Abstract
A cost-effective and efficacious influenza vaccine for use in commercial poultry farms would help protect against avian influenza outbreaks. Current influenza vaccines for poultry are expensive and subtype specific, and therefore there is an urgent need to develop a universal avian influenza vaccine. We have constructed a live bacterial vaccine against avian influenza by expressing a conserved peptide from the ectodomain of M2 antigen (M2e) on the surface of Lactococcus lactis (LL). Chickens were vaccinated intranasally with the lactococcal vaccine (LL-M2e) or subcutaneously with keyhole-limpet-hemocyanin conjugated M2e (KLH-M2e). Vaccinated and nonvaccinated birds were challenged with high pathogenic avian influenza virus A subtype H5N2. Birds vaccinated with LL-M2e or KLH-M2e had median survival times of 5.5 and 6.0 days, respectively, which were significantly longer than non-vaccinated birds (3.5 days). Birds vaccinated subcutaneously with KLH-M2e had a lower mean viral burden than either of the other two groups. However, there was a significant correlation between the time of survival and M2e-specific serum IgG. The results of these trials show that birds in both vaccinated groups had significantly (P < 0.05) higher median survival times than non-vaccinated birds and that this protection could be due to M2e-specific serum IgG.
1. Introduction
The US Poultry industry annually produces over 43 billion pounds of high-quality broiler chickens and turkeys and over 90 billion eggs, which in 2010 had a market value of $34.7 billion [1, 2]. Avian diseases are a constant threat to the industry. Viruses are of particular concern because antibiotics cannot control them, although vaccines can control some avian viral infections. Avian vaccines are an important component of protecting the value of commercial poultry. However, many commercial birds are not vaccinated because of the cost, labor, and difficulty in differentiating infected from vaccinated animals.
Avian influenza virus is an important concern to the poultry industry both in the USA and worldwide. It is highly contagious and causes two levels of disease [3]. Low pathogenic strains cause a disease that is seldom fatal but results in slower growth and lower egg production. The highly pathogenic form of the disease results in systemic morbidity and a high mortality rate (90–100%). Highly pathogenic avian influenza (HPAI) is a significant public health concern because of recent highly pathogenic H5N1 avian influenza outbreaks causing human deaths in Asia, Europe, Middle East, and Africa. According to the world health organization (WHO) update, since 2003 until February 2013, there were 620 confirmed cases of human infection with H5N1, of which 367 died due to disease complications. Although there are avian influenza vaccines approved in the USA for use in commercial poultry, they are subtype specific and costly to administer because they require parenteral delivery (intramuscular or subcutaneous).
Lactococcus lactis (LL) is a nonpathogenic, Gram-positive bacterium that is being developed as a delivery vehicle for vaccines. Various heterologous bacterial and viral antigens have been expressed from L. lactis, and antigen-specific immune responses have been reported [4–12]. The efficacy of lactococcal vaccines has been validated in many reports that have shown protection from infectious challenge of vaccinated animals [4, 12–15]. In mammals, L. lactis does not colonize the oral cavity or gastrointestinal tract but remains metabolically active and survives passage after oral administration [16–19]. It is thought that noncolonizing bacteria may be preferred over commensal bacteria for vaccine delivery because they may avoid antigen tolerance [20]. Little is known about L. lactis in chickens, but the closely related Streptococcus genus is abundant in chicken gastrointestinal contents [21, 22].
The M2 protein of avian influenza virus is one of three proteins with domains exposed outside the virus particle. The ectodomain of M2 (M2e) includes a peptide region that is conserved among all subtypes and therefore has been a main focus for the development of a universal influenza vaccine. In the intact virion, M2e is not the dominant immunogen [23–26]. However, antibodies to the M2e peptide increase survival and reduce disease upon infectious challenge in mice and chicken [27–32].
In this report, live L. lactis that expresses M2e (LL-M2e) or keyhole-limpet-hemocyanin- (KLH-) conjugated M2e (KLH-M2e) was used to vaccinate chickens. Immune responses were measured, and the vaccinated and nonvaccinated birds were challenged with highly pathogenic avian influenza virus. The results of these trials show that birds in both vaccinated groups had significantly (P < 0.05) higher median survival times than nonvaccinated birds and that this protection could be due to M2e-specific serum IgG.
2. Material and Methods
2.1. Vaccine Construction
DNA encoding (Figure 1) 10 tandem copies (10xM2e) of M2e (SLLTEVETLTRNGWECKCSDSSD) was prepared commercially (Blue Heron Biotechnology) using codons preferred by L. lactis. BspE1 restriction sites were included at each end for cloning. 10xM2e was cloned into the lactococcal expression vector pP16pip as described [8] using standard methods, creating pBG-10xM2e, and transformed [33] into the plasmid-free strain L. lactis LM2301 [34].
Figure 1.

Coding region of 10xM2e used for expression in L. lactis. (a) Protein and nucleic acid sequence of M2e from A/chicken/Pennsylvania/1370/1983/H5N2 is shown. (b) Nucleic acid sequence of coding region of 10xM2e used for expression in L. lactis is shown. The bases in italics were added to the ends of the coding region for 10xM2e. BspE1 sites (underlined) were used to clone the sequence in-frame into the unique BspE1 site in pip.
