Abstract
BACKGROUND
Metastases to the brain represent a feared complication and contribute to the morbidity and mortality of breast cancer. Despite improvements in therapy, prognostic factors for development of metastases are lacking. KISS1 is a metastasis suppressor that demonstrates inhibition of metastases formation in several types of cancer. The purpose of this study was to determine the importance of KISS1 expression in breast cancer progression and the development of intracerebral lesions.
METHODS
In this study, we performed a comparative analysis of 47 brain metastases and 165 primary breast cancer specimens by using the antihuman KISS1 antibody. To compare KISS1 expression between different groups, we used a 3-tier score and the automated score computer software (ACIS) evaluation. To reveal association between mRNA and protein expression, we used quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. Significance of immunohistochemistry stainings was correlated with clinicopathological data.
RESULTS
We identified that KISS1 expression is significantly higher in primary breast cancer compared with brain metastases (P < .05). The mRNA analysis performed on 33 selected ductal carcinoma brain metastatic lesions and 36 primary ductal carcinomas revealed a statistically significant down-regulation of KISS1 protein in metastatic cases (P = .04). Finally, we observed a significant correlation between expression of KISS1 and metastasis-free survival (P = .04) along with progression of breast cancer and expression of KISS1 in primary breast cancer specimens (P = .044).
CONCLUSIONS
In conclusion, our study shows that breast cancer expresses KISS1. Cytoplasmic expression of KISS1 may be used as a prognostic marker for increased risk of breast cancer progression.
Keywords: KISS1, breast cancer, brain metastases, mediator, suppressor, IHC, RNA
INTRODUCTION
Patients who develop brain metastases from any primary tumor survive, on average, <2 years from diagnosis.1–3 The incidence of brain metastases from breast adenocarcinoma is rising.4,5 Although recent improvements in chemotherapy and radiotherapy have been remarkable, advanced metastatic disease, especially in the brain, remains incurable.6 Therefore, novel strategies for early diagnosis, prognostic factors, and better therapies are urgently needed.
Different groups of cellular proteins participate in metastases. To verify their expression, high throughput arrays have been proposed.7 In breast cancer, this method has revealed hundreds of different genes that may impact development of metastases to the bone and brain.8,9 However, the high variability of these results has required additional verification steps to compare back-to-back expression of metastatic lesions and matched primary specimens7,9–12 In addition, mRNA levels do not necessarily correlate with actual protein expression, which mediates the biology. To date, prognostic factors in treatment of breast cancer include the expression of estrogen receptors (ER), progesterone receptors (PR), as well as amplification of the human epidermal growth factor receptor 2 (HER2) gene.13 However, more studies are needed to determine the predictors of metastatic development. The majority of deaths in breast cancer are associated with metastases. It is therefore important to understand mechanisms and factors involved in regulation of metastases. Among metastases suppressors, KISS1 is one of the inhibitors that has demonstrated significant inhibition of metastases formation in breast cancer and pancreatic carcinoma models.14,15 In the literature, it has been shown that loss of KISS1 expression is associated with cancer progression in gastric, ovarian, bladder, esophageal, and hepatocellular carcinoma.16–21 In vitro treatment of breast cancer cells with the product of KISS1 gene, KISSPEPTIN 10, has been shown to reduce the invasive properties of breast cancer cells.22 Recent clinical studies conducted by several groups indicate a correlation between KISS1 expression and poor prognosis.23 The relationship between development of metastases and KISS1 expression in primary breast cancer specimens has not been well established.
The aim of this study was to define the relationship between KISS1 expression and development of brain metastases. To address this question, we used immunohistochemistry (IHC) to evaluate expression of KISS1 in cancer and non-cancer breast specimens. The present study evaluated the expression profiles of KISS1 in primary invasive ductal carcinoma (IDC), ductal carcinoma in situ (DCIS), brain metastasis from human breast (BMHB), metastatic lymph nodes (LN), and non-neo-plastic breast parenchyma (NBreast) from a total of 389 patients.
MATERIALS AND METHODS
Reagents
We used a monoclonal antibody recognizing the KISS1 protein that was developed at the UAB Epitope Recognition Immunoreagent Shared Resource (Dr. M.A. Accavitti-Loper). Antibody specificity was validated by mass spectrometry and sequencing of immunoprecipitated peptides. TMA sections (BR801, BR8011, BR812) obtained from BIOMAX (Rockville, Md) were stained with antibodies that recognize human HER2 (A0485, DAKO), ER (RM901-01, Thermo-Fisher Scientific, Pittsburgh, Penn), and PR (RM-9102, Thermo-Fisher) antigens.
Patient Information and Study Design
The study was approved by the institutional review board of the University of Chicago. The eligible specimens were collected based on inclusion and exclusion criteria. Inclusion criteria: 1) brain lesion along with primary cancer specimens with pathological diagnosis, 2) informed consent obtained or waiver of consent, 3) follow-up information available. Exclusion criteria: 1) no patient information or follow-up, 2) lack of enough tissue to perform TMA. A panel of 47 assessable intracerebral lesions that were resected at the University of Chicago Medical Center were examined and divided between experimental and validation sets. Expression of metastatic suppressor KISS1 was evaluated based on automated computer evaluation software (ACIS). To validate the ACIS findings, evaluation of KISS1 immunoreactivities was performed by using a 3-tier score system.
Medical records were reviewed for pertinent patient information. The analyzed data included birth date, date of diagnosis, date of operation, TNM stage, and date of relapse/metastasis. The time of metastases-free survival was defined as the time of primary surgical therapy to the time of developing brain lesions.
