Abstract
We demonstrate the use of capillary zone electrophoresis with an electrokinetic sheath-flow electrospray interface coupled to a triple quadrupole mass spectrometer for the accurate and precise quantification of leu-enkaphalin in a complex mixture using multiple-reaction monitoring (MRM). Assay time is <6 minutes, with no re-equilibration required between runs. A standard curve of Leu-enkephalin was performed in the presence of a background tryptic digest of bovine albumin. We demonstrate reasonably reproducible peak heights (21% relative standard deviation), retention times (better than 1% relative standard deviation), and robust electrospray quality. Our limit-of detection (3σ) was 60 pM, which corresponds to the injection of 115 zeptomole of peptide. This is a 10–20-fold improvement in mass sensitivity than we have obtained by nano HPLC/MRM and substantially better than reported for LC/MS/MS. Further quantification was performed in the presence of stable-isotope labeled versions of the peptides; under these conditions, linearity was observed across nearly four orders of magnitude. The concentration detection limit was 240 pM for the stable-isotope labeled quantification.
Nanoscale reversed-phase liquid chromatography is routinely used as the primary separation technique in bottom-up proteomics (HPLC/MS/MS). 1–3 It has advantages in sensitivity and peak shape, and is amenable to in-line automation and low-volume injections. Efforts to improve the depth of protein identification have largely relied on multi-dimensional gradient separations and longer, shallower gradients.1–3 More recently, these techniques have been coupled to triple-quadrupole and hybrid linear ion-trap triple quadrupole (QqQ) mass spectrometers employing Multiple/Selected Reaction Monitoring (MRM/SRM) acquisitions to perform targeted detection and relative and absolute quantification of peptide mixtures in a complex biological matrix.4–6 One of the first studies to employ this technique in a relevant biological background was Anderson et al., in which they used targeted acquisition to monitor 47 proteins from high-abundance protein-depleted human plasma digested with trypsin.7
MRM is a gas-phase separation technique by which the first quadrupole (Q1) isolates a parent m/z, which is passed to the second quadrupole (Q2) for fragmentation, and the third quadrupole stabilizes a single fragment ion, which is allowed to pass to the detector. The combination of parent and fragment m/z is called a transition, which is tuned for each peptide of interest. This process can be accomplished in a few milliseconds, and generates a linear ion response enabling absolute-quantification with appropriate calibration curves and standards.7–9 Despite being performed at modest resolution, typically 0.7 amu, the specificity of a single transition in biologically relevant mixtures can be quite high. MRM differs from conventional tandem mass spectrometry because the third quadrupole is not scanned but instead tuned to the preselected ion of interest.
Efforts to provide open-access to to MRM data have resulted in the availability of large MRM repositories where specific transitions have been curated for many compounds and proteins of interest. The predominant information required to define a MRM transition is parent ion m/z, and one or more fragment m/z ion masses. Further refinement is provided by accurate retention times. This information is portable, and easily compared between different labs, experiments, tissue, and even mass-spectrometers. The Clinical Proteomic Tumor Analysis Consortium (CPTAC) initiative and the Institute for Systems Biology (ISB) have spent considerable effort in validating the portability of MRM transitions.10–12
Recently, our group and others have employed novel CE/CZE interfaces for ESI-MS/MS analysis to approach bottom-up proteomics analysis.13,14 The utility of this separation technique has been demonstrated on proteome samples of intermediate complexity, including bacterial secretomes and nuclear histone fractions.13,14 Since its development by Olivares et al., 15 on a single quadrupole instrument, CE/CZE-MS analyses of peptides and proteins have historically been acquired on high-resolution instrumentation and to our knowledge, have never been coupled to a high-sensitivity triple quadrupole for CE-ESI-MRM. CE has been employed as a separation technique utilizing MRM/SRM before, but this was limited to small-molecule analysis including antibiotics and nucleoside adducts.16–18 SRM experiments on peptides reported previously have all been performed on ion-trap style instrumentation, which approximates a MRM transition through the use of selective ion extraction.16,17 Preparative CE as a prelude to traditional LC/MS/MS (MRM) was used in a multi-dimensional approach by Kuzyk et al. but as a method of assessing peptide purity and not as the direct CE-ESI interface.8
Our objective here was to couple the higher separation resolution and mass-sensitivity of CZE-ESI to compound the sensitivity and specificity gains afforded by MRM on a modern triple quadrupole instrument. The versatility of MRM has been exploited to provide relative and absolute quantification across a myriad of biological contexts, and samples. This includes bacterial lysates, SDS-PAGE bands, blood plasma, and others.4,7–9,19,20 Recently Aebersold’s group demonstrated that MRM is highly adaptable and even preferred for non internal standard absolute quantification using the "best-flyer" hypothesis for bottom-up quantification.10,21 To this end, we developed and implemented an electrokinetically pumped sheath-flow CZE-ESI for the MRM analysis of Leu-enkephalin, a opioid neurotransmitter in the presence of a biological background. Leu-enkephalin has been separated using CZE, MEKC previously, and thus represents an ideal peptide from which to establish MRM-based quantitative CZE-ESI separations.22,23 We demonstrate that the CZE-ESI MRM response is linear at low concentrations and that it produces reproducible migration times. We furthermore demonstrate that CZE-ESI MRM is amenable to normalization both against the biological background and by using stable isotopic dilution via chemical labeling (mTRAQ).
