condition per experiment: *P<0.05, **P<0.01, two-tailed unpaired t-test. Scale bar, 2.5 μm. ctrl shRNA, non-targeting shRNA control; DCF, 2,7-dichlorofluorescin; Drp1, dynamin-related protein 1; EA, ethacrynic acid; GDAP1, ganglioside-induced differentiation associated protein 1; GSH, glutathione; NS, not significant; ROS, reactive oxygen species; s.e.m., standard error of the mean; shDrp1, shRNA against Drp1 (dynamin related protein 1); shGDAP1, shRNA against GDAP1; shRNA, short hairpin RNA.
To further test the impact of GDAP1 on fission, we made use of transient Pex11β–myc overexpression, which elongates peroxisomes independent of other stimuli [6, 30, 31]. In N1E-115 cells the strongest elongation effect can be observed 7 h after transfection. Subsequently, the endogenous fission machinery of the cell is activated; in consequence 24 h after Pex11β–myc transfection cells predominantly show a spherical peroxisomal morphology, which represents the new equilibrium of forced elongation and endogenous fission events (supplementary Fig S3 online). Also in this paradigm, knockdown of GDAP1 resulted in less cells with spherical peroxisomes 24 h after Pex11β–myc transfection, as did knockdown of the well-established fission factor Drp1 (supplementary Fig S4A,B online), supporting that GDAP1 acts as peroxisomal fission factor.
Missense disease mutants promote peroxisomal fission
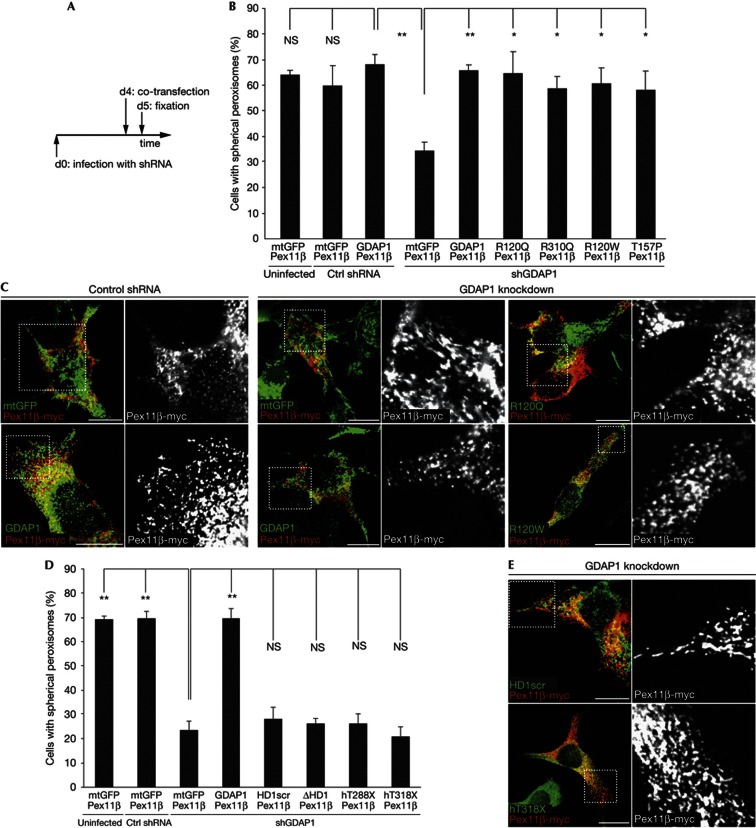
We have previously rescued mitochondrial fission after GDAP1 knockdown by re-expressing GDAP1 enabling us to examine also disease-linked mutants [17]. This experimental paradigm was adapted for the analysis of peroxisomal fragmentation. Four days after lentiviral infection with control or GDAP1-targeted shRNAs, N1E-115 cells were transiently co-transfected with Pex11β–myc and expression vectors encoding mitochondrially targeted eGFP (mtGFP) as control, GDAP1 wildtype, dmGDAP1 or rmGDAP1 missense mutants (Fig 3A). Mutants covering different domains of the protein were selected (supplementary Fig S5A online). Twenty-four hours after transfection, when tubulation by Pex11β–myc-overexpression is counterbalanced by active fission (supplementary Fig S3 online), cells were fixed and morphologically analysed. Uninfected and control infected mtGFP and Pex11β–myc expressing cells showed predominantly spherical peroxisomes. GDAP1 knockdown resulted in more cells with elongated peroxisomes as fission is decreased (Fig 3B,C). Re-introduction of wildtype GDAP1 into these knockdown cells restored the peroxisomal morphology to control levels, validating the experimental setup and GDAP1’s role as fission factor. Overexpression of all tested missense disease mutants in GDAP1 knockdown cells also reconstituted the peroxisomal morphology (Fig 3B,C). Wildtype GDAP1, tested dmGDAP1s or rmGDAP1s showed no significant differences in their ability to induce peroxisomal fragmentation. This was unexpected, as disease-causing mutant forms of GDAP1 influence mitochondrial dynamics (supplementary Table SI online; [16, 17]).