Construction of Tissue Array-Experimental Set
For immunohistochemistry studies, primary invasive ductal breast cancer and brain metastases specimens, from 1980 to 2008, were obtained from University of Chicago Tissue Bank. All specimens used in this study were previously stained via hematoxylin and eosin (H&E) and then reviewed by study-assigned pathologists (PP and HS). The experimental set of 51 patients included 20 primary cancer specimens (invasive ductal carcinoma), 20 human metastatic brain lesions of ductal carcinoma and 14 cases of nontumorigenic breast. All were examined via tissue microarray (TMA). Of note, the recipient block also included 3 cores from primary breast cancer cases and their matched brain lesions.
TMA recipient blocks were constructed from paraffin- embedded specimens previously fixed in 10% formaldehyde. From each donor block, 2 cylinders of 0.6-mm diameter tissue were taken from representative areas based on H&E staining and transferred to a paraffin recipient block with 0.8-mm distance between the samples, using a Tissue Arrayer (Beecher Instruments, Gene Micro-Array Technologies, Silver Spring, Md). Sections of the resulting tumor tissue microarray block (4 mm thick) were transferred to the Superfrost/Plus microscope glass slides (Fisher Scientific).
Validation Set
Given the exploratory nature of the study, an independent series of 145 primary IDCs, 27 metastatic brain lesions (ductal carcinoma), 46 samples from DCIS, 113 samples of NBreast, and 7 LN metastases (ductal carcinoma) from 338 patients were used to validate the IHC data. Unstained tissue microarray slides (TMA) BR801, BR8011, BR812, CIHDR2A, and CIHDR2B (with no overlapping cases) were received from BIOMAX (Rockville, Md) and Dr. Olopade (University of Chicago), respectively. The features of the patient samples are summarized in Table 1. Validation was carried out by calculation of a 3-tier score performed by 2 pathologists (PP and HS).
Table 1.
Patient Characteristics of Brain Metastatic Lesions and Primary Casesa
| Characteristics | N(%) of Cases 47 |
N(%) of Cases 165b |
P Valuec |
|---|---|---|---|
| Age at diagnosis | |||
| Mean (SD) | 51.38 ± 10.40 | 51.66 ± 11.80 | .883 |
| <35 | 2 (4.3%) | 9 (5.45%) | .815 |
| 35–55 | 27 (57.5%) | 102 (61.8%) | |
| >55 | 18 (38.3%) | 54 (32.7%) | |
| Stages | .738 | ||
| T0 | 1 (0.6%) | ||
| T1 | 11 (23.4%) | 49 (29.7%) | |
| T2 | 30 (63.8%) | 87 (52.8%) | |
| T3 | 4 (8.6%) | 20 (12.1%) | |
| T4 | 2 (4.2%) | 8 (4.8%) | |
| ER status | .562 | ||
| Positive | 18 (38.3%) | 55 (43.7%) | |
| Negative | 26 (55.31%) | 76 (52.4%) | |
| Unknown | 3 (6.4%) | 4 (3.9%) | |
| PR status | .027 | ||
| Positive | 13 (27.6%) | 77 (46.7%) | |
| Negative | 31 (66.0%) | 84 (50.9%) | |
| Unknown | 3 (6.4%) | 4 (2.4%) | |
| Her2 tumor status | .626 | ||
| Positive | 17 (36.2%) | 72 (43.6%) | |
| Negative | 27 (57.4%) | 82 (49.7%) | |
| Unknown | 3 (6.4%) | 11 (7.4%) | |
| LN removal | .029 | ||
| N0 | 20 (42.6%) | 83 (50.3%) | |
| N1 | 14 (29.9%) | 65 (39.4%) | |
| N2 | 8 (17.0%) | 10 (6.1%) | |
| N3 | 5 (10.7%) | 7 (4.24%) |
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor; LN, lymph node; PR, progesterone receptor.
Histology of brain metastases is based on patient’s primary tumor;
Information about patient cases obtained from only University of Chicago Archive directory and used to create the TMA: CIHDR2A and CIHDR2B; TMA obtained from BIOMAX has limited or no information about patients. To characterize the expression of patient samples used for TMA (BIOMAX), we stained sections for HER2, PR and ER expressions followed by evaluation evaluation of IHC scoring.
P values between IDC samples with no (n = 165) or with records of intracerebral metastases (n = 47) were determined by t test for continuous variables (age), or chi-square tests or Fisher exact tests for categorical variables.
IHC
Sections of 4 µm obtained from the tissue microarray were deparaffinized in xylene and then rehydrated. After deparaffinization, heat-induced epitope retrieval was conducted by immersing slides in Coplin jars filled with 10 mMol/L citrate buffer (pH 6.0) or ET buffer (pH = 9.0) and then blocked with 1% H2O2 in phosphate-buffered saline (PBS) for 15 minutes (room temperature [RT]) after treatment with Tris buffer saline Tween 20 (TBST) buffer and 5% nonfat dry milk reagent. For IHC staining, slides were incubated first with mouse antihuman KISS1 antibodies (clone 6A4.27; dilution 1:250) and then sections were incubated with a secondary antibody conjugated to a peroxidase-labeled polymer (DAKO REAL Envision System (DAKO, Glostrup, Denmark). Each incubation step was followed by 3 washes for 5 minutes in TBST buffer. Reaction products were developed with DAB and counterstained with hematoxylin. Negative controls were obtained by omitting the primary antibody.