2.2. Polyclonal Antiserum
M2e peptide was prepared commercially (Global Peptide Services) and covalently linked to keyhole limpet hemocyanin (KLH) using a commercial kit according to the manufacturer's instructions (Thermo-Fisher Scientific). Polyclonal antiserum was prepared by injecting subcutaneously New Zealand white rabbits with 50 μg (M2e-equivalent) conjugate (KLH-M2e) mixed with adjuvant (TiterMax). M2e-specific titer was measured by ELISA as described [35].
2.3. L. lactis Surface Expression of M2e
M2e expression was measured using a modification of the indirect cellular ELISA [35]. Exponential phase cultures of L. lactis (pBG-10xM2e) were centrifuged (5,000 ×g, 5 min), washed, and resuspended in PBS to a final cell optical density (600 nm) = 1.0. Serial 1 : 2 dilutions of washed cells were added to a 96-well microtiter plate. Plates were further processed as described using polyclonal rabbit anti-M2e serum and preimmune serum, goat-anti-rabbit IgG-horseradish peroxidase conjugate (Santa Cruz Biotechnology), and chemiluminescent substrate (Thermo-Fisher Scientific). Plates were read in a Tecan Infinite F500 plate reader.
2.4. Virus Stocks
Highly Pathogenic Avian influenza A/chicken/Pennsylvania/1370/1983 (H5N2) virus was obtained from United States Department of Agriculture. Viral stocks were grown in day 10 embryonated chicken eggs for 3–5 days. Allantoic fluid was collected, tested by hemagglutination assay [36], and stored at −85°C. Egg infectious dose (50%) was calculated by the formula of Reed and Muench [37].
2.5. Serum and Fecal Analysis
Serum and fecal samples were analyzed for M2e antibodies by ELISA [35] using microtiter plates coated with 2 μg/mL M2e peptide. A standard curve was included on each plate by making a serial dilution of chicken IgG (Rockland). The plates were developed with goat antichicken IgG-horseradish peroxidase conjugate (Bethyl Laboratories) and chemiluminescent substrate (Thermo-Fisher Scientific). Luminescent signal was converted to IgG concentration by extrapolation from the graph of luminescence versus IgG concentration of the standards, and corrected for sample dilution. Each sample was analyzed at three dilutions in duplicate.
2.6. Cell-Mediated Immune Response
A lymphocyte proliferation assay was performed as described [38]. Peripheral lymphocytes were isolated from blood collected 14 days after the final vaccination, stained with carboxyfluorescein diacetate succinimidyl ester, and stimulated for 72 h at 39°C with 20 μg/mL M2e peptide, 20 μg/mL nonspecific peptide, or 10 μg/mL concanavalin A (Sigma). Stimulated and nonstimulated lymphocytes were stained with mouse antichicken CD4-R-phycoerythrin clone CT-4 (Thermo-Fisher Scientific) and analyzed by flow cytometry on a Cytomics FC 500 instrument (Becton Dickinson) and FlowJo software (Tree Star).
2.7. Vaccination and Challenge
Two groups of six seventeen-day-old Neo Brown chickens were vaccinated intranasally on three consecutive days with 4 × 1010 cfu LL-M2e in 100 μL PBS. The regimen was repeated 2 and 4 weeks later. Six seventeen-day-old Neo Brown chickens were each vaccinated subcutaneously on the back of the neck with a 1 : 1 mixture of Titermax Gold adjuvant and 50 μg M2e equivalent KLH-M2e conjugate in a total volume of 400 μL. The subcutaneous vaccination was repeated 2 and 4 weeks later. A negative control group of six seventeen-day-old Neo Brown chickens was not vaccinated. L. lactis control was not used in the experiment because of two reasons: (1) previous data in our lab [8, 14] and previous publications have suggested that L. lactis does not elicit acquired immune response [4–15], and (2) the HEPA-filtered primary containment cage within the animal biosafety laboratory-3+ (ABSL-3+) could accommodate only 12 birds at a time and it was decided by investigators and Oregon State ethical committee members to use vaccinated and nonvaccinated groups only. Serum was collected before vaccination and 1 week after the last vaccination. Fecal samples were collected before and after vaccination, mixed with 0.3 mL 0.5% bovine serum albumin, 0.02% NaN3, and 1x protease inhibitor (Boehringer Mannheim) in PBS, and stored at −20°C until assayed.
Birds were challenged intranasally 2 weeks after vaccination with 1 × 104 egg infectious dose of highly pathogenic avian influenza virus A/chicken/Pennsylvania/1370/1983 (H5N2) in 100 μL PBS using a micropipettor to deliver the virus into the nasal opening. Tracheal swabs were collected prior to infection and on day 3 after infection. Body weight and cloacal temperature were recorded daily.
