The transcription factor Foxo1 is required for the differentiation of memory CD8+ T cells, and its absence hinders clearance of secondary infections.
Abstract
The forkhead O transcription factors (FOXO) integrate a range of extracellular signals, including growth factor signaling, inflammation, oxidative stress, and nutrient availability, to substantially alter the program of gene expression and modulate cell survival, cell cycle progression, and many yet to be unraveled cell type–specific responses. Naive antigen-specific CD8+ T cells undergo a rapid expansion and arming of effector function within days of pathogen exposure. In addition, by the peak of expansion, they form precursors to memory T cells capable of self-renewal and indefinite survival. Using lymphocytic choriomeningitis virus Armstrong to probe the response to infection, we found that Foxo1−/− CD8+ T cells expand normally with no defects in effector differentiation, but continue to exhibit characteristics of effector T cells long after antigen clearance. The KLRG1lo CD8+ T cells that are normally enriched for memory-precursor cells retain Granzyme B and CD69 expression, and fail to up-regulate TCF7, EOMES, and other memory signature genes. As a correlate, Foxo1−/− CD8+ T cells were virtually unable to expand upon secondary infection. Collectively, these results demonstrate an intrinsic role for FOXO1 in establishing the post-effector memory program that is essential to forming long-lived memory cells capable of immune reactivation.
Intracellular infectious agents stimulate several thousand antigen-specific naive CD8+ T cells to expand up to 10,000-fold resulting in lymphocytosis and lymphadenopathy (Wirth and Harty, 2009). Within this expanded T cell population, there exist several distinct subsets that can be characterized by both function and phenotype. Cells exhibiting strong cytotoxicity to the instigating agent express high levels of perforin, granzymes, and the killer cell lectin–like receptor G1 (KLRG1). With sterilizing immunity, many of these terminally differentiated effector cells die at a high rate over a 2-wk period after the peak of the expansion. In contrast, a subset of T cells does not express KLRG1, displays a relatively reduced rate of cell death, and preferentially contributes to indelible antigen-specific immune memory (Sarkar et al., 2008; Parish and Kaech, 2009). Experiments with single-cell transfers show that these diverse populations arise from a common precursor (Stemberger et al., 2007; Gerlach et al., 2010), and this commitment may be influenced early in the process of naive T cell activation (Celli et al., 2008; Beuneu et al., 2010).
The differentiation and expansion of CD8+ effector T cells depends on co-stimulation, growth factors such as IL-2 (Williams et al., 2006; Bachmann et al., 2007; Obar et al., 2010; Pipkin et al., 2010), and inflammatory cytokines, especially IL-12, that promote the expression of TBX21 (Curtsinger et al., 2003; Takemoto et al., 2006; Joshi et al., 2007; Pearce and Shen, 2007). Further studies have shown that IL-2 acts, in part, through the transcriptional repressor BLIMP1 (encoded by Prdm1); accordingly, Prdm1−/− T cells display a defect in effector cell differentiation and more readily form memory-precursor cells. The pro-memory transcriptional profile that includes Bcl6, Tcf7, and Eomes is inhibited by BLIMP1, whereas a transcription factor associated with effector T cells, TBX21, is enhanced by BLIMP1 (Kallies et al., 2009; Rutishauser et al., 2009; Shin et al., 2009; Ji et al., 2011; Yang et al., 2011).
Studies have shown that the AKT signaling pathway promotes effector cell differentiation at the expense of memory cell precursors (Hand et al., 2010; Kim et al., 2012). In addition, the mammalian target of rapamycin, a downstream target of AKT, is a major regulator of memory CD8+ T cell differentiation (Araki et al., 2009; Pearce et al., 2009). Thus treatment with rapamycin or metformin enhanced both the quantity and quality of memory CD8+ T cells. Because inhibition of the FOXO1 transcription factor is a major conduit of AKT-mediated signaling, we set out to determine whether FOXO1 broadly affects the contingency of effector versus memory-precursor differentiation, and to what extent FOXO1 determines the program of memory T cell gene expression. Here, we show that the loss of Foxo1 has little effect on the expansion and survival of antigen-stimulated CD8+ T cells, but causes them to maintain an activated effector phenotype. These persisting Foxo1-null T cells were unable to expand upon reactivation, and this phenotype correlated with a extensive pattern of gene expression that favors the formation of effector T cells. We conclude that FOXO1 regulates the fate of effector versus memory-precursor T cells, and this has implications for the manner with which the physiological state of the organism impacts the outcome of an immune response.
RESULTS
Normal expansion of Foxo1−/− CD8+ T cells after viral infection
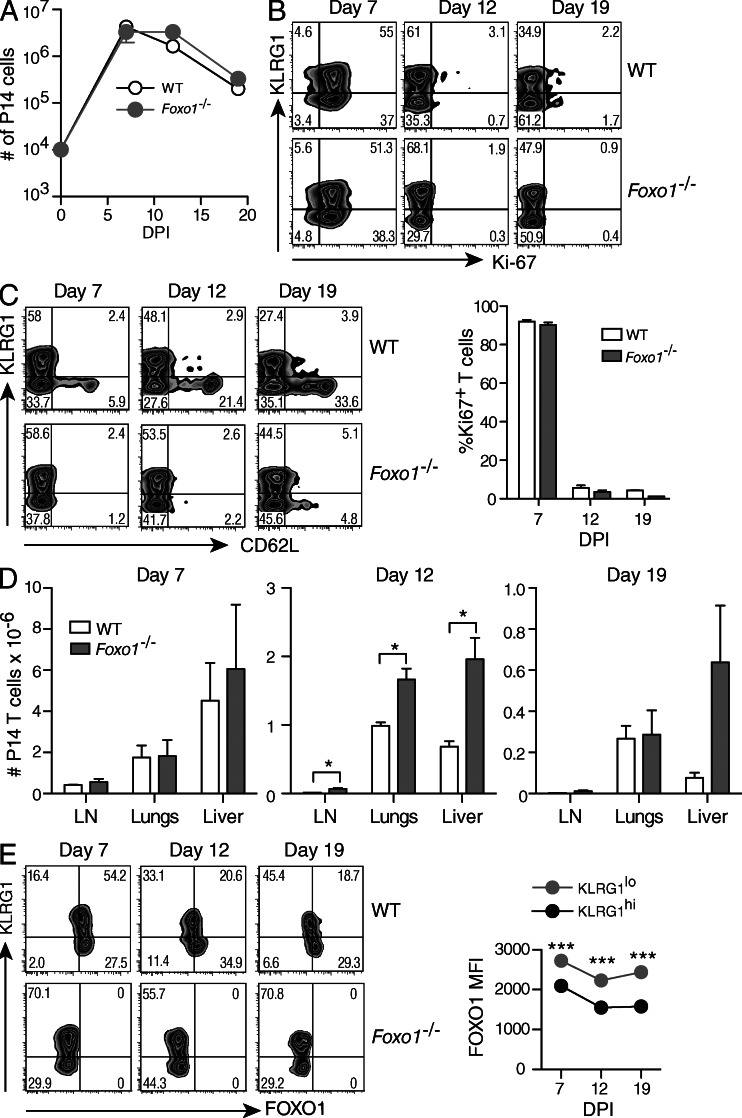
To study the role of FOXO1 in CD8+ T cells responding to a viral infection in vivo, we took advantage of mice in which exon 2 of Foxo1 is flanked by loxP sites (Foxo1f/f mice; Paik et al., 2007). Because crossing Foxo1f/f mice with mice carrying the Tg(Cd4-cre)1Cwi transgene (Cd4Cre mice) results in autoimmunity caused by a defect on regulatory T cells (Kerdiles et al., 2010), we produced Foxo1f/f mice with the Cre recombinase under the control of the human Granzyme B promoter (GZMBCre). Recombination was thus restricted to activated CD8+ T cells (Jacob and Baltimore, 1999; Rutishauser et al., 2009). These mice were further crossed to include the P14 T cell antigen receptor transgenes (P14) endowing T cells with specificity for the immunodominant epitope of lymphocytic choriomeningitis virus (LCMV) gp33 in association with H2Db. Equal numbers of spleen cells from Foxo1f/fGZMBCre+P14+ (Foxo1−/− P14) and Foxo1f/fGZMBCre−P14+ (WT P14) mice were co-transferred into naive host mice, and the T cells were followed by expression of CD45.2 or CD45.1/2, respectively. Recipient mice were then infected with LCMV Armstrong (LCMV-Arm; Oldstone, 2002). The analysis of splenic T cells revealed an equivalent expansion of Foxo1−/− and WT P14 T cells upon viral infection with little or no difference in the rate of contraction (Fig. 1 A). Although there appears to be a slight delay in the contraction of Foxo1−/− T cells in the experiment presented, this was not consistently seen in three other independent experiments. In addition, similar percentages of Ki-67+ cells in WT and Foxo1−/− P14 cells were found at the peak of the response (7 d after infection) with virtually no Ki-67+ cells present at days 12 and 19 (Fig. 1 B, top panels and bottom graph). Since FOXO1 is important for the expression of the homing molecules, l-selectin (CD62L) and CCR7 (Fabre et al., 2008; Kerdiles et al., 2009), we examined CD62L expression in the spleen and the distribution of WT and Foxo1−/− P14 cells after viral infection in lymph nodes (LN), lungs, and liver. As expected, expression of CD62L was low on Foxo1−/− P14 cells compared with WT P14 cells (Fig. 1 C). However, Foxo1−/− T cells were at least as abundant as WT P14 T cells in all the organs analyzed, excluding a defect in the homing capacity of Foxo1−/− P14 cells due to low CD62L expression (Fig. 1 D). Thus, loss of FOXO1 had no effect on the overall proliferation of virus-specific CD8+ T cells in response to LCMV infection.