To detect HER2, ER, and PR expressions, tissue sections were deparaffinized, rehydrated through xylenes and serial dilutions of ethyl alcohol (EtOH) with distilled water followed by incubation with antigen retrieval buffer (DAKO, S1699) in steamer at over 97°C for 20 minutes. Either HER-2 antibody (A0485, 1:100 dilution, DAKO), ER (RM901-01, 1:50 dilution, Thermo-Fisher Scientific), or PR (RM-9102, 1:50 dilution, Thermo-Fisher Scientific) were applied on tissue sections (1 hour, RT). After TBS wash, tissue sections were incubated with either biotinylated antirabbit IgG (7.5 µg/mL, BA-1000, Vector Laboratories, Burlingame, Calif), Bond Polymer Refine Detection (DS9800, Leica Microsystems, Buffalo Grove, Ill) or combination of Envision+ system (DAKO, K4003) and DAB+ chromogen (DAKO, K3468) for 30 minutes at RT. The antigen-antibody binding was detected by an Elite kit (PK-6100, Vector Laboratories) and a DAB (DAKO, K3468) system. Tissue sections were briefly immersed in hematoxylin for counterstaining and were covered with cover glasses.
Scoring Systems
Immunoassaying intensity was evaluated by two pathologists (coauthors PP and HS) and scored using either 3 tier score (the staining levels 0, 1+, 2+, and 3+) or Automated Cellular Imaging System (ACIS, Clarient, Calif). Measurements by ACIS were performed based on 3 criteria: the color defined by hue, the “darkness” defined as luminosity, and density defined as saturation.24 The positive score was detected as a cytoplasmic expression and later defined as viability at low magnification (10×) and was presented as Brown IOD per 10 µM.2 The expression of markers detected by ACIS was validated by using validation test.
RNA Extraction of Tissue Bank and OCT-Frozen Specimens
Total RNA was extracted from breast cancer specimens containing at least 80% of tumor cells. Sections (5 µm) of formalin-fixed, paraffin-embedded tissue, were deparaffinized and subjected to RNeasy FFPE kit (Qiagen, Valencia, Calif). From OCT-embedded sections, the total RNA was isolated according to a published protocol.25 The RNA concentration was determined using Nanodrop Spectrophotometer (NanoDrop Technologies, Wilmington, Del). The quality of isolated mRNAs was assessed by using Agilent 2100 Bioanalyzer (Agilent technologies, Santa Clara, Calif) together with the reagents in the RNA6000 Nano LabChip kit. All samples were within a range of 5 to 500 ng/µL.
Reverse Transcription
RNA extraction from formalin-fixed paraffin-embedded or OCT-embedded tissue sections was performed by using RNeasy isolation kit (Qiagen). Next, the isolated RNA was reverse-transcribed in a final volume of 20 µL using a 1-step iScript Synthesis Kit (Bio-Rad, Hercules, Calif) according to the manufacturer’s instructions, with the following conditions: 5× iScript reaction mix, iScript reverse transcriptase, nuclease-free water and 1 µg of total RNA. The reactions were performed at 42°C for 30 min, followed by inactivation of the enzyme at 85°C for 15 minutes. The cDNA was stored at −20°C.
Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Real-time RT-PCR analyses of mRNAs were performed using the OPTICON 2 Detection System instrument and software (Bio-Rad). These primers were synthesized by IDT (Coralville, Iowa). Real-time RT-PCR was performed with the SYBR GreenER Super Mix (Invitrogen, Carlsbad, Calif) using 1 µL of diluted cDNA and 300 nmol/L of the primers in 10 µL of final reaction mixture. The ratios of KISS1 to GAPDH mRNAs were calculated for each sample.
The PCR program was 95°C for 4 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 45 seconds. The relative expression was calculated by 2−ΔΔCT (CT = fluorescence threshold value; ΔCT = CT of the target gene-CT of the reference gene (GAPDH); ΔΔCT = ΔCT of the tumor −ΔCT of the reference sample). The sequences of the PCR primers for the study of KISS1 were described before.26
Statistical Analysis
Immunohistochemistry scoring and analyses were performed blindly. Patient characteristics between experimental and validation specimen sets (Table 1) were compared using t-test for continuous variables and Pearson chi-square/Fisher exact tests for categorical variables. Associations between KISS1 expression in BMHB, IDC, DCIS, NBreast, and LN were tested using contingency methods (Fisher exact test). Brown IOD data in BMHB, IDC, NBreast, and LN groups were compared using Kruskal-Wallis analysis of variance (ANOVA) with Dunn multiple comparisons post-test.27 The KISS-1 RNA expression in unmatched/matched samples was compared using either Mann-Whitney test or 2-way ANOVA model with repeated measures. The repeated measurements of 2-way ANOVA assume normality distribution of the data, fixed effects for the 2 factors (measure time and sample group) and random effects for patient variability.28 Metastases-free survival was defined as the time from mastectomy to the date of clinically documented intracerebral metastatic disease attributed to breast cancer. Fisher exact test was used to compare ACIS grades of protein expressions among different tissues. Spearman correlation is a nonparametric measurement of statistical correlations. Spearman correlation was used to test for an association between HER2, ER, PR, and KISS1 in the 2 cohorts of patients. A P value of <.05 was considered statistically significant. All statistical analyses were carried out using STATA 11.1 version (StataCorp LP, College Station, Tex) and GraphPad Prism 4.02 version (GraphPad, San Diego, Calif) softwares.