EXPERIMENTAL SECTION
Chemicals and Materials
Bovine serum albumin, Leu-enkephalin, ammonium acetate, ammonium bicarbonate, and triethyl-ammonium bicarbonate (TEAB) were purchased from Sigma (St. Louis, MO). Sequencing-grade trypsin was purchased from Promega (Madison, WI). LC/MS grade solvents (water, acetonitrile, methanol) were purchased from Burdick and Jackson (B&J) (Morristown, NJ), Formic acid was purchased from Fisher (Waltham, MA). Fused capillaries were purchased from Polymicro Technologies (Phoenix, AZ). mTRAQ reagents Δ0 Da. and Δ8 Da. were purchased from ABSciex (Foster City, CA).
Sample Preparation
Bovine serum albumin (BSA, 2.2 mg/mL) was prepared in ammonium bicarbonate buffer (50 mM, pH 8.2) and then reduced at 95 °C for 5 minutes with 16 mM dithiothreitol. Iodoacetamide was added to a final concentration of 48 mM, and alkylation was performed at room temperature for 20 minutes in the dark. 50 µg of trypsin was added to 1 mL (2,200 µg) of BSA and digestion was performed for 10 hours at 37 °C. The BSA tryptic-digest was diluted to a final concentration of 66 nM in ammonium acetate buffer (10 mM, pH 5.5). A stock solution of Leu-enkephalin was made in water at a concentration of 50 µM, then diluted by the BSA tryptic-digest (66 nM) to a concentration range of 1 nM to 10 µM as working standards.
Compound Tuning
MRM source and fragment ion conditions were optimized while infusing a Leu-enkephalin solution. Briefly, 100 fmol/µL of N-YGGFL-OH was infused at 500 nL/min via syringe pump into a 5500 QTrap (AB Sciex). Automatic compound tuning (Analyst Compound Optimization V1.5.1) was used to determine Q1, Q3, and CE (collision energy, eV) for multiple fragment ions for the peptide. The [M+H]1+ ion at 556.20 m/z was the only observed parent ion for Leu-enkaphalin, and 397.20 m/z was the most intense fragment ion.
The transitions used in this study and their optimized parameters are summarized in Table 1. Unlike small molecule MRM, peptides typically exhibit a broad range of optimized values for most instrument parameters and constant values of DP, EP, CXP were used (Declustering Potential, Entrance Potential, Collision Exit Potential respectively) in this study. This set-up is consistent with that reported by the MacCoss group.24 MRM transitions for the SIS (Stable-Isotope-Standard) coded versions of Leu-enkephalin were determined by adding the appropriate mTRAQ masses (140, 148 Da.) and increasing th collision energy by 8 eV. MRM transitions to common peptides found within the biological matrix itself used for normalization were previously determined and are part of a common collection of MRM transitions used to QA/QC (Quality Assurance, Quality Control) QqQ instruments within our and other laboratories.7,12,20
Table 1.
