Key Points
An NES within CALM is necessary and sufficient for CALM-AF10–mediated transformation.
Presence of the CALM NES confers transformation potential to AF10 through perturbation of H3K79 methylation and Hoxa cluster expression.
Abstract
The t(10;11) chromosomal translocation gives rise to the CALM-AF10 fusion gene and is found in patients with aggressive and difficult-to-treat hematopoietic malignancies. CALM-AF10–driven leukemias are characterized by HOXA gene up-regulation and a global reduction in H3K79 methylation. DOT1L, the H3K79 methyltransferase, interacts with the octapeptide/leucine zipper domain of AF10, and this region has been shown to be necessary and sufficient for CALM-AF10–mediated transformation. However, the precise role of CALM in leukemogenesis remains unclear. Here, we show that CALM contains a nuclear export signal (NES) that mediates cytoplasmic localization of CALM-AF10 and is necessary for CALM-AF10–dependent transformation. Fusions of the CALM NES (NESCALM-AF10) or NES motifs from heterologous proteins (ABL1, Rev, PKIA, APC) in-frame with AF10 are sufficient to immortalize murine hematopoietic progenitors in vitro. The CALM NES is essential for CALM-AF10–dependent Hoxa gene up-regulation and aberrant H3K79 methylation, possibly by mislocalization of DOT1L. Finally, we observed that CALM-AF10 leukemia cells are selectively sensitive to inhibition of nuclear export by Leptomycin B. These findings uncover a novel mechanism of leukemogenesis mediated by the nuclear export pathway and support further investigation of the utility of nuclear export inhibitors as therapeutic agents for patients with CALM-AF10 leukemias.
Introduction
The t(10;11)(p13;q14) translocation gives rise to the CALM-AF10 fusion gene and was originally identified in the U937 human monocytic cell line.1 Subsequently, CALM-AF10 translocations have been identified in patients with acute myeloid leukemia (AML), T-cell acute lymphoblastic leukemia and malignant lymphoma and are generally associated with poor prognoses.2,3 Although several breakpoints have been identified, CALM-AF10 fusion proteins consistently include most of the CALM coding sequence in-frame with a portion of AF10 that contains the octapeptide/leucine zipper (OM-LZ) protein interaction domain.4-6 Retroviral transduction of CALM-AF10 causes acute leukemia in a murine bone marrow transplantation model.7 Similarly, mice that express a CALM-AF10 transgene develop acute leukemia at a median age of 12 mo.8 CALM-AF10 leukemias are characterized by up-regulated expression of the HOXA homeobox genes, including HOXA5, HOXA7, HOXA9, and HOXA10.8-10
AF10 (also known as MLLT10) was first identified as a fusion partner for the MLL gene in patients with AML.11 AF10 encodes a nuclear protein that is a putative transcription factor and contains LAP/PHD-finger domains and nuclear localization sequences.12,13 The AF10 carboxy-terminus contains an OM-LZ domain, which has previously been shown to be necessary and sufficient for CALM-AF10–mediated leukemogenesis.5,14 The OM-LZ domain of AF10 interacts with various proteins, including the H3K79 methyltransferase, DOT1L.14 CALM-AF10 leukemias are marked by global hypomethylation of H3K79, whereas the Hoxa locus is H3K79 hypermethylated.15,16 Therefore, aberrant recruitment of DOT1L by the OM-LZ domain of AF10 is thought to be critical for CALM-AF10–mediated leukemogenesis.16 However, the precise mechanism by which this occurs has not yet been elucidated.
The clathrin assembly lymphoid myeloid leukemia (CALM; also known as PICALM) gene encodes a 652-aa protein. CALM predominantly localizes to the cytoplasm and has been shown to be necessary for the orderly progression of clathrin-mediated endocytosis.17 Structurally, CALM contains domains that are involved in endocytosis, including an epsin N-terminal homology domain18 and a clathrin-binding domain in the carboxy-terminus.17,19 CALM has also been shown to shuttle between the cytoplasm and the nucleus, where it may activate transcription, although this remains poorly understood.20 Although perturbation of endocytosis as a result of CALM gene translocations has been hypothesized to play a role in leukemogenesis,5,21 the specific contributions of CALM to CALM-AF10–dependent leukemogenesis remain unclear.
Here, we performed structure-function analyses to elucidate the contributions of CALM to CALM-AF10–mediated leukemogenesis. We determined that the CALM carboxy-terminus (aa 544-553) contains a nuclear export signal (NES) that mediates cytoplasmic localization of CALM-AF10. Using in vitro bone marrow clonogenic assays and in vivo transplantation experiments, we discovered that the CALM NES is both necessary and sufficient for CALM-AF10–mediated leukemogenesis. These findings reveal a novel oncogenic mechanism by which an NES within a leukemogenic fusion protein mediates transformation.
Methods
Generation of CALM-AF10 mutant constructs
The bicistronic murine stem cell virus-internal ribosome entry site-enhanced green fluorescent protein (MSCV-IRES-eGFP) retroviral vector encoding CALM-AF10 was a kind gift from Eric Delabesse. The CALM-AF10 chimera fuses all but the last 4 aa of CALM (NCBI accession NP_001008660; also missing 8 aa from exon 17a) to exon 11 (aa 234) of AF10 (NCBI accession NP_001182555). For clarity and consistency with previous publications, CALM amino acid numbering is in reference to the longest CALM isoform (NCBI accession NP_009097.2), which is a total of 652 aa. Truncation mutants were generated by polymerase chain reaction (PCR) amplification of CALM domains using primers containing a BamHI site and Flag tag at the 5′ end and an NsiI site at the 3′ end. These CALM products were cloned in-frame with AF10 using an intermediate plasmid in which an NsiI site was introduced at the 5′ end of exon 11. Point mutations within the CALM NES were generated using a Site Directed Mutagenesis kit (Agilent Technologies). NES fusion constructs were generated by cloning double-stranded oligonucleotides in-frame with AF10 (aa 234) using BamHI and NsiI overhangs. The CALM-AF10ΔOMLZ construct was made by deleting AF10 nucleotide 2128-2349 (coding for aa 710-783) by PCR. All constructs were verified by sequencing (Duke DNA Analysis Facility).
CalmNULL hematopoietic cell lines
CALM-deficient cells were obtained from Picalmfit1 (herein referred to as CalmNULL) mice.19,22 CalmNULL CALM-AF10 leukemias were generated by transplanting CALM-AF10–transduced CalmNULL E14 fetal liver cells into lethally irradiated B6(Cg)-TyrC-2J/J mice. Upon development of symptoms, mice were sacrificed, and bone marrow cells were cultured with interleukin (IL)-3. These cells were used to detect CALM-AF10 by immunofluorescence using an anti-CALM antibody. CalmNULL hematopoietic cell lines were also generated by in vitro immortalization of E14 fetal livers with MLL-ENL.23 CALM-AF10 or CALM(mutNES1+2)-AF10 were ectopically expressed by retroviral transduction in these cells and their subcellular localization was assessed using an anti-CALM antibody.
Infection of hematopoietic progenitors and methylcellulose assay
Mice were bred and maintained at the Duke Animal Facility. All in vivo and euthanasia procedures in this study were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal studies have been approved by the Duke University Institutional Animal Care and Use Committee (protocol no. A029-10-02). All efforts were made to minimize animal suffering. Primary bone marrow cells were isolated from 4- to 8-wk-old B6(Cg)-TyrC-2J/J mice (B6-albino mice, Jackson Laboratory) injected 4-5 d previously with 5-fluorouracil (150 mg/kg, tail vein injection). Hematopoietic precursor cells (HPs) were enriched from hind leg long bones by lineage depletion and were infected as previously described,23 with the following modifications: antibodies used for lineage depletion were limited to those against CD11b, Gr1, and B220 (eBioscience). Transduced HPs were plated in methylcellulose medium (HSC-CFU media; Miltenyi Biotec) containing IL-3, IL-6, granulocyte macrophage–colony stimulating factor (all at 10 ng/mL; PeproTech), and stem cell factor (100 ng/mL; PeproTech) at a concentration of 1000 cells/mL per well (in a 6-well plate). Seven days later, colonies consisting of >100 cells were counted. Cells harvested from the pooled colonies were serially replated under identical conditions at a concentration of 10 000 cells/mL to generate secondary and tertiary colonies. Cytospin preparations and immunophenotypic analyses (anti-Mac-1, anti-Gr-1, and anti-c-Kit antibodies from eBioscience) were performed using cells from tertiary colonies.
