EMBO Reports (2013) 14 6, 568–576 doi:; DOI: 10.1038/embor.2013.53
Among their many implications in plant and metazoan biology, microRNAs (miRNAs) control developmental patterning by facilitating acquisition and maintenance of cell fates. Whilst the core mechanisms of miRNA biogenesis and action are relatively well understood, how such processes might be modulated is only starting to emerge. Evidence in plants and human cells indicates that autophagy, a regulated process that delivers cytosolic material to lysosomes for degradation, is important to maintain miRNA homeostasis [1,2]. In this issue of EMBO reports, Zhang and Zhang report a role for autophagy in the miRNA pathway of the worm Caenorhabditis elegans [3]. They describe how ablation of some autophagy components rescues the developmental defects of genetically sensitized animals partly compromised for miRNA biogenesis or action. Autophagy seems to regulate the effector step of the worm miRNA pathway by selectively targeting AIN-1—an orthologue of the fly and mammalian GW182/TNRC6 that mediates silencing of miRNA target transcripts. Modulation of miRNA action through autophagy is, therefore, a conserved theme across phylae and kingdoms.
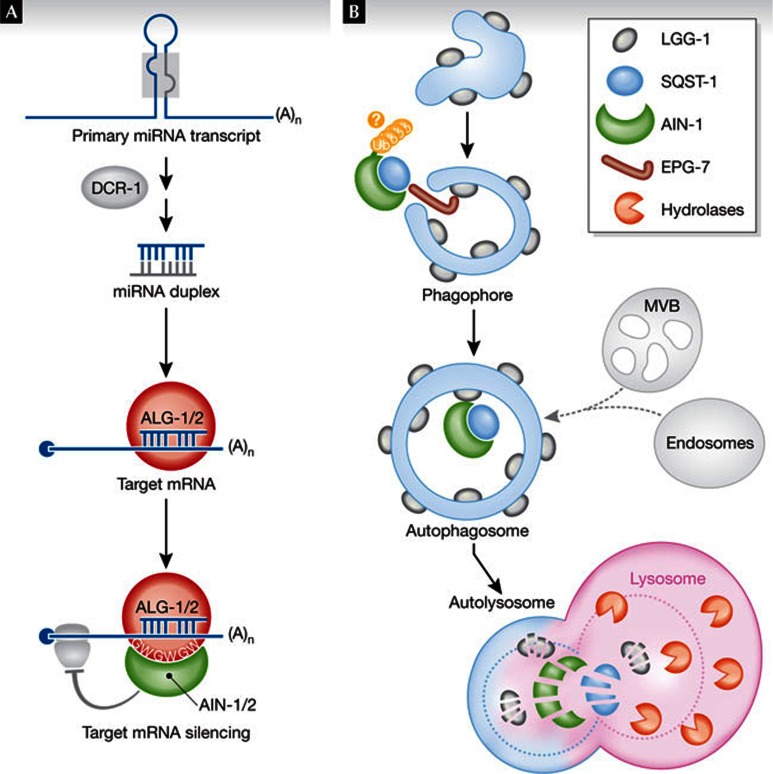
Metazoan miRNAs are encoded by stem–loop-containing, non-coding primary transcripts, which on nuclear maturation, are processed by the cytosolic RNase-III Dicer into 21-nt, double-stranded miRNA duplexes. One strand of the duplex is transferred into an Argonaute (AGO) protein, which scans the cellular content for mRNAs with partial or complete miRNA complementarity. Target transcripts are then silenced through translation inhibition and accelerated decay mediated by GW182/TNRC6—a component of the miRNA-induced silencing complex (miRISC) that interacts with AGO through repeated glycine–tryptophan (GW) residues (Fig 1A). Whilst this mechanistic scheme has been largely established biochemically, forward genetics and cell biology in various organisms are uncovering unsuspected regulatory layers in miRNA biogenesis and action, including an apparently ubiquitous link to membrane biogenesis and functions [4,5,6]. These findings have shed light on possible mechanisms of miRISC disassembly and recycling during the formation of intraluminal vesicles in late endosomes known as ‘multivesicular bodies’ (MVBs; [7,8]). As MVBs share features and regulators with lysosomes and autophagosomes—the vacuoles that fuse with lysosomes to carry out enzymatic digestion of cytosolic material—the possibility has emerged that the turnover of some miRISC components could be modulated by selective autophagy [9]. Selectivity in autophagy is enabled by specialized receptors, chiefly those in the SQST-1/P62 family, which recognize substrates for autophagy and target them to nascent autophagosome membranes through their interaction with autophagy-related (atg) and ectopic PGL granule (epg) family proteins.