RESULTS
To investigate whether brain metastases, breast cancer specimens, LNs, and NBreast exhibit different KISS1 immunoreactivities, we stained TMA slides with antibodies that recognize KISS1 protein. Representative images are shown in Figure 1 to demonstrate examples of negative (no expression), mild (1+) and moderate (2+) expressions. The primary breast cancer specimen serves as a positive control for KISS1 expression and represents high expression (3+). To analyze KISS1 expression, we created an experimental set with specimens that represent brain metastases, primary cancer, LN metastases, and NBreast cases. To evaluate expression of KISS1 marker, we used ACIS algorithm. Intensity of KISS1 expression was measured by ACIS software and is presented as Brown IOD/µM2. The results of expression profiling are presented in detail in Figure 2A. Kruskal-Wallis tests indicated a significant difference in expression of KISS1 suppressor (P = .014) among BMHB, IDC (primary cases), NBreast, and LN metastatic samples.
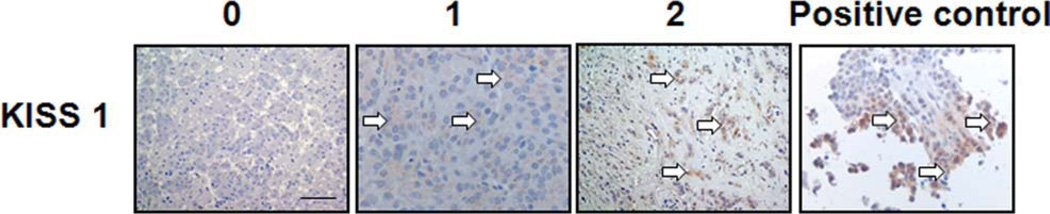
Figure 1.
Localization of KISS1 staining in tissue microarray (TMA) sections containing brain metastatic lesions (BMHB). Representative staining results of KISS1 expressed in BMHB are shown at 40× original magnification. Sections of formalin-fixed paraffin-embedded specimens were stained for KISS1 antibodies. Positive staining for the KISS1 antibody (clone 6A4, UAB) is shown, illustrating 0 to 3+ staining scale. Primary human breast carcinoma was used as positive control. Of note, we did not observe (3+) staining in brain metastases. Arrows point to the stained cells.
Figure 2.
The automated computer software (ACIS) score and immunohistochemistry (IHC) intensities of KISS1 stainings in tumor cells of patient samples. Evaluation of KISS1 immunoreactivity was assessed by ACIS (A) and intensities of cytoplasmic staining were calculated manually in each core at magnification (200×) by pathologists and plotted as percentage of cases having mild (1+), moderate (2+), and strong (3+) intensities of expression or no expression (0) (B). Statistical analyses for ACIS score, conducted by using Kruskal-Wallis test, indicated a significant difference in expression of KISS1 among BMHB, IDC, DCIS, NBreast, and LN samples (P = .014). Dunn multiple comparison test (2 sided) was applied to determine the difference between groups. P values <.05 were observed between IDC samples and LN along with difference between IDC and intracerebral metastases (BMHB). In the validation set (B), statistical significance between IDC, NBreast, BMHB, and LN samples was revealed by using the Fisher test (P < .001). The pairwise comparison Fisher exact test: IDC versus DCIS, P = .11; IDC versus BMHB, P < .001; IDC versus NBreast, P < .001; IDC versus LN, P = 013.
Dunn multiple comparison post-test (2 sided) demonstrated a significantly higher expression of KISS1 in IDC specimens (64.67 ± 21.47) than in BMHB (3.71 ± 0.92, P <.01) and LN metastases (1.72 ± 0.28, P < 0.01). Of note, no significant difference was observed between KISS1 expression in IDC sample and NBreast (9.47 ± 2.45, P > .05).
To validate the TMA findings, we performed an independent analysis of expression using ACIS grading (Fig. 2B). We used a validation set that contains a panel of IDC samples (n = 145) and brain metastatic lesions (n = 27), and stained with antibodies against KISS1 protein. In validation experiments, expression of KISS1 in breast cancer (IDC) was found to be strong (3+) and moderate (2+) in 16.6% (24 of 145) and 39.3% (57 of 145) specimens, whereas mild (1+) in 24.8% (36 of 145) or no expression was detected in 19.3% (28 of 145) cases. In contrast, brain lesions (BMHB) demonstrated either no expression (0) or weak (1+) expression in 92.6% cases (25 of 27) and moderate/ strong immunoreactivity (2 of 27) in 7.4% of cases. Fisher exact test indicated a significant difference in expression of KISS1 among BMHB, IDC, DCIS, NBreast, and LN samples (P < .001). For the pairwise comparison Fisher exact test: IDC versus BMHB, P < .001; IDC versus NBreast, P < .001; IDC versus DCIS, P = .11; IDC versus LN, P = .013.
Finally, to understand the correlation between HER2, PR, ER, and KISS1 markers, we performed evaluation of breast cancer specimens without dividing them into the grade or stages. Overall, we observed the Spearman correlation between KISS1 expression with PR staining (Spearman rho = 0.28, P = .047) and no statistical correlation was observed between KISS1 and HER2 or ER stains (Spearman rho = .152, P = .29 and Spearman rho = .200, P = .165, respectively).