Figure 3.
Peroxisomal fission is dependent on the GDAP1-targeting domain and the HD1, but unaffected by CMT-disease associated N-terminal missense mutations. (A) N1E-115 cells were infected with lentiviruses encoding shRNA against GDAP1 or a non-targeting control (ctrl) shRNA or left uninfected (d0). After 4 days (d4), cells were co-transfected with Pex11β–myc and GDAP1 wildtype, mutant forms of GDAP1, or mtGFP. Twenty-four hours later (d5), cells were fixed and stained (C,E). Peroxisomal morphology of double-positive cells was quantified in a blinded counting (B,D). (GDAP1 with scrambled HD1 (HD1 scr), GDAP1 with deleted HD1 (ΔHD1), C-terminal truncated human GDAP1 (hT288X, hT318X)). Values represent means and s.e.m. of at least three independent experiments, 100 cells were counted per condition per experiment: *P<0.05, **P<0.01, two-tailed unpaired t-test. Scale bar, 10 μm. CMT, Charcot-Marie-Tooth disease; ctrl shRNA, non-targeting shRNA control; GDAP1, ganglioside-induced differentiation associated protein 1; HD1, hydrophobic domain 1; mtGFP, mitochondria-localized green fluorescent protein; NS, not significant; s.e.m., standard error of the mean; shGDAP1, shRNA against GDAP1; shRNA, short hairpin RNA.
To confirm this finding, we expressed Pex11β–myc in N1E-115 cells overexpressing GDAP1 wildtype, dmGDAP1, or rmGDAP1 missense mutants and analysed peroxisomal morphology 7 h after start of transfection (supplementary Fig S5B online). In this setting, an active fission factor will counterbalance the Pex11β peroxisome elongation effect, which is maximal at this time point in control cells (supplementary Fig S3 online). All tested disease-associated missense mutants were able to induce fragmentation of Pex11β–myc-stimulated peroxisomes with comparable efficiency as GDAP1 wildtype, resulting in an overall spherical morphology (supplementary Fig S5C online). In summary, in both experimental approaches (Fig 3B,C and supplementary Fig S5C online) we found that the disease-associated missense mutations do not impair the peroxisomal fission capacity of GDAP1. As mitochondrial fission activity of rmGDAP1 missense mutants is reduced [16, 17], our results further indicate that mutations in N-terminal domains differently affect GDAP1’s activity at mitochondria versus peroxisomes.
GDAP1-induced fission depends on its HD1
Two domains are required for mitochondrial fission: the HD1 and the C-terminal TA-domain, which mediates mitochondrial targeting (supplementary Fig S5A and supplementary Table SI online) [18]. To examine whether mitochondrial and peroxisomal fission induction by GDAP1 depend on the integrity of the same domains, we used the experimental setting as described before in Fig 3A. We tested GDAP1 constructs coding for a protein with a scrambled HD1 (HD1scr), a deleted HD1 (ΔHD1) or C-terminal truncated GDAP1. The mutant hT288X corresponds to the largest GDAP1 truncation described in CMT patients lacking the HD1 and TA-domain, and hT318X is an artificial truncation mutant lacking only the TMD and the C-terminal residues [14, 15]. None of these mutants was able to induce peroxisomal fragmentation (Fig 3D,E). These results were confirmed using the experimental settings as described in supplementary Fig S5B and (supplementary Fig S5D online). Thus, the TA-domain and the HD1 are essential for fission at peroxisomes as well as at mitochondria. Mutations in GDAP1 interfering with targeting are associated with more severe clinical phenotypes than point mutations leading to amino acid exchanges in the N-terminal part of GDAP1 [32]. Our results correlate with the different severities and imply that disease-associated missense mutations in the N-terminal part of GDAP1 affect exclusively mitochondrial dynamics, while loss or C-terminal truncation of GDAP1 additionally affect peroxisomal dynamics (Supplementary Table SI online).
