Key Points
miRNAs activate NK cells through a TLR–NF-κB signaling pathway and may have therapeutic applications in cancer.
Abstract
MicroRNAs (miRNAs) bind to complementary sequences of target mRNAs, resulting in translational repression or target degradation and thus gene silencing. miRNAs are abundant in circulating blood, yet it is not known whether, as a class of regulatory molecules, they interact with human natural killer (NK) cells. Here we found that the treatment of human NK cells with several mature miRNAs in the presence of a low concentration of interleukin-12 induced CD69 expression, interferon-γ production, and degranulation marker CD107a expression. In vivo, infusion of several miRNAs alone in murine peripheral blood also resulted in comparable NK-cell activation, but not T-cell activation. Furthermore, miRNA administration significantly protected mice from tumor development in an NK cell–dependent manner. Mechanistically, we found that miRNA stimulation led to downstream activation of nuclear factor κB (NF-κB), an effect that was blunted by a block in Toll-like receptor 1(TLR1) signaling and attenuated in lymphoma patients. Knockdown of TLR1 resulted in less activation by miRNAs. Collectively, we show that miRNAs have a capacity to selectively activate innate immune effector cells that is, at least in part, via the TLR1–NF-κB signaling pathway. This may be important in the normal host defense against infection and/or malignant transformation.
Introduction
MicroRNAs (miRNAs) are small, noncoding RNAs first described in Caenorhabditis elegans in 1993.1 Later, it was found that their deletion or deregulation was associated with cancer development.2 Starting with about 70 to 100 nucleotides (nt), the pre-miRNA are processed into an 18- to 25-nt, mature, single-stranded RNA by the ribonuclease Dicer and the RNA-induced silencing complex.3 The mature miRNAs bind to 3′ untranslated regions of target mRNAs, either inducing degradation of mRNAs in the presence of the RNA-induced silencing complex or blocking protein translation processes.4 Computational analysis has predicted that more than one-third of human protein-coding genes (including some tumor suppressor genes and oncogenes) are regulated by miRNAs.5 miRNAs play essential and pleiotropic roles in both physiologic and disease processes, including normal cell development as well as malignant transformation and metastasis.6,7
More recent evidence shows that miRNAs reside and circulate in the blood and other body fluids of both healthy donors and patients with diseases including cancer.8 These miRNAs, which are detectable in serum and plasma, can serve as potential markers for cancer diagnosis, prognosis, and even targets for treatment.9 For example, levels of miR-155 increase in patients with breast cancer and lung cancer in comparison with levels found in healthy donors, and changes in miR-155 expression levels also correlate with the metastasis of breast cancer.10,11 Similarly, significantly higher levels of circulating miR-21 are found in patients with hepatocellular carcinoma and breast cancer in comparison with healthy donors.12,13 Interestingly, expression of miR-15b in the cerebrospinal fluid is strikingly increased in patients with glioma compared with healthy donors.14 miR-122 is the most dominant miRNA found in the liver and constitutes 70% of the cloned hepatic miRNA in adult mice.15 In patients with chronic hepatitis or hepatocellular carcinoma, levels of circulating miR-122 were demonstrated to be higher in comparison with levels in healthy donors.16,17 Some miRNAs are also found to be downregulated in cancer patients.18 However, the biological functions of the circulating miRNAs remain largely unknown in both normal and disease states.
Natural killer (NK) cells are a critical component of innate immunity in that they often provide the first line of defense against malignant transformation and viral infection. We and others found that miRNAs are expressed by NK cells and intrinsically regulate their function and development.19-22 However, to the best of our knowledge, it has not been determined whether extrinsic or circulating miRNAs are able to activate NK cells. In this study, using both in vitro and in vivo approaches, we found that synthetic circulating miRNAs and miRNA-containing exosomes freshly isolated from healthy donors have a capacity to activate NK cells. This appears to occur via a Toll-like receptor (TLR) signaling pathway. Our study suggests that we have identified a novel role for miRNAs in the innate immune response.
Methods
Cell culture
Primary human NK cells, human peripheral blood mononuclear cells (PBMCs), and mouse spleen cells were cultured in complete RPMI 1640 media (Invitrogen) containing 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). Cells were cultured at 37°C and supplemented with 5% carbon dioxide. The human interleukin (IL)-2–dependent NK cell line NK-92, a generous gift of Dr Hans G. Klingemann (Rush University Medical Center, Chicago, IL),23 was cultured similarly, except that 20% FBS was used.
Mice
Eight-week-old C57BL/6 and athymic nude mice were obtained from the Jackson Laboratory. The Ohio State University Animal Care and Use Committee approved all animal work.
Human NK-cell isolation
Human NK cells were first enriched from the peripheral blood of healthy donors (American Red Cross) with the RosetteSep NK-cell enrichment mixture (StemCell Technologies) and Ficoll-Paque Plus (Amersham) centrifugation. The enriched NK cells were further purified by positive selection using anti-CD56 magnetic-activated cell sorting beads (Miltenyi Biotec). NK cells with purity greater than 99%, which was confirmed by flow cytometry, were used (supplemental Figure 1). PBMCs from lymphoma patients were first stained with CD3-PE and CD19-PE and subjected to negative selection for NK cells using magnetic-activated cell sorting LS columns (Miltenyi Biotec). The enriched NK cells were then further purified by fluorescence-activated cell sorting (FACS) after being stained with CD3–fluorescein isothiocyanate (FITC) and CD56-allophycocyanin (APC) antibodies (Abs). For healthy donors, PBMCs were directly stained with CD3-FITC and CD56-APC Abs and subjected to sorting for NK cells. The purity of sorted NK cells immediately lysed for real-time reverse transcriptase polymerase chain reaction (RT-PCR) analysis is greater than 97%. The Ohio State University Institutional Review Board approved all human work.