Multiple Reaction Monitoring (MRM) Transitions and parameters used in this study.
| Name | Q1 | Q3 | Dwell time (msec) | Collision energy (eV) |
|---|---|---|---|---|
| Unlabeled Leu-enkephalin | 556.20 | 397.20 | 100 | 28.00 |
| [C][C]TESLVNR (BSA) | 569.75 | 818.44 | 100 | 33.90 |
| LVNELTEFAK (BSA) | 582.32 | 951.48 | 100 | 34.40 |
| AEFVEVTK (BSA) | 461.75 | 722.41 | 100 | 29.14 |
| Q1 | Q3 | Dwell time (msec) | Collision energy(eV) | |
| Δ0 labeled Leu-enkephalin | 696.37 | 537.28 | 50 | 46.00 |
| Δ0 labeled Leu-enkephalin | 696.37 | 418.21 | 50 | 49.00 |
| Δ0 labeled Leu-enkephalin | 696.37 | 565.27 | 50 | 39.00 |
| Δ8 labeled Leu-enkephalin | 704.37 | 426.21 | 50 | 49.96 |
| Δ8 labeled Leu-enkephalin | 704.37 | 573.27 | 50 | 40.65 |
| Δ8 labeled Leu-enkephalin | 704.37 | 545.40 | 50 | 45.69 |
| Unlabel Leu-enkephalin | 556.39 | 279.17 | 50 | 28.50 |
| [C][C]TESLVNR (BSA) | 569.75 | 818.44 | 50 | 33.90 |
| LVNELTEFAK (BSA) | 582.32 | 951.48 | 50 | 34.40 |
| AEFVEVTK (BSA) | 461.75 | 722.41 | 50 | 29.14 |
Shown here are the Q1, Q3, Dwell and CE eV values utilized in the unlabeled (Top) and Stable Isotope Standard (SIS) experiments performed.
Compound Labeling
Leu-enkephalin (N-YGGFL-OH) 1.2 mg was dissolved in 240 µL triethylammonium bicarbonate buffer (TEAB pH8.0) (0.5 M in Burdick & Jackson water). 1-unit vials of mTRAQ reagent Δ0 and Δ8 were allowed to reach room temperature and briefly centrifuged. 100 µL of anhydrous-isopropanol was added to each tube and mixed. 20 µL of the Leu-enkephalin solution in an additional 2 mM ammonium bicarbonate was added to lower the rate of spurious tyrosine hydroxyl labeling.25 The tube was allowed to sit at room temperature for 1 hour and the reaction was quenched by adding 100 µL of 0.5% formic acid. The solutions were dried in an Eppendorf Vacuum concentrator and reconstituted in 100 µL of 0.1% formic acid (1 mM). Labeling efficiency of both the Δ0 and Δ8 were determined by infusion of the solution through an ESI Q-TOF (Bruker-Daltronics, Billerica, MA) and determined to be approximately 50% by comparison of the 596.20 m/z and 696.20 m/z peak intensities. Previous labeling in the absence of free primary amine resulted in heterogeneous labeling of the N-YGGFL-OH producing singly, and doubly labeled species (Data not shown). The Leu-enkephalin labeled with mTRAQ reagent Δ8 (1 mM) was added to the BSA whole digest to a final concentration of 50 nM, which was used as the biological matrix. The Leu-enkephalin labeled with mTRAQ reagent Δ0 (1mM) was diluted by the matrix to the concentration range of 1.5 nM to 10 µM, as working standards.
CZE Acquisition
High voltages were provided by two Spellman CZE 1000R high-voltage power supplies. Electrospray was generated using an electrokinetically pumped sheath flow through a nanospray emitter.14,26,27 The emitter was borosilicate glass capillary (o.d. 1.0 mm, i.d. 0.75 mm, 10 cm length) pulled with a Sutter instrument P-1000 flaming/brown micropipette puller. The size of the emitter opening was 5–10 µm. Voltage programming was controlled by LabView software.
The separation capillary (i.d. 50 µm, o.d. 149 µm, length 30.0 cm) was uncoated. Samples were injected by applying 8 kV to the sample reservoir and 2 kV to the sheath flow electrospray reservoir for 3 seconds. For separation, 8 kV was applied on the injection end of the capillary and 2 kV on the sheath flow reservoir for 5 minutes. The separation buffer was 10 mM ammonium acetate (pH 5.5). The electrospray sheath flow liquid contained 50% v/v methanol, 50% v/v water, and 0.1% formic acid.
MS/MS (MRM) Parameters
MRM data were acquired in manual acquisition (tune) mode on a QTrap 5500 hybrid linear-ion trap triple quadrupole instrument (AB Sciex) with the Nanospray III DCI interface heater installed and set to 160°C. A custom electrokinetically-pumped nanospray source was attached via a nanospray source ring.26 Ion spray voltage was provided either through an external power supply or via a conductive clip from the internal power supply of the instrument. GS1 (Sheath gas) was disabled on the instrument and the Curtain gas was set within the software parameters to (5) arbitrary units, which is lower than the (10) setting allowed by default. Ion spray voltage was set to 2.2 kV, DP at 100 V, and CXP at 35 V. Resolution for all transitions was performed at Q1,Q3 Unit resolution (0.7 amu FWHM). All other parameters are set according to the default calibration under which the instrument was operating. Dwell times varied depending on the number of transitions in the method, but were typically 50–100 ms in order to acquire 8–12 points across each peak profile. Inter transition pause times were set to 4 ms; no period or MRM scheduling were used.