Real-Time PCR Data Correlates with Protein Expression Detected by IHC
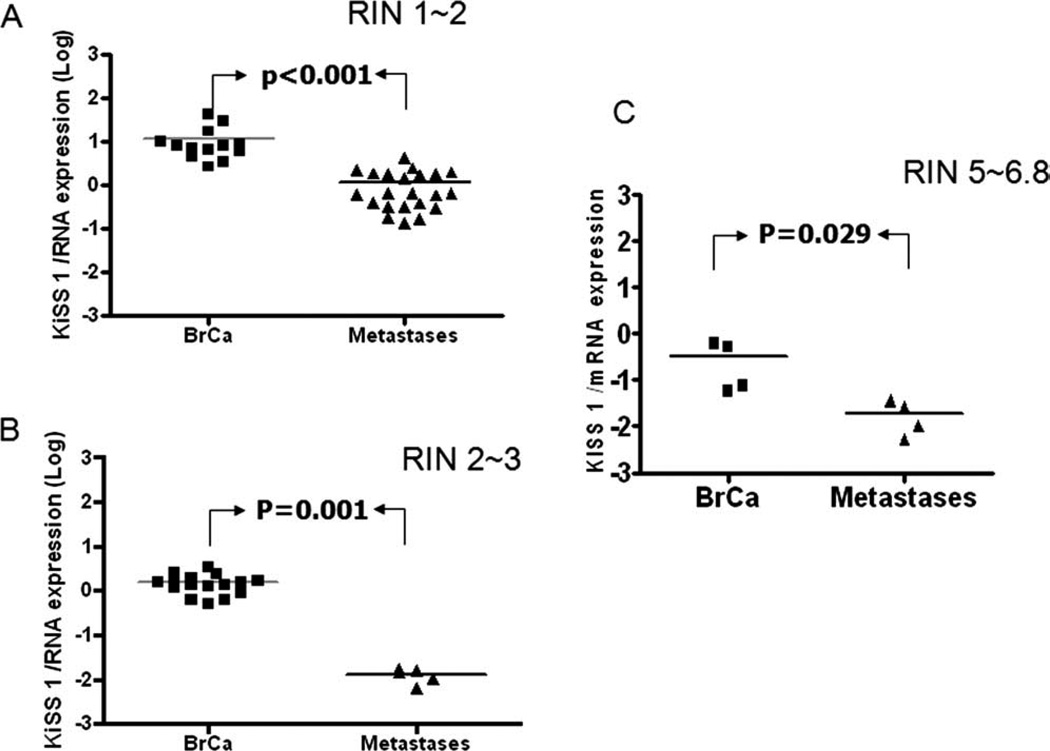
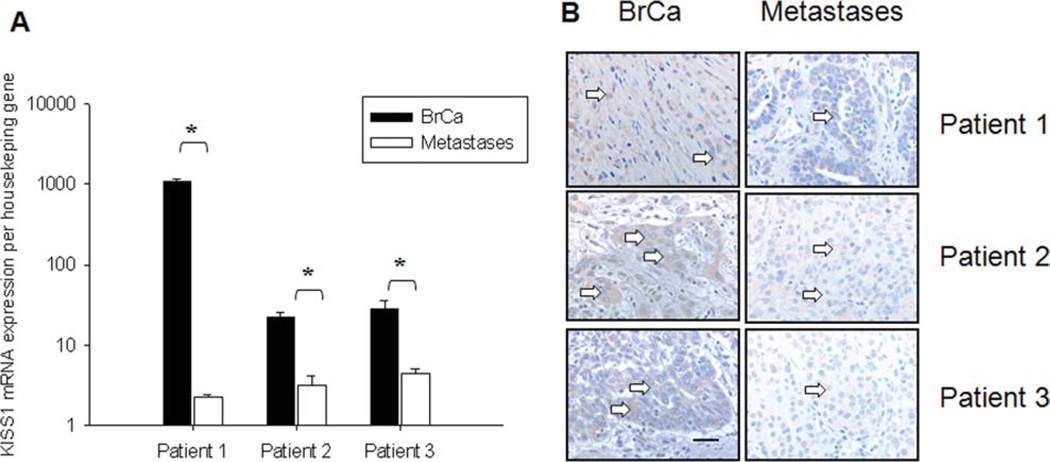
To confirm the differential expression of KISS1 protein in metastatic lesions and primary cases, we measured the relative level of mRNA. It is well documented that quality of mRNA isolated from paraffin embedding tissue is very poor. In our experiments, the RNA integrity number (RIN) varied from 1.1 to 3.7 (average ~2.2, for RNA isolated from formalin-fixed paraffin-embedded tissue) and 5.1 to 6.8 (average ~6.2) for the frozen sections. The relative expression was determined as Log difference relative to the normalized reference sample (2 sets of normal samples) as presented in Figure 3. Materials to perform mRNA analyses from the IDC (breast cancer, BrCa) samples and brain metastases were available from 67 patients (including 3 matched cases). Real-time PCR data revealed that regardless of RIN number, KISS1 expression was lower in BMHB compared with IDC samples (Fig. 3A and B). The same trend was observed for the OCT frozen sections (Fig. 3C). Thus, we observed significant down-regulation of KISS1 expression in metastatic samples having RIN 1–2, RIN 2–3, and RIN numbers >5 (Mann-Whitney test; P < .001, P = .001, and P = .029, respectively). To determine the status of KISS1 expression in matched samples (primary IDC and their intracerebral metastases, n = 3) we performed a quantitative analysis of mRNA expression on patient samples at day 1, day 2, and day 3. As seen in Figure 4A, KISS1 mRNA is significantly higher in all IDC specimens compared with the matched brain lesions (2-way ANOVA with repeated measurements on log-transformed data, F-test P < .001). Importantly, for this set of the samples we observed a correlation between mRNA expression level and IHC stainings (Fig. 4B). Therefore, this data support the hypothesis of the biological significance of KISS1 expression in brain metastases (BMHB).