Figure 1.
Autophagy degrades AIN-1. (A) Snapshot of the Caenorhabditis elegans miRNA pathway. The unique worm Dicer enzyme DCR-1 produces miRNAs that load into one of two AGO proteins, ALG-1 and ALG-2. On retrieval of miRNA target transcripts, the GW182/TNRC6 paralogues AIN-1 and AIN-2 interact with ALG-1 and ALG-2 through specialized GW residues to direct silencing through translational inhibition and accelerated mRNA decay. (B) A simplified model for autophagy-mediated regulation of AIN-1 steady-state levels. It is unclear if SQST-1–AIN-1 interaction requires prior ubiquitination of AIN-1 (uuuu), a modification often, albeit not always, displayed by autophagy receptor cargoes. Note that MVBs and late endosomes, putative sites of miRISC unloading and recycling, can also intercept this degradation pathway. AGO, Argonaute; AIN-1/2, Argonaute-interacting protein 1/2; ALG-1/2, Argonaute-like gene 1; DCR-1, Dicer; EPG-7, ectopic PGL granules 7; GW, glycine–tryptophan; miRISC, miRNA-induced silencing complex; mRNA, messenger RNA; miRNA, microRNA; MVB, multivesicular body; SQST-1, sequestosome 1; TNRC6, trinucleotide repeat containing 6.
The recent demonstration, in both human cells and plants, that autophagy is indeed an integral part of miRNA homeostasis [1,2], prompted Zhang and Zhang to explore whether the same was true in C. elegans. They first used a reverse genetic approach based on well-characterized developmental defects shown by animals defective for miRNA biogenesis or action, including the reiteration of stage-specific cell proliferation programmes known as ‘heterochronic’ defects. The authors found that heterochronic defects in loss-of-function mutants of alg-1 (one of two worm miRNA-effector AGOs; Fig 1A) and ain-1 (one of two worm GW182/TNRC6 paralogues; Fig 1A) were partly suppressed by secondary null mutations in several epg genes. Similarly, heterochronic defects caused by a hypomorphic mutation in the single Dicer gene of C. elegans (DCR-1; Fig 1A) were rescued in animals carrying a second atg-2 loss-of-function mutation.
To rule out that these results were not confounded by general autophagy-dependent regulations of larval development, the authors used animals expressing GFP ‘sensor’ constructs based on fusions to hbl-1 and lin-41—two genetically verified targets of the C. elegans let-7 miRNA family. In both cases, loss of GFP, monitored at the cognate let-7 expression stages, was significantly stronger in autophagy mutants than in wild-type animals. This difference in silencing efficacy probably resulted from enhanced let-7 activity because it was not observed in let-7 mutant worms. Conversely, increasing autophagy by starvation or by inhibiting signalling from the key autophagy regulatory kinase mTOR, increased GFP production from the let-7 sensors. Abnormal vulval development due to altered silencing of let-60/RAS, another target of the let-7 family, was also corrected in autophagy mutants, in an AIN-1-dependent manner. To test if their findings were restricted to the let-7 family members or, instead reflected a general effect of autophagy on miRNA action, Zhang and Zhang explored the genetic modification of neuronal fate specification defects caused by a hypomorphic mutation in the let-7-unrelated miRNA lsy-6. As with let-7, mutations in selected epg and atg genes could partly rescue the neuronal specification defects, which conversely, were exacerbated by mTOR signalling mutations. Therefore, loss-of-autophagy activity suppresses the developmental and regulatory defects caused by inefficient miRNA activity in C. elegans.