Figure 1.
Kinetics of CD8+ T cell expansion in response to LCMV-Arm infection. A 1:1 mix of 104 CD45.1+/2+ WT (Foxo1f/fGZMBCre−) and CD45.2+ Foxo1−/− (Foxo1f/fGZMBCre+) P14 cells were transferred to CD45.1+ WT host mice. Mice were then infected with LCMV-Arm, and lymphoid and nonlymphoid organs were harvested and analyzed at day 7, 12, and 19 after infection. (A) WT and Foxo1−/− P14 cells were identified in the spleen by their expression of CD45.1, CD45.2, CD8, and Vα2 and total numbers of P14 cells were determined. DPI, days post infection. (B) Ki67 staining of spleen cells from WT and Foxo1−/− P14 cells at different time points. (C) Expression of KLRG1 and CD62L on WT and Foxo1−/− P14 cells isolated from the spleen. (D) Total numbers of WT and Foxo1−/− P14 cells in the indicated organs. (E) Expression of KLRG1 and FOXO1 on WT and Foxo1−/− P14 cells isolated from the spleen. Data representative of 1 out of 3 independent experiments each, with n = 3,4.
FOXO transcription factors are subject to complex post-translational regulation that includes nuclear egress and cytoplasmic localization followed by 14–3-3-mediated degradation. As such, we sought to determine how the amounts of FOXO1 might change during the course of CD8+ T cell expansion and contraction. The results showed there was heterogeneous FOXO1 expression that was inversely correlated with KLRG1 expression (Fig. 1 E, WT). We also note that FOXO1 expression was not detected in the GZMBCre+ T cells. These results are consistent with the possibility that FOXO1 is active in the precursors to memory cells, and plays less of a role in KLRG1hi cells. To address this issue, we focused our analysis on memory CD8+ T cell differentiation with or without the deletion of Foxo1.
KLRG1lo CD8+ memory-precursor T cells display an enhanced effector phenotype
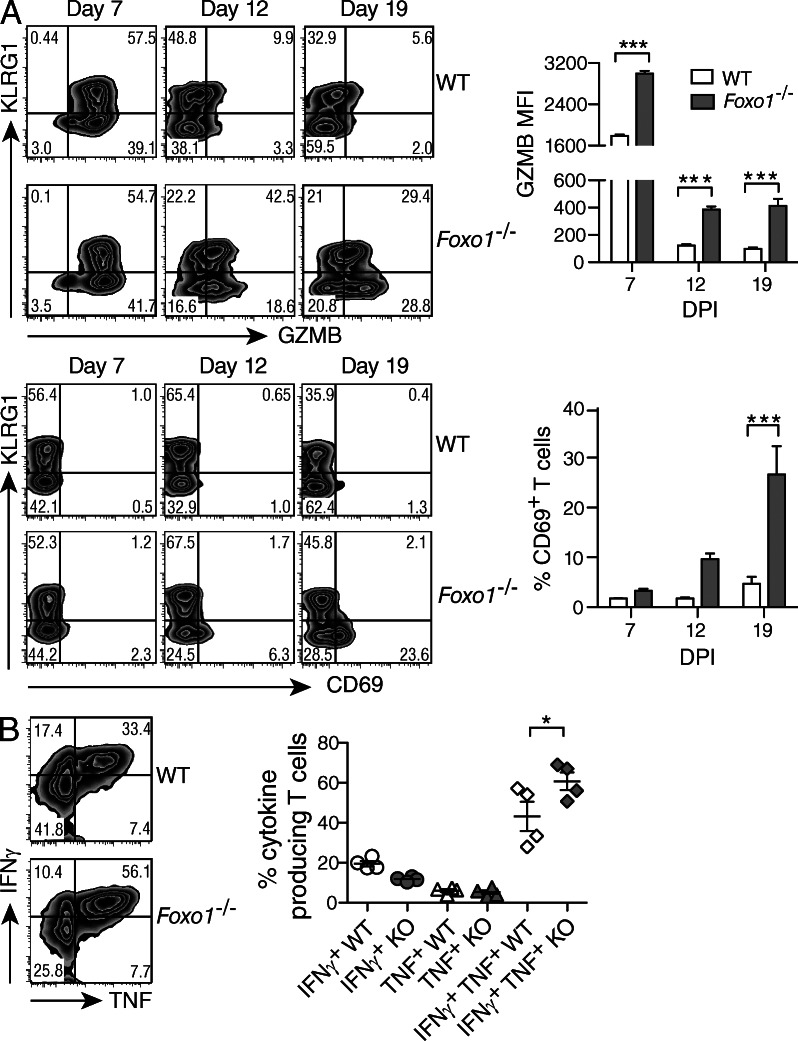
After the peak expansion of CD8+ T cells in response to virus infection, effector cells selectively die, but the surviving effector cells and memory-precursor cells become quiescent, and those within the secondary lymphoid organs eventually extinguish the expression of cytotoxic effector molecules such as GZMB (Kaech et al., 2002). To examine how the deletion of Foxo1 affected the resolution of the response, we examined CD8+ T cells for the expression of molecules that play a role in effector function. Although the percentage of GZMB+ cells among WT and Foxo1−/− P14 cells was similar at the peak of the response (Fig. 2 A, top left), the mean fluorescence intensity (MFI) in Foxo1−/− P14 cells was higher (Fig. 2 A, top right). In addition, WT P14 cells rapidly lost GZMB during the contraction phase (d12-19), whereas Foxo1−/− P14 cells retained GZMB in reduced, though substantial amounts. In fact, the KLRG1lo cells increased GZMB expression at d19 against a constant expression within the KLRG1hi population. Similarly, the KLRG1lo Foxo1−/− T cells regained expression of CD69 during the contraction phase (Fig. 2 A, bottom), and comparable results for the expression of GZMB and CD69 were found for liver T cells (not depicted). These data suggest that in the absence of FOXO1, CD8+ T cells fail in their transition from effector to memory cells, showing a sustained effector phenotype. How the CD8+ T cells reexpress effector molecules in the absence of antigen is unclear. In accord with the enhanced and extended effector function, there was a greater number of Foxo1−/− multicytokine-producing (TNF and IFN-γ) effector T cells compared with WT (Fig. 2 B). To ensure that Foxo1−/− T cells were functional, we infected WT (Foxo1f/fGZMBCre−) and Foxo1−/− (Foxo1f/fGZMBCre+) mice and found no detectable LCM virus by day 7 (unpublished data).
Figure 2.