Bone marrow transplantation and assessment of mice
Recipient B6-albino mice were lethally irradiated (10 Gy in 2 split doses, 3 h apart) using an X-RAD 320 irradiator 24 h prior to transplantation. Mice were injected (tail vein) with 50 000-200 000 infected, lineage–depleted HPs (CALM-AF10, NESCALM-AF10, CALM(520-583)-AF10, or Hoxa9/Meis1; described previously24) along with 200 000 freshly harvested bone marrow cells to ensure radioprotection. Hematopoietic engraftment of GFP-positive cells was assessed by flow cytometry of peripheral blood leukocytes. Mice were monitored for signs of disease and sacrificed when moribund by CO2 euthanasia. Spleens were weighed, and the morphology of leukemia cells was analyzed by peripheral blood and bone marrow smears using the Diff-Quik Stain (Dade Behring, Inc). The concentration of leukocytes in the blood was determined by flow cytometry (Accuri C6) following lysis of red cells with ammonium chloride solution (StemCell Technologies). The percentage of blasts (GFP-positive) and bone marrow immunophenotype (all antibodies from eBioscience) were analyzed by flow cytometry (Accuri C6).
Results
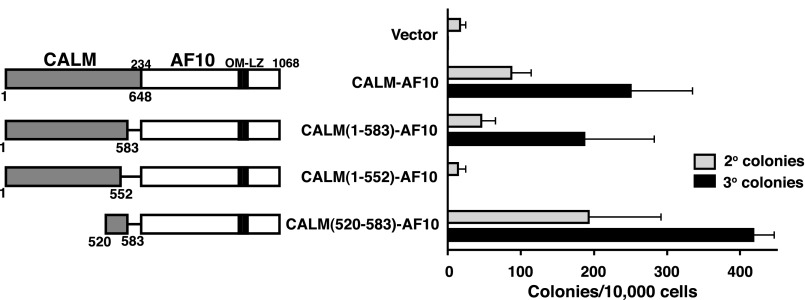
CALM aa 520-583 are sufficient for CALM-AF10–mediated immortalization
It has previously been determined that a region in the CALM carboxy-terminus (aa 400-648) is sufficient for CALM-AF10–mediated transformation.5 To identify specific domains within this region that contribute to transformation, we performed structure-function analysis of the CALM-AF10 fusion protein. Two CALM-AF10 deletion mutants were generated [CALM(1-583)-AF10 and CALM(1-552)-AF10; Figure 1], and we tested the ability of these truncation mutants to immortalize primary murine HPs using an in vitro bone marrow clonogenic assay.23 HPs were retrovirally transduced with an empty MSCV-IRES-eGFP vector, full-length CALM-AF10, or the CALM-AF10 truncation mutants. Cells were seeded in methylcellulose, and colony formation was scored upon subsequent replatings; the presence of colonies following the third passage is indicative of extended self-renewal, herein referred to as “immortalization.” As shown in Figure 1, full-length CALM-AF10 consistently immortalizes HPs in this assay. The secondary and tertiary colonies are compact hypercellular blast-like colonies and consist of immature Mac-1+/Gr-1+ myeloid cells and macrophages (supplemental Figure 1A-C).
Figure 1.
The region between CALM aa 520-583 is essential for CALM-AF10–mediated immortalization. Murine bone marrow progenitor cells were transduced with retroviral constructs expressing CALM-AF10 proteins schematically shown on the left. Bar graph (right) indicates the number of colonies generated per 10 000 cells seeded in second and third passage methylcellulose cultures. The mean ± SEM are shown from duplicate samples analyzed in 3 [CALM(1-552)-AF10], 4 [CALM(520-583)-AF10], or 5 [Vector, CALM-AF10, and CALM(1-583)-AF10] independent experiments.
Similar to full-length CALM-AF10, CALM(1-583)-AF10–transduced progenitors gave rise to hundreds of colonies in the third round of plating (Figure 1). However, CALM(1-552)-AF10–transduced cells lost their colony-forming potential in secondary passage, similar to those transduced with the empty vector control (Figure 1). In addition, we created a CALM-AF10 mutant that includes CALM aa 520-583 fused to AF10 [CALM(520-583)-AF10]. As shown in Figure 1, CALM(520-583)-AF10–transduced cells gave rise to hundreds of secondary and tertiary colonies, comparable with those seen with full-length CALM-AF10. Expression of the appropriately sized CALM-AF10 mutant proteins was verified by western blot (supplemental Figure 2B). From these results, we conclude that aa 520-583 of CALM are sufficient for CALM-AF10–mediated immortalization.
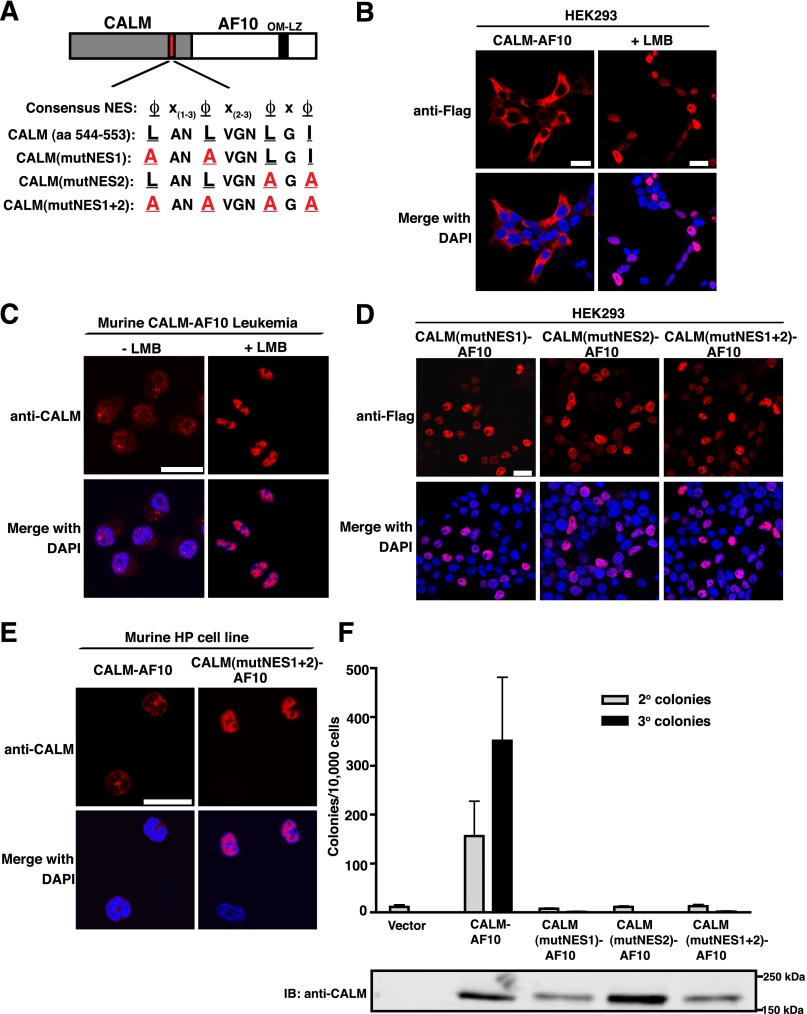
An NES is located within CALM aa 520-583
Sequence analysis of CALM aa 520-583 revealed the presence of a putative NES located between aa 544 and 553 (Figure 2A).16 An NES is a hydrophobic leucine-rich amino acid sequence that is recognized by the nuclear export receptor, CRM1 (also known as XPO1). CRM1 mediates export of NES-containing proteins from the nucleus to the cytoplasm through the nuclear pore complex.25 Leptomycin B (LMB) is a nuclear export inhibitor (NEI) that covalently modifies CRM1 in the NES-binding pocket.26,27 Of note, it has previously been reported that treatment with LMB causes nuclear accumulation of CALM.20 Because the CALM-AF10 protein localizes primarily to the cytoplasm,16 the identification of an NES motif in CALM raises the possibility that localization of the fusion protein is dependent on CRM1-mediated nuclear export. Likewise, it has been reported that fluorescent protein-fused CALM-AF10 becomes nuclear in the presence of LMB.21,28 In accord with these findings, we observed that CALM-AF10 localizes to the cytoplasm of transiently transfected HEK293 cells (Figure 2B left panels), and following treatment with LMB, there is a dramatic redistribution of CALM-AF10 to the nucleus (Figure 2B right panels). We next analyzed the localization of CALM-AF10 in a murine CALM-AF10 leukemia cell line. As shown in Figure 2C, CALM-AF10 localizes to both the nucleus and the cytoplasm of the leukemic cells. Upon addition of LMB, CALM-AF10 becomes exclusively nuclear (Figure 2C). These results indicate that CALM-AF10 undergoes nucleocytoplasmic shuttling and is exported out of the nucleus in a CRM1-dependent manner.
Figure 2.