Peak Processing, Integration
Peaks were integrated and fit using Multiquant v2.0.2 (AB Sciex) with the MQ4 algorithm and a 3-point Gaussian fit. Data were fit to a linear calibration curve corrected by a MRM transition from bovine albumin Q1569.70--> Q3818.40 m/z or the heavy labeled mTRAQΔ8-YGGFL peak area added in at constant concentration. Linear calibration curves were weighted with a 1/X bias (X = concentraton of YGGFL) to reduce the influence of higher concentrations on the slope-intercept formula. Linear Data fits for the unlabeled N-YGGFL-OH were similar to the linear fits presented in S-3 for the Stable-Isotope-Dilution data. Data presented within Figure 2 were treated with a 3-point median filter and then convoluted with a 2 point (1.05s) Gaussian filter, then fit with a Gaussian function
| (1) |
where amplitude is the peak height, t is time, t0 is the peak center, σ is the peak standard deviation, and offset is the average background signal.
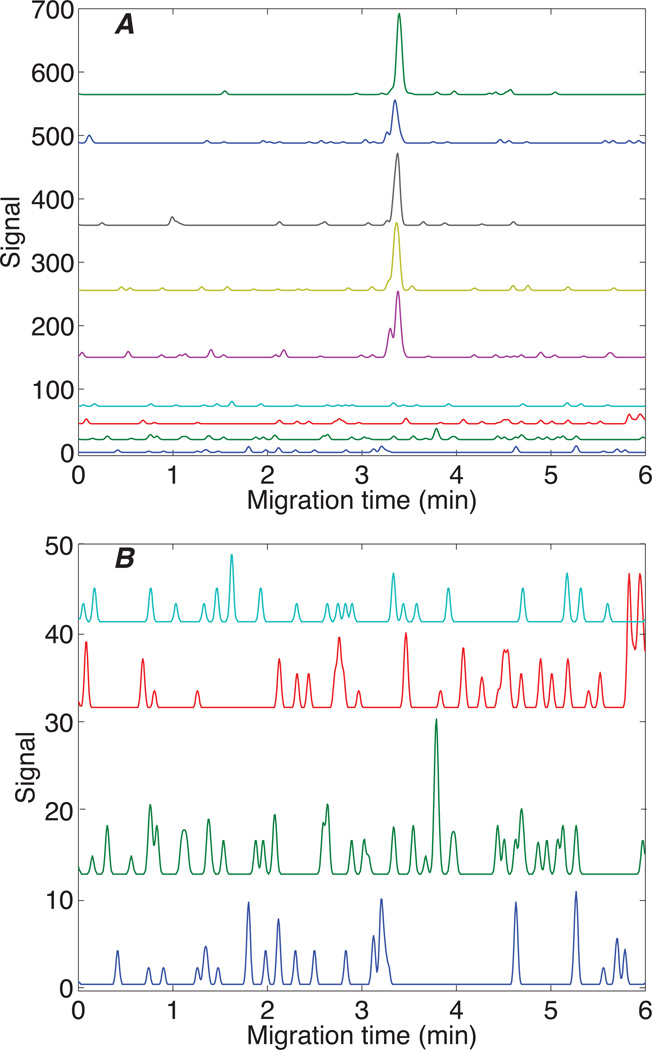
Figure 2.
Five replicate analysis of 1 nM (2 amol) leu-enkaphalin transition (top curves) and four blanks (bottom curves) using the 556.2 → 397.1 transition. Curves offset for clarity.