Phenotype of virus-specific CD8+ T cells. A 1:1 mix of 104 CD45.1+/2+ WT (Foxo1f/fGZMBCre−) and CD45.2+ Foxo1−/− (Foxo1f/fGZMBCre+) P14 cells were transferred to CD45.1+ WT host mice. Mice were then infected with LCMV-Arm, and spleen cells were harvested and analyzed at different time points. (A) The expression of KLRG1, GZMB, and CD69 was determined in WT and Foxo1−/− P14 cells at different time points (left), and the amount of GZMB (MFI) and percentage of CD69 are depicted (right). Data are representative of 3 independent experiments, with n = 3–4. (B) At day 7 after infection, spleen cells were stimulated with PMA and ionomycin and the frequency of IFN-γ– and TNF-producing cells was determined. Data are representative of 2 independent experiments each, with n = 4.
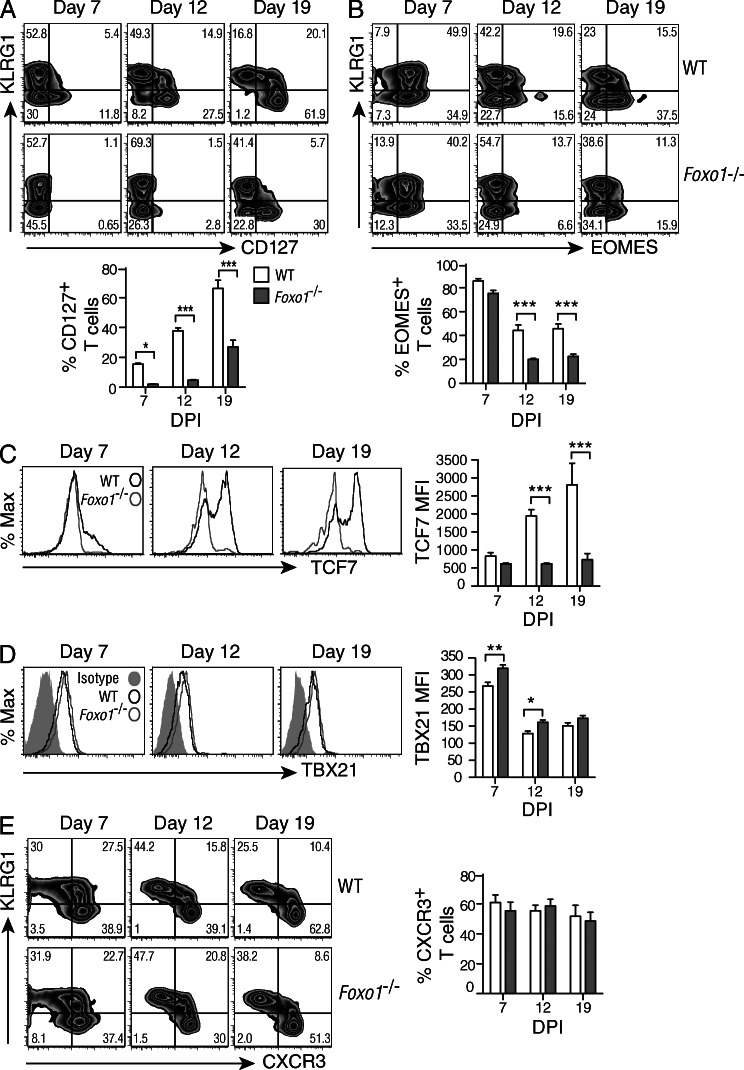
At the peak of the expansion phase, T cell diversity is evident based on the differential expression of several diverse cell surface receptors and transcription factors. For example, the KLRG1hiCD127lo population is enriched for committed effector cells, whereas the KLRG1loCD127hi population is enriched for the precursors of memory T cells (Joshi et al., 2007). A complication in the present analysis is that FOXO1 directly controls the expression of CD127 at least in naive and acutely activated T cells (Kerdiles et al., 2009), and as such may not accurately reflect the state of CD8+ T cell differentiation. This is depicted in Fig. 3 A, at 7 and 12 d, where an emerging CD127+ population is visible in the WT but not the Foxo1−/−. However, we note that by 19 d, CD127 expression has been reestablished on a proportion of the KLRG1lo cells, and this suggests that at least a subset of activated T cells can express CD127 in the absence of FOXO1.
Figure 3.
Enhanced effector phenotype in the absence of FOXO1. A 1:1 mix of 1 × 104 CD45.1+/2+ WT (Foxo1f/fGZMBCre−) and CD45.2+ Foxo1−/− (Foxo1f/fGZMBCre+) P14 cells were transferred to CD45.1+ WT host mice. Mice were then infected with LCMV-Arm, and spleen cells were harvested and analyzed at different time points. (A–E) WT, open bars; Foxo1−/−, filled bars. (A) The expression of KLRG1 and CD127 was determined in WT and Foxo1−/− P14 cells, and the percentage of CD127+ cells is depicted (bottom). (B) The expression of KLRG1 and EOMES was determined in WT and Foxo1−/− P14 cells, and the percentage of EOMES+ cells is depicted (bottom). (C and D) WT and Foxo1−/− P14 cells were analyzed for their expression of TCF7 (C) and TBX21 (D), and the amount of both are presented as mean fluorescence intensity (MFI, right). (E) The expression of KLRG1 and CXCR3 was determined in WT and Foxo1−/− P14 cells (left), and the percentage of CXCR3+ cells depicted (right). Data are representative of 3 independent experiments each, with n = 3–4.
The T-box transcription factors EOMES (eomesodermin) and TBX21 (T-bet) are required for the formation and function of effector and memory CD8+ T cells (Kaech and Cui, 2012). TBX21 is required for the formation of KLRG1hi effector cells, and its overexpression skews the population toward terminal effector cell differentiation. In contrast, EOMES is required for T cells to survive in the long term as memory cells expressing IL2Rβ (CD122; Intlekofer et al., 2005; Banerjee et al., 2010). An examination of co-transferred WT and Foxo1−/− P14 cells showed that within the WT T cell population there was a remaining KLRG1loEOMEShi population at 19 d after infection that was largely missing in the Foxo1−/− P14 cells (Fig. 3 B). This correlates with the absence of the high-mobility group protein, TCF7, a Wnt-activated transcription factor shown to be important for Eomes expression and maintenance of CD8+ memory T cells (Zhou et al., 2010; Fig. 3 C). Thus, there was a substantial deficit in the expression of two transcription factors shown to be required for the maintenance and function of CD8+ memory T cells.
Collectively, these results demonstrate an important role for FOXO1 in regulating essential characteristics associated with the lineage of precursor cells destined to become self-renewing, long-lived memory T cells. A recent study showed that IL-12 promotes Tbx21 expression by inactivating FOXO1, and Tbx21 expression can be inhibited indirectly by FOXO1 overexpression (Rao et al., 2012). Furthermore, FOXO1 appears to directly target Eomes, and thus constitutes a second pathway that converges on Eomes expression to promote a memory cell phenotype (Rao et al., 2012). In line with these results, Foxo1−/− P14 cells displayed higher TBX21 expression compared with WT P14 cells (Fig. 3 D), and this correlated with lower levels of EOMES (Fig. 3 B), supporting a critical role for FOXO1 in regulating both Tbx21 and Eomes expression.
Finally, CXCR3 is up-regulated on CTLs after activation and is thought to facilitate colocalization of T cells with viral antigens in the spleen. Whereas KLRG1hi P14 cells were shown to exhibit a heterogeneous expression pattern, KLRG1lo T cells remained uniformly CXCR3hi throughout a viral response (Hu et al., 2011). This was seen for both WT and Foxo1−/− P14 cells (Fig. 3 E), suggesting that Foxo1−/− CD8+ T cells have equal access to antigen, and that this aspect of CTL differentiation appears to be unaffected by Foxo1 deletion.