2.8. Tracheal Swab Analysis
Tracheal swabs were collected by inserting a calcium alginate fiber-tipped applicator swab (Fisher Scientific) into the trachea and moving the swab 10 times up and down about 1 cm. The swab was immediately placed in 1 mL minimal essential medium (Life Technologies) plus streptomycin (100 μg/mL), penicillin (100 units/mL), and amphotericin B (0.25 μg/mL) and stored at −80°C. Viral content of the tracheal swabs was assessed by plaque assay. Briefly, the swabs were thawed and mixed by vortexing. A 1 : 10 dilution series of each sample was prepared in PBS and then added to tissue cultures of Madin-Darby canine kidney cells (MDCK). After 72 h at 37°C + 5% CO2, plates were fixed with formalin and stained with crystal violet, and plaques were counted.
2.9. Statistical Analysis
Data were analyzed using GraphPad software, Prism 4.0, and InStat 3.0. Serum means and tracheal plaques were compared using two-tailed, unpaired Student's t-test with Welch's correction. Correlation analysis between serum response and survival was done using Pearson's analysis (two-tailed), and a line was plotted using linear regression analysis. Survival was analyzed by the method of Kaplan and Meier using the logrank test. Temperature and weight change was analyzed using nonparametric ANOVA (Kruskal-Wallis test).
2.10. Animals
All procedures using animals complied with all state and federal laws and were approved by the Oregon State University Institutional Animal Care and Use Committee (approval number 3682). All experiments involving high pathogenic avian influenza virus were conducted in CDC/APHIS-USDA approved ABSL-3+ high containment facility at VMAIL, Oregon State University.
3. Results
3.1. Vaccine Design and Construction
The M2e sequence from HPAI strain A/Chicken/Pennsylvania/1370/1983/H5N2 was selected for construction of the vaccine (Figure 1). Ten tandem repeats of the coding region for the M2e were cloned in-frame into the coding region for a surface protein from L. lactis. The genetic fusion was cloned into an expression vector with a strong promoter for L. lactis and transformed into L. lactis (LL-M2e).
3.2. Expression of M2e
Expression of M2e protein on the surface of LL-M2e was measured by ELISA assay using preimmune and rabbit polyclonal anti-M2e antibodies. The results show that the level of expression of M2e was proportional to the amount of LL-M2e bound to the ELISA plate (Figure 2). Preimmune serum did not show any signal. The expression of M2e protein on the surface of LL vector with no M2e gene was not detected using preimmune and polyclonal antibodies suggesting that the antibody specific to M2e is not binding to any protein from the LL vector with no M2e gene (data not shown). Three independent experiments were conducted.
Figure 2.
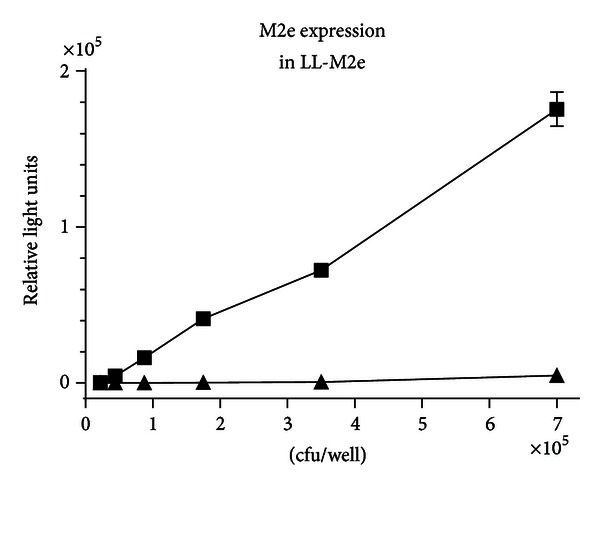
Expression of M2e from L. lactis. A 1/2 serial dilution of LL-M2e washed cells was bound to a 96-well microtiter plate and analyzed by ELISA. Cell surface-bound M2e was detected by adding a constant amount/well of preimmune (▲) or postimmune (■) rabbit polyclonal M2e antiserum. Expression of M2e protein on the surface of LL vector with no M2e gene was not detected using preimmune and polyclonal antibodies (data not shown). The signal was developed with secondary antibody anti-rabbit IgG-horseradish peroxidase conjugate and chemiluminescent substrate. Error bars indicate standard deviation. Three independent experiments were conducted (n = 3).
3.3. Immune Response
Intranasal vaccination with LL-M2e was tested in 12 chickens. In addition, another group of 6 chickens was vaccinated subcutaneously with M2e peptide conjugated to keyhole limpet hemocyanin (KLH-M2e). Six birds were nonvaccinated, which formed a negative control group. One week after the final dose of vaccine, blood was collected, and the M2e-specific serum IgG response was measured by ELISA.
The results indicate that 8 of 12 birds vaccinated with LL-M2e had a measurable humoral response, and the group mean was significantly (P < 0.05) higher than that of the nonvaccinated group (Figure 3). All birds vaccinated with KLH-M2e had a measurable M2e-specific response that was significantly (P < 0.05) higher than responses in either of the other groups. None of the birds in the nonvaccinated group showed an M2e-specific humoral response.
Figure 3.