To bolster their developmental and reverse genetic approach, the authors embarked on biochemical and cell biology analyses of the key miRISC components AIN-1 and AIN-2, and ALG-1 and ALG-2 during embryonic development. An AIN-1::GFP reporter diffusively distributed in the cytoplasm in wild-type embryos clearly accumulated at the protein but not at the mRNA level, and formed many aggregates in several autophagy mutant embryos. This aggregation process, typical of autophagy-targeted proteins under conditions of altered autophagosome maturation, was not seen with AIN-2–GFP. ALG-1 and ALG-2 also accumulated in aggregates in mutant embryos, although this effect was clearer under certain stress conditions. Confirming AIN-1 as a direct target of autophagy under normal developmental conditions, AIN-1::GFP aggregates co-localized in atg-3 mutants with SQST-1/P62—both a selective receptor and a target of autophagy (Fig 1B). Furthermore, in atg-2 or epg-6—mutants defective in autophagosome maturation—endogenous AIN-1 aggregates co-localized with punctae formed of LGG-1, the worm paralogue of the autophagosome membrane-associated protein ATG-8 (Fig 1B). In vivo co-immunoprecipitaton and in vitro glutathione S-transferase pulldowns confirmed direct interactions between AIN-1 and LGG-1. Nonetheless, the AIN-1::GFP pattern was unchanged in single sqst-1 or double sqst-1 lgg-1 mutant worms compared with wild-type worms, suggesting the involvement of at least one additional factor in recruiting AIN-1 to autophagosomes. A probable candidate was the recently characterized EPG-7 scaffold protein, which links cargo and receptor complexes with the autophagic assembly machinery in C. elegans [10]. EPG-7 is selectively required for interactions between SQST-1/P62 aggregates and LGG-1 punctae, and for their subsequent degradation (Fig 1B). A null mutation in epg-7 but not in sqst-1, indeed caused accumulation of AIN-1::GFP aggregates and suppressed the neuronal fate specification defects of lsy-6 hypomorphic animals.
Whilst the results of Zhang and Zhang clearly implicate autophagy as a negative regulator of the worm miRNA pathway, they are also interesting in several other respects. First, they emphasize the value of genetically sensitized backgrounds in the discovery of pathways that modulate rather than directly affect miRNA biogenesis and action. Indeed, none of the developmental defects associated with miRNA dysfunction were observed in worms carrying single mutations in autophagy components. The modulatory role of autophagy was only evident when animals that were partly defective in specific or generic miRNA functions were secondarily subjected to the ablation of autophagy components. Intrinsic to the potency of these ‘genetic modifier’ approaches is the primary availability of hypomorphic alleles of individual miRNA or miRNA pathway components. These alleles are nearly always recovered through forward genetics, which until recently relied on tedious gene-mapping procedures. However, given the present advent of powerful genome re-sequencing methodologies the prospect of generating large allelic series seems much more favourable. In fact, this endeavour should be considered a priority if the various tenants of miRNA biology are to be fully understood through modifier gene identification but also interactome studies. A second interesting aspect emerging from the work of Zhang and Zhang, and others [1,2], is the notion that autophagy is part of a global homeostatic regulatory mechanism that prevents overt perturbations to the miRNA pathway. Whilst such perturbations might be artificially induced genetically, as in the present study, they will probably arise naturally through various stresses of abiotic or biotic origin. It is noteworthy that the role of autophagy in the plant miRNA pathway was discovered through the study of the viral virulence factor P0, which suppresses antiviral gene silencing by increasing the turnover of AGO1 through autophagy [2]. Unveiling the mode of action of P0 revealed that selective AGO1 degradation is, in fact, part of a normal regulatory process. As with viruses, other intracellular parasites, including important bacterial pathogens in human, establish intimate interactions with the endomembrane system of their hosts [11]; some of these pathogens also profoundly modify cellular miRNA biogenesis and action in ways that probably contribute to disease. Whether these effects are mediated through alterations of autophagy or other membrane-based miRNA regulatory processes, and whether pathogen virulence factors can be used as molecular probes of the underlying mechanisms, are interesting prospects for future investigation.
Footnotes
The author declares that he has no conflict of interest.
References
- Gibbings D et al. (2012) Nat Cell Biol 14: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B et al. (2012) Proc Natl Acad Sci USA 109: 15942–15946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang H (2013) EMBO Rep [Epub ahead of print] doi:; DOI: 10.1038/embor.2013.53 [DOI] [Google Scholar]
- Brodersen P et al. (2012) Proc Natl Acad Sci USA 109: 1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Ruvkun G (2012) Proc Natl Acad Sci USA 109: 4568–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L et al. (2013) EMBO J 32: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ et al. (2009) Nat Cell Biol 11: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Lee YS et al. (2009) Nat Cell Biol 11: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D, Voinnet O (2010) Trends Cell Biol 20: 491–501 [DOI] [PubMed] [Google Scholar]
- Lin L et al. (2013) J Cell Biol 201: 113–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cossart P (2012) Trends Cell Biol 22: 283–291 [DOI] [PubMed] [Google Scholar]