RESULTS AND DISCUSSION
Adaptation of CZE-ESI-QqQ Interface
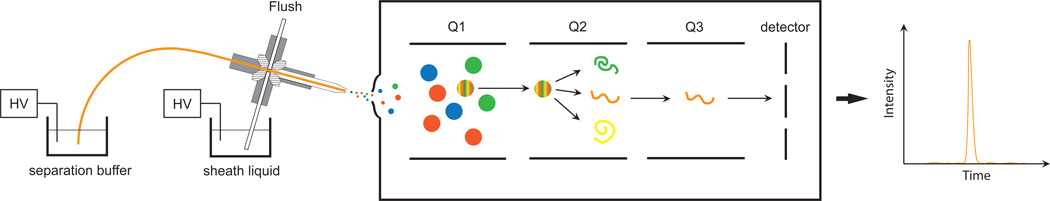
The electrophoresis system was assembled from components previously reported, with the following modifications.14,26 The existing CZE-ESI interface was moved to a 15° angle with respect to the source, and it was coupled the existing commercial nano-spray interface collar from AB Sciex. The separation buffer reservoir was made from a 20 mL scintillation vial molded to the source stage using Sugru putty (Sugru, UK). A schematic of the interface is illustrated as Scheme 1.
Scheme 1.
Schematic of CZE-ESI QqQ interface. Shown here is diagram of the modified CZE interface as in Wojcik et al.24 The CZE needle is positioned 15° offset with respect to the X axis of the orifice. The interface to the triple quadrupole (QqQ is a modified form of the nanopray III Sciex source, with the laminar-flow particle discriminator installed and heated to 160°C.
Quantitative-response Curves
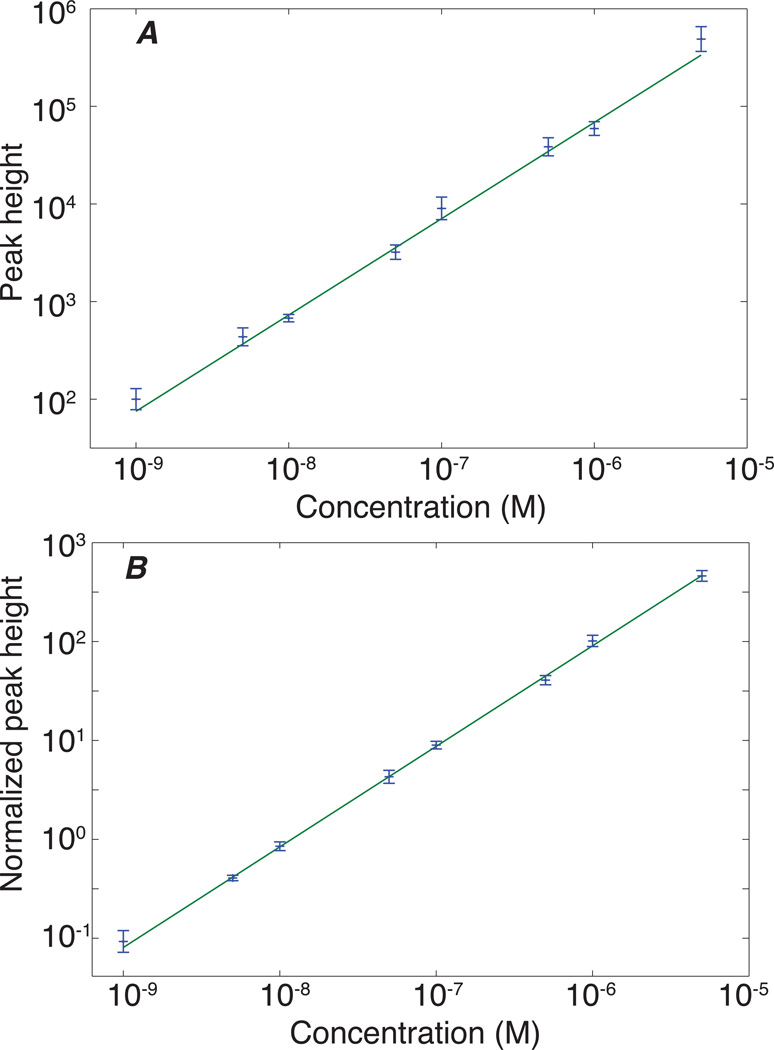
Calibration curves were generated for the Q1 556.20, --> Q3 397.20 m/z transition of Leu-enkephalin. This transition was treated as the quantifier transition, the remaining transitions were used as qualifiers. Five replicates were obtained for analyte concentrations ranging from 10−9 to 5×10−6 M, spanning 3.5 orders of magnitude. Testing for linearity across a wide dynamic range is not straightforward. A linear least squares fit to a plot of peak amplitude versus concentration will be dominated by the highest concentration signal. Instead, analysis of the log-log plot of the calibration curve can provide useful insight into the data’s linearity. A linear fit that produces a slope of one in the log-log plot confirms the original data’s linearity. Table 2 summarizes the results. The slope of the log-log plot is within experimental error of 1.0, which confirms a linear relationship between the signal and peptide concentration across the concentration range. However, the values of the reduced chi-squared statistic were much larger than expected, which suggests that there is a source of systematic error.