Cell stimulation by miRNAs
Purified human primary NK cells, PBMCs, or freshly isolated mouse splenocytes were resuspended at 1 × 106 cells/100 μL in complete RPMI 1640 media, then plated on a 96-well plate in the presence of recombinant human (rh) IL-12 (Genetics Institute Inc). miRNAs were placed in complex with DOTAP, a cationic liposomal formulation (Roche), according to the manufacturer’s instruction. Briefly, 2 μg miRNAs were dissolved in 25 μL hepes-buffered saline, combined with 10 μg DOTAP solution in 25 μL hepes-buffered saline, and incubated for 15 minutes. Next, 50 μL of complete RPMI 1640 media were added to the miRNAs–DOTAP mixture and mixed well before being added to each well preseeded with cells. The final concentration of DOTAP was 50 μg/mL, and the final concentration of miRNA was 10 μg/mL. Vehicle control for all experiments consisted of 50 μg/mL of DOTAP. RNA control for all experiments consisted of 50 μg/mL of DOTAP complexed with 10 μg/mL of a nonspecific, single-stranded control RNA called RNA41, which is similar in size to miRNA.24 The cells were stimulated for 36 hours (unless specified) with miRNAs and 2.5 ng/mL rhIL-12. The dose of 2.5 ng/mL rhIL-12 is less than what is typically used for stimulation of NK cells (10 ng/mL). For the TLR blocking assay, the aforementioned cells were preincubated with TLR1 (InvivoGen), TLR3 (Hycult Biotech), or TLR6 (InvivoGen) blocking Abs or control immunoglobulin G (IgG; Equitech Bio) at a concentration of 10 μg/mL for 1.5 hours prior to stimulation with miRNAs. The blocking Abs were also kept in the culture during the stimulation with miRNAs.
Flow cytometric analysis
The stimulated cells were stained with monoclonal antibodies (mAbs) at 4°C for 20 minutes, washed with phosphate-buffered saline (PBS) and fixed with 1% formalin, followed by FACS analysis using an LSRII (BD Bioscience) to detect surface expression of each antigen. Human NK cells were gated as CD56+CD3- and mouse NK cells were gated as NK1.1+CD3− cells within the lymphocyte gate. The following anti-human mAbs used were: CD3-FITC, CD56-APC, CD107a-PE, and CD69-PE. The following anti-mouse mAbs used were: CD3-FITC, NK1.1-APC, CD69-PE, and CD107a-PE. All mAbs were purchased from BD Bioscience.
Real-time reverse-transcriptase and enzyme-linked immunosorbent assay
Total RNA was extracted and reverse transcribed into cDNA. The interferon (IFN)-γ mRNA expression level was determined by real-time RT-PCR using Taqman PCR Master Mix (Applied Biosystems), while TLRs, p65, and IRAK1 mRNA expression levels were assessed using SYBR Green Master Mix (Applied Biosystems). Expression levels were normalized to an 18S or β-actin internal control and then analyzed by the ΔΔCt method. The sequences of primers and/or probes are available upon request. To detect secreted IFN-γ protein, cells were stimulated with miRNAs and IL-12 as described above, and cell-free supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA) as previously described.25
CD107a degranulation assay
To detect the capacity of NK cells for cytotoxic activity, human NK cells and murine splenocytes were stimulated in vitro and treated in vivo with miRNAs, respectively. A total of 0.5 million miRNA-stimulated human and murine NK cells were then incubated at the ratio of 1:1 with K562 and YAC-1 target cells, respectively. However, only human NK cells were cocultured with a low concentration (2.5 ng/mL) of IL-12. Subsequently, 2.5 μL anti-human or 1 μL anti-mouse CD107a antibody was added to this coculture for 1 hour. Then 1 μL/mL of the secretion inhibitor, monensin (eBioScience), was added for an additional incubation of 3 hours. The cells were washed with PBS and stained with CD3 (human and mouse) and CD56 (human) or NK1.1 mAbs (mouse) and then analyzed via flow cytometry using an LSRII.
In vivo miRNA stimulation
The complexes of miRNAs and vehicle, Lipofectamine 2000 (Invitrogen), were prepared according to the manufacturer’s instruction. Briefly, 30 μL of Lipofectamine 2000 was mixed with 20 μg RNA-Ctl, miR-122, or miR-15b dissolved in 170 μL PBS. The liposome complexes were administered intravenously (200 μL/mouse) into mice through tail veins. Four days later, the injected mice were sacrificed, and total splenocytes were isolated for degranulation assay or stained with aforementioned mAbs for flow cytometric analysis.
Immunoblotting
Total protein lysates were prepared with T-Per (tissue protein extraction reagent; ThermoScientific) supplemented with proteinase and phosphatase inhibitors. Proteins were resolved on a 4% to 20% SDS-PAGE gel and transferred onto PVDF membranes (Amersham). The Abs used for blotting were TLR1 (Cell Signaling), phospho (p)-p65 (Cell Signaling), p65 (Cell Signaling), and β-actin (Santa Cruz).
TLR1 shRNA knockdown
A TLR1 short hairpin RNA (shRNA) plasmid was constructed by inserting RNA interference sequences into pSUPER-retrovirus vector expressing green fluorescent protein (GFP). Viruses were prepared by transfecting the shRNA plasmid and packaging plasmids into phoenix cells. Infection was performed as previously described.26 Briefly, NK-92 cells were cocultured with virus-containing media and centrifuged at 1800 rpm at 32°C for 45 minutes, then incubated for 2 to 4 hours at 32°C. This infection cycle was repeated twice. Upon completion of this infection, GFP-positive cells were sorted on a FACSAria II cell sorter (BD Bioscience). Knockdown of TLR1 in the sorted NK-92 cells was confirmed by immunoblotting.