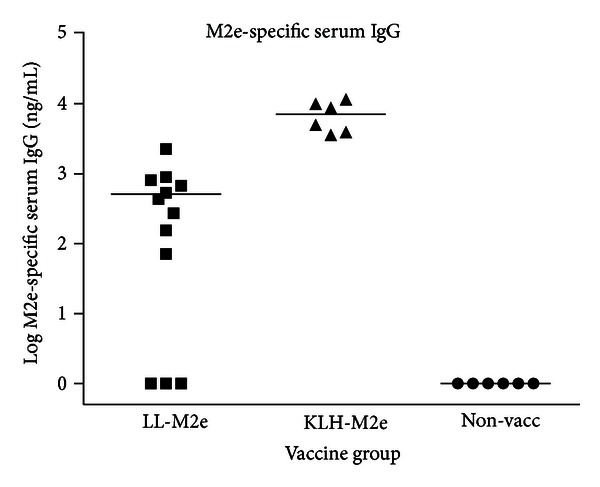
Serum response. Serum was collected 1 week after final vaccination and analyzed for M2e-specific serum IgG by ELISA using microtiter plates coated with 2 μg/mL M2e peptide. The plates were developed with goat antichicken IgG-horseradish peroxidase conjugate and chemiluminescent substrate. Luminescent signal from each sample was converted to concentration of chicken IgG by extrapolation from the graph of luminescence versus IgG concentration of the standards and corrected for sample dilution. Each sample was analyzed at two or three different dilutions, and each dilution was analyzed in duplicate. Group mean M2e-specific IgG concentration is shown.
3.4. Infectious Challenge
Two weeks after the final dose of vaccine, the vaccinated (LL-M2e and KLH-M2e vaccination groups) and nonvaccinated chickens were challenged with the highly pathogenic strain A/chicken/Pennsylvania/1370/H5N2.
Body temperature was monitored, and the results show that all groups responded similarly (Figure 4(a)). Temperatures rose with an average of 1.04°C from day 0 to day 3 and then declined with an average of 1.38°C by day 10 after infection. There was no statistically significant difference in temperatures among all groups at any time after infection.
Figure 4.
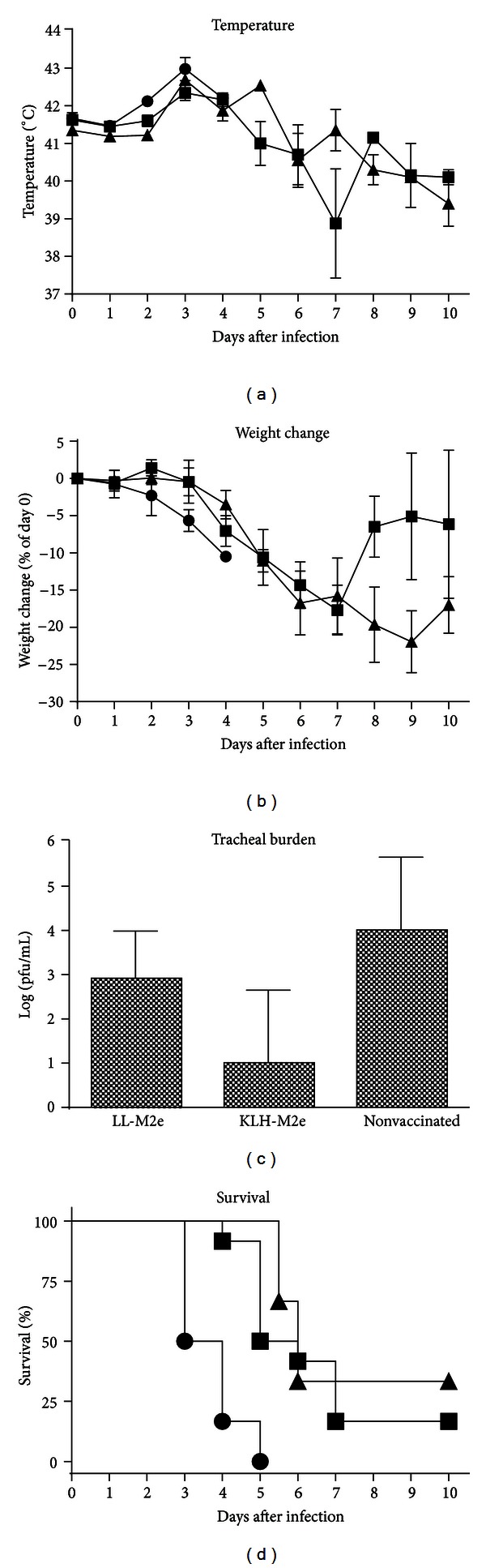
Weight loss, body temperature, tracheal burden, and survival. Groups of vaccinated (intranasal LL-M2e (■), subcutaneous KLH-M2e (▲)) or nonvaccinated (⚫) birds were challenged with high pathogenic avian influenza H5N2 (A/chicken/Pennsylvania/1370/1983). n = 12 (LL-M2e), n = 6 (KLH-M2e and nonvaccinated). Error bars indicate standard deviation. (a) Body temperature was measured for each bird, and the group mean is shown. (b) Weight loss for each bird was calculated as a difference compared to day 0, and the group mean is shown. (c) Tracheal swabs were collected 3 days after infection and analyzed for viral plaques. (d) Survival was monitored for 10 days after infection.