GDAP1-induced fission depends on Drp1 and Mff
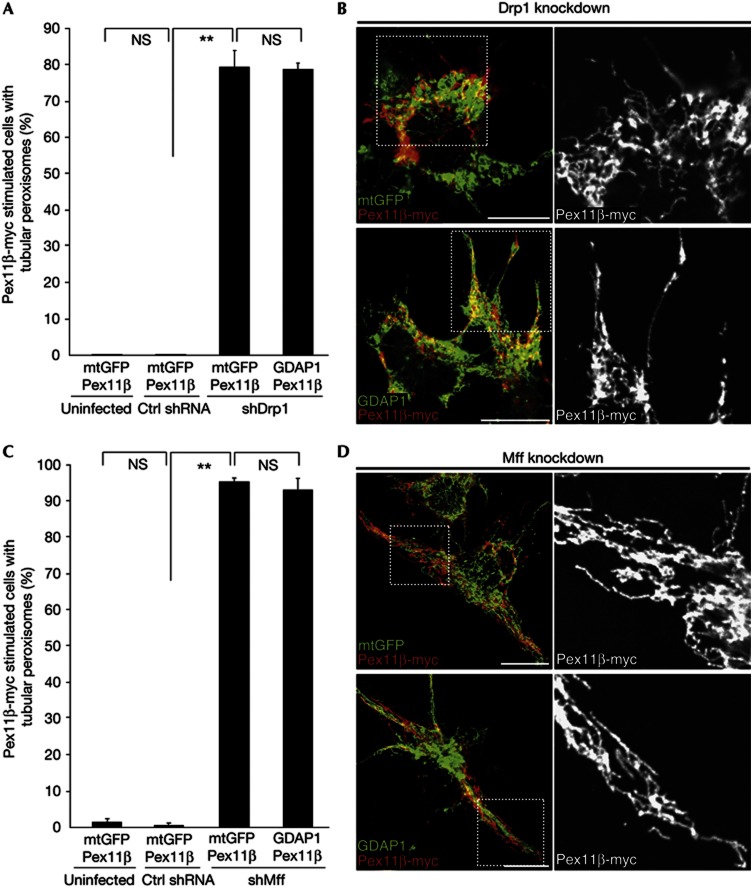
The previous data confirm that GDAP1-dependent fragmentation requires the intact HD1 and proper protein targeting to the peroxisomal membrane as for mitochondrial fission. Next, we investigated whether GDAP1-induced peroxisomal fission is dependent on Drp1, the executive GTPase in mitochondrial and peroxisomal fission [14, 15] and on Mff, the adaptor protein for Drp1 at peroxisomes and mitochondria in mammalian cells [10, 12]. We used Drp1 or Mff knockdown (supplementary Fig S2C,D online) in combination with Pex11β–myc and GDAP1 overexpression, as we did before in GDAP1 knockdown cells (Fig 3A). N1E-115 cells were infected with lentiviruses encoding non-targeting control shRNA, Drp1, or Mff shRNA constructs, followed by co-transfection with expression vectors coding for Pex11β–myc and mtGFP as control, or for Pex11β–myc and GDAP1. The next day, cells were fixed and morphologically analysed. As expected, Pex11β–myc expression together with control mtGFP led to highly tubular peroxisomes in Drp1 knockdown cells (Fig 4A,B). Overexpression of combined Pex11β–myc and GDAP1 in Drp1 knockdown cells resulted in an identical tubular peroxisomal morphology. Similarly, knockdown of Mff led to highly tubular peroxisomes (Fig 4C,D). In the absence of Mff, GDAP1 expression could not alter the peroxisomal morphology, demonstrating that GDAP1 cannot substitute for Mff in peroxisomal fission. We also analysed the mitochondrial morphology in N1E-115 cells with Mff knockdown and found that GDAP1-induced mitochondrial fission is as well dependent on Mff (supplementary Fig S6 online). Thus our data show that GDAP1-induced peroxisomal fission, like mitochondrial fission, is dependent on Mff and Drp1 [17].
Figure 4.
GDAP1-induced peroxisomal fragmentation depends on Drp1 and Mff. The experiment was performed as illustrated in Fig 3A. Knockdown of Drp1 (A,B) or Mff (C,D) in combination with co-expression of Pex11β–myc and GDAP1 or mtGFP result in tubular peroxisomes. Values represent means and s.e.m. of three independent experiments, 100 cells were counted per condition per experiment: **P<0.01, two-tailed unpaired t-test. Scale bar, 10 μm. Ctrl shRNA, non-targeting shRNA control; Drp1, dynamin-related protein 1; GDAP1, ganglioside-induced differentiation associated protein 1; Mff, mitochondrial fission factor; mtGFP, mitochondria-localized green fluorescent protein; s.e.m., standard error of the mean; shMff, shRNA against Mff (mitochondrial fission factor); shRNA, short hairpin RNA.