Foxo1−/− CD8+ T cells fail to up-regulate memory-associated signature genes
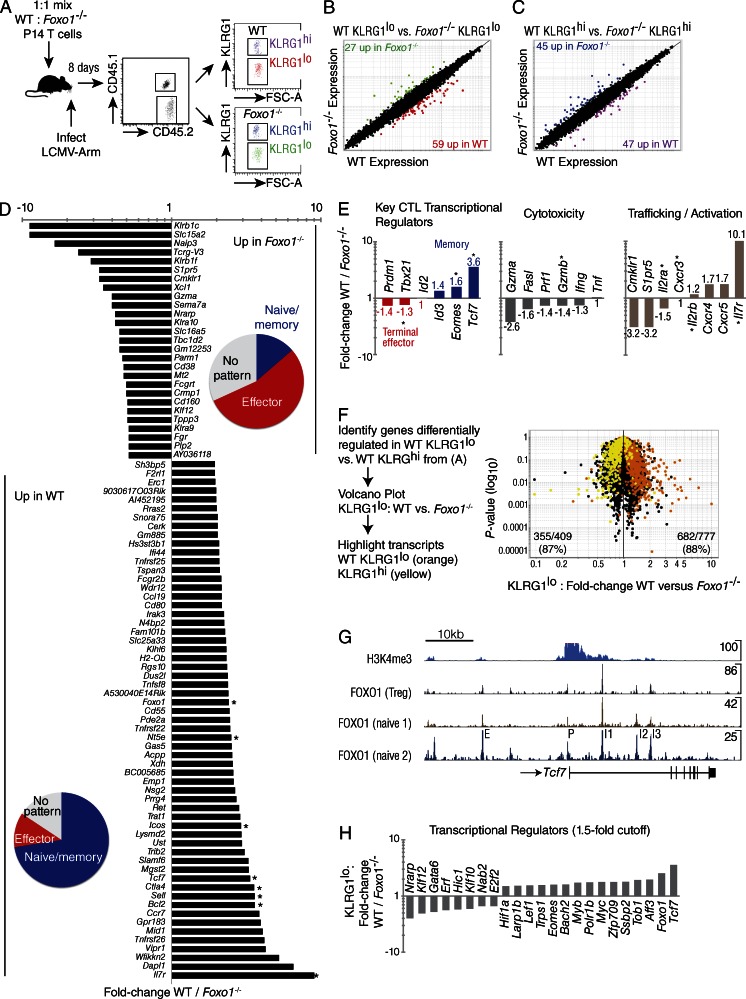
To obtain an unbiased analysis of genes differentially expressed in antigen-specific Foxo1−/− CD8+ T cells responding to infection, we performed microarray analysis on KLRG1lo and KLRG1hi FACS-sorted, congenically marked WT and Foxo1−/− P14 cells 8 d after infection with LCMV-Arm (Fig. 4). For this experiment, we performed gene deletion in Rosa26Cre-ERT2 Foxo1f/f (Foxo1−/−) P14 mice just before adoptive transfer (Kerdiles et al., 2009). A virtually complete deletion of Foxo1 was obtained before adoptive transfer and maintained throughout the duration of the experiment (unpublished data). We used a replicate mixed transfer experimental design to sort Foxo1f/f (WT) and Foxo1−/− P14 cells of the same TCR specificity from the same infected animals, thus controlling for cytokine environment and antigen load on CTL gene expression (Fig. 4 A). We compared the gene expression profile between WT versus Foxo1−/− P14 CD8+ T cells in KLRG1lo (Fig. 4 B) and KLRG1hi (Fig. 4 C) sorted cells. The data showed that FOXO1 regulates gene expression in both populations. We then focused our analysis on sorted KLRG1lo T cells known to seed the memory population and display abundant differences in both effector molecules and key transcription factors when compared with KLRG1hi T cells (Figs. 2 and 3, respectively; Kaech and Cui, 2012). We identified genes passing a twofold up or down cut-off in KLRG1lo cells, identifying 27 genes down and 59 genes up in WT relative to Foxo1−/− P14 CD8+ T cells (Fig. 4, B and D). As expected, and as a positive control for our analysis, we observed decreased abundance of mRNAs of known FOXO1 targets in Foxo1−/− P14 CD8+ T cells: Sell (3.9 fold); Ctla4 (3.8 fold); Il7r (10.1 fold); and Foxo1 itself (2.5 fold; Fig. 4 D). We grouped the twofold list of regulated genes into naive/memory, effector, or no pattern based on their expression during the CD8+ T cell response to infection as reported by the Immgen project (Best et al., 2013). Interestingly, Foxo1−/− P14 CD8+ T cells were enriched for genes associated with effector T cells, whereas WT P14 CD8+ T cells genes were enriched for genes associated with T cell naive/memory (Fig. 4 D, pie charts).
Figure 4.
Deletion of Foxo1 prevents the development of a memory program of gene expression. A 1:1 mix of 104 WT (Foxo1f/f Rosa26) and Foxo1−/− (Foxo1f/f Rosa26Cre-ERT2) P14 cells were prepared from tamoxifen-treated mice and transferred to CD45.1+ WT host mice. Microarray gene-expression analysis was performed by sorting WT and Foxo1−/− KLRG1lo P14 cells 8 d after infection with LCMV-Arm. (A) Experimental scheme for mixed transfer and FACS of WT and Foxo1−/− P14 T cells on day 8 of LCMV-Armstrong infection. The four double-sorted populations were analyzed on the same microarray chip. (B) KLRG1lo WT versus Foxo1−/− reveals 59 genes up and 27 genes down with a twofold cutoff. (C) KLRG1hi WT versus Foxo1−/− reveals 47 genes up and 45 genes down with a twofold cutoff. (D) Identification of genes KLRG1lo WT / Foxo1−/− with a twofold cutoff. Pie charts indicate portion of genes with an expression pattern tied to naive/memory (blue), effector (red), or no pattern (gray). Asterisk indicates expression was verified by flow cytometry. (E) Classification of a subset of the genes in (D) by function. (F) Genes differentially regulated in WT KLRG1lo versus WT KLRG1hi, were overlaid with genes from KLRG1lo WT versus KLRG1lo Foxo1−/− CD8+ T cells, showing nearly a 90% concordance of transcripts. (G) Naive CD4+ ChIP-Seq data showing several FOXO1 binding sites proximal to and within Tcf7. Data were analyzed from T reg cells and two different datasets analyzing naive CD4 T cells. Peaks are labeled as putative: intergenic enhancer (E), promoter (P), and intronic enhancers 1, 2, and 3 (I1, I2, I3). Peak sequences are listed in Table I. (H) Transcription factors known to play a role in the formation of memory cells, summarizing data presented in D with the addition of other transcription factors differentially regulated in WT vs. Foxo1−/− P14 cells. Data are from 1 experiment with n = 3.
Although 8 d after infection has been found to be the peak of cytotoxic T cell expansion, virus has been cleared by this point, and subsets of CTL can already be identified as terminally differentiated effector cells or memory cell precursors. Memory precursors are associated with expression of Id3, Eomes, and Tcf7, whereas terminally differentiated effectors are associated with expression of Prdm1 (BLIMP1), Id2, and elevated Tbx21 (Kaech and Cui, 2012). As shown, these signature transcription factors are differentially regulated in KLRG1lo Foxo1−/− versus WT P14 cells (Fig. 4 E, left); furthermore, these results are consistent with aforementioned results (Fig. 3, B–D) showing that TCF7 and EOMES were markedly reduced in Foxo1−/− T cells, whereas Tbx21 was increased. In addition, CTLs exert their effector function through the granule exocytosis pathway, FAS, and cytokine production. We found elevated granzymes, perforin, Fas, and Ifng in KLRG1lo Foxo1−/− P14 cells. We further analyzed KLRG1lo cells for activation and trafficking molecules (Fig. 4 E, right). Altogether, these data show a bias for Foxo1−/− CD8+ T cells toward terminal differentiation, increased effector function, and altered trafficking/activation (Fig. 4 E, middle and right).