Bioluminescent imaging
Balb/C mice–derived A20 B-cell lymphoma cells were retrovirally transduced with a Pinco-pGL3-luciferase (luc)/GFP plasmid, and the GFP-positive cells were sorted by a FACSAria II cell sorter (BD Biosciences). Then 1 × 105 luc-expressing A20 cells were injected into each athymic nude mice via tail veins. miRNAs were administered via a tail-vein injection at the following 3 time points: 3 days prior to, 4 days after, and 18 days after A20 implantation. For NK depletion, each mouse was administered 200 μg TM-β1 (IL-2/15Rβ) mAb intraperitoneally on day 6 prior to and day 3 after A20 cells implantation, 200 μm per time. Three weeks after A20 implantation, mice were injected with luciferin (150 mg/kg body weight; Gold Biotechnology), anesthetized with isoflurane, and imaged using an IVIS-100 imaging system (Xenogen).
Statistics
Data were compared by Student 2-tailed t test. P < .05 was considered statistically significant.
Results
miRNAs enhance surface expression of the activation marker, CD69, on human NK cells
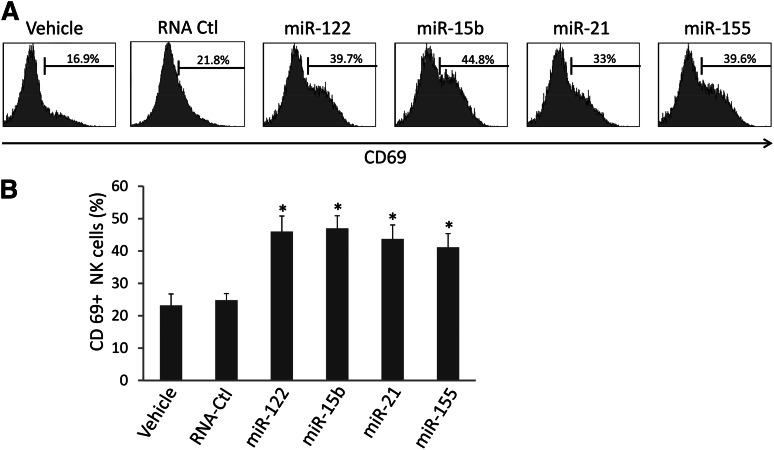
We synthesized several miRNAs previously demonstrated to be present in the circulation, including miR-122, miR-15b, miR-21, and miR-155, and placed them in culture with highly purified human NK cells (supplemental Figure 1). Alone, each miRNA was unable to activate NK cells (data not shown). However, when placed in culture for 36 hours in the presence of a low concentration of IL-12 (2.5 ng/mL), we observed a twofold to threefold increase in the surface expression of CD69 for each miRNA (Figure 1A-B). In contrast, a nonspecific, single-stranded control RNA termed RNA41,24 which is of similar size to miRNAs, prepared in an identical fashion, and incubated for an equal amount of time at an identical concentration with human NK cells, did not significantly induce CD69 surface expression (Figure 1A-B). Similar results were also obtained with either a longer (72 hours) or shorter (24 hours) exposure (not shown). To further verify whether this effect was sequence specific, we mutated miR-122 and miR-15b sequences by substituting uridine (U) with guanosine (G) in the miRNA sequence. We found that these 2 mutated miRNAs completely lost their capability to activate NK cells (supplemental Figure 2).
Figure 1.
miRNAs increase CD69 surface expression on NK cells. (A) Highly purified (≥99%) human NK cells were treated with miRNAs, DOTAP vehicle control, or nonspecific, single-stranded RNA, RNA41 (RNA-Ctl),24 for 36 hours in the presence of a low concentration of IL-12 (2.5 ng/mL). The treated cells were then subjected to flow cytometric analysis to determine the percentage of CD69+ cells. Representative data from 1 of 6 donors with similar results are shown. (B) Summary of data from 4 donors obtained in 1 experiment. *P < .05 and error bars represent standard deviation.
Consistent with these data obtained with synthetic miRNAs, we also found that CD-9–expressing exosomes isolated from healthy donor serum and containing relatively high concentrations of miRNAs, such as miR-122 and miR-21, were able to significantly activate NK cells purified from the corresponding (autologous) donors (supplemental Figure 3).
Treatment with miRNAs augments human NK-cell IFN-γ production and degranulation
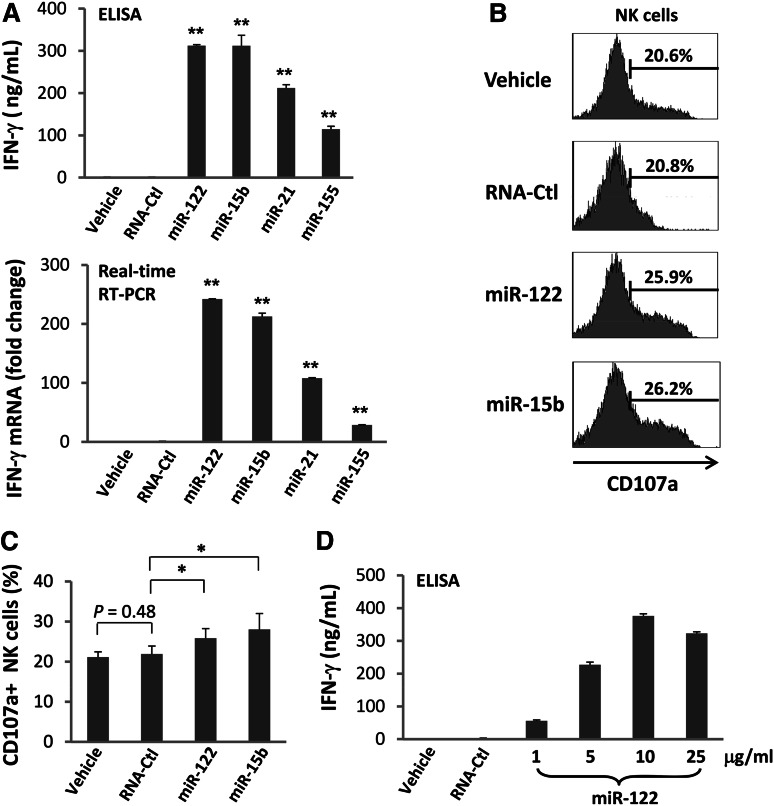
High expression of activation marker CD69 on NK cells is usually coupled with functional activation.27 We therefore assessed primary human NK cells for secretion of IFN-γ following miRNA stimulation. We observed variable but consistently significant increases of IFN-γ protein secretion in each instance (Figure 2A, upper panel). The data were confirmed by real-time RT-PCR (Figure 2A, lower panel) and were also found to be dependent of the concentration of miRNAs (Figure 2D). Similar results were observed in the NK cell line NK-92 (supplemental Figure 4). Although the extent of NK-92 activation was less, most likely due to IL-2 prestimulation of this IL-2–dependent cell line, these data exclude the possibility that the activation of NK cells by miRNAs was caused by contamination of other immune cell subsets. Consistent with the results of CD69 surface expression, the control RNA41 did not induce IFN-γ expression in NK cells (Figure 2A). Hereafter, we included only miR-122 and miR-15b for our experimental conditions in assessing NK-cell activation because of their stronger stimulation of IFN-γ expression when compared with the other 2 miRNAs (miR-21 and miR-155; Figure 2A).