Concluding remarks
Existing peroxisomes can elongate and are subsequently fragmented by the peroxisomal fission machinery to generate new peroxisomes. Here we show that the mitochondrial fission factor GDAP1 is also a peroxisomal fission factor. Loss of GDAP1 results in elongated peroxisomal morphologies, as the fission capacity is decreased, whereas overexpression promotes peroxisomal fragmentation. GDAP1-induced fission relies on the presence of Mff and Drp1, demonstrating that GDAP1 influences fission upstream of the conserved basic fission machinery of mitochondria and peroxisomes. GDAP1 is not ubiquitously expressed, however, its accessory function is essential for myelinated peripheral nerves as mutations in GDAP1 lead to CMT. To induce fission, the integrity of HD1 is essential in GDAP1. Consistently, truncations lacking this domain or the targeting TA domain have lost the ability to promote peroxisomal fission. All other disease mutations in the N-terminal cytosolic GDAP1 domain are still able to promote peroxisomal fragmentation. These findings stand in contrast to our previous observations at the MOM, where the fission-induction function of these recessively inherited missense mutants is reduced [16, 17], suggesting different regulatory mechanisms for GDAP1-induced fission at both organelles. Furthermore, our results reveal a difference in the cell biology of CMT-associated N-terminal missense mutations and the more severe C-terminal truncation mutations of GDAP1.
Methods
Constructs
All constructs for transient transfections and lentiviral vectors and production of viruses have been described [16–18, 27, 31]. Lentiviruses expressing shRNA targeting Mff or Pex19 were purchased from Sigma (clone ID: NM_029409.2-484s1c1, NM_029409.2-693s1c1; NM_002857.2-88s1c1, NM_002857.2-90s1c1). MtDsRed and GFP–SKL were inserted into a modified pSicoR backbone (Addgene) containing the CMV promoter and multiple cloning sites from the pcDNA3.1 vector (Invitrogen).
Cell culture
Hippocampi of P1 C57BL/6J mice (Janvier, France) were dissected, trypsinized, and triturated. In total, 50,000–100,000 cells/ml were seeded on Matrigel (BD Bioscience) coated cover slips in Neurobasal medium (Gibco) supplemented with B27 (Gibco), 50 ng/ml NGF (Harlan), 2 mM L-glutamine (Gibco), 4 mg/ml D-glucose (Sigma). Mouse primary hippocampal neurons were infected with lentiviruses 1 day after preparation and kept in culture for 4–6 days. COS7 cells stably expressing GFP–SKL, HEK-293T cells, and N1E-115 cells were maintained, transfected and infected as described previously [17].
Immunocytochemistry
Immunocytochemistry was described [16]. For co-staining of GDAP1 and Pex11β–myc, cells were permeabilized with Digitonin (2.5 μg/ml, 5 min) and stained using antibodies against myc, followed by a permeabilization with 0.2% Triton X-100 for 10 min and subsequently stained for GDAP1. Antibodies: anti-GDAP1 [16], anti-human GDAP1 [32], anti-Pex14 (Proteintech Europe), anti-cytochrome c (Pharmingen), anti–myc (clone 9E10).
Biochemical Methods
Expression and purification of recombinant proteins and native polyacrylamide gel electrophoresis were performed as described [27]. The measurement of GSH levels and DCF fluorescence in cells has been performed according to [17, 29]. Western blotting, detection and quantification was performed as described [16]. Antibodies: anti-GDAP1 [16], anti-Dlp1 (BD Bioscience), anti-Pex19 (Sigma), anti-Mff (A. van der Bliek), anti-GAPDH (HyTest), and anti-β-actin (Sigma).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
The authors thank Alexander van der Bliek for kindly providing reagents, Michèle Telorack for help in performing the GSH measurement, Claudia Grou and Manuel Pinto (IBMC, Porto) for support in in vitro translation assay and Ned Mantei for critically reading the manuscript. We thank the Light Microscopy Centre of the ETH Zürich. This work was supported by the Swiss National Science Foundation and the National Center for Competence in Research (NCCR) Neural Plasticity and Repair (to U.S.), BBSRC (BB/K006231/1), and the Portuguese Foundation for Science and Technology (FCT) and Fundo Europeu De Desenvolvimento Regional (FEDER) (PTDC/SAU-OSM/103647/2008; PTDC/BIA-BCM/118605/2010 to M.S.; SFRH/BD/73532/2010 to S.G.).