Figure 3.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of KISS1 mRNA expression levels in the paraffin-embedded or OCT frozen samples from IDC and metastatic lesions. Real-time PCR was performed on cDNA obtained from mRNA having low (A), middle (B), and high quality by using primer sets for KISS1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). mRNAs from samples having renewal identification number (RIN)1-2, RIN2-3, and RIN5 (and more) were designated as low-, mild-, and high-quality groups. Transcription levels of target proteins are represented as log gene expression of target gene in BMHB versus IDC samples. Down-regulation of KISS1 expression was observed in BMHB compared with IDC samples. (Mann-Whitney test, for RIN1-2, RIN2-3, and RIN numbers more than 5, P < .001, P = .001, and P = .029, respectively).
Figure 4.
The characterization of KISS1 expression in matched cases. Analysis of KISS1 expression was performed based on mRNA expression (A) or via IHC (B) staining in the 3 pairs of matched paraffin-embedded samples from breast cancer (IDC) and intracerebral metastatic lesions (metastases). Real-time PCR was performed using primer sets for KISS1, and GAPDH. Transcription levels of target proteins are calculated based on Ct method (*2-way ANOVA with repeated measurements, F-test P < .001). Arrows point to the stained cells.
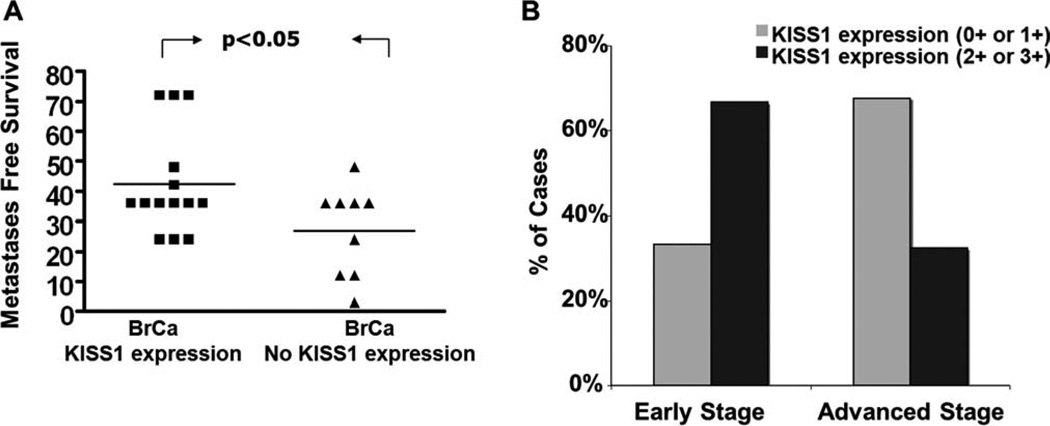
KISS1 Expression Correlates with Metastases-Free Survival
To evaluate the potential associations between KISS1 expression in the brain lesions and clinicopathological data, we compared cases that exhibit negative expression with those that exhibit positive (0 or 1+ compared to 2+ or 3+) cytoplasmic stainings in IDC. To analyze the significance of KISS1 expression, the cohort of IDC patients with brain metastases was re-analyzed. As shown in Figure 5A, the group of patients with positive expression of KISS1 protein (1+ or more) was found to have a trend toward slower disease progression. These patients developed brain metastases 42.4 ± 4.6 (95% confidence interval [CI], 32.4, 52.5) months after primary tumor resection via lumpectomy or mastectomy, whereas the group of the patients with negative KISS1 expression had brain metastases after only 27.0 ± 5.0 (95% CI, 15.5, 38.5) months. Finally, we compared the level of KISS1 expression between patients with different stages of breast cancer disease. Because different complications such as brain metastases occur during stages 2 and 3, we considered them together as group “Advanced cancer”. As seen in Figure 5B, weak KISS1 expression (0 or 1+) was more frequent in patients with “Advanced cancer” stage than early stage (68% in “Advanced stage” while 33% in “Early stage”, P = .005). Overall, this data suggests that expression of KISS1 correlates with stage of breast cancer and may suggest that lack of KISS1 contributes to the systemic spread of breast cancer throughout the body and formation of distant metastases at sites such as the brain.
Figure 5.
The biological importance of KISS1 protein expression for brain dissemination and progression of breast cancer. (A) Impact of KISS1 expression on occurrence of brain metastasis. Median months from detection of breast cancer to dissemination to the brain were evaluated. KISS1 negative (N9) and KISS1 positive (N14) (t test, P = .04). (B) Dependence of breast cancer progression on expression of KISS1 marker in primary breast cancer patients. KISS1 expression was evaluated based on IHC staining and scored by using a 3-tier score system. All samples were divided between early stages (T0 and T1) and advanced (T2, T3, and T4) groups. Percentage of cases expressing differential levels of KISS1 in primary IDC samples is shown. Nonparametric Fisher exact test P = .044.
DISCUSSION
The aim of this study was to identify the association of KISS1 with development of brain metastatic disease in breast cancer patients. Development of metastases depends on a complex interplay between extracellular environment, cell surface, and cell propensity to proliferate in the new environment. Because each metastatic lesion represents a clone from the primary tumor that expresses the necessary proteins for seeding, we decided to compare protein expression between the primary tumors and metastases. Using a tissue microarray, tumor sections were stained with a KISS1 biomarker. We studied the expression of that biomarker in a large number of specimens including many validation steps. First, to create TMA, we used duplicate of each core to avoid nonfavorable manipulation during IHC. Second, in the TMA, we incorporated a panel of normal breast parenchyma to see nonspecific staining of normal tissue. Finally, each TMA staining was evaluated by staining of additional set of human specimens.