Figure 2.
miRNAs increase IFN-γ production by NK cells. (A) Highly purified (≥99%) human NK cells were treated with miRNAs, DOTAP vehicle control, or nonspecific, single-stranded RNA, RNA41 (RNA-Ctl),24 for 36 hours in the presence of a low concentration of IL-12 (2.5 ng/mL). Supernatants were harvested for ELISA and cells were harvested for real-time RT-PCR analysis to determine the levels of IFN-γ secretion (upper panel) and gene expression (lower panel), respectively. Gene expression of the vehicle was normalized to 1. Experimental values are each presented as fold change compared with that of the vehicle. Data shown represent 1 of 3 donors with similar results. (B-C) Highly purified human NK cells were treated with miRNAs or DOTAP vehicle control for 36 hours in the presence of a low concentration of IL-12 (2.5 ng/mL). The cells were then incubated with K562 tumor cells at a ratio of 1:1 (effector: target). After 4 hours, CD107a expression was assessed by flow cytometric analysis. Shown in C are representative data from 1 of 5 donors with similar results. (C) Summary data of 5 donors. In all panels, error bars represent standard deviation. *P < .05 and **P < .01. (D) Cells were treated and data were collected as described in (A), with the exception that concentrations of miR-122 were varied, as indicated on the X-axis. Data suggest that miR-122 activates NK cells in a dose-dependent fashion.
Upon activation with cytokines, human CD56bright NK cells secrete abundant IFN-γ; in contrast, CD56dim NK cells produce negligible amounts of IFN-γ in response to cytokine stimulation.28,29 Interestingly, upon miRNA stimulation, both CD56bright and CD56dim NK cells become activated, resulting in higher expression of CD69 and IFN-γ secretion when compared with parallel cultures treated with vehicle alone (supplemental Figure 5A-B).
To determine whether miRNAs also stimulate NK-cell cytotoxic effector functions, we measured CD107a expression by a flow cytometric assay to quantify degranulation of primary human NK cells upon contact with K562 tumor cell targets in the presence of IL-12. Compared with the controls, we observed that CD107a expression underwent a moderate but statistically significant increase following stimulation with miR-122 and miR-15b (Figures 2B-C). A 51Cr-release cytotoxicity assay with K562 targets was also performed; however, it did not reach statistical significance because of interdonor variation in cytotoxic activity (not shown).
miRNAs activate NK cells in vivo
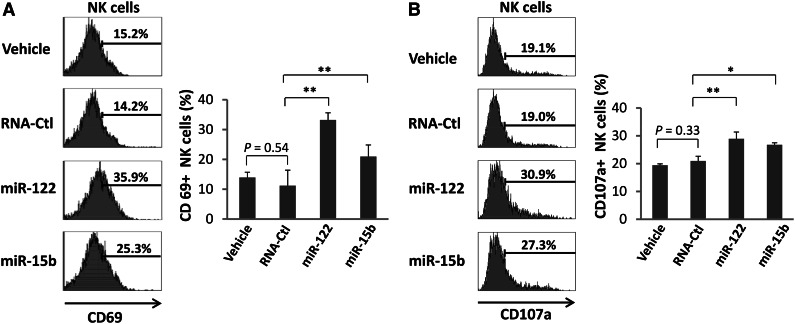
To support the physiological relevance for our findings, we treated normal wild-type mice with miR-122 and miR-15b by injecting 1 dose of miRNAs, without administration of exogenous IL-12, and sacrificed the mice 4 days later. Flow cytometric analysis indicated that NK cells from miRNA-treated mice have twofold to threefold higher surface expression of the activation marker CD69 in comparison with that from either vehicle- or RNA-Ctl sequence-treated mice (Figure 3A). In vivo NK-cell activation by miRNAs was also evidenced by ex vivo coculture with YAC tumor cells in the absence of any exogenous IL-12, which resulted in a statistically significant increase in CD107a expression (Figure 3B).