Weight decreased to a maximum loss of between 18 and 22% over the first 7 to 9 days and then gradually increased (Figure 4(b)). There was no statistical difference in weight loss among the groups at any time after infection.
Viral burden in tracheal swabs was measured 3 days after infection (Figure 4(c)). Mean tracheal burden varied among groups from 101 to 104 pfu/mL, and there was a significantly (P < 0.05) lower mean value for birds vaccinated with KLH-M2e than either of the other two groups. There was no significant (P > 0.05) difference between the group vaccinated with LL-M2e and the nonvaccinated group.
Birds in both vaccinated groups had significantly (P < 0.05) higher median survival times than nonvaccinated birds (Figure 4(d)). Birds vaccinated with LL-M2e or KLH-M2e had median survival times of 5.5 and 6.0 days, respectively. Nonvaccinated birds had a median survival time of 3.5 days. Two of 12 birds from the LL-M2e group survived and 2 of 6 birds from the KLH-M2e group survived, whereas none of the six nonvaccinated birds survived. There was no statistical difference in survival between the two vaccinated groups.
An analysis of M2e-specific serum IgG as a function of survival showed a significant (P < 0.01, R 2 = 0.9197) correlation for the group vaccinated with LL-M2e (Figure 5). An analysis of the group vaccinated with KLH-M2e showed a similar trend, but the correlation was not statistically significant (P > 0.05, data not shown).
Figure 5.
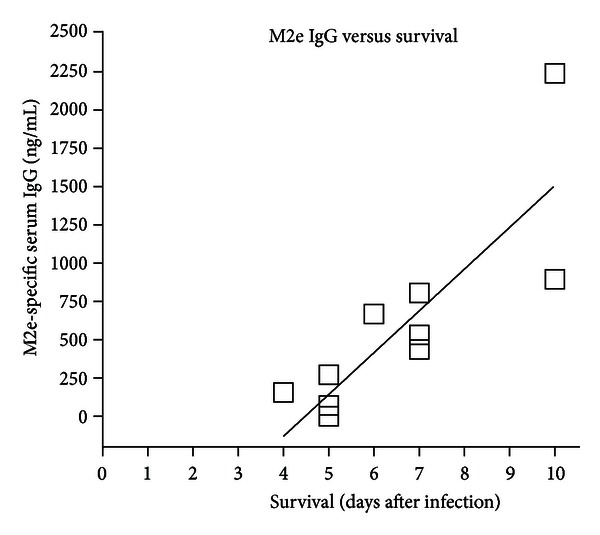
Correlation analysis. M2e-specific serum IgG from birds vaccinated with LL-M2e (n = 12) was plotted as a function of survival following infectious challenge. The line represents a linear regression analysis.
Although there was a correlation between M2e-specific serum response and survival in the group vaccinated with LL-M2e, additional immune responses were analyzed. CD4+ T lymphocytes from 6 LL-M2e-vaccinated birds were analyzed for M2e-specific proliferation. In addition, fecal samples for detection of IgA were collected 2 weeks after final vaccination and just prior to infection. In all of the birds tested, M2e-specific CD4+ lymphocyte response or fecal IgA was below a threshold limit of our assay in vaccinated birds (data not shown).
4. Discussion
The ectodomain of the M2 protein is an attractive choice for a cross-subtype vaccine. Its amino acid sequence is not only highly conserved, but also nonglycosylated, which is essential for antigens expressed from bacteria. Although M2e is only weakly antigenic in the context of an infection or whole virus vaccine, antibodies against M2e reduce infection and protect against viral replication and death, at least in mammalian models [25, 28–32, 39].
The strategy of using M2e as a vaccine is different from the conventional one currently used for human seasonal vaccines. Current vaccines and natural infection induce a humoral response to two immunodominant viral surface antigens, hemagglutinin (HA) and neuraminidase (NA). However, HA and NA undergo intense selective pressure due to the host immune response and are constantly changing. The use of M2e as antigen would avoid the problem of genetic drift and shift that characterizes both HA and NA because M2e is genetically stable. Even under prolonged selective pressure, only a single amino acid (proline to leucine or histidine at position 9) has been found to change in M2e [40]. It is likely that the genetic drift in the highly conserved M2e would be low [26, 41], which suggests that M2e vaccines would be universally effective against many subtypes and would not require seasonal modification like the current influenza vaccines.
L. lactis has been used as a vaccine delivery vehicle for various types of antigens, including those from bacteria and viruses [7, 13, 25, 42, 43]. Our expression system in L. lactis has been used previously to display an antigen (M6c) from Streptococcus pyogenes [8]. Mice vaccinated intranasally with the L. lactis-M6c developed a significant humoral response to M6c that correlated with protection from infectious challenge [14].