CALM contains a functional NES that is necessary for CALM-AF10–mediated immortalization. (A) Alignment of the NES within CALM (aa 544-553) and the consensus sequence of CRM1-dependent NES, where ϕ represents any hydrophobic residue and x represents any amino acid. The hydrophobic aa of the CALM NES were point-mutated to alanines (A) to create 3 mutants: CALM(mutNES1)-AF10, CALM(mutNES2)-AF10, and CALM(mutNES1+2)-AF10. (B) Confocal immunofluorescence (IF) analysis of HEK293 cells transiently transfected with Flag-tagged CALM-AF10 and analyzed in the absence (left) or presence (right) of LMB (10 nM, 1 h). (C) Confocal IF analysis of murine CalmNULL CALM-AF10 leukemia cells grown in the absence (left) or presence (right) of LMB (0.1 nM, 12 h). (D) Confocal IF of HEK293 cells transiently transfected with CALM(mutNES)-AF10 mutants. (E) Confocal IF of MLL-ENL–immortalized CalmNULL hematopoietic precursors retrovirally infected with Flag-tagged CALM-AF10 (left panels) or CALM(mutNES1+2)-AF10 (right panels). Cell nuclei were visualized with DAPI (blue). Bars represent 20 μm for all panels. (F) Colony-forming assay of murine HPs infected with empty vector, CALM-AF10, or NES point mutants. Bars represent the number of colonies generated per 10 000 cells seeded in second and third passage cultures. The mean ± SEM are shown from duplicate samples analyzed in 2 [CALM(mutNES1)-AF10], 3 [CALM(mutNES2)-AF10], and CALM[mutNES1+2)-AF10] or 6 (Vector and CALM-AF10) independent experiments. Bottom panel is a western blot of HEK293 cells transfected with empty vector, CALM-AF10, and CALM(mutNES)-AF10 point mutants.
To determine the functional role of the putative CALM NES, we point-mutated conserved hydrophobic residues of the NES within full-length CALM-AF10. As shown in Figure 2A, 2 or 4 leucine/isoleucine residues were replaced with alanines [L544A and L547A in CALM(mutNES1)-AF10; L551A and I553A in CALM(mutNES2)-AF10, and L544A, L547A, L551A, and I553A in CALM(mutNES1+2)-AF10]. Unlike wild-type CALM-AF10, CALM(mutNES1)-AF10, CALM(mutNES2)-AF10, and CALM(mutNES1+2)-AF10 localize exclusively to the nuclei of HEK293 cells (Figure 2D). Similarly, when retrovirally expressed in murine hematopoietic cells, CALM-AF10 localizes predominantly to the cytoplasm, whereas CALM(mutNES1+2)-AF10 is exclusively nuclear (Figure 2E). From these findings, we conclude that CALM contains a functional NES between aa 544 and 553 that mediates the cytoplasmic localization of CALM-AF10.
The CALM NES is necessary for CALM-AF10–mediated immortalization in vitro
Because aa 520-583 of CALM are sufficient for immortalization (Figure 1) and include a CRM1-dependent NES (Figure 2A), we hypothesized that the immortalizing ability of CALM-AF10 may be dependent on the presence of the CALM NES. To test this hypothesis, CALM(mutNES)-AF10 constructs (Figure 2A) were retrovirally transduced into murine HPs and seeded in methylcellulose. Upon serial replatings, full-length CALM-AF10 immortalized bone marrow cells, whereas none of the NES point mutants resulted in colony formation (Figure 2F). Expression of the appropriately sized CALM-AF10 and NES point mutant proteins (180 kDa) was verified by western blot (Figure 2F bottom). These results suggest that a functional NES within CALM-AF10 is necessary for immortalization. Of note, the subcellular localization of the CALM-AF10 truncation mutants corroborates the link between immortalizing potential and cytoplasmic localization. When expressed in HEK293 cells, CALM(1-583)-AF10 and CALM(520-583)-AF10 are predominantly cytoplasmic, whereas CALM(1-552)-AF10 localizes to the nucleus (supplemental Figure 2A). Therefore, we conclude that the CALM NES is necessary for the cytoplasmic localization of CALM-AF10 and for in vitro immortalization.
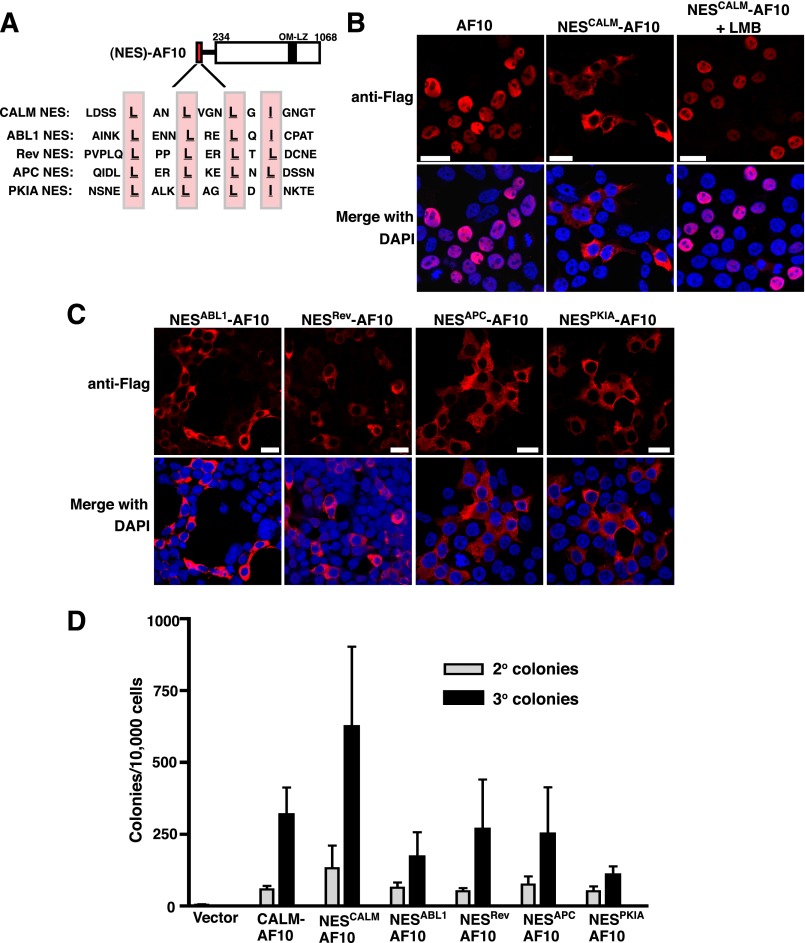
The CALM NES is sufficient for CALM-AF10–mediated immortalization in vitro
To determine whether the CALM NES is sufficient for CALM-AF10–mediated immortalization, we fused CALM aa 540-557 in-frame with AF10 (NESCALM-AF10; Figure 3A). CALM aa 540-557 include the conserved CRM1-dependent NES (Figure 2A), with 4 aa flanking each side to maintain the structure of the NES.29 When transiently transfected into HEK293 cells, NESCALM-AF10 localizes predominantly to the cytoplasm, whereas AF10 (aa 234-1068) displays a nuclear distribution (Figure 3B). Following treatment with LMB, NESCALM-AF10 becomes exclusively nuclear (Figure 3B). Therefore, fusion of the CALM NES to AF10 efficiently mediates CRM1-dependent nuclear export of AF10.
Figure 3.
Fusion of conserved NES protein motifs to AF10 confers cytoplasmic localization and in vitro immortalization potential. (A) Alignment of the CALM NES (aa 540-557) and the NES motifs from heterologous proteins, ABL1 (aa 1086-1103), Rev (aa 70-87), APC (aa 63-79), and PKIA (aa 33-50), fused in-frame with AF10 (at aa 234). Key hydrophobic residues within each NES are highlighted. (B) Confocal IF analysis of HEK293 cells transfected with Flag-tagged AF10 (aa 234-1068) or the NESCALM-AF10 fusion in the absence (middle) or presence (right) of LMB (10 nM, 1 h). (C) Confocal IF of HEK293 cells transfected with the heterologous NES-AF10 fusions. Cell nuclei were stained with DAPI (blue). Bars represent 20 μm. (D) Colony-forming assay of the NES-AF10 constructs. Bars represent the number of colonies generated per 10 000 cells seeded in second and third round cultures. The mean ± SEM are shown from duplicate samples analyzed in 3 (NESABL1-AF10), 4 (NESCALM-AF10, NESPKIA-AF10, NESAPC-AF10, NESRev-AF10), or 5 (Vector and CALM-AF10) independent experiments.
To test whether the CALM NES is sufficient to impart immortalizing potential to AF10, we examined the ability of NESCALM-AF10 to immortalize murine HPs in vitro. It has previously been demonstrated that expression of AF10 alone does not lead to immortalization.30 However, similar to CALM-AF10, NESCALM-AF10 transduction into HPs gives rise to hundreds of secondary and tertiary colonies (Figure 3D) with characteristics similar to CALM-AF10 colonies (supplemental Figure 1A-C). Therefore, the CALM NES fused to AF10 is sufficient for myeloid immortalization in vitro.