Figure 3.
miRNAs activate NK cells in vivo. (A) Mice were treated in vivo with vehicles or 20 μg RNA-Ctl, miR-122, or miR-15b for 4 days. The treated mice were then sacrificed, and total splenocytes were isolated for flow cytometric analysis to measure CD69 surface expression after gating on CD3-NK1.1+ NK cells. Representative data from 1 of 6 mice with similar results (left) as well as summary data from 3 mice (right) in 1 experiment are shown. (B) The prepared splenocytes from A were cocultured with YAC-1 tumor cells for 3 hours without any exogenous IL-12 and subjected to flow cytometric analysis of CD107a expression after gating on CD3-NK1.1+ NK cells. For (A) and (B), *P < .05 and **P < .01, respectively, and error bars represent standard deviation.
miRNAs do not activate T cells in vivo or in vitro
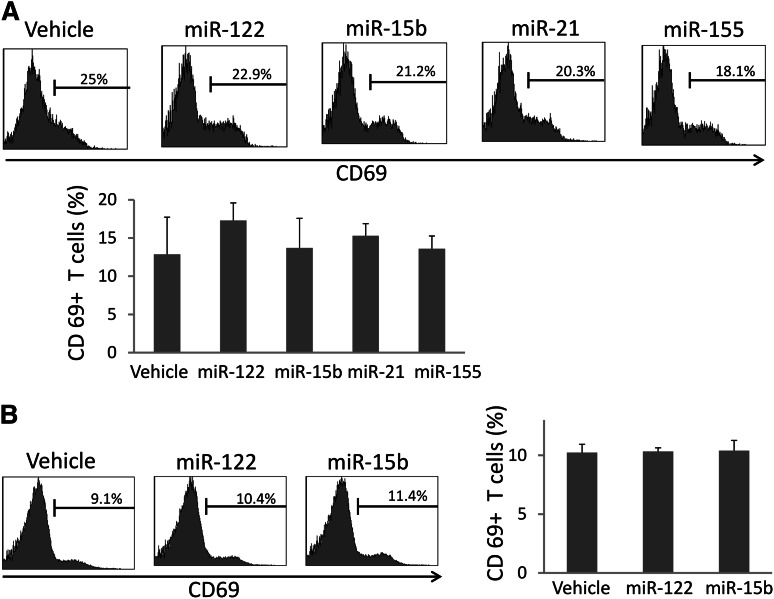
In contrast to NK cells, human T cells were not activated by miRNAs when assessed in culture of whole PBMCs (Figure 4A). Moreover, mouse T cells were not found to be activated following the in vivo infusion of miRNA (Figure 4B). These data suggest that miRNAs may selectively activate the innate immune response without activating adaptive (T-cell) immune response.
Figure 4.
miRNAs do not activate T cells in vivo. (A) Human PBMCs were stimulated with miRNAs as described in Figure 1A and subjected to flow cytometric analysis of CD69 surface expression within CD3+ T cells. Depicted are representative data from 1 of 6 donors (top), as well as summary data from 3 donors in 1 experiment (bottom). Data suggest that miRNAs do not significantly change human T-cell surface expression of CD69. (B) Mice were treated in vivo, and cells were prepared for flow cytometric analysis as described in Figure 3A. Depicted are representative data from 1 of 6 mice with similar data (left), as well as summary data from 3 mice in 1 experiment (right), suggesting that miRNAs do not significantly change murine T-cell surface expression of CD69 in vivo. For (A) and (B), error bars represent standard deviation.
Activation of NK cells by miRNAs occurs via the TLR signaling pathway
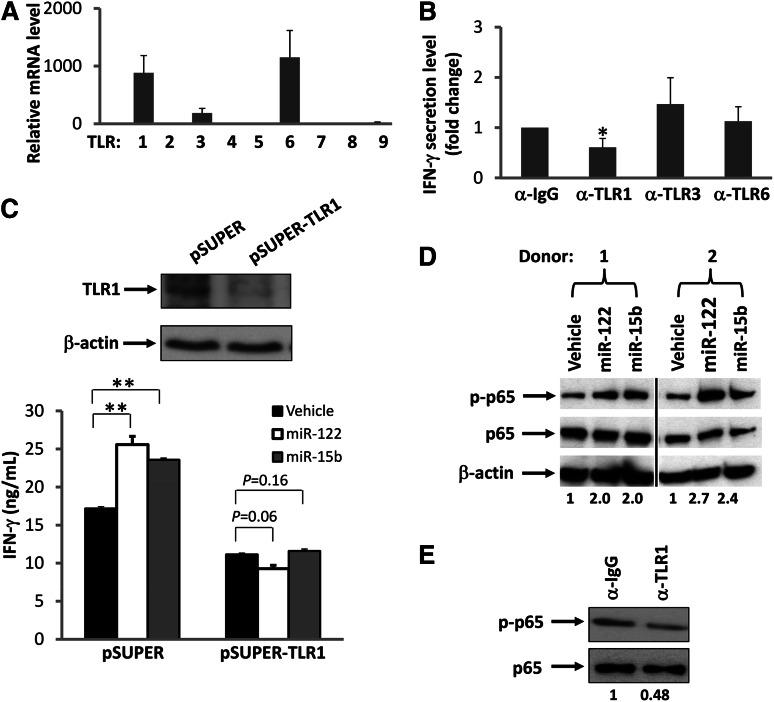
TLR9 has been shown to recognize and respond to bacterial and viral or synthetic deoxyoligonucleotides that contain unmethylated CpG dinucleotide motifs.30 Murine TLR7 and human TLR8 have also been shown to recognize viral RNA.24,31 We therefore speculated that miRNAs might be activating NK cells through the TLR8 or TLR9 pathway. We first determined mRNA expression levels of TLR8 and TLR9, as well as other TLRs, in resting human NK cells as well as cells costimulated with IL-12 and miRNAs. Surprisingly, we found that TLR8 and TLR9 are negligibly expressed in both resting and activated human NK cells, suggesting that activation of NK cells by miRNAs is unlikely to occur through these receptors. In contrast, we found that resting human NK cells express relatively higher transcript levels of TLR1, TLR3, and TLR6 (Figure 5A). Accordingly, we disrupted TLR signaling by blocking each of these 3 TLRs with their respective blocking Abs. For TLR1, we used an antibody with a capability to block signaling activated by its ligand, Pam3CSK4 (supplemental Figure 6). We found that blockade of TLR1 in primary NK cells significantly reduced miRNA-mediated induction of IFN-γ production by ∼50%, while blockade of TLR3 and TLR6 had no significant effect on NK-cell activation (Figure 5B). To further demonstrate that TLR1 may participate in miRNA-induced NK-cell activation, an shRNA approach was taken. We first confirmed that TLR1, but not TLR3, was successfully knocked down in NK-92 cells (Figure 5C and supplemental Figure 7). We then found that TLR1 knockdown caused NK-92 cells to lose their capability for miRNA-mediated activation as no increase in IFN-γ production was observed, while the empty vector (pSUPER)–transduced control cells remained responsive to miRNAs (Figure 5C). A confocal microscopy study of HEK293T cells cotransfected with miRNAs and TLR1-YFP fusion plasmid indicated that miRNAs and TLR1 protein colocalize with each other within these cells (supplemental Figure 8). Collectively, these data suggest that NK-cell activation by miRNAs occurs, at least in part, via the TLR1 signaling pathway; however, this might be different in other types of immune cells, as recently reported.32
Figure 5.