In the present report, we have similarly expressed 10 tandem copies of M2e on the surface of L. lactis and measured immune responses. Most of the vaccinated chickens developed an M2e-specific serum IgG response. The lack of robust M2e-specific fecal IgA or CD4+ T lymphocyte response could be due to differential processing of antigen by the immune system, which needs to be explored further. This indicates that LL-M2e induced mainly humoral response, but less significant cellular response.
The infectious challenge results show that chickens vaccinated intranasally with LL-M2e or vaccinated subcutaneously with KLH-M2e survived infectious challenge longer than nonvaccinated birds. Birds vaccinated with LL-M2e or KLH-M2e had median survival times of 5.5 and 6.0 days, respectively. Nonvaccinated birds had a median survival time of 3.5 days. Two of 12 birds from the LL-M2e group survived and 2 of 6 birds from the KLH-M2e group survived, whereas none of the six nonvaccinated birds survived. Birds in both vaccinated groups had significantly (P < 0.05) higher median survival times than nonvaccinated birds.
Weight loss or body temperature did not differ much among treatment groups, and therefore we believe that apparently these measures of health were not predictive of survival.
Viral burden was also measured. Previous experiments (not shown) indicated that viral burden in the tracheal swabs peaked at day 3 after infection. Birds vaccinated subcutaneously with KLH-M2e had a lower mean viral burden than either of the other two groups. Perhaps this is a reflection of the higher M2e-specific serum IgG. We found no statistical significance in the amount of virus in tracheal swabs at day 3 after infection between birds vaccinated with LL-M2e and nonvaccinated birds.
An analysis of our data showed that protection may be due to M2e-specific serum IgG. Birds with higher M2e-specific IgG tended to survive longer. Previous studies present conflicting results on the mechanism of protection provided by M2e vaccines. Some studies show that antibodies to M2e provide protection [32, 44, 45]. Another study showed no correlation between M2e-specific titer and protection from infectious challenge [46]. Some reports show that M2e vaccines can induce an M2e-specific CD4+ T-cell response that may contribute to protection [47, 48]. Still other reports suggest that M2e-specific antibodies may bind to infected cells and direct natural killer T cells, macrophages, or other host immune cells to kill the infected cell [26, 49–51]. A recent article by El Bakkouri et al. [52] has suggested that alveolar macrophages and Fc receptor-dependent elimination of influenza A virus-infected cells are essential for immune protection by anti-M2e IgG. Our results are consistent with a mechanism of protection that depends on an M2e-specific serum IgG response.
5. Conclusion
In conclusion, the data show that intranasal vaccination of chickens with LL-M2e or subcutaneous vaccination of chickens with KLH-M2e provided a significant increase in survival compared to nonvaccinated birds. Survival and protection could be due to serum M2e-specific IgG response to the vaccine.
Conflict of Interests
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper. Further, none of the authors have any financial conflict of interests with Becton Dickinson for using Cytomics FC 500 instrument and Tree Star for using FlowJo software in this paper.
Acknowledgments
This work was supported by grants from the United States Department of Agriculture AICAP (2008-55204-18863) and NIFA (2011-85204-30046) programs. The authors thank Fred Schnell for assisting with flow cytometry.
References
- 1.USDA Economic Research Service. 2012, http://www.ers.usda.gov/
- 2.US Poultry and Egg Association. 2012, http://www.uspoultry.org/economic_data/
- 3.Londt BZ, Banks J, Alexander DJ. Highly pathogenic avian influenza viruses with low virulence for chickens in in vivo tests. Avian Pathology. 2007;36(5):347–350. doi: 10.1080/03079450701589134. [DOI] [PubMed] [Google Scholar]
- 4.Bermúdez-Humarán LG, Cortes-Perez NG, Lefèvre F, et al. A novel mucosal vaccine based on live lactococci expressing E7 antigen and IL-12 induces systemic and mucosal immune responses and protects mice against human papillomavirus type 16-induced tumors. Journal of Immunology. 2005;175(11):7297–7302. doi: 10.4049/jimmunol.175.11.7297. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain L, Wells JM, Robinson K, Schofield K, LePage R. Mucosal immunization with recombinant Lactococcus lactis . In: Pozzi G, Wells JM, editors. Gram-Positive Bacteria as Vaccine Vehicles for Mucosal Immunization. Austin, Tex, USA: Landes Bioscience; 1997. pp. 83–106. [Google Scholar]
- 6.Dieye Y, Hoekman AJW, Clier F, Juillard V, Boot HJ, Piard JC. Ability of Lactococcus lactis to export viral capsid antigens: a crucial step for development of live vaccines. Applied and Environmental Microbiology. 2003;69(12):7281–7288. doi: 10.1128/AEM.69.12.7281-7288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enouf V, Langella P, Commissaire J, Cohen J, Corthier G. Bovine rotavirus nonstructural protein 4 produced by Lactococcus lactis is antigenic and immunogenic. Applied and Environmental Microbiology. 2001;67(4):1423–1428. doi: 10.1128/AEM.67.4.1423-1428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geller BL, Wade N, Gilberts TD, Hruby DE, Johanson R, Topisirovic L. Surface expression of the conserved C repeat region of streptococcal M6 protein within the Pip bacteriophage receptor of Lactococcus lactis . Applied and Environmental Microbiology. 2001;67(12):5370–5376. doi: 10.1128/AEM.67.12.5370-5376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MH, Roussel Y, Wilks M, Tabaqchali S. Expression of Helicobacter pylori urease subunit B gene in Lactococcus lactis MG1363 and its use as a vaccine delivery system against H. pylori infection in mice. Vaccine. 2001;19(28-29):3927–3935. doi: 10.1016/s0264-410x(01)00119-0. [DOI] [PubMed] [Google Scholar]