Table 2.
Results of a linear least squares fit to the log-log calibration curve (Fig 1).
| transition | Q1556.20 → Q3397.20 m/z |
| slope of log-log plot | 0.987 ± 0.027 |
| reduced chi-squared | 5.5 |
| Mean RSD peak height | 21% |
| Mean RSD peak area | 21% |
| Migration time | 3.408 ± 0.038 min |
| Peak width | 0.0305 ± 0.0036 min (1.8 ± 0.2 s) |
| transition | Q1 556.20 → Q3 397.20 m/z normalized with Q1 569.80→ Q3 818.40 m/z |
| slope of log-log plot | 1.017 ± 0.015 |
| reduced chi-squared | 0.342 |
| Mean RSD peak height | 29% |
| Mean RSD peak area | 28% |
| concentration detection limit | 60 pM |
| mass detection limit | 115 zmol |
The obvious source of that systematic error is uncontrolled variation in injection volume. We also measured three transitions associated with the BSA digest (Q1 569.70, --> Q3 818.40; Q1 582.32, --> Q3 951.48; and Q1 461.75, --> Q3 722.40 m/z). We normalized the peak height obtained from Leu-enkephalin with the peak height from the Q1 569.75, --> Q3 818.44 m/z transition. The normalized data are under control; the reduced chi squared statistic for the log-log plot is close to 1 (p~0.90), and the slope is within experimental error of unity. Figure 1 presents log-log plot of the calibration curve for both the pre-normalized and normalized data. Normalizing the data to invariant proteins within the biological matrix is one of the best strategies to correct for errors in injection in both CZE and LC/MS/MS data. This strategy has been successfully employed to correct for theses variances in other LC/MS/MS MRM studies including both label-free and labeled quantification.7,8,20,21 In order to assess the quality of the electrospray plume, we monitored the peak-shape of the underlying MRM transition itself. Since this work utilized a beam-line style instrument, heterogenity in the spray quality itself might be observed as large swings in counts/s−1, signal drop out, etc. None of these spray artifacts were not observed. An example of the raw signal from a single transition of Leu-enkephalin is shown as S-4.
Figure 1.
Log-log calibration curve for Lue enkephalin in 66 nM BSA digest for the 556.2 → 397.1 transition. Propagation of errors was used to estimate the uncertainty in the log(signal). The line is the result of a linear least squares fit of a first order polynomial to the data. The slope of the line and the reduced chi-squared for the fit are included in Table 1. A – raw data. B – normalized to the 569.75→ 818.44 transition from a BSA peptide, which corrects for variations in injection volume.
Reproducibility of Migration Time
Migration times were determined using the peak center (t0) and peak widths using the sigma value (σ) from the Gaussian fit using equation 1. The migration time was remarkably reproducible. The average migration time was 3.408 ± 0.038 minutes, corresponding to a 1% relative standard deviation. A major source of uncertainty in the migration time was the peak width itself, which averaged 0.0305 ± 0.0036 min. This highly reproducible migration time will be valuable in the analysis of complex samples, where an ion’s transition is probed only when the parent ion is migrating from the capillary. This precision provides an enhanced level of confidence in discriminating between correct from incorrect MRM transitions.5,12,21,24 Precise migration times also enable the use of period-based and interleaved dwell time approaches that can increase the number of transitions monitored into the hundreds or even thousands while providing improved ion-statistics by increasing the effective time spent per peptide transition.28,29
Detection Limit
The noise in the baseline was estimated as the standard deviation of a set of four filtered blanks. The concentration detection limit was 35 pM for the Q1 556.20 --> Q3 397.20 m/z transition. Figure 2 presents five electropherograms generated by injection of a 1 nM (8 amol) sample injection. Four blanks are also presented in the figure. This sensitivity compares very favorably to those observed in nano-HPLC- MRM studies in comparable biological background. Our detection and quantification limits are as much as 1.5 -- 2 orders of magnitude superior to those reported for peptides from similar complexity in SDS-PAGE extracted protein digests.19,30 Sinnaeve et al. reported capillary LC/MS (MRM) LOQ values for Leu-enkephalin of 0.5–1 pmol/mL and mass detection limits of 500 amol.17,31 Our absolute mass detection limit (3 σ) for Leu-Enkephalin was three orders of magnitude superior, 335 zeptomoles, (Table 2).