From our sorted cells, we made an expression list of genes up or down in WT KLRG1lo versus WT KLRG1hi. This established a list of genes expressed at higher levels in terminal-effector cells relative to those T cells with increased memory potential. When we overlaid the genes up or down 1.5-fold from this list into a volcano plot of WT versus Foxo1−/− KLRG1lo CD8+ T cells, there was an ∼90% concordance of transcripts; genes associated with terminal effector cells were increased in Foxo1−/− T cells, whereas genes associated with memory-precursor cells were increased in WT (Fig. 4 F). Specifically, the results show that Foxo1−/− KLRG1lo cells are enriched for terminally differentiated transcripts (87% of KLRG1hi-biased transcripts are higher in KLRG1lo Foxo1−/− versus KLRG1lo WT), whereas WT KLRG1lo cells are enriched for memory transcripts (88% of the KLRG1lo-biased transcripts are up in WT KLRG1lo versus Foxo1−/− KLRG1lo). A trivial explanation for this analysis would be that there was a bias for KLRG1hi cell differentiation caused by the absence of FOXO1, and the microarray data simply reflects this while not revealing a direct role for Foxo1 in driving memory cell differentiation. However, at the peak of the response there are approximately equivalent proportions of KLRG1hi and KLRG1lo cells in WT and Foxo1−/− T cells, and yet TCF7, EOMES, TBX21, and GZMB on KLRG1lo cells are differentially expressed within these populations throughout infection (Figs. 2, 3). We infer that the effect is not simply a population bias but an intrinsic FOXO1-dependent effect on key memory and effector molecules. These results suggest a failure of Foxo1−/− CD8+ T cells at the peak of the response to differentiate into memory-precursor cells.
The fact that so many genes associated with the memory state are of decreased abundance in Foxo1−/− T cells suggests two nonmutually exclusive possibilities. One, FOXO1 is essential to terminate the effector state allowing genes required for differentiated memory cells to be expressed under the control of other factors. Two, FOXO1 is essential for driving the memory program through direct induction of known targets Sell, Il7r, and of master transcription factors of memory such as Tcf7 and Eomes (Rao et al., 2010; Zhou et al., 2010). Tcf7 is expressed in both CD4+ and CD8+ T cells from early thymic developmental stages and in peripheral cells, it is notably absent in effector cells and then once again expressed in high amounts in memory T cells (Yu et al., 2010; Best et al., 2013).
In support of a direct role for FOXO1 in memory cell formation, we have determined, using a chromatin immunoprecipitation sequencing (ChIP-Seq) approach, that FOXO1 binds directly to multiple sites close to, and within the Tcf7 gene in naive CD4+ T cells, and these binding sites were also found in two other T cell-specific ChIP-Seq datasets (Fig. 4 G). Five of the peaks were tentatively labeled as E (intergenic enhancer), P (promoter), I1 (intronic enhancer-1), I2 (intronic enhancer-2), and I3 (intronic enhancer-3). For each of these elements, we list the nucleotide sequences corresponding to each ChIP-Seq peak (Table I). As shown, each has one or more tandem consensus forkhead binding sequences at or surrounding the peak, and this is consistent with specific FOXO1 binding. In addition, we show the H3K4me3 histone marks at the transcription start site indicative of an open locus (Pekowska et al., 2011). This apparently direct regulation of Tcf7 suggests that memory cell differentiation after TCR and cytokine stimulation requires FOXO1 nuclear localization.
Table 1.
Nucleotide sequences found at FOXO1-binding peaks within the Tcf7 gene
| Elementa | Peak sequences |
| E | 5′-TATCTTGTTTTGCTC-3′ |
| P | 5′-GGAGTAAACAGACCC-3′ |
| I1 | 5′-ACTGTTGTTTCCTGC-3′ |
| 5′-TGTACAAACAAGGCT-3′ | |
| 5′-GGAGGAAACAGGTGT-3′ | |
| l2 | 5′-CAGGGTGTTTGTAGT-3′ |
| 5′-TCTAAAAACATCCTG-3′ | |
| 5′-AAGGAAAACACAAGC-3′ | |
| l3 | 5′-GACTGTGTTTATTTT-3′ |
| 5′-CTCTGAAACAGAGAC-3′ |
Sequences in bold correspond to a forkhead consensus site.
See Fig. 4 G.
Finally, for clarity, we summarized the data focusing on transcription factors differentially regulated in WT versus Foxo1−/− P14 T cells (Fig. 4 H). This list includes other transcription factor genes known to play a role in the formation of memory, such as the Tcf7-related Lef1 (Zhou and Xue, 2012). It also may point to a role for FOXO1-regulated transcription factors and DNA binding proteins as of yet not recognized as being involved in CD8+ T cell memory formation.
Collectively, these analyses suggest a basic and essential role for FOXO1 in programming fundamental aspects of the memory state in CD8+ T cells after response to infection in vivo. We therefore tested whether Foxo1−/− T cells can expand in response to a secondary antigenic challenge as a measure of the requirement for Foxo1 in an immune recall response.
Foxo1−/− CD8+ T cells fail to make a memory response
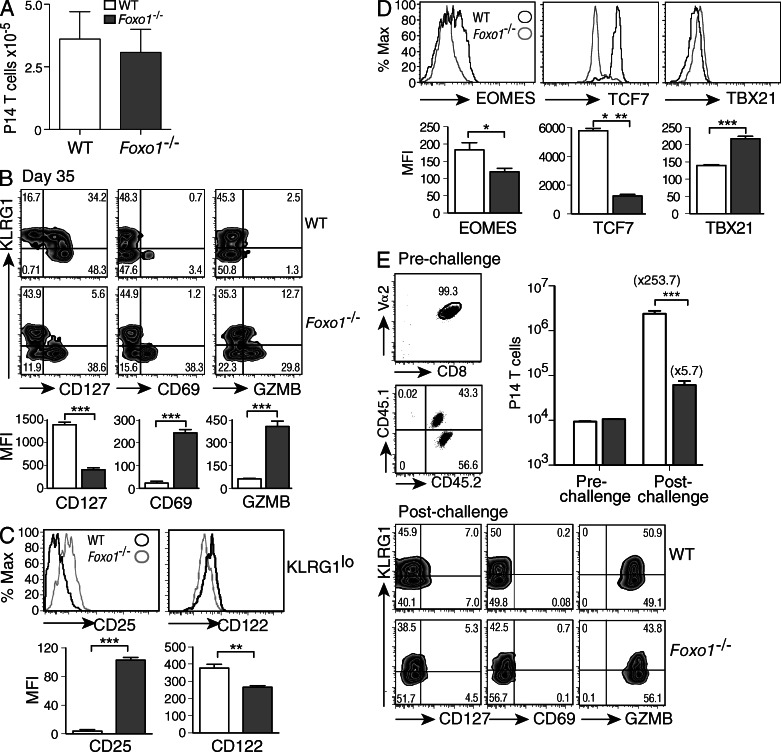
The data presented demonstrate that, in the absence of FOXO1, virus-specific CD8+ T cells display a prolonged effector phenotype and fail to up-regulate a multitude of key memory-associated genes. To better understand the role of FOXO1 in CD8+ T cell differentiation after viral infection, we characterized Foxo1−/− P14 spleen cells from LCMV-Arm infected mice at later time points (35 d after infection). As seen for early time points (Fig. 1 A), the numbers of P14 cells were comparable between WT and Foxo1−/− P14 cells (Fig. 5 A). Even at this later time, Foxo1−/− P14 cells exhibit an enhanced effector-like phenotype (Fig. 5 B) characterized by low expression of the memory-associated marker CD127, and high expression of CD69 and GZMB. As shown for early time points (Fig. 2 A; and Fig. 3, A and B), this phenotype was only observed in KLRG1lo memory-like cells (Fig. 5, B and C). This KLRG1lo population also expressed high amounts of the high-affinity IL-2 receptor CD25 and low levels of the IL-2 receptor β-chain CD122 (Fig. 5 C), consistent with the microarray data (Fig. 4 E, right). Early in infection, high CD25 expression was shown to promote terminal-effector CD8+ T cell differentiation in vivo (Kalia et al., 2010), while high CD122 expression is associated with CD8+ memory T cells (Intlekofer et al., 2005). This supports the concept that FOXO1 is essential for the transition from effector to long-lived memory CD8+ T cells. In addition, the expression of memory-related transcription factors EOMES and TCF7 was still low, whereas the expression of TBX21 was higher in KLRG1lo Foxo1−/− P14 cells compared with WT at this late time-point (Fig. 5 D). These phenotypic changes are consistent with an extended effector-like phenotype in Foxo1−/− T cells (Fig. 5 B).
Figure 5.