- 10.Pei H, Liu J, Cheng Y, et al. Expression of SARS-coronavirus nucleocapsid protein in Escherichia coli and Lactococcus lactis for serodiagnosis and mucosal vaccination. Applied Microbiology and Biotechnology. 2005;68(2):220–227. doi: 10.1007/s00253-004-1869-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez CA, Eichwald C, Burrone O, de Mendoza D. Rotavirus vp7 antigen produced by Lactococcus lactis induces neutralizing antibodies in mice. Journal of Applied Microbiology. 2005;99(5):1158–1164. doi: 10.1111/j.1365-2672.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- 12.Xin KQ, Hoshino Y, Toda Y, et al. Immunogenicity and protective efficacy of orally administered recombinant Lactococcus lactis expressing surface-bound HIV Env. Blood. 2003;102(1):223–228. doi: 10.1182/blood-2003-01-0110. [DOI] [PubMed] [Google Scholar]
- 13.Lei H, Xu Y, Chen J, Wei X, Lam DMK. Immunoprotection against influenza H5N1 virus by oral administration of enteric-coated recombinant Lactococcus lactis mini-capsules. Virology. 2010;407(2):319–324. doi: 10.1016/j.virol.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Mannam P, Jones KF, Geller BL. Mucosal vaccine made from live, recombinant Lactococcus lactis protects mice against pharyngeal infection with Streptococcus pyogenes . Infection and Immunity. 2004;72(6):3444–3450. doi: 10.1128/IAI.72.6.3444-3450.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou P, Liu W, Chen YH. The epitope recognized by a monoclonal antibody in influenza A virus M2 protein is immunogenic and confers immune protection. International Immunopharmacology. 2005;5(4):631–635. doi: 10.1016/j.intimp.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Corthier G, Delorme C, Ehrlich SD, Renault P. Use of luciferase genes as biosensors to study bacterial physiology in the digestive tract. Applied and Environmental Microbiology. 1998;64(7):2721–2722. doi: 10.1128/aem.64.7.2721-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drouault S, Corthier G, Ehrlich SD, Renault P. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Applied and Environmental Microbiology. 1999;65(11):4881–4886. doi: 10.1128/aem.65.11.4881-4886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klijn N, Weerkamp AH, de Vos WM. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Applied and Environmental Microbiology. 1995;61(7):2771–2774. doi: 10.1128/aem.61.7.2771-2774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vesa T, Pochart P, Marteau P. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Alimentary Pharmacology and Therapeutics. 2000;14(6):823–828. doi: 10.1046/j.1365-2036.2000.00763.x. [DOI] [PubMed] [Google Scholar]
- 20.Bermúdez-Humarán LG, Langella P, Miyoshi A, et al. Production of human papillomavirus type 16 E7 protein in Lactococcus lactis . Applied and Environmental Microbiology. 2002;68(2):917–922. doi: 10.1128/AEM.68.2.917-922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czerwiński J, Højberg O, Smulikowska S, Engberg RM, Mieczkowska A. Influence of dietary peas and organic acids and probiotic supplementation on performance and caecal microbial ecology of broiler chickens. British Poultry Science. 2010;51(2):258–269. doi: 10.1080/00071661003777003. [DOI] [PubMed] [Google Scholar]
- 22.Olsen KN, Henriksen M, Bisgaard M, Nielsen OL, Christensen H. Investigation of chicken intestinal bacterial communities by 16S rRNA targeted fluorescence in situ hybridization. Antonie van Leeuwenhoek. 2008;94(3):423–437. doi: 10.1007/s10482-008-9260-0. [DOI] [PubMed] [Google Scholar]
- 23.Feng J, Zhang M, Mozdzanowska K, et al. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virology Journal. 2006;3:102–113. doi: 10.1186/1743-422X-3-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson BE, Moran TM, Bona CA, Popple SW, Kilbourne ED. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin: II. Sequential infection of mice simulates human experience. Journal of Immunology. 1987;139(6):2010–2014. [PubMed] [Google Scholar]
- 25.Liu W, Li H, Chen YH. N-terminus of M2 protein could induce antibodies with inhibitory activity against influenza virus replication. FEMS Immunology and Medical Microbiology. 2003;35(2):141–146. doi: 10.1016/S0928-8244(03)00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schotsaert M, de Filette M, Fiers W, Saelens X. Universal M2 ectodomain-based influenza A vaccines: preclinical and clinical developments. Expert Review of Vaccines. 2009;8(4):499–508. doi: 10.1586/erv.09.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babapoor S, Neef T, Mittelholzer C, et al. A novel vaccine using nanoparticle platform to present immunogenic M2e against avian influenza infection. Influenza Research and Treatment. 2011;2011:12 pages. doi: 10.1155/2011/126794.126794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Filette M, Jou WM, Birkett A, et al. Universal influenza A vaccine: optimization of M2-based constructs. Virology. 2005;337(1):149–161. doi: 10.1016/j.virol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Fan J, Liang X, Horton MS, et al. Preclinical study of influenza virus a M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine. 2004;22(23-24):2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 30.Frace AM, Klimov AI, Rowe T, Black RA, Katz JM. Modified M2 proteins produce heterotypic immunity against influenza A virus. Vaccine. 1999;17(18):2237–2244. doi: 10.1016/s0264-410x(99)00005-5. [DOI] [PubMed] [Google Scholar]