Stable Isotope labeling
In order to further characterize potential sources of peak-shape error, we also acquired comparable calibration curves of an isotopically-light labeled (mTRAQ) Leu-enkephalin in the presence of a constant background Internal Standard (IS) of a heavy isotope form of the mTRAQ label using stable isotope dilution to normalize the data.32,33 This compound represents a nearly ideal internal-standard as the heavy and light forms of the peptide differ only due to the addition of stable isotopes, and thus have identical chemical, ionization and migration-time properties. As indicated in Table 2, detection limits for this analysis (35 pM) were degraded vs. the unlabeled peptide mixture (240 pM) with a mass detection limit of 2 amol. As expected, the ideal internal standard had weighted linear concentration curves with R2 values of 0.9997 (weighted ln(x)) Slope =0.0168x+/−0.98598). In particular the RSD for these corrected data was < 4% for all concentrations and 0.48% for the lowest concentration of Leu-enkephalin observed (1.5 fmol/µL) when plotted as log data as in Fig 1 (slope not shown). In order to represent the data typical for that observed in LC/MS-MRM analysis we re-plotted the data against a linear slope, weighted 1/x for concentration. The summary of these data is shown as S-1, S-2. This includes SIS (Stable Isotope Standard) normalized curve and integrated peaks for one concentration point (S-1) and the summary table for the integrated data of triplicate injections over 3.5 orders of magnitude (S-2). Linearity for these data were also excellent with a slope (weighted 1/x =0.020x+(−0.017). r=0.99933. RSD for the linear plotting ranged from 0.85 -- 23.8% , demonstrating excellent inter-run reproducibility. Accuracy was good, with an average accuracy of 99.85% (σ=3). The lowest quantified point has poor accuracy overall (179.95%). Removal of the lowest quantified point, demonstrates excellent accuracy for all points with an average accuracy of 89.8% and a RSD of 16%. Area response [ratio] Analyte peak area / IS peak area S-3 was excellent with a RSD of 0.9% at 3σ.
There are two obvious sources for the difference in sensitivity between the stable isotope and unlabeled forms of the analysis. First, these data were not corrected for the approximately 50% labeling efficiency engineered into the mTRAQ standard. In labeling a solution containing just a single peptide, tyrosine hydroxyl-mediated NHS ester (N-hydroxysuccinimide) formation occurred at a high rate generating a mixture of unlabeled, singly, and doubly labeled forms of the N-YGGFL-OH peptide.25 This observation confounded attempts to accurately assess an absolute detection and quantification limit using the completely labeled, but heterogeneous peptide; thus the partially labeled, but quantifiable fraction was used. Labeling efficiency accounts for a factor of two degradation in detection limit. In addition, since there are two chemically identical peptides eluting simultaneously in the labeled experiment, one could expect an approximately 2-fold effect from ion-suppression. These two potential sources of error may explain all of the quantitative differences observed from the labeled and unlabeled forms of the quantification.
SUMMARY AND CONCLUSIONS
The application of capillary electrophoresis as a separation technique to complement existing MRM-based peptide quantification has been demonstrated with high reproducibility and sensitivity, using an apparatus that can be easily interfaced to multiple mass spectrometers. We observe highly sensitive detection and quantification at the pM level of Leu-enkephalin, a common opioid neurotransmitter peptide, in the presence of a large excess of bovine albumin digested with trypsin. This sample simulates the biological context under which most bottom-up proteomics based quantification is performed. The relatively robust accuracy in peak height (RSD 21%), and excellent reproducibility in migration time (RSD <1%) in a contextual background suggest a broad applicability for bottom-up and endogenous peptide quantification by CZE-ESI MRM based approaches. Further quantitative accuracy was demonstrated utilizing stable-isotope dilution, which is the de facto method for high precision and accurate LC/MS/MS peptide quantification. This method is well suited to implementing CE/CZE as a complementary method to LC-based approaches for biological quantification.34
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Elliott Jones, Loren Olsen and Dr. Sahana Mollah (AB Sciex) for donation of the nanospray source ring. We also thank Dr. Bill Boggess in the Notre Dame Mass Spectrometry and Proteomics Facility. We also acknowledge funding from the National Institutes of Health R01GM096767 (NJD).