Foxo1−/− CD8+ T cells fail to expand upon secondary viral challenge. (A–E) WT, open squares; Foxo1−/− filled squares. A 1:1 mix of 104 CD45.1+/2+ WT (Foxo1f/fGZMBCre−) and CD45.2/2+ Foxo1−/− (Foxo1f/fGZMBCre+) P14 cells were prepared and transferred to CD45.1+ WT host mice. Mice were then infected with LCMV-Arm, and spleen cells were analyzed 35 d after infection. (A) Number of cells recovered per spleen at day 35. (B) WT and Foxo1−/− P14 cells were analyzed by their expression of KLRG1 and CD127, CD69, and GZMB (top), and the MFI of these molecules was quantified on the KLRG1lo population (bottom). (C) Expression of CD25 and CD122 on KLRG1lo P14 cells. (D) WT and Foxo1−/− P14 cells were analyzed for expression of transcription factors important for CD8+ memory and effector T cells, and the amount of protein expressed on the KLRG1lo population graphed as MFI. (E) P14 (CD8+Vα2+) WT (CD45.1+/2+) and Foxo1−/− (CD45.1+) cells from hosts 35 d after LCMV-Arm infection were sorted and transferred at a 1:1 ratio into naive WT mice. Mice were then infected with LCMV-Arm and the expansion of WT and Foxo1−/− P14 cells determined at d 5 after infection (top right). The phenotype of P14 T cells 5 d after secondary challenge (bottom). Data are representative of 2 independent experiments for a total n = 11.
Finally, we tested the ability of these cells to respond to a second challenge with LCMV-Arm. To do this, WT and Foxo1−/− P14 cells were sorted from mixed adoptively transferred mice at 35 d after infection and transferred as a 1:1 mixture into naive recipients. These mice were challenged with LCMV-Arm and analyzed for T cell expansion 5 d post-infection (Fig. 5 E). The results showed that in the absence of FOXO1, CD8+ T cells failed to accumulate, and were at a 45-fold disadvantage with respect to the WT CD8+ T cells. Of note, the phenotype of the cells post-challenge (Fig. 5 E) was similar to the one found at the peak of the response during the primary infection (Figs. 2 and 3), with higher expression of GZMB in Foxo1−/− P14 cells compared with WT P14 cells. Collectively, the results of the experiments described demonstrate an intrinsic role for FOXO1 in regulating the transition from effector cells to long-lived memory cells capable of reactivation.
DISCUSSION
Infection by an intracellular pathogen almost invariably results in the expansion of antigen-specific CD8+ T cells that participate in killing the pathogen-infected cells. These T cells exhibit characteristics of terminal differentiation in that they have a finite life span and they maintain a highly differentiated program of gene expression associated with cytotoxicity and inflammation. Although competing models of CTL memory exist, available evidence favors the concept that memory-precursor cells differentiate from a subset of this effector population, ultimately becoming a cell-type perhaps like no other in the immune system. These memory T cells are self-renewing, with a population turnover of 2–3 mo, and their progeny can be shown to persist for the life of the individual (Parretta et al., 2008; Choo et al., 2010). Yet, they are poised to respond to a reinfection by rapid growth, progression through the cell cycle, and expression of a cytotoxic, proinflammatory program of gene expression (Tanchot et al., 1997; Murali-Krishna et al., 1999; Zhang and Bevan, 2011).
The control of the effector versus memory-precursor lineage decision has been illuminated over the past decade, including a role for stimulatory and inflammatory cytokines as well as a description of several transcription factors necessary for the distinct programs of gene expression (Rutishauser and Kaech, 2010; Best et al., 2013). In this study, we describe experiments that demonstrate a high order control over this fate decision mediated by the forkhead transcription factor, FOXO1. With the deletion of Foxo1, CD8+ T cells fail to acquire the characteristics of memory-precursor cells 7 d post-infection, they retain many aspects of effector T cells for an extended period, and they do not exhibit the vast expansion characteristic of a recall immune response. Given its extensive range of posttranslational modifications (Hedrick et al., 2012), we infer that the lineage decision to acquire characteristics of effector vs. memory-precursor cells would appear to center, to an extent, on the integration of extrinsic signals that are manifest in the cellular localization and target specificity of FOXO1.
These results are in concert with studies showing that activation of AKT, a major pathway to FOXO inhibition, is key to the differentiation of effector cells at the expense of memory-precursory cells (Hand et al., 2010; Kim et al., 2012). In addition, the IL-12–mediated inactivation of FOXO1 was shown to promote TBX21–expressing effector T cells, and the rapamycin-mediated inhibition of TBX21 expression was FOXO1 dependent. Also, this same study established a role for FOXO1 in switching T cells from an effector to a memory-like phenotype through an increase in EOMES expression (Rao et al., 2012). Here, we establish that FOXO1, in the context of rapidly expanding T cells, directly and indirectly refines a program of gene expression to produce the phenotypic characteristics of memory cells. Of particular importance may be a direct role for FOXO1 in controlling the expression of Tcf7, encoding a Wnt, β-catenin-regulated transcription factor. This might be viewed as contrary to studies showing that FOXO factors compete with β-catenin for TCF interactions, and thus suppress the expression of TCF target genes; however, these studies examined FOXO3 and FOXO4. We surmise that this effect may not apply to FOXO1, but rather, FOXO1 may transcriptionally promote the expression of Tcf7 (Almeida et al., 2007; Hoogeboom et al., 2008). In this regard, the present results closely parallel those analyzing Tcf7 mutant T cells in that such T cells maintain expression of GZMB, and exhibit reduced expression of EOMES, CD62L, CCR7, and IL2β (Zhou et al., 2010). Our conclusion is that FOXO1 is required for the expression of Tcf7 as part of an extended program of gene expression that gives rise to memory T cells.
FOXO1 has been found to regulate pluripotency and self-renewal in several contexts. In embryonic stem cells (ES cells) in mouse and human beings, it does so by direct control of Oct4 and Sox2—encoding two of the factors necessary to maintain pluripotency in ES cells or induce pluripotency in differentiated cell types (Zhang et al., 2011). It is necessary for the maintenance of tumor stem cells in acute myelogenous leukemia (AML) cells, which explains the paradoxical finding that AKT activation or the inactivation of multiple Foxo genes can inhibit tumorigenicity in many examples of AML (Sykes et al., 2011). In addition, FOXO factors are essential for the maintenance of hematopoietic stem cells, although largely through the detoxification of reactive oxygen species (Tothova et al., 2007). Although we might be tempted to draw parallels between the self-renewal capacity of stem cells and memory T cells, we note that FOXO transcription factors have also been shown to control acne (Melnik, 2011), bone homeostasis (Almeida, 2011), liver metabolism (Kousteni, 2012), muscle mass (Goodman et al., 2011), and the trade-off between aging and cancer (Kloet and Burgering, 2011). In fact, there may be few physiological processes that are not guided, at least in part, by FOXO transcription factors. Perhaps what all these processes have in common is a requirement for integrating characteristics defining the physiological state of the organism to effect an adaptively advantageous program of gene expression different for each and every tissue. A corollary to this thesis is that gene expression is not determined by any one transcription factor; rather, a transcription factor exerts activity in a highly context-dependent manner (Barolo and Posakony, 2002).
These results imply that AKT-mediated inhibition of FOXO1 ensures the memory program is not prematurely engaged, as early in the response, the cytokine or TCR-driven cytoplasmic sequestration of FOXO1 may promote high TBX21 and an effector phenotype so long as antigen is present. How this pathway plays into T cell exhaustion in chronic infection or continued formation of memory T cells in viral latency will be a topic of future studies. An implication is that the magnitude and effectiveness of a recall immune response may depend on the physiological state of the individual, where high levels of growth factors or superabundant nutrients may well blunt the formation or function of memory T cells. In addition, the role of memory T cells in persistent or latent viral infections is only beginning to be understood (Youngblood et al., 2012), and the course of such an infection could be readily affected by the activity of FOXO1 in antigen-specific T cells. The delicate balance between latency and recrudescence may be controlled in part through inactivation of FOXO caused by physiological stress.
MATERIALS AND METHODS
Mice and tamoxifen treatment.