Conserved NES protein motifs mediate nuclear export and confer immortalization potential to AF10
To determine whether the sole structural contribution of CALM is an NES, we fused NES motifs from other proteins to AF10 and assessed the immortalizing ability of the resulting chimeric proteins. Based on previous studies that characterized the functionality of multiple nuclear export consensus sequences, we chose to fuse the NES protein motifs of ABL1, Rev (from HIV-1), APC, and PKIA to AF10 (Figure 3A).29,31 As shown in Figure 3C, NESABL1-AF10, NESRev-AF10, NESAPC-AF10, and NESPKIA-AF10 all localize to the cytoplasm when expressed in HEK293 cells. Therefore, each of these conserved NES protein motifs mediates efficient nuclear export of AF10.
Next, we examined the ability of the NES-AF10 constructs to immortalize murine HPs in vitro. Primary murine HPs were retrovirally transduced with NESABL1-AF10, NESRev-AF10, NESAPC-AF10, or NESPKIA-AF10 and seeded in methylcellulose. As shown in Figure 3D, transduction by each of the NES-AF10 fusions resulted in secondary and tertiary colonies, indicative of immortalization. These results emphasize that the fusion of an NES to AF10 is critical for the acquisition of oncogenic properties.
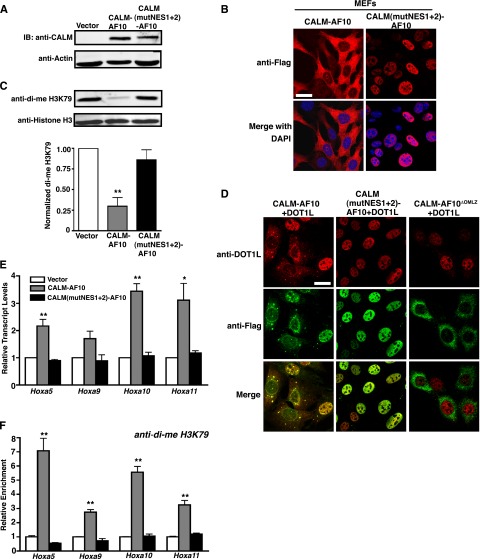
The CALM NES is necessary for CALM-AF10–dependent aberrant H3K79 methylation and increased Hoxa gene expression
To explore molecular mechanisms by which nuclear export of CALM-AF10 contributes to leukemogenesis, we analyzed the epigenetic and transcriptional status of cells expressing cytoplasmic or nuclear CALM-AF10. Murine embryonic fibroblasts (MEFs) were retrovirally transduced with empty vector, CALM-AF10, or CALM(NESmut1+2)-AF10, and protein expression was verified by western blot (Figure 4A). CALM-AF10 localizes primarily to the cytoplasm of MEFs. However, it should be noted that some punctate nuclear staining is detectable in CALM-AF10–expressing cells, likely reflecting the fact that CALM-AF10 shuttles between the nucleus and the cytoplasm. As expected, CALM(NESmut1+2)-AF10 localizes exclusively to the nuclei of MEFs (Figure 4B).
Figure 4.
The CALM NES is necessary for aberrant H3K79 methylation and elevated Hoxa cluster expression. (A-B) Western blot (A) and confocal IF (B) of MEFs stably infected with empty vector, CALM-AF10, and CALM(mutNES1+2)-AF10. Data are representative of one set of MEFs (out of 3) stably infected with the respective constructs. (C) Representative western blot of di-me H3K79 and histone H3 levels in stably infected MEFs. Quantification data (bottom) represent the mean ± SEM of di-me H3K79 values normalized to actin from separately generated MEF lines for each construct (n = 3). Statistical analysis was performed by one-way ANOVA followed by Dunnett’s multiple comparison test using empty vector-transduced MEFs as the control; **P < .01 (D) Confocal IF of MEFs coexpressing DOT1L (red) and Flag-tagged CALM-AF10 (green, left), CALM(NESmut1+2)-AF10 (green, middle), or CALM-AF10ΔOMLZ (green, right). Bar represents 20 μm. (E) Hoxa transcript levels were measured by real-time reverse transcription-PCR and normalized to housekeeping genes GAPDH and β2M and then to empty vector-infected MEFs by the ΔΔCt method. (F) Chromatin immunoprecipitation analysis of di-me H3K79 in the promoter regions of the Hoxa cluster genes. Hoxa amplification was measured by real-time PCR as a percent of input and then normalized to the vector control. For panels E-F, results are shown as mean ± SEM from separately generated MEF lines for each construct (n = 3). White bars represent empty vector, gray bars are CALM-AF10, and black bars are CALM(mutNES1+2)-AF10. Statistical analyses were performed using one-way ANOVA for each Hoxa gene followed by Dunnett’s multiple comparison test. Only CALM-AF10 was found to be significantly different from the vector control: **P < .01, *P < .05.
AF10 interacts with the histone H3K79 methyltransferase DOT1L through the OM-LZ motif,14 which is necessary for transformation. CALM-AF10 leukemia cells display a global reduction in H3K79 dimethylation, a possible result of mistargeting DOT1L away from chromatin.15,21 To determine whether this phenotype is dependent on nuclear export of CALM-AF10, we analyzed levels of dimethylated (di-me) H3K79 in MEFs expressing cytoplasmic or nuclear CALM-AF10. In agreement with published results,15,21 stable expression of CALM-AF10 resulted in a 70% decrease in di-me H3K79 compared with the vector control (Figure 4C middle lane, quantified in lower graph). However, expression of CALM(mutNES1+2)-AF10 did not affect H3K79 methylation (Figure 4C right lane, quantified in lower graph). These results suggest that nuclear export of CALM-AF10 may be required for its ability to interfere with DOT1L activity.
Exclusion of DOT1L from the nucleus could explain the global loss of H3K79 methylation observed in CALM-AF10–expressing cells. To test whether CALM-AF10 directly mislocalizes DOT1L from the nucleus, DOT1L was coexpressed with CALM-AF10 or CALM(NESmut1+2)-AF10 in MEFs. As shown in Figure 4D, CALM-AF10 and DOT1L co-localize. The distribution is variable, with 56% of cells expressing CALM-AF10 and DOT1L in the cytoplasm and 44% of cells displaying predominantly nuclear distribution of CALM-AF10 and DOT1L (Figure 4D left panels). In contrast, CALM(NESmut1+2)-AF10 and DOT1L co-localize exclusively in the nucleus (100% of the cells) (Figure 4D middle panels). As previously reported,16 DOT1L remains nuclear in the presence of a CALM-AF10 mutant missing the OM-LZ domain (AF10 aa 710-783; CALM-AF10ΔOMLZ) (Figure 4D right panels). These results emphasize that full-length CALM-AF10 and DOT1L co-localize, and in the presence of CALM-AF10, DOT1L becomes cytoplasmic in approximately one-half of the cells.
In addition to reduced H3K79 methylation, the transforming potential of CALM-AF10 has been linked to transcriptional up-regulation of Hoxa cluster genes.8-10 As shown in Figure 4E, stable expression of CALM-AF10 in MEFs correlates with elevated transcript levels of Hoxa5 (2-fold), Hoxa9 (1.7-fold), Hoxa10 (3.5-fold), and Hoxa11 (3-fold) compared with empty vector. In contrast, expression of CALM(mutNES1+2)-AF10 did not significantly change Hoxa expression levels (Figure 4E). It has been shown that H3K79 is locally hypermethylated at the Hoxa5 locus in CALM-AF10 leukemia cells. To assess whether the Hoxa locus is hypermethylated in CALM-AF10–expressing MEFs, we performed chromatin immunoprecipitation (ChIP). H3K79 dimethylation is enriched at the promoters of the Hoxa genes in CALM-AF10 cells (Figure 4F), whereas CALM(mutNES1+2)-AF10 cells exhibit a H3K79 methylation pattern similar to that of empty vector-expressing cells (Figure 4F).
The observation that H3K79 is aberrantly methylated in an NES-dependent manner led us to examine whether LMB prevents DOT1L targeting to the Hoxa locus in CALM-AF10 cells. Vector or CALM-AF10–expressing MEFs were incubated with 1 nM LMB for 24 h, and dimethyl H3K79 ChIP was performed. LMB incubation reduced H3K79 methylation on the Hoxa promoter regions in CALM-AF10 cells by ∼40% (supplemental Figure 3A). However, LMB did not significantly alter H3K79 methylation in vector expressing cells (supplemental Figure 3B). These results suggest that a CRM1-NES interaction is critical for DOT1L targeting and/or activity at the Hoxa locus in CALM-AF10 cells. Together, these data support the importance of the CALM NES in inducing the perturbed epigenetic and transcriptional state observed in CALM-AF10 leukemias.
