A new polymorphism near the IL28B locus negatively affects induction of IL28B and exhibits strong predictive value for HCV treatment response and spontaneous resolution.
Abstract
Approximately 3% of the world population is chronically infected with the hepatitis C virus (HCV), with potential development of cirrhosis and hepatocellular carcinoma. Despite the availability of new antiviral agents, treatment remains suboptimal. Genome-wide association studies (GWAS) identified rs12979860, a polymorphism nearby IL28B, as an important predictor of HCV clearance. We report the identification of a novel TT/-G polymorphism in the CpG region upstream of IL28B, which is a better predictor of HCV clearance than rs12979860. By using peripheral blood mononuclear cells (PBMCs) from individuals carrying different allelic combinations of the TT/-G and rs12979860 polymorphisms, we show that induction of IL28B and IFN-γ–inducible protein 10 (IP-10) mRNA relies on TT/-G, but not rs12979860, making TT/-G the only functional variant identified so far. This novel step in understanding the genetic regulation of IL28B may have important implications for clinical practice, as the use of TT/G genotyping instead of rs12979860 would improve patient management.
Hepatitis C virus (HCV) is a positive-strand RNA virus that belongs to the Flaviviridae family. Among seven distinct genotypes, genotype 1 is the most frequent in the United States and in Western Europe (Lauer and Walker, 2001). 30% of infected individuals spontaneously clear the virus, whereas the others develop chronic infection. Morbidity associated with chronic hepatitis C mainly results from the development of liver fibrosis and cirrhosis, with complications such as hepatocellular carcinoma and liver failure. Treatment of chronic hepatitis C with pegylated IFN-α and ribavirin (PEG-IFN-α/RBV) results in a sustained virological response (SVR) in ∼50% of patients infected with genotype 1/4, and in 70–90% of those infected with genotype 2/3 (Zeuzem et al., 2009). Although new antiviral agents such as telaprevir (Zeuzem et al., 2011) and boceprevir (Bacon et al., 2011; Poordad et al., 2011) improve cure rates, treatment remains long, burdened with adverse effects, costly, with limited availability to many areas, and may lead to the emergence of resistance.
Recent genome-wide association studies (GWAS) showed that single nucleotide polymorphisms (SNPs) nearby IL28B (mainly rs12979860) are strongly associated with spontaneous (Rauch et al., 2010) and treatment-induced clearance of HCV infection (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Rauch et al., 2010). IL28B encodes type III IFN-λ3, a cytokine which has antiviral properties both in vitro and in vivo (Kotenko et al., 2003; Sheppard et al., 2003; Meager et al., 2005; Robek et al., 2005; Ank et al., 2006; Hou et al., 2009). Numerous investigators confirmed the importance of IL28B SNPs in the natural course and treatment of HCV infection (Mangia et al., 2010; Rallón et al., 2010; McCarthy et al., 2010; Lange and Zeuzem, 2011).
It remains unknown whether rs12979860 exerts biological effects or whether it is in linkage disequilibrium (LD) with other functional polymorphisms. The expression of IL28B was reported to be lower in whole blood or PBMCs from individuals carrying the mutant alleles than those carrying the WT alleles (Suppiah et al., 2009; Tanaka et al., 2009), but these observations were not unequivocally confirmed in liver biopsies from patients with chronic hepatitis C (Honda et al., 2010; Urban et al., 2010; Dill et al., 2011). The functional effect was proposed to result from rs8103142, a nonsynonymous SNP in LD with rs12979860, which induces a K-to-R substitution at amino acid position 70 of IL28B (Ge et al., 2009). However, this SNP was not among the most significantly associated with clearance in GWAS. Furthermore, K70 and R70 protein variants did not induce different levels of IFN-stimulated gene (ISG) expression or different inhibition of HCV replication in Huh-7.5 cells (Urban et al., 2010). Other investigators performed gene mapping but failed to detect new SNPs with a stronger genetic effect (di Iulio et al., 2011) or with a clear functional mechanism.
RESULTS AND DISCUSSION
By sequencing the putative regulatory region upstream of IL28B, we identified a T deletion (previously described as rs67272382 NT_011109.16:g.12007372delT) adjacent to a T-to-G substitution (previously described as rs74597329 NT_011109.16:g.12007373T>G), located at position 39739154 and 39739155 in chromosome 19 (Genome Build 37.1). The presence of the TT/-G substitution, as well as the previously known rs12979860, was explored in a cohort of 540 Caucasian patients with chronic HCV infection and in 93 patients with spontaneous HCV clearance (Table S1). The rs12979860 and TT/-G polymorphisms were in strong LD (R2 = 0.91) and both had a minor allele frequency of 0.38