C57BL/6 CD45.1+ mice were maintained in pathogen-free conditions. Rosa26Cre-ERT2 mice were provided by T. Ludwig (The Ohio State University, Columbus, OH; Guo et al., 2007). Foxo1f/f and Tg(GZMB-cre)1Jcb/J (GZMBCre) mice have been previously described (Jacob and Baltimore, 1999; Paik et al., 2007). Foxo1f/f mice (backcrossed to C57BL/6, n > 12) were crossed to GZMBCre or Rosa26Cre-ERT2 mice. In addition, mice were crossed with Tg(TcrLCMV)327Sdz (P14) mice to generate Foxo1f/f GZMBCre+P14 or Foxo1f/f Rosa26Cre-ERT2 P14 CD45.2+ mice. CD45.1+/2+ WT P14 mice were produced by a further cross with CD45.1+ C57BL/6 mice. Rosa26Cre-ERT2-mediated deletion of floxed alleles was induced by intraperitoneal injection of 1 mg of tamoxifen (Sigma-Aldrich) emulsified in 200 ml of sunflower seed oil (Sigma-Aldrich) every day for 5 d, followed by 5 d of rest (ERcre+, Foxo1−/−; ERcre−, WT). All procedures were approved by the Animal Care and Use Committee of the University of California, San Diego.
Adoptive transfer and viral infection.
Spleen cells were harvested from mice as described in the previous section. P14 cells were identified as CD8+Vα2+ and numbers were adjusted to obtain a 1:1 mix of 104 WT:Foxo1−/− cells. P14 cells were transferred intravenously to 8–10-wk-old CD45.1+ WT host mice. Mice were then infected with 2 × 105 pfu LCMV-Armstrong i.p. Virus was grown, identified, and quantified as previously described (de la Torre and Oldstone, 1992).
Flow cytometry.
Cell suspensions isolated from the indicated organs were incubated for 15 min at 4°C in PBS containing 1% FCS (Omega), 2 mM EDTA, 0.01% NaN3, and the indicated fluorochrome-conjugated antibodies. All intracellular staining was done with FoxP3 Fix/Perm (eBioscience). Antibodies against FOXO1 and TCF7 (rabbit polyclonal) were purchased from Cell Signaling Technology. Alexa Fluor 647 anti–rabbit secondary antibody and PE-conjugated anti–human Granzyme B (GZMB) antibodies were purchased from Invitrogen. The rest of the antibodies used were purchased from eBioscience or BD. Data were collected on a Fortessa (BD) and analyzed with FlowJo software (Tree Star).
Cell sorting and microarray.
Splenocytes were harvested and the CD8+ fraction enriched by negative selection using biotinylated antibodies against CD4, B220, CD11c, and DX5 (all from eBioscience), followed by streptavidin microbeads using MACS system in a 4°C cold room (Miltenyi Biotec). The negative fraction was then stained with antibodies against CD45.1, CD45.2, KLRG1, CD8, and Vα2. KLRG1lo donor WT (CD45.1+/2+) and Foxo1−/− (CD45.2+) P14 (CD8+Vα2+) splenocytes were sorted from host mice on day 7 after LCMV Armstrong infection using a BD ARIA (BD). Total RNA was extracted from sorted cells using RNeasy Microkit (QIAGEN), and RNA was labeled with biotin with the BioArray High Yield RNA Transcript Labeling kit (Enzo Diagnostics) and purified with an RNeasy Mini kit (QIAGEN). The resulting cRNA was hybridized to GeneChip Mouse Gene 1.0 ST arrays at the UCSD VA/VMRF microarray and NGS core. The raw CEL files were obtained, and the data were normalized and analyzed with the GenePattern software suite.
ChIP and ChIP-seq.
Methods for ChIP-Seq are described previously (Lin et al., 2010). Sequences were mapped to the mouse genome mm9 assembly (NCBI) using the Bowtie alignment tool and unique tags were visualized by preparing custom tracks for the UCSC Genome Browser where the total number of tags was normalized to 107. DNA sequence analysis was performed using HOMER. Data are have been deposited in the the Gene Expression Omnibus database under accession no. GSE46025. Foxo1 binding was referenced for T reg cells (Samstein et al., 2012) and two datasets for naive CD4 T cells (Naive 1; Ouyang et al., 2012; Naive2; this study). H3K4me3 histone marks were analyzed as previously described (Lin et al., 2010).
Statistical analysis.
Differences between datasets were analyzed by unpaired two-tailed Student’s t test or two-way ANOVA, as indicated.
Acknowledgments
This work was supported by National Institutes of Health grant 5R01AI103440 to SMH.
The authors declare that they have no conflicting interests.
Footnotes
Abbreviations used:
- KLRG1
- killer cell lectin–like receptor G1
- LCMV
- lymphocytic choriomeningitis virus
References
- Almeida M. 2011. Unraveling the role of FoxOs in bone—insights from mouse models. Bone. 49:319–327 10.1016/j.bone.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M., Han L., Martin-Millan M., O’Brien C.A., Manolagas S.C. 2007. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J. Biol. Chem. 282:27298–27305 10.1074/jbc.M702811200 [DOI] [PubMed] [Google Scholar]
- Araki K., Turner A.P., Shaffer V.O., Gangappa S., Keller S.A., Bachmann M.F., Larsen C.P., Ahmed R. 2009. mTOR regulates memory CD8 T-cell differentiation. Nature. 460:108–112 10.1038/nature08155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M.F., Wolint P., Walton S., Schwarz K., Oxenius A. 2007. Differential role of IL-2R signaling for CD8+ T cell responses in acute and chronic viral infections. Eur. J. Immunol. 37:1502–1512 10.1002/eji.200637023 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., Reiner S.L. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S., Posakony J.W. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167–1181 10.1101/gad.976502 [DOI] [PubMed] [Google Scholar]
- Best J.A., Blair D.A., Knell J., Yang E., Mayya V., Doedens A., Dustin M.L., Goldrath A.W.; Immunological Genome Project Consortium 2013. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14:404–412 10.1038/ni.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuneu H., Lemaître F., Deguine J., Moreau H.D., Bouvier I., Garcia Z., Albert M.L., Bousso P. 2010. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 33:412–423 10.1016/j.immuni.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Celli S., Garcia Z., Beuneu H., Bousso P. 2008. Decoding the dynamics of T cell-dendritic cell interactions in vivo. Immunol. Rev. 221:182–187 10.1111/j.1600-065X.2008.00588.x [DOI] [PubMed] [Google Scholar]
- Choo D.K., Murali-Krishna K., Anita R., Ahmed R. 2010. Homeostatic turnover of virus-specific memory CD8 T cells occurs stochastically and is independent of CD4 T cell help. J. Immunol. 185:3436–3444 10.4049/jimmunol.1001421 [DOI] [PubMed] [Google Scholar]
- Curtsinger J.M., Johnson C.M., Mescher M.F. 2003. CD8 T cell clonal expansion and development of effector function require prolonged exposure to antigen, costimulation, and signal 3 cytokine. J. Immunol. 171:5165–5171 [DOI] [PubMed] [Google Scholar]
- de la Torre J.C., Oldstone M.B. 1992. Selective disruption of growth hormone transcription machinery by viral infection. Proc. Natl. Acad. Sci. USA. 89:9939–9943 10.1073/pnas.89.20.9939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre S., Carrette F., Chen J., Lang V., Semichon M., Denoyelle C., Lazar V., Cagnard N., Dubart-Kupperschmitt A., Mangeney M., et al. 2008. FOXO1 regulates l-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J. Immunol. 181:2980–2989 [DOI] [PubMed] [Google Scholar]
- Gerlach C., van Heijst J.W., Swart E., Sie D., Armstrong N., Kerkhoven R.M., Zehn D., Bevan M.J., Schepers K., Schumacher T.N. 2010. One naive T cell, multiple fates in CD8+ T cell differentiation. J. Exp. Med. 207:1235–1246 10.1084/jem.20091175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C.A., Mayhew D.L., Hornberger T.A. 2011. Recent progress toward understanding the molecular mechanisms that regulate skeletal muscle mass. Cell. Signal. 23:1896–1906 10.1016/j.cellsig.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K., McMinn J.E., Ludwig T., Yu Y.H., Yang G., Chen L., Loh D., Li C., Chua S.J., Jr, Zhang Y. 2007. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology. 148:3987–3997 10.1210/en.2007-0261 [DOI] [PubMed] [Google Scholar]
- Hand T.W., Cui W., Jung Y.W., Sefik E., Joshi N.S., Chandele A., Liu Y., Kaech S.M. 2010. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc. Natl. Acad. Sci. USA. 107:16601–16606 10.1073/pnas.1003457107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick S.M., Hess Michelini R., Doedens A.L., Goldrath A.W., Stone E.L. 2012. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12:649–661 10.1038/nri3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogeboom D., Essers M.A., Polderman P.E., Voets E., Smits L.M., Burgering B.M. 2008. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J. Biol. Chem. 283:9224–9230 10.1074/jbc.M706638200 [DOI] [PubMed] [Google Scholar]