REFERENCES
- 1.Washburn MP, Wolters D, Yates JR., 3rd Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 2.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Fröhlich F, Cox J, Mann M. Mol. Cell Proteomics. 2011;10:M110.003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolters DA, Washburn MP, Yates JR., 3rd Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 4.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Mol. Cell Proteomics. 2005;4:1134–1144. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths JR, Unwin RD, Evans CA, Leech SH, Corfe BM, Whetton AD. J. Am. Soc. Mass Spectrom. 2007;18:1423–1428. doi: 10.1016/j.jasms.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Anderson L, Hunter CL. Mol. Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Mol. Cell Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CY, Picotti P, Huettenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R, Vitek O. Molecular & Cellular Proteomics: MCP. 2011 doi: 10.1074/mcp.M111.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham A-JL, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faserl K, Sarg B, Kremser L, Lindner H. Anal. Chem. 2011;83:7297–7305. doi: 10.1021/ac2010372. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Champion MM, Sun L, Champion PAD, Wojcik R, Dovichi NJ. Anal. Chem. 2012;84:1617–1622. doi: 10.1021/ac202899p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivares JA, Nguyen NT, Yonker CR, Smith RD. Anal. Chem. 1987;59:1230–1232. [Google Scholar]
- 16.Soto-Chinchilla JJ, García-Campaña AM, Gámiz-Gracia L. Electrophoresis. 2007;28:4164–4172. doi: 10.1002/elps.200600848. [DOI] [PubMed] [Google Scholar]
- 17.Sinnaeve BA, Storme ML, Van Bocxlaer JF. J Sep Sci. 2005;28:1779–1784. doi: 10.1002/jssc.200500114. [DOI] [PubMed] [Google Scholar]
- 18.Lee ED, Mück W, Henion JD, Covey TR. J. Chromatogr. 1988;458:313–321. doi: 10.1016/s0021-9673(00)90575-2. [DOI] [PubMed] [Google Scholar]
- 19.Llarrull LI, Toth M, Champion MM, Mobashery S. J. Biol. Chem. 2011;286:38148–38158. doi: 10.1074/jbc.M111.288985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiGiuseppe Champion PA, Champion MM, Manzanillo P, Cox JS. Mol. Microbiol. 2009;73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig C, Claassen M, Schmidt A, Aebersold R. Mol. Cell Proteomics. 2012;11:M111.013987. doi: 10.1074/mcp.M111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beijersten I, Westerlund D. Anal. Chem. 1993;65:3484–3488. doi: 10.1021/ac00071a025. [DOI] [PubMed] [Google Scholar]
- 23.Banks JF, Jr, Dresch T. Anal. Chem. 1996;68:1480–1485. doi: 10.1021/ac9509824. [DOI] [PubMed] [Google Scholar]
- 24.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, Carr SA, Maccoss MJ. Anal. Chem. 2010;82:10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalkhof S, Sinz A. Anal Bioanal Chem. 2008;392:305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- 26.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Commun. Mass Spectrom. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 27.Wojcik R, Li Y, Maccoss MJ, Dovichi NJ. Talanta. 2012;88:324–329. doi: 10.1016/j.talanta.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertsch A, Jung S, Zerck A, Pfeifer N, Nahnsen S, Henneges C, Nordheim A, Kohlbacher O. J. Proteome Res. 2010;9:2696–2704. doi: 10.1021/pr1001803. [DOI] [PubMed] [Google Scholar]
- 29.Dresen S, Ferreirós N, Gnann H, Zimmermann R, Weinmann W. Anal Bioanal Chem. 2010;396:2425–2434. doi: 10.1007/s00216-010-3485-2. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Gruidl M, Remily-Wood E, Liu RZ, Eschrich S, Lloyd M, Nasir A, Bui MM, Huang E, Shibata D, Yeatman T, Koomen JM. J. Proteome Res. 2010;9:4215–4227. doi: 10.1021/pr1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinnaeve BA, Van Bocxlaer JF. J Chromatogr A. 2004;1058:113–119. [PubMed] [Google Scholar]
- 32.DeSouza LV, Taylor AM, Li W, Minkoff MS, Romaschin AD, Colgan TJ, Siu KWM. J. Proteome Res. 2008;7:3525–3534. doi: 10.1021/pr800312m. [DOI] [PubMed] [Google Scholar]
- 33.Kirkpatrick DS, Gerber SA, Gygi SP. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Ban E, Park SH, Kang MJ, Lee HJ, Song EJ, Yoo YS. Electrophoresis. 2012;33:2–13. doi: 10.1002/elps.201100344. [DOI] [PubMed] [Google Scholar]