- 31.Mozdzanowska K, Feng J, Eid M, et al. Induction of influenza type A virus-specific resistance by immunization of mice with a synthetic multiple antigenic peptide vaccine that contains ectodomains of matrix protein 2. Vaccine. 2003;21(19-20):2616–2626. doi: 10.1016/s0264-410x(03)00040-9. [DOI] [PubMed] [Google Scholar]
- 32.Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nature Medicine. 1999;5(10):1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 33.Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Applied and Environmental Microbiology. 1989;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh PM, McKay LL. Recombinant plasmid associated with cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. Journal of Bacteriology. 1981;146(3):937–944. doi: 10.1128/jb.146.3.937-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ausubel FM, Brent R, Kingston RE, et al. Current Protocols in Molecular Biology. New York, NY, USA: John Wiley & Sons; 1998. [Google Scholar]
- 36.Donald HB, Isaacs A. Counts of influenza virus particles. Journal of General Microbiology. 1954;10(3):457–464. doi: 10.1099/00221287-10-3-457. [DOI] [PubMed] [Google Scholar]
- 37.Swayne DE, Senne DA, Beard CW. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. Kennett Square, Pa, USA: American Association of Avian Pathologists; 1998. [Google Scholar]
- 38.Sitz K, Birx D. Lymphocyte proliferation assay. Methods in Molecular Medicine. 1999;17:343–353. doi: 10.1385/0-89603-369-4:343. [DOI] [PubMed] [Google Scholar]
- 39.Grandea AG, III, Olsen OA, Cox TC, et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(28):12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zharikova D, Mozdzanowska K, Feng J, Zhang M, Gerhard W. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. Journal of Virology. 2005;79(11):6644–6654. doi: 10.1128/JVI.79.11.6644-6654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerging Infectious Diseases. 2006;12(4):569–574. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei H, Sheng Z, Ding Q, et al. Evaluation of oral immunization with recombinant avian influenza virus HA1 displayed on the Lactococcus lactis surface and combined with the mucosal adjuvant cholera toxin subunit B. Clinical and Vaccine Immunology. 2011;18(7):1046–1051. doi: 10.1128/CVI.00050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YJ, Ma GP, Li GW, et al. Oral vaccination with the porcine rotavirus VP4 outer capsid protein expressed by Lactococcus lactis induces specific antibody production. Journal of Biomedicine and Biotechnology. 2010;2010:9 pages. doi: 10.1155/2010/708460.708460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessa J, Schmitz N, Hinton HJ, Schwarz K, Jegerlehner A, Bachmann MF. Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design. European Journal of Immunology. 2008;38(1):114–126. doi: 10.1002/eji.200636959. [DOI] [PubMed] [Google Scholar]
- 45.Denis J, Acosta-Ramirez E, Zhao Y, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26(27-28):3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 46.Mozdzanowska K, Zharikova D, Cudic M, Otvos L, Gerhard W. Roles of adjuvant and route of vaccination in antibody response and protection engendered by a synthetic matrix protein 2-based influenza A virus vaccine in the mouse. Virology Journal. 2007;4:118–131. doi: 10.1186/1743-422X-4-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eliasson DG, El Bakkouri K, Schön K, et al. CTA1-M2e-DD: a novel mucosal adjuvant targeted influenza vaccine. Vaccine. 2008;26(9):1243–1252. doi: 10.1016/j.vaccine.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 48.Mozdzanowska K, Furchner M, Zharikova D, Feng J, Gerhard W. Roles of CD4+ T-cell-independent and -dependent antibody responses in the control of influenza virus infection: evidence for noncognate CD4+ T-cell activities that enhance the therapeutic activity of antiviral antibodies. Journal of Virology. 2005;79(10):5943–5951. doi: 10.1128/JVI.79.10.5943-5951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. Journal of Immunology. 2004;172(9):5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 50.Tompkins SM, Zhao ZS, Lo CY, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerging Infectious Diseases. 2007;13(3):426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang R, Song A, Levin J, et al. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Research. 2008;80(2):168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 52.El Bakkouri K, Descamps F, de Filette M, et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. Journal of Immunology. 2011;186(2):1022–1031. doi: 10.4049/jimmunol.0902147. [DOI] [PubMed] [Google Scholar]