NESCALM-AF10 is sufficient to induce leukemogenesis in vivo
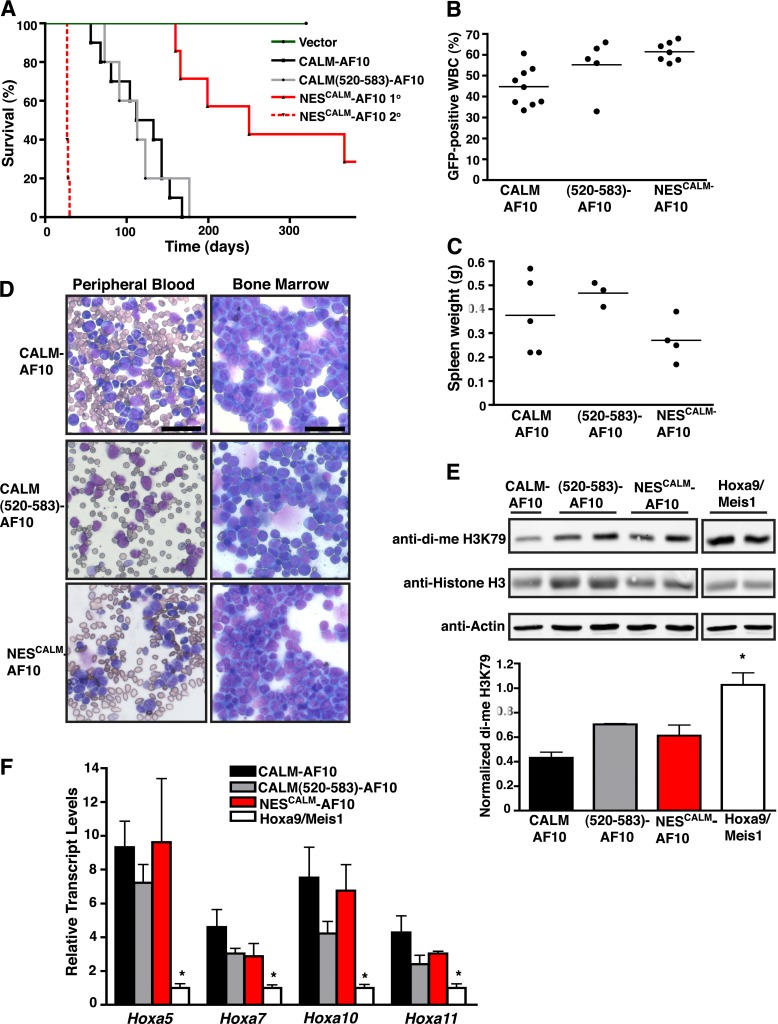
To analyze the in vivo leukemogenic potential of NESCALM-AF10, we injected CALM-AF10–, CALM(520-583)-AF10–, or NESCALM-AF10–transduced progenitors into lethally irradiated syngeneic mice. As shown in Figure 5A, expression of full-length CALM-AF10 caused acute leukemia in 100% of mice (n = 10), with a median survival of 122 d. CALM(520-583)-AF10 also induced leukemogenesis in 100% of mice (n = 5), with a latency similar to that of CALM-AF10 (median survival of 113 d). Importantly, expression of NESCALM-AF10 was sufficient to cause acute leukemia in 5 of 7 recipient mice, albeit with a prolonged latency (median survival of 250 d) (Figure 5A). Engraftment, as reflected by the percentage of GFP-expressing leukocytes in the peripheral blood 24-27 d posttransplant, occurred in all recipients (30%-70% GFP; Figure 5B). Leukemias from mice in each cohort displayed similar characteristics, including enlarged spleens (Figure 5C), massive bone marrow infiltration by myeloblasts, and leukocytosis (Figure 5D). The vast majority of bone marrow blasts coexpressed the myeloid antigens Mac-1 and Gr-1, whereas the expression of cKit and B220 were absent or seen on a small fraction of the cells (supplemental Figure 4). Because the NESCALM-AF10 leukemias had a longer latency, we assessed their transplantability into secondary recipients. The transplanted mice died of aggressive leukemias, with a median latency of 28 d (Figure 5A). From these results, we conclude that CALM(520-583)-AF10 and NESCALM-AF10 induce AMLs similar to full-length CALM-AF10.
Figure 5.
CALM(520-583)-AF10 and NESCALM-AF10 are sufficient to induce leukemias in vivo with characteristics similar to CALM-AF10 leukemias. (A) Kaplan-Meier survival curve of mice transplanted with empty vector (n = 4), CALM-AF10 (n = 10), CALM(520-583)-AF10 (n = 5), or NESCALM-AF10 (n = 7) transduced bone marrow cells. All vector-transduced mice remained leukemia-free through the course of the study and were sacrificed at 320 d posttransplantation. The survival curve of secondary transplant mice (n = 5) injected with NESCALM-AF10 primary leukemias (2 primary leukemias injected to 2 or 3 mice) is depicted by a dashed red line. (B) Percentage of GFP-positive white blood cells (WBC) in the peripheral blood of mice measured at 27 d (CALM-AF10), 25 d [CALM(520-583)-AF10], or 24 d (NESCALM-AF10) posttransplantation. (C) Spleen weights from mice with CALM-AF10, CALM(520-583)-AF10, and NESCALM-AF10 leukemias measured at time of death. A normal murine spleen weighs approximately 0.09 g. (D) Representative peripheral blood and bone marrow smears from mice with CALM-AF10, CALM(520-583)-AF10, and NESCALM-AF10 leukemias. Bars represent 40 μm for all panels. (E) Representative western blots of di-me H3K79, histone H3, and Actin protein levels in separate CALM-AF10 (shown is 1 sample of 3), CALM(520-583)-AF10 (shown are 2 samples of 3), NESCALM-AF10 (shown are 2 samples of 3), and Hoxa9/Meis1 (shown are 2 samples of 3) leukemias. Quantification data represent the mean ± SEM of di-me H3K79 values normalized to histone H3 from separate leukemias (n = 3 for each). Statistical analysis was performed by one-way ANOVA followed by Dunnett’s multiple comparison test. Only Hoxa9/Meis1 was found to be statistically different from CALM-AF10: *P < .05. (F) Hoxa transcript levels were obtained by real-time RT-PCR and normalized to housekeeping genes GAPDH and β2M and then to Hoxa9/Meis1 leukemic cells by the ΔΔCt method. Results are shown as mean ± SEM compiled from 3 CALM(520-583)-AF10, 3 NESCALM-AF10, 4 CALM-AF10, or 4 Hoxa9/Meis1 separate leukemias. Statistical analysis was performed by one-way ANOVA for each Hoxa gene followed by Dunnett’s multiple comparison test. Only Hoxa9/Meis1 was found to be statistically different from CALM-AF10: *P < .05.
To further characterize leukemias induced by CALM-AF10, CALM(520-583)-AF10, and NESCALM-AF10, H3K79 methylation was quantified and compared with AMLs induced by co-transduction of Hoxa9 and Meis1 (Hoxa9/Meis1). These leukemias serve as a control, because retroviral expression of the Hoxa9 and Meis1 effector genes recapitulates the disease while bypassing the upstream epigenetic alterations regulating their expression. As shown in Figure 5E, CALM-AF10, CALM(520-583)-AF10, and NESCALM-AF10 leukemic cells all display reduced H3K79 dimethylation levels in comparison with Hoxa9/Meis1 leukemic cells (reduction of 55%, 30%, and 40%, respectively).