KISS1 first was identified as suppressor gene in a hybridization assay between metastatic and nonmetastatic cell lines and their derivates.29 Furthermore, in vivo inoculation of KISS1 expressing human breast carcinoma MDA-MB435 into mice led to suppression of lung metastases by 95%.14 The KISS1 gene codes several regulatory peptides called kisspeptins (KPs).30 Later studies found that of the 3 KPs, the largest cleavage product of KISS1 protein, KP-54, had metastatic suppressive potential. KP-54, KP-13, and KP-10 are ligands for the KISS1 receptor (previously GPR54, AXOR12, or hOT7T175). KPs and their receptors are expressed in normal placenta and are thought to play a role in controlling invasion of placenta into the endometrium. Recently, it has been shown that KISS1 regulates hormonal secretion and puberty by secretion in the hypothalamus and pituitary gland.31 Although there is some evidence that points toward an anti-invasive mechanism of KISS1 activity via down-regulation of MMP2 and MMP9, the major mechanism involves suppression of colonization.32
In our IHC study, KISS1 expression was different between the brain metastases, primary IDC, LN metastases, and normal tissue (P < .05). The most significant difference was down-regulation of KISS1 expression in BMHB compared with IDC (P = .007), or to normal parenchyma (P = .002). Although Stark et al33 detected low level of KISS1 mRNA expression in metastatic specimens which correlates with our IHC results, we believe that real-time PCR method is not sufficient for the characterization of KISS1 expression. Because mRNA of KISS1 does not necessarily reflect the post-translational modification and the amount of secreted protein, we therefore went one step further by also showing changes in protein expression.
Despite convincing preclinical models of metastases suppression by KISS1 not only in breast cancer, but also in ovarian cancer and melanoma, clinical translation is missing. In our study, we confirmed findings published by Martin et al,34 who showed the correlation between KISS1 expression and TNM grading. In addition, we illustrate the Spearman correlation between KISS1 and PR protein expressions for the clinical samples.
Histological staging will continue to be an integral part of clinical approach as we learn more of the metastasis process. Actually, in ovarian carcinoma and human hepatocelluar carcinoma, Prentice et al,35 and later Shengbing et al,21 found KISS1 expression to be an independent favorable prognostic marker. Unfortunately, in our study we are unable to provide Kaplan-Meier survival curve due to the fact that 95% of our patients with brain metastatic lesions remain alive. However, by measuring metastases free survival, we found a clear trend in prolongation of this interval with high KISS1 expression. Whether patients with low KISS1 expression need more frequent screening for recurrence is still too early to say, but a strong argument could be made that the use of such markers to customize cancer treatments could become more routine.
Acknowledgments
We thank Lori Loftis, Deidre Anderson, Zakiya Moton, and Tatyana Grushko (Department of Surgery, Pathology and Medicine, University of Chicago) for their precious help in obtaining clinicopathological data and technical assistance with the IRB application. The authors also extend thanks to Leslie Martin and LeiAnn Arceneaux (Pathology Core, University of Chicago) for their help with the tissue microarray database and experimental design. Finally, we are very grateful to the Drs. Theodore Karrison and Dingcai Cao (Department of Health Studies and Surgery, University of Chicago) for the help with statistical analysis.
FUNDING SUPPORT
This research was supported by the NCI (R01CA122930-MSL, R01CA138587-MSL, R01CA134981-DRW, P50CA125182-OIO), the American Cancer Society (RSG-07-276-01-MGO-MSL), and the National Foundation for Cancer Research (DRW).
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
REFERENCES
- 1.Kirsch DG, Ledezma CJ, Mathews CS, et al. Survival after brain metastases from breast cancer in the trastuzumab era. J Clin Oncol. 2005;23:2114–2116. doi: 10.1200/JCO.2005.05.249. author reply 2116–2117. [DOI] [PubMed] [Google Scholar]
- 2.Gori S, Rimondini S, De Angelis V, et al. Central nervous system metastases in HER-2 positive metastatic breast cancer patients treated with trastuzumab: incidence, survival, and risk factors. Oncologist. 2007;12:766–773. doi: 10.1634/theoncologist.12-7-766. [DOI] [PubMed] [Google Scholar]
- 3.Tham YL, Sexton K, Kramer R, Hilsenbeck S, Elledge R. Primary breast cancer phenotypes associated with propensity for central nervous system metastases. Cancer. 2006;107:696–704. doi: 10.1002/cncr.22041. [DOI] [PubMed] [Google Scholar]
- 4.Lassman AB, DeAngelis LM. Brain metastases. Neurol Clin. 2003;21:1–23. doi: 10.1016/s0733-8619(02)00035-x. vii. [DOI] [PubMed] [Google Scholar]
- 5.Chang EL, Lo S. Diagnosis and management of central nervous system metastases from breast cancer. Oncologist. 2003;8:398–410. doi: 10.1634/theoncologist.8-5-398. [DOI] [PubMed] [Google Scholar]
- 6.Guarneri V, Conte PF. The curability of breast cancer and the treatment of advanced disease. Eur J Nucl Med Mol Imaging. 2004;31(Suppl 1):S149–S161. doi: 10.1007/s00259-004-1538-5. [DOI] [PubMed] [Google Scholar]