- Hu J.K., Kagari T., Clingan J.M., Matloubian M. 2011. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc. Natl. Acad. Sci. USA. 108:E118–E127 10.1073/pnas.1101881108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Jacob J., Baltimore D. 1999. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 399:593–597 10.1038/21208 [DOI] [PubMed] [Google Scholar]
- Ji Y., Pos Z., Rao M., Klebanoff C.A., Yu Z., Sukumar M., Reger R.N., Palmer D.C., Borman Z.A., Muranski P., et al. 2011. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12:1230–1237 10.1038/ni.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., Kaech S.M. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Cui W. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Hemby S., Kersh E., Ahmed R. 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 111:837–851 10.1016/S0092-8674(02)01139-X [DOI] [PubMed] [Google Scholar]
- Kalia V., Sarkar S., Subramaniam S., Haining W.N., Smith K.A., Ahmed R. 2010. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 32:91–103 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kallies A., Xin A., Belz G.T., Nutt S.L. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kerdiles Y.M., Beisner D.R., Tinoco R., Dejean A.S., Castrillon D.H., DePinho R.A., Hedrick S.M. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10:176–184 10.1038/ni.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles Y.M., Stone E.L., Beisner D.R., McGargill M.A., Ch’en I.L., Stockmann C., Katayama C.D., Hedrick S.M. 2010. Foxo transcription factors control regulatory T cell development and function. Immunity. 33:890–904 10.1016/j.immuni.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.H., Sullivan J.A., Plisch E.H., Tejera M.M., Jatzek A., Choi K.Y., Suresh M. 2012. Signal integration by Akt regulates CD8 T cell effector and memory differentiation. J. Immunol. 188:4305–4314 10.4049/jimmunol.1103568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet D.E., Burgering B.M. 2011. The PKB/FOXO switch in aging and cancer. Biochim. Biophys. Acta. 1813:1926–1937 10.1016/j.bbamcr.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Kousteni S. 2012. FoxO1, the transcriptional chief of staff of energy metabolism. Bone. 50:437–443 10.1016/j.bone.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Jhunjhunwala S., Benner C., Heinz S., Welinder E., Mansson R., Sigvardsson M., Hagman J., Espinoza C.A., Dutkowski J., et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik B.C. 2011. Isotretinoin and FoxO1: A scientific hypothesis. Dermatoendocrinol. 3:141–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali-Krishna K., Lau L.L., Sambhara S., Lemonnier F., Altman J., Ahmed R. 1999. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science. 286:1377–1381 [DOI] [PubMed] [Google Scholar]
- Obar J.J., Molloy M.J., Jellison E.R., Stoklasek T.A., Zhang W., Usherwood E.J., Lefrançois L. 2010. CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8+ T cells in primary and secondary responses. Proc. Natl. Acad. Sci. USA. 107:193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone M.B. 2002. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Curr. Top. Microbiol. Immunol. 263:83–117 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Liao W., Luo C.T., Yin N., Huse M., Kim M.V., Peng M., Chan P., Ma Q., Mo Y., et al. 2012. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 491:554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., et al. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 128:309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish I.A., Kaech S.M. 2009. Diversity in CD8(+) T cell differentiation. Curr. Opin. Immunol. 21:291–297 10.1016/j.coi.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parretta E., Cassese G., Santoni A., Guardiola J., Vecchio A., Di Rosa F. 2008. Kinetics of in vivo proliferation and death of memory and naive CD8 T cells: parameter estimation based on 5-bromo-2′-deoxyuridine incorporation in spleen, lymph nodes, and bone marrow. J. Immunol. 180:7230–7239 [DOI] [PubMed] [Google Scholar]
- Pearce E.L., Shen H. 2007. Generation of CD8 T cell memory is regulated by IL-12. J. Immunol. 179:2074–2081 [DOI] [PubMed] [Google Scholar]
- Pearce E.L., Walsh M.C., Cejas P.J., Harms G.M., Shen H., Wang L.S., Jones R.G., Choi Y. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 460:103–107 10.1038/nature08097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J.C., Ferrier P., Spicuglia S. 2011. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30:4198–4210 10.1038/emboj.2011.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 32:79–90 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.R., Li Q., Odunsi K., Shrikant P.A. 2010. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 32:67–78 10.1016/j.immuni.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.R., Li Q., Gubbels Bupp M.R., Shrikant P.A. 2012. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. 36:374–387 10.1016/j.immuni.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser R.L., Kaech S.M. 2010. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol. Rev. 235:219–233 [DOI] [PubMed] [Google Scholar]
- Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., Kaech S.M. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein R.M., Arvey A., Josefowicz S.Z., Peng X., Reynolds A., Sandstrom R., Neph S., Sabo P., Kim J.M., Liao W., et al. 2012. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 151:153–166 10.1016/j.cell.2012.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., Ahmed R. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Blackburn S.D., Intlekofer A.M., Kao C., Angelosanto J.M., Reiner S.L., Wherry E.J. 2009. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 31:309–320 10.1016/j.immuni.2009.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemberger C., Huster K.M., Koffler M., Anderl F., Schiemann M., Wagner H., Busch D.H. 2007. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 27:985–997 10.1016/j.immuni.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Sykes S.M., Lane S.W., Bullinger L., Kalaitzidis D., Yusuf R., Saez B., Ferraro F., Mercier F., Singh H., Brumme K.M., et al. 2011. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 146:697–708 10.1016/j.cell.2011.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto N., Intlekofer A.M., Northrup J.T., Wherry E.J., Reiner S.L. 2006. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177:7515–7519 [DOI] [PubMed] [Google Scholar]
- Tanchot C., Lemonnier F.A., Pérarnau B., Freitas A.A., Rocha B. 1997. Differential requirements for survival and proliferation of CD8 naïve or memory T cells. Science. 276:2057–2062 10.1126/science.276.5321.2057 [DOI] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Williams M.A., Tyznik A.J., Bevan M.J. 2006. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 441:890–893 10.1038/nature04790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Harty J.T. 2009. Initial TCR transgenic precursor frequency alters functional behaviour of CD8 T cells responding to acute infection. Adv. Exp. Med. Biol. 633:71–80 10.1007/978-0-387-79311-5_7 [DOI] [PubMed] [Google Scholar]
- Yang C.Y., Best J.A., Knell J., Yang E., Sheridan A.D., Jesionek A.K., Li H.S., Rivera R.R., Lind K.C., D’Cruz L.M., et al. 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12:1221–1229 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B., Wherry E.J., Ahmed R. 2012. Acquired transcriptional programming in functional and exhausted virus-specific CD8 T cells. Curr. Opin. HIV AIDS. 7:50–57 10.1097/COH.0b013e32834ddcf2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Sharma A., Sen J.M. 2010. TCF1 and beta-catenin regulate T cell development and function. Immunol. Res. 47:45–55 10.1007/s12026-009-8137-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bevan M.J. 2011. CD8(+) T cells: foot soldiers of the immune system. Immunity. 35:161–168 10.1016/j.immuni.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yalcin S., Lee D.F., Yeh T.Y., Lee S.M., Su J., Mungamuri S.K., Rimmelé P., Kennedy M., Sellers R., et al. 2011. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 13:1092–1099 10.1038/ncb2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Xue H.H. 2012. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J. Immunol. 189:2722–2726 10.4049/jimmunol.1201150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Yu S., Zhao D.M., Harty J.T., Badovinac V.P., Xue H.H. 2010. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 33:229–240 10.1016/j.immuni.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]