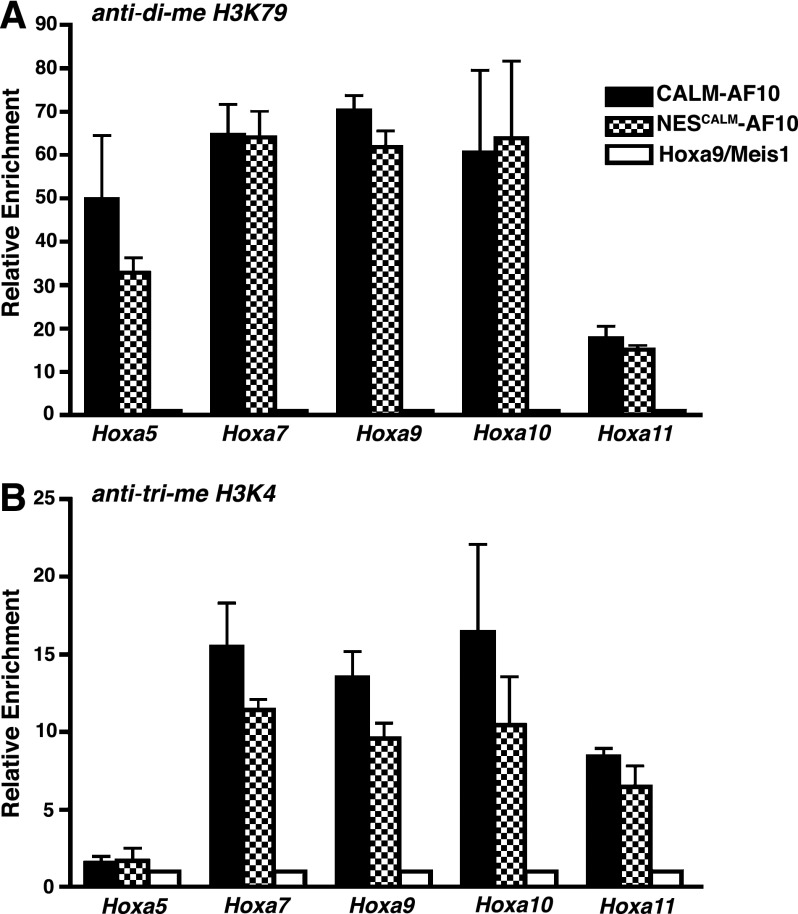
In contrast to Hoxa9/Meis1 leukemic cells, CALM-AF10, CALM(520-583)-AF10, and NESCALM-AF10 leukemic cells all display elevated Hoxa5, Hoxa7, Hoxa10, and Hoxa11 transcript levels (Figure 5F). Therefore, the CALM NES fused to AF10 is sufficient to up-regulate Hoxa cluster expression. We next assessed the epigenetic status of the Hoxa locus in the CALM-AF10 and NESCALM-AF10 leukemic cells. As observed in CALM-AF10–expressing MEFs (Figure 4F), both CALM-AF10 and NESCALM-AF10 leukemic cells exhibit enriched H3K79 dimethylation on Hoxa gene promoters (Figure 6A). We observed a similar pattern of H3K4 trimethylation, a mark of active transcription, on the Hoxa promoters in CALM-AF10 and NESCALM-AF10 cells (Figure 6B). Therefore, the fusion of the CALM NES to AF10 phenocopies the aberrant epigenetic and transcriptional profile observed in CALM-AF10 leukemias.
Figure 6.
H3K79 and H3K4 are hypermethylated on the Hoxa locus in CALM-AF10 and NESCALM-AF10 leukemias. ChIP analysis of di-me H3K79 (A) and trimethylated H3K4 (B) in the promoter regions of the Hoxa cluster genes. Hoxa amplification was measured by real-time PCR as a percent of input then normalized to Hoxa9/Meis1 control leukemic cells. Black bars represent CALM-AF10, checkered bars are NESCALM-AF10, and white bars are Hoxa9/Meis1 leukemias. Results are shown as mean ± SEM compiled from 3 separate leukemias.
Inhibition of CRM1 impairs viability of human CALM-AF10 leukemia cells
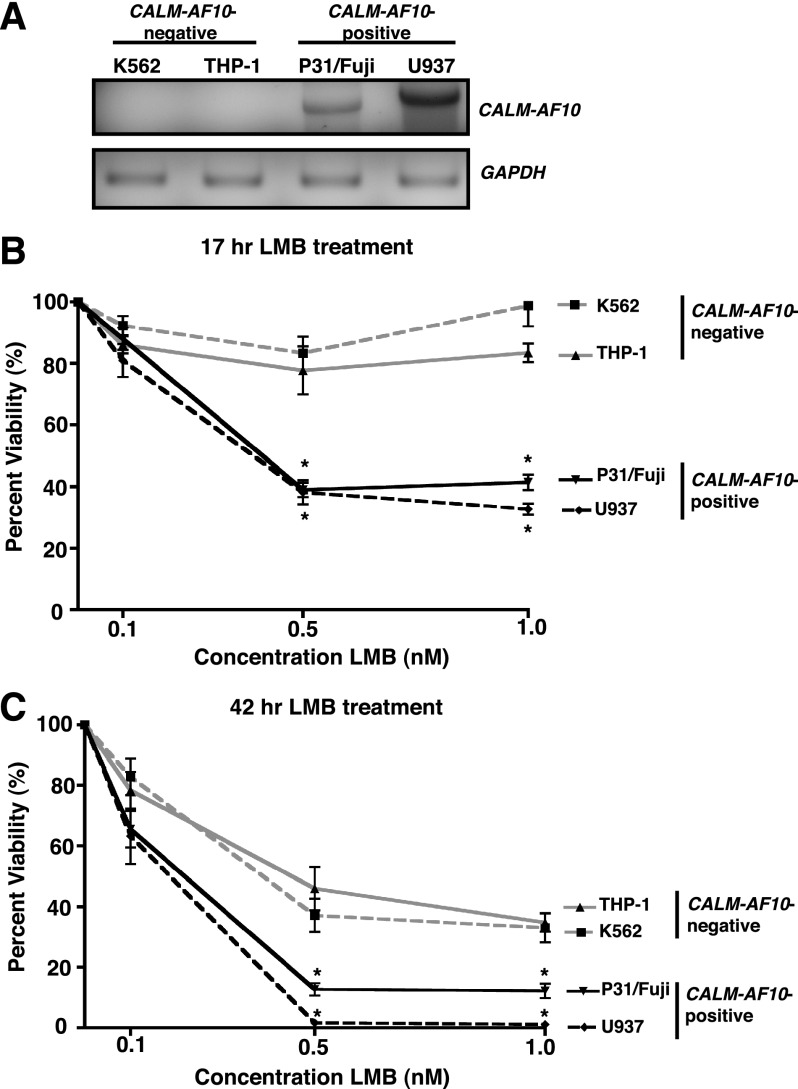
We have shown that mutation of the CALM NES results in nuclear accumulation of CALM-AF10 and impairs leukemogenic transformation. We therefore hypothesized that treatment with a NEI could prevent CALM-AF10 from interacting with the nuclear export receptor CRM1 and potentially hinder cell proliferation. To test the sensitivity of CALM-AF10–expressing cells to NEIs, we compared the viability of CALM-AF10–positive (P31/Fujioka6 and U937) and CALM-AF10–negative (K562 and THP-1) human leukemia cell lines in the presence of LMB. As determined by reverse transcription-PCR, P31/Fujioka and U937 cells both express the CALM-AF10 transcript, whereas THP-1 and K562 cells do not (Figure 7A). Of note, transcripts of differing sizes were amplified from the P31/Fujioka and U937 cells, consistent with the alternate CALM-AF10 fusion breakpoints in these cell lines.6 Cells were seeded at fixed densities, and cell viability was measured in the presence of varying concentrations of LMB. CALM-AF10–positive cells showed a greater sensitivity to 0.5 and 1 nM LMB after 17 h of treatment, with ∼40% viable cells remaining compared with 80%-100% CALM-AF10–negative cells (Figure 7B). The same effect was observed after 42 h of LMB treatment; 1%-11% of the CALM-AF10–positive cells were viable, whereas 35%-45% of the CALM-AF10–negative cells were viable in the presence of 0.5 or 0.1 nM LMB (Figure 7C). These results indicate that CALM-AF10–expressing human leukemia cells display increased sensitivity to inhibition of CRM1 and support the investigation of the therapeutic utility of NEIs in CALM-AF10 leukemias.
Figure 7.
Blocking nuclear export decreases viability of human CALM-AF10 leukemia cell lines. (A) Confirmation of expression of the CALM-AF10 transcript in P31/Fujioka (P31/Fuji) and U937 cells by RT-PCR. GAPDH serves as a control for equal input of RNA for RT-PCR. (B-C) CALM-AF10–positive (P31/Fujioka and U937) and CALM-AF10–negative (K562 and THP-1) cell lines were grown in the presence of increasing concentrations of LMB (0, 0.1, 0.5, 1 nM) for 17 h (B) or 42 h (C). The number of viable cells was determined by flow cytometry and is shown as a percent of untreated for each cell line. Results are shown as mean ± SEM from 3-6 independent experiments. The viabilities of both P31/Fujioka and U937 cell lines are statistically different from either K562 or THP-1 at 0.5 and 1 nM LMB, as determined by two-way ANOVA followed by Bonferroni’s Multiple Comparison Test: *P < .05.