Interaction of TLRs and miRNAs. (A) Real-time RT-PCR was used to determine the level of TLR mRNA expressed in highly purified human NK cells. The mRNA level of TLR5 was found to be lowest and was normalized to 1. The mRNA level of other TLRs was presented as relative to that of TLR5. Data are shown as the average of 3 donors. (B) Purified human NK cells were preincubated for 1.5 hours with a nonspecific IgG or anti-TLR1, anti-TLR3, or anti-TLR6–blocking antibody (α). Cells were then stimulated with miR-122 as described in Figure 1A in the presence of the blocking antibody, and supernatants were harvested to measure IFN-γ protein via ELISA. The concentration of IFN-γ in the purified NK cells incubated with IgG and miR-122 was arbitrarily set at 1. Data were averaged from 3 donors. *P < .05 and error bars represent standard deviation (SD). (C) NK-92 cells were infected with pSUPER-TLR1-GFP retroviruses, and stably transfected cells were sorted based on GFP expression. Both the vector-transduced cells (pSUPER) and the TLR1 knockdown cells (pSUPER-TLR1, confirmed by immunoblotting; upper panel) were stimulated with miR-122 or miR-15b as described in Figure 1A, and cell-free supernatants were collected to assess IFN-γ secretion via ELISA (lower panel). **P < .01 and error bars represent SD. (D) Purified human NK cells were stimulated with miR-122 or miR-15b as described in Figure 1A and subsequently subjected to immunoblotting using p65 and phospho-p65 (p-p65) Abs. β-actin immunoblotting was included to demonstrate equal loading of total protein. Data shown represent 2 of 4 donors with similar results. Numbers beneath each lane represent quantification of p-p65 by densitometry, normalized by p65. (E) The experiment was performed as in D except that anti-TLR1 blocking mAb or its control IgG was included in the culture in the presence of miR-122. Data shown represent 1 of 3 donors with similar results. Numbers beneath each lane represent quantification of p-p65 by densitometry, normalized by p65.
Nuclear factor (NF)-κB represents a signaling pathway downstream of several TLRs, including TLR1.33 To further test our hypothesis that miRNAs activate NK cells via TLR signaling, we assessed NF-κB activation in NK cells after treatment with miRNAs. We found that although the total level of p65 protein, a transactivation component of NF-κB signaling, was unchanged, treatment with miRNAs induced an increase in the phosphorylation of p65 in primary NK cells primed with IL-12 (Figure 5D). To further confirm that miRNA-induced activation of NK cells requires TLR1-mediated NF-κB signaling, we blocked TLR1 with its blocking antibody and found that the signaling blockade inhibited phosphorylation of p65 (Figure 5E). Importantly, when we treated purified primary human NK cells with miR-122 and miR-15b in the same way as we did to determine p65 activation, we did not observe any phosphorylation of IRF3, an activation event specifically mediated via TLR3 (data not shown).
miRNAs enhance antitumor activity of NK cells in vivo to control tumor development
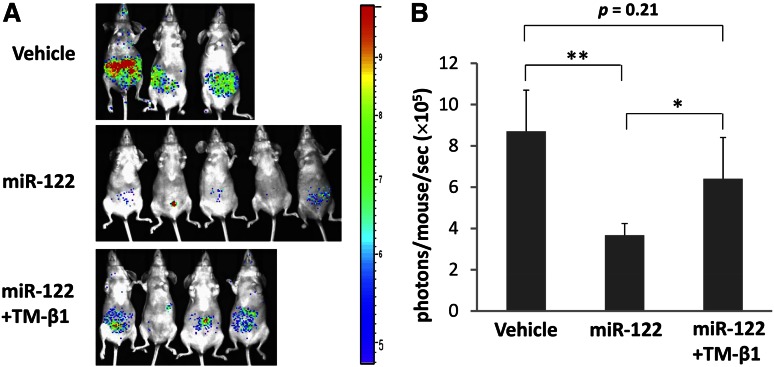
A hallmark of NK-cell function is to kill tumor cells or virally infected cells. To further validate the physiological relevance of NK activation by miRNAs, we treated mice with miR-122 prior to and post implantation of A20 lymphoma cells in the presence or absence of NK-cell depletion. To exclude the possible T-cell effect, athymic nude mice were used. As shown in Figure 6, after 3 doses of miR-122, the growth of A20 tumor cells in mice was significantly suppressed when compared with vehicle-administered mice. However, this effect was largely abrogated when the NK cells were depleted by TM-β1 mAb treatment, suggesting that miRNA suppressed tumor growth at least in part through NK-cell activation. More robust inhibition of tumor growth was observed when we repeated our experiment with more frequent miRNA injections and a longer duration of treatment (supplemental Figure 9).