Author contributions: N.H. was the primary experimentalist, contributed to the study design and writing of the manuscript. S.G. and A.N. contributed experimental work. M.S., U.S. and A.N. conceived and directed experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Wanders RJ, Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75: 295–332 [DOI] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Rachubinski RA (2007) Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol 23: 321–344 [DOI] [PubMed] [Google Scholar]
- Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J (2006) The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol 173: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD (2006) Growth and division of peroxisomes. Int Rev Cytol 255: 237–290 [DOI] [PubMed] [Google Scholar]
- van der Zand A, Gent J, Braakman I, Tabak HF (2012) Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 [DOI] [PubMed] [Google Scholar]
- Koch A, Schneider G, Luers GH, Schrader M (2004) Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci 117: 3995–4006 [DOI] [PubMed] [Google Scholar]
- Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M (2005) A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 16: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gould SJ (2003) The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem 278: 17012–17020 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Tanaka A, Fujiki Y (2007) Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp Cell Res 313: 1675–1686 [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S, Erdmann R (2005) Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. Febs J 272: 5169–5181 [DOI] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Yoon Y (2007) Mitochondria and peroxisomes: are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays 29: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Camoes F, Bonekamp NA, Delille HK, Schrader M (2009) Organelle dynamics and dysfunction: A closer link between peroxisomes and mitochondria. J Inherit Metab Dis 32: 163–180 [DOI] [PubMed] [Google Scholar]
- Delille HK, Alves R, Schrader M (2009) Biogenesis of peroxisomes and mitochondria: linked by division. Histochem Cell Biol 131: 441–446 [DOI] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U (2005) Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol 170: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann A, Wagner KM, Ruegg M, Suter U (2009) GDAP1 mutations differ in their effects on mitochondrial dynamics and apoptosis depending on the mode of inheritance. Neurobiol Dis 36: 509–520 [DOI] [PubMed] [Google Scholar]
- Wagner KM, Ruegg M, Niemann A, Suter U (2009) Targeting and function of the mitochondrial fission factor GDAP1 are dependent on its tail-anchor. PLoS One 4: e5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Cuesta A, Pedrola L, Palau F, Marin I (2004) Evolutionary and structural analyses of GDAP1, involved in Charcot-Marie-Tooth disease, characterize a novel class of glutathione transferase-related genes. Mol Biol Evol 21: 176–187 [DOI] [PubMed] [Google Scholar]
- Noack R et al. (2011) Charcot-Marie-Tooth disease CMT4A: GDAP1 increases cellular glutathione and the mitochondrial membrane potential. Hum Mol Genet 21: 150–162 [DOI] [PubMed] [Google Scholar]
- Niemann A, Berger P, Suter U (2006) Pathomechanisms of mutant proteins in Charcot-Marie-Tooth disease. Neuromol Med 8: 217–242 [DOI] [PubMed] [Google Scholar]
- Cassereau J, Chevrollier A, Bonneau D, Verny C, Procaccio V, Reynier P, Ferre M (2011) A locus-specific database for mutations in GDAP1 allows analysis of genotype-phenotype correlations in Charcot-Marie-Tooth diseases type 4A and 2K. Orphanet J Rare Dis 6: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Brambillasca S, Colombo S (2007) How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Schrader M (2001) Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J Histochem Cytochem 49: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Delille HK, Schrader M (2008) Targeting of hFis1 to peroxisomes is mediated by Pex19p. J Biol Chem 283: 31107–31115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A, Landgraf C, Lorenzen S, Rosenkranz K, Volkmer-Engert R, Erdmann R, Rottensteiner H (2006) Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci 119: 2508–2517 [DOI] [PubMed] [Google Scholar]
- Pinto MP, Grou CP, Alencastre IS, Oliveira ME, Sa-Miranda C, Fransen M, Azevedo JE (2006) The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J Biol Chem 281: 34492–34502 [DOI] [PubMed] [Google Scholar]
- Yagita Y, Hiromasa T, Fujiki Y (2013) Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J Cell Biol 200: 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman I, Kode A, Biswas SK (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc 1: 3159–3165 [DOI] [PubMed] [Google Scholar]
- Li X, Gould SJ (2002) PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol 156: 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ (1998) Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem 273: 29607–29614 [DOI] [PubMed] [Google Scholar]
- Kabzinska D, Niemann A, Drac H, Huber N, Potulska-Chromik A, Hausmanowa-Petrusewicz I, Suter U, Kochanski A (2011) A new missense GDAP1 mutation disturbing targeting to the mitochondrial membrane causes a severe form of AR-CMT2C disease. Neurogenetics 12: 145–153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.