Discussion
Fusion proteins have been shown to mediate hematopoietic oncogenesis by a variety of mechanisms. Some chimeric fusion proteins involve DNA-binding transcription factors (eg, AML1-ETO or NUP98-HOXA9),32,33 cofactors (eg, MOZ-rearrangements or MN1-TEL),34,35 or chromatin-modifying proteins (eg, MLL-fusions),36 and co-opting of the properties of the fusion partners leads to aberrant transcriptional activity. Other fusion partners provide homo-oligomerization domains that enable dimerization of the fusion protein, resulting in enhanced or novel activities (such as PML-RARα or some MLL-fusions).37-39 Several endocytic proteins (eg, CALM, EPS15, or CLTC) are targeted by chromosomal translocations, and perturbation of endocytosis has been proposed as a mechanism of leukemogenesis (reviewed in Crosetto et al40 and Lanzetti et al41). The well-documented involvement of CALM in endocytosis17,42 and its potential transcriptional role in the nucleus20 led us to assume that either (or both) of these functions could be involved in the oncogenicity of CALM-AF10. However, our structure-function analysis of CALM revealed an unexpected and novel mechanism of leukemic transformation that is dependent on the presence of an NES.
Using bone marrow immortalization and transplantation assays, we have demonstrated that an NES within CALM (aa 544-553) is both necessary and sufficient for CALM-AF10–mediated leukemogenesis. These findings complement the observations of Deshpande et al,5 who reported that the carboxy-terminus of CALM (aa 400-648) is sufficient for CALM-AF10–driven transformation. These authors attributed their findings to the presence of a clathrin-binding domain in the carboxy-terminus of CALM and concluded that disruption of endocytosis may be a mechanism by which CALM-AF10 is oncogenic.5 We have recently determined that the CALM clathrin-binding domain spans aa 583-652 at the extreme C terminus, which is distinct from the NES.19 Similarly, Stoddart et al21 have shown that endocytosis is not perturbed in CALM-AF10 cells. Instead, these authors suggested that homo-oligomerization of CALM-AF10 may mediate its transformation potential, based on the ability of the last 55 aa of CALM within CALM-AF10 (aa 593-648) to facilitate dimerization.21 Our observation that CALM aa 540-557 or NES motifs from heterologous proteins are sufficient to confer transforming potential to AF10 led us to conclude that a CRM1-dependent NES represents the functional contribution of CALM in CALM-AF10–mediated leukemogenesis and is distinct from the motifs of CALM involved in endocytosis or oligomerization.
We have shown that fusion of the 18-aa CALM NES in-frame with AF10 (NESCALM-AF10) is sufficient to immortalize murine progenitor cells in vitro and induce leukemogenesis in vivo. It is important to note that NESCALM-AF10 leukemias display prolonged latencies, whereas a larger portion of CALM spanning the NES fused to AF10 [CALM(520-583)-AF10] fully recapitulates the rapid onset of CALM-AF10 leukemias (Figure 5A). It is possible that the longer CALM(520-583)-AF10 allows for proper folding of the NES and/or contributes to its overall stability. It is also possible that CALM(520-583)-AF10 contains an additional domain(s) that contributes to leukemogenesis. For example, CALM contains a putative transcriptional activation domain (TAD) that spans aa 408-572.28 Additional residues of the TAD present in CALM(520-583)-AF10 could recruit transcriptional complexes or directly interact with DNA regulatory regions. Because the function of the CALM TAD remains unknown, additional studies are required to elucidate its potential contribution to CALM-AF10–dependent leukemogenesis. Importantly, our demonstration that NESCALM-AF10 induces leukemogenesis in vivo suggests that the CALM NES is sufficient for CALM-AF10–mediated immortalization.
CALM-AF10 leukemias are characterized by a global reduction in H3K79 methylation, and this phenotype may contribute to leukemogenesis by increasing chromosomal instability.15 Here, we demonstrate that export of AF10 out of the nucleus correlates with this perturbed epigenetic state. It has previously been proposed that CALM-AF10 acts in a dominant negative manner on endogenous AF10 to regulate global H3K79 hypomethylation.15 Because the OM-LZ domain of AF10 is necessary and sufficient for CALM-AF10–mediated transformation, CALM-AF10 could alter the subcellular localization of the OM-LZ binding partner, DOT1L. The relative exclusion of DOT1L from the nucleus may explain the global loss of H3K79 methylation observed in CALM-AF10–expressing cells. Indeed, we observed that CALM-AF10 and DOT1L co-localize in both the cytoplasm and nucleus (Figure 4D). Alternatively, CALM-AF10 has also been shown to alter the localization and function of another OM-LZ interactor, IKAROS, but the consequences of this mislocalization have not been elucidated.43,44 Finally, GAS41 is another OM-LZ binding protein that functions as a component of the SWI/SNF complex, and its mislocalization could potentially result in transcriptional deregulation.45 Further investigations are necessary to determine the effects of nuclear export of CALM-AF10 on endogenous AF10 and its known OM-LZ binding partners.
The up-regulation of effector Hoxa homeobox genes has been shown to be mediated by DOT1L-dependent H3K79 methylation in both MLL-rearranged and CALM-AF10 leukemias.14,16 Our finding that both H3K79 hypermethylation and Hoxa gene up-regulation are dependent on the CALM-derived NES (Figure 4E-F) is novel and intriguing. Because the CALM-derived NES mediates nuclear export of CALM-AF10, the effects on the Hoxa loci may be indirect, possibly by mislocalizing important transcriptional/epigenetic regulators to the cytoplasm. However, Okada et al16 showed that CALM-AF10 binds the Hoxa5 locus by ChIP. Although we have not been able to replicate these findings (data not shown), it is possible that CALM-AF10 directly binds chromatin as it undergoes nucleocytoplasmic shuttling. Likewise, the nuclear pore has been characterized as a site of active gene transcription in eukaryotes.46 Therefore, it is conceivable that the CALM-derived NES targets CALM-AF10 to the nuclear periphery (via CRM1), where Hoxa locus-specific gene activation may occur. Although the precise mechanism(s) by which CALM-AF10 mediates its transcriptional and epigenetic effects is complex, our LMB studies implicate the necessity of a CRM1 interaction. Intriguingly, another leukemic fusion protein, SET-NUP214, was found to bind both CRM1 and DOT1L.47 SET-NUP214 leukemic cells also have up-regulated Hoxa gene expression and local H3K79 hypermethylation, supporting a potential oncogenic mechanism involving DOT1L and CRM1.47 In addition, new AF10 fusion partners, NAP1L1, HNRNPH1, and DDX3X, have recently been identified in T-cell acute lymphoblastic leukemias.48,49 Similar to CALM, NAP1L1 and DDX3X contain characterized CRM1 interaction domains,50,51 and hnRNP H1 has a putative NES. Therefore, CRM1 interaction may be a recurrent feature of leukemic fusion proteins, emphasizing its potential role in mediating leukemogenesis. Further mechanistic studies aimed at understanding the role played by CRM1 interaction in leukemogenesis are needed.
Aggressive hematopoietic malignancies harboring CALM-AF10 translocations are seen in both pediatric and adult patients and are associated with a poor prognosis. Improving the outcome of CALM-AF10 leukemias depends on the development of targeted therapies with improved efficacy and reduced toxicity. Based on our discovery that immortalization by CALM-AF10 is dependent on a CALM NES, we hypothesize that NEIs represent an innovative approach to selectively target these malignancies. Importantly, we have shown that human CALM-AF10–expressing leukemic cells are more sensitive to treatment with LMB than CALM-AF10–negative lines (Figure 7B-C). Additionally, blocking nuclear export has been proposed as a therapeutic strategy against many types of cancer based on diverse mechanisms of action, such as preventing signaling through p53 or nuclear factor κB.52,53 In fact, LMB was tested as an anticancer agent in phase 1 clinical trials, but due to dose-limiting toxicity, it was not pursued further.54 Despite concerns about toxicity, targeting nuclear export remains an attractive possibility, and new, more potent, and less toxic agents are being tested.55 Therefore, blocking CRM1-dependent nuclear export is a potential therapeutic possibility for patients with CALM-AF10 leukemias. In addition, it may be beneficial to develop a small molecule inhibitor specifically directed against the NES of CALM to selectively block nuclear export of the CALM-AF10 oncoprotein.
In summary, a CALM-derived NES confers transformation potential to AF10 through perturbation of the epigenetic and transcriptional state. Nuclear export of AF10 mislocalizes a fraction of DOT1L to the cytoplasm, which correlates with a global loss of H3K79 methylation. In contrast, H3K79 hypermethylation is present at the Hoxa locus and is also dependent on the presence of a CALM-derived NES. From these results, we hypothesize that CRM1 inhibitors may represent a novel therapeutic approach for patients with CALM-AF10 leukemias.
Supplementary Material
Acknowledgments
The authors thank Michael Armstrong, Oren Becher, Lisa Crose, Chi Dang, Jonathan Haldeman, Corinne Linardic, and Rob Wechsler-Reya for thoughtful comments and suggestions, Eric Delabesse for providing the CALM-AF10 construct, and Yi Zhang for providing the hDOT1L construct.