Figure 6.
miRNA stimulation enhances antitumor activity of NK cells in vivo. (A) Ventral bioluminescence imaging of mice bearing A20 lymphoma. Athymic nude mice were injected with 1 × 105 luc-expressing A20 cells via tail veins and subjected to miRNA stimulation alone or combined with TM-β1 treatment according to the schedule described in the Materials and Methods section. The pseudo color indicates the relative signal strength for tumor growth, with strongest in red and weakest in purple. (B) Quantification summary of units of photons per second per mouse from (A). Data are shown as mean ± standard deviation from each group of mice. *P < .05; **P < .01.
Components of the TLR signaling pathway are downregulated in NK cells from cancer patients
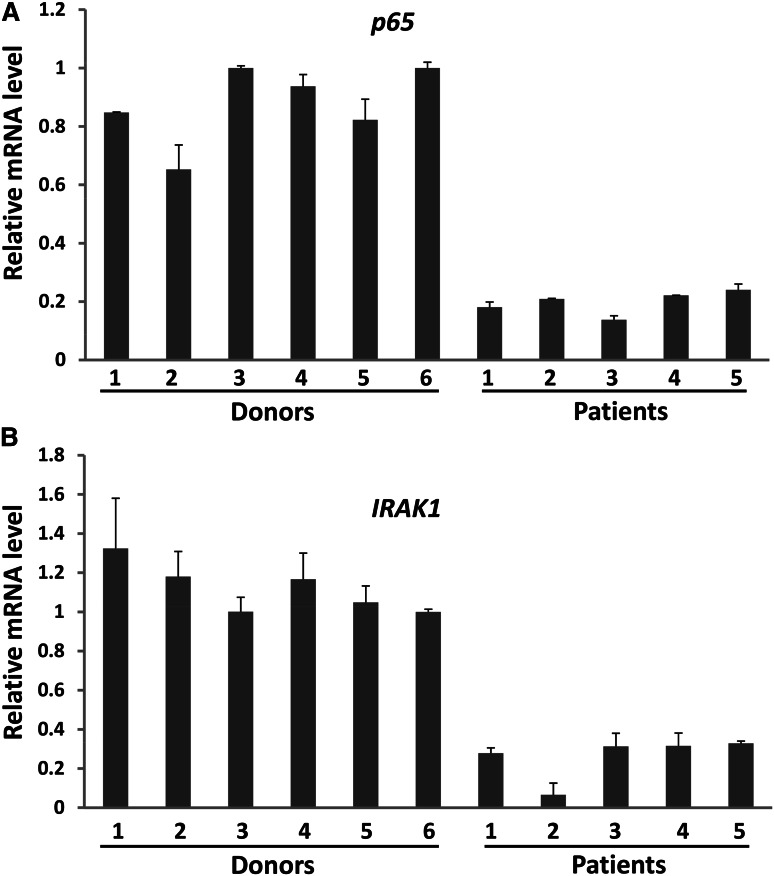
The data presented provide strong evidence that miRNAs have a role in combating tumors by directly activating NK cells. Yet, some cancer patients with disease progression have high levels of circulating miRNAs, which suggests that there may be a mechanism(s) by which tumors escape surveillance by miRNA-activated NK cells. To investigate this, we purified NK cells from PBMCs of lymphoma patients; these patients are reported to have elevated expression of miRNAs such as miR-155.18 We found that although there was no significant difference of TLR1 expression between healthy donors and cancer patients (supplemental Figure 10), p65 and IRAK1, the 2 main components in the TLR1–NF-κB signaling pathway, were consistently and significantly downregulated in NK cells from lymphoma patients (Figure 7A-B). We speculate that higher miRNAs in cancer patients may therefore be unable to activate NK cells in order to control tumor progression. We will look to validate this in future studies.
Figure 7.
Downregulation of the NF-κB signaling pathway components in NK cells from lymphoma patients. (A,B) NK cells were isolated from PBMCs of both healthy donors and lymphoma patients as described in the Materials and Methods section. The purified NK cells were then immediately subjected to RNA extraction and cDNA synthesis. The expression levels of p65 (A) and IRAK1 (B) were determined by SYBR Green real-time RT-PCR assay.
Discussion
miRNAs exist in the peripheral blood of healthy donors as well as in patients with cancer.8 As supported by our in vivo antitumor data, it is conceivable that miRNAs boost NK cells’ ability to participate in surveillance of malignant transformation or infectious insult in otherwise healthy individuals. On the other hand, the prevalence of circulating miRNAs in some cancer patients also implies that other factors secreted by malignant tumors, such as transforming growth factor-β (TGF-β), might somehow serve to disarm this form of innate immune activation. In addition, in lymphoma patients we found that both p65 and IRAK1 have consistently lower mRNA expression in NK cells when compared with those in healthy donors, suggesting that downregulation of NF-κB signaling in NK cells might be a potential underlying mechanism by which tumors evade immune surveillance by miRNAs. However, this should not imply that modulation of miRNAs in cancer patients is irrelevant. In fact, high levels of miR-21 expression are associated with a more favorable clinical outcome for diffuse large B-cell lymphoma.34 During manuscript preparation, a report was published that showed that systemic mi-RNA-34a delivery abrogates in vivo tumor growth of this disease.35
We found the activation induced by miRNAs to be relatively specific for innate immune effector cells (ie, NK cells) and absent in T cells, both in vitro and in vivo. This finding may have application when selective activation of these relatively distinct arms of the immune response is desired. For example, in the setting of blood and marrow transplantation, the NK-cell innate immune response can function to kill activated T cells, thereby contributing to the suppression of donor T-cell–mediated graft-versus-host disease.36,37
We worked to ensure that the NK-cell activation noted in the presence of miRNAs was not due to nonspecific binding and activation by contaminants. Our high-performance liquid chromatography–purified RNA negative control was prepared and handled in an identical fashion to our miRNAs, making it unlikely that NK-cell activation in response to the miRNAs was the result of endotoxin or other contaminants. With regard to the RNA control, we used a nonspecific, single-stranded RNA called RNA41, which is similar in size to miRNAs,24 in the majority of in vitro experiments and did not see induction of IFN-γ or CD107a degranulation. Further, to rule out the potential bias due to the use of a single RNA (RNA41) as a control, we also mutated several nt of miR-122 and miR-15b and found that all NK-cell activation was lost.