- 7.Nishizuka I, Ishikawa T, Hamaguchi Y, et al. Analysis of gene expression involved in brain metastasis from breast cancer using cDNA microarray. Breast Cancer. 2002;9:26–32. doi: 10.1007/BF02967543. [DOI] [PubMed] [Google Scholar]
- 8.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 9.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmieri D, Fitzgerald D, Shreeve SM, et al. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Mol Cancer Res. 2009;7:1438–1445. doi: 10.1158/1541-7786.MCR-09-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaitan D, Sankpal UT, Weksler B, et al. Role of KCNMA1 gene in breast cancer invasion and metastasis to brain. BMC Cancer. 2009;9:258. doi: 10.1186/1471-2407-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge X, Yamamoto S, Tsutsumi S, et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Palmieri D, Bronder JL, Herring JM, et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007;67:4190–4198. doi: 10.1158/0008-5472.CAN-06-3316. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 15.McNally LR, Welch DR, Beck BH, et al. KISS1 overexpression suppresses metastasis of pancreatic adenocarcinoma in a xenograft mouse model. Clin Exp Metastasis. 2010;27:591–600. doi: 10.1007/s10585-010-9349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid K, Wang X, Haitel A, et al. KiSS-1 overexpression as an independent prognostic marker in hepatocellular carcinoma: an immunohistochemical study. Virchows Arch. 2007;450:143–149. doi: 10.1007/s00428-006-0352-9. [DOI] [PubMed] [Google Scholar]
- 17.Ikeguchi M, Yamaguchi K, Kaibara N. Clinical significance of the loss of KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:1379–1383. doi: 10.1158/1078-0432.ccr-1519-02. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Carbayo M, Capodieci P, Cordon-Cardo C. Tumor suppressor role of KiSS-1 in bladder cancer: loss of KiSS-1 expression is associated with bladder cancer progression and clinical outcome. Am J Pathol. 2003;162:609–617. doi: 10.1016/S0002-9440(10)63854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhar DK, Naora H, Kubota H, et al. Downregulation of KiSS-1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int J Cancer. 2004;111:868–872. doi: 10.1002/ijc.20357. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SL, Yu Y, Jiang T, Lin B, Gao H. Expression and significance of KiSS-1 and its receptor GPR54 mRNA in epithelial ovarian cancer. Zhonghua Fu Chan Ke Za Zhi. 2005;40:689–692. [PubMed] [Google Scholar]
- 21.Shengbing Z, Feng LJ, Bin W, Lingyun G, Aimin H. Expression of KiSS-1 gene and its role in invasion and metastasis of human hepatocellular carcinoma. Anat Rec (Hoboken) 2009;292:1128–1134. doi: 10.1002/ar.20950. [DOI] [PubMed] [Google Scholar]
- 22.Olbrich T, Ziegler E, Turk G, Schubert A, Emons G, Grundker C. Kisspeptin-10 inhibits bone-directed migration of GPR54-positive breast cancer cells: Evidence for a dose-window effect. Gynecol Oncol. 2010;119:571–578. doi: 10.1016/j.ygyno.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Yi X, Li CY, Zhang SH, Wang XH, Li ZQ, Yang F. Relationship and clinical significance of KiSS-1, nuclear factor kappa B (NF-kappaB), p50, and matrix metalloproteinase 9 expression in breast cancer. Zhonghua Bing Li Xue Za Zhi. 2008;37:238–242. [PubMed] [Google Scholar]
- 24.Ma PC, Tretiakova MS, MacKinnon AC, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008;47:1025–1037. doi: 10.1002/gcc.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strand C, Enell J, Hedenfalk I, Ferno M. RNA quality in frozen breast cancer samples and the influence on gene expression analysis--a comparison of 3 evaluation methods using microcapillary electrophoresis traces. BMC Mol Biol. 2007;8:38. doi: 10.1186/1471-2199-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeguchi M, Hirooka Y, Kaibara N. Quantitative reverse transcriptase polymerase chain reaction analysis for KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003;129:531–535. doi: 10.1007/s00432-003-0469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn O. Multiple Comparisons Using Rank Sums. Technometrics. 1964;6:241–252. [Google Scholar]
- 28.Glantz SA, Slinker BK. Primer of applied regression and analysis of variance. 2nd ed. New York, NY: McGraw-Hill; 1990. [Google Scholar]
- 29.Lee JH, Welch DR. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer. 1997;71:1035–1044. doi: 10.1002/(sici)1097-0215(19970611)71:6<1035::aid-ijc20>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 30.Kotani M, Detheux M, Vandenbogaerde A, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 31.Beck BH, Welch DR. The KISS1 metastasis suppressor: a good night kiss for disseminated cancer cells. Eur J Cancer. 2010;46:1283–1289. doi: 10.1016/j.ejca.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nash KT, Phadke PA, Navenot JM, et al. Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99:309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol. 2005;131:191–198. doi: 10.1007/s00432-004-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin TA, Watkins G, Jiang WG. KiSS-1 expression in human breast cancer. Clin Exp Metastasis. 2005;22:503–511. doi: 10.1007/s10585-005-4180-0. [DOI] [PubMed] [Google Scholar]
- 35.Prentice LM, Klausen C, Kalloger S, et al. Kisspeptin and GPR54 immunoreactivity in a cohort of 518 patients defines favourable prognosis and clear cell subtype in ovarian carcinoma. BMC Med. 2007;5:33. doi: 10.1186/1741-7015-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]