This work was supported by NCI R01 CA 109281 (D.S.W.), a St. Baldrick’s Research Grant (D.S.W.), a V Foundation/Applebee’s Grant (D.S.W.), and a Hyundai Hope Grant (C.P.L.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.E.C. and C.P.L. designed the research, performed the research, and wrote the manuscript; P.B.S. performed the research; and D.S.W. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel S. Wechsler, 397 Hanes House, DUMC 102382, Duke University Medical Center, Durham, NC 27710; e-mail: dan.wechsler@duke.edu.
References
- 1.Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA. 1996;93(10):4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohlander SK, Muschinsky V, Schrader K, et al. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia. 2000;14(1):93–99. doi: 10.1038/sj.leu.2401614. [DOI] [PubMed] [Google Scholar]
- 3.Dreyling MH, Schrader K, Fonatsch C, et al. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood. 1998;91(12):4662–4667. [PubMed] [Google Scholar]
- 4.Silliman CC, McGavran L, Wei Q, Miller LA, Li S, Hunger SP. Alternative splicing in wild-type AF10 and CALM cDNAs and in AF10-CALM and CALM-AF10 fusion cDNAs produced by the t(10;11)(p13-14;q14-q21) suggests a potential role for truncated AF10 polypeptides. Leukemia. 1998;12(9):1404–1410. doi: 10.1038/sj.leu.2401109. [DOI] [PubMed] [Google Scholar]
- 5.Deshpande AJ, Rouhi A, Lin Y, et al. The clathrin-binding domain of CALM and the OM-LZ domain of AF10 are sufficient to induce acute myeloid leukemia in mice. Leukemia. 2011;25(11):1718–1727. doi: 10.1038/leu.2011.153. [DOI] [PubMed] [Google Scholar]
- 6.Narita M, Shimizu K, Hayashi Y, et al. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br J Haematol. 1999;105(4):928–937. doi: 10.1046/j.1365-2141.1999.01433.x. [DOI] [PubMed] [Google Scholar]
- 7.Deshpande AJ, Cusan M, Rawat VP, et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 2006;10(5):363–374. doi: 10.1016/j.ccr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Caudell D, Zhang Z, Chung YJ, Aplan PD. Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res. 2007;67(17):8022–8031. doi: 10.1158/0008-5472.CAN-06-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dik WA, Brahim W, Braun C, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19(11):1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 10.Caudell D, Aplan PD. The role of CALM-AF10 gene fusion in acute leukemia. Leukemia. 2008;22(4):678–685. doi: 10.1038/sj.leu.2405074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaplin T, Bernard O, Beverloo HB, et al. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood. 1995;86(6):2073–2076. [PubMed] [Google Scholar]
- 12.Linder B, Jones LK, Chaplin T, et al. Expression pattern and cellular distribution of the murine homologue of AF10. Biochim Biophys Acta. 1998;1443(3):285–296. doi: 10.1016/s0167-4781(98)00226-7. [DOI] [PubMed] [Google Scholar]
- 13.Saha V, Chaplin T, Gregorini A, Ayton P, Young BD. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc Natl Acad Sci USA. 1995;92(21):9737–9741. doi: 10.1073/pnas.92.21.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Lin YH, Kakadia PM, Chen Y, et al. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood. 2009;114(3):651–658. doi: 10.1182/blood-2009-03-209395. [DOI] [PubMed] [Google Scholar]
- 16.Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006;8(9):1017–1024. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tebar F, Bohlander SK, Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell. 1999;10(8):2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford MG, Pearse BM, Higgins MK, et al. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291(5506):1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 19.Scotland PB, Heath JL, Conway AE, et al. The PICALM protein plays a key role in iron homeostasis and cell proliferation. PLoS ONE. 2012;7(8):e44252. doi: 10.1371/journal.pone.0044252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vecchi M, Polo S, Poupon V, van de Loo JW, Benmerah A, Di Fiore PP. Nucleocytoplasmic shuttling of endocytic proteins. J Cell Biol. 2001;153(7):1511–1517. doi: 10.1083/jcb.153.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoddart A, Tennant TR, Fernald AA, Anastasi J, Brodsky FM, Le Beau MM. The clathrin-binding domain of CALM-AF10 alters the phenotype of myeloid neoplasms in mice. Oncogene. 2012;31(4):494–506. doi: 10.1038/onc.2011.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klebig ML, Wall MD, Potter MD, Rowe EL, Carpenter DA, Rinchik EM. Mutations in the clathrin-assembly gene Picalm are responsible for the hematopoietic and iron metabolism abnormalities in fit1 mice. Proc Natl Acad Sci USA. 2003;100(14):8360–8365. doi: 10.1073/pnas.1432634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16(14):4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgado E, Albouhair S, Lavau C. Flt3 is dispensable to the Hoxa9/Meis1 leukemogenic cooperation. Blood. 2007;109(9):4020–4022. doi: 10.1182/blood-2006-01-039586. [DOI] [PubMed] [Google Scholar]
- 25.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 26.Kudo N, Wolff B, Sekimoto T, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242(2):540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 27.Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324(5930):1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 28.Archangelo LF, Gläsner J, Krause A, Bohlander SK. The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene. 2006;25(29):4099–4109. doi: 10.1038/sj.onc.1209438. [DOI] [PubMed] [Google Scholar]
- 29.Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256(1):213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 30.DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99(10):3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 31.Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat Cell Biol. 2000;2(9):653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- 32.Gardini A, Cesaroni M, Luzi L, et al. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008;4(11):e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yassin ER, Sarma NJ, Abdul-Nabi AM, et al. Dissection of the transformation of primary human hematopoietic cells by the oncogene NUP98-HOXA9. PLoS ONE. 2009;4(8):e6719. doi: 10.1371/journal.pone.0006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsumoto T, Yoshida N, Kitabayashi I. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci. 2008;99(8):1523–1527. doi: 10.1111/j.1349-7006.2008.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buijs A, van Rompaey L, Molijn AC, et al. The MN1-TEL fusion protein, encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity. Mol Cell Biol. 2000;20(24):9281–9293. doi: 10.1128/mcb.20.24.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2011;152(2):141–154. doi: 10.1111/j.1365-2141.2010.08459.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5(5):821–830. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 38.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4(2):99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 39.Martin ME, Milne TA, Bloyer S, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4(3):197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 40.Crosetto N, Tikkanen R, Dikic I. Oncogenic breakdowns in endocytic adaptor proteins. FEBS Lett. 2005;579(15):3231–3238. doi: 10.1016/j.febslet.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 41.Lanzetti L, Di Fiore PP. Endocytosis and cancer: an ‘insider’ network with dangerous liaisons. Traffic. 2008;9(12):2011–2021. doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- 42.Miller SE, Sahlender DA, Graham SC, et al. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147(5):1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greif PA, Tizazu B, Krause A, Kremmer E, Bohlander SK. The leukemogenic CALM/AF10 fusion protein alters the subcellular localization of the lymphoid regulator Ikaros. Oncogene. 2008;27(20):2886–2896. doi: 10.1038/sj.onc.1210945. [DOI] [PubMed] [Google Scholar]
- 44.Greif PA, Bohlander SK. Up a lymphoid blind alley: does CALM/AF10 disturb Ikaros during leukemogenesis? World J Biol Chem. 2011;2(6):115–118. doi: 10.4331/wjbc.v2.i6.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debernardi S, Bassini A, Jones LK, et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99(1):275–281. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- 46.Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17(2):100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Van Vlierberghe P, van Grotel M, Tchinda J, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111(9):4668–4680. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandimarte L, Pierini V, Di Giacomo D, et al. Blood. 2012;120.. MLLT10 gene promiscuity unravels involvement of RNA processing genes in pediatric T-acute lymphoblastic leukemia [abstract] Abstract 1431. [Google Scholar]
- 50.Miyaji-Yamaguchi M, Kato K, Nakano R, Akashi T, Kikuchi A, Nagata K. Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol Cell Biol. 2003;23(18):6672–6684. doi: 10.1128/MCB.23.18.6672-6684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma D, Bhattacharya J. Evolutionary constraints acting on DDX3X protein potentially interferes with Rev-mediated nuclear export of HIV-1 RNA. PLoS ONE. 2010;5(3):e9613. doi: 10.1371/journal.pone.0009613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeda A, Sarma NJ, Abdul-Nabi AM, Yaseen NR. Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins. J Biol Chem. 2010;285(21):16248–16257. doi: 10.1074/jbc.M109.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mutka SC, Yang WQ, Dong SD, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69(2):510–517. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74(4):648–649. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranganathan P, Yu X, Na C, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120(9):1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.