In addition to the potential of using miRNAs for cancer diagnosis and prognosis, miRNAs are also promising for therapeutic applications, and they will be evaluated in clinical trials. Our findings suggest that those miRNAs that function as tumor suppressors, eg, miR-12238 included in the current study, may be promising agents for treatment of cancer because, on the one hand, they may directly and specifically target oncogene expression, yet on the other hand, they are also able to activate innate immune effector cells against tumor cells. Our discovery may also shed light on the manner in which the host mounts an immune response against infectious pathogens and/or malignant transformation, as well as the manner by which tumors or pathogens edit the immune response to escape immune activation by circulating miRNAs.
In summary, we provide what we believe to be the first evidence that miRNAs, as a class of regulatory molecules, directly activate both human and mouse NK cells and that this NK-cell activation occurs, at least in part, via the TLR1 signaling pathway. This identifies a new function of miRNAs with physiological relevance and their potential for applications in preventing or treating cancer and infections either alone or as an adjuvant.
Supplementary Material
Acknowledgments
This project was supported by grants from the National Cancer Institute (CA155521 to J.Y. and CA095426 and CA068458 to M.A.C.), a 2012 scientific research grant from the National Blood Foundation (J.Y.), an institutional research grant (IRG-67-003-47) from the American Cancer Society (J.Y.), and The Ohio State University Comprehensive Cancer Center Pelotonia grant (J.Y.).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.H. designed research, performed experiments, and wrote the paper; J.C., L.-C.W., H.M., Y.P., T.H., M.W., S.Y., and S.S. performed experiments; C.A.A.-B., S.V., D.M.B., C.C.H., X.H., K.G., and S.M.D. designed the research; J.Z. analyzed data; M.A.C. designed the research and edited the manuscript; and J.Y. devised the concept, designed research, performed experiments, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jianhua Yu, The Ohio State University, Biomedical Research Tower 816, 460 West 12th Ave, Columbus, OH 43210; e-mail: jianhua.yu@osumc.edu; and Michael A. Caligiuri, The Ohio State University, Biomedical Research Tower 816, 460 West 12th Ave, Columbus, OH 43210; e-mail: michael.caligiuri@osumc.edu.
References
- 1.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 7.Rovira C, Güida MC, Cayota A. MicroRNAs and other small silencing RNAs in cancer. IUBMB Life. 2010;62(12):859–868. doi: 10.1002/iub.399. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 10.Roth C, Kasimir-Bauer S, Pantel K, Schwarzenbach H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol Oncol. 2011;5(3):281–291. doi: 10.1016/j.molonc.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth C, Rack B, Müller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 13.Tomimaru Y, Eguchi H, Nagano H, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56(1):167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro-oncol. 2012;14(1):29–33. doi: 10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen CJ, Lee MH. Early diagnosis of hepatocellular carcinoma by multiple microRNAs: validity, efficacy, and cost-effectiveness. J Clin Oncol. 2011;29(36):4745–4747. doi: 10.1200/JCO.2011.39.0054. [DOI] [PubMed] [Google Scholar]
- 17.Waidmann O, Bihrer V, Pleli T, et al. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J Viral Hepat. 2012;19(2):e58–e65. doi: 10.1111/j.1365-2893.2011.01536.x. [DOI] [PubMed] [Google Scholar]
- 18.Fang C, Zhu DX, Dong HJ, et al. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol. 2012;91(4):553–559. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]
- 19.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187(12):6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehniger TA, Wylie T, Germino E, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20(11):1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TD, Lee SU, Yun S, et al. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 2011;118(20):5476–5486. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trotta R, Chen L, Ciarlariello D, et al. miR-155 regulates IFN-γ production in natural killer cells. Blood. 2012;119(15):3478–3485. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong JH, Maki G, Klingemann HG. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8(4):652–658. [PubMed] [Google Scholar]
- 24.Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 25.Yu J, Wei M, Becknell B, et al. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity. 2006;24(5):575–590. doi: 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Becknell B, Trotta R, Yu J, et al. Efficient infection of human natural killer cells with an EBV/retroviral hybrid vector. J Immunol Methods. 2005;296(1-2):115–123. doi: 10.1016/j.jim.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Borrego F, Peña J, Solana R. Regulation of CD69 expression on human natural killer cells: differential involvement of protein kinase C and protein tyrosine kinases. Eur J Immunol. 1993;23(5):1039–1043. doi: 10.1002/eji.1830230509. [DOI] [PubMed] [Google Scholar]
- 28.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper MA, Fehniger TA, Turner SC, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97(10):3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 30.Takeshita F, Gursel I, Ishii KJ, Suzuki K, Gursel M, Klinman DM. Signal transduction pathways mediated by the interaction of CpG DNA with Toll-like receptor 9. Semin Immunol. 2004;16(1):17–22. doi: 10.1016/j.smim.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 32.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 34.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 35.Craig VJ, Tzankov A, Flori M, Schmid CA, Bader AG, Müller A. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia. 2012;26(11):2421–2424. doi: 10.1038/leu.2012.110. [DOI] [PubMed] [Google Scholar]
- 36.Olson JA, Leveson-Gower DB, Gill S, Baker J, Beilhack A, Negrin RS. NK cells mediate reduction of GVHD by inhibiting activated, alloreactive T cells while retaining GVT effects. Blood. 2010;115(21):4293–4301. doi: 10.1182/blood-2009-05-222190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7(5):340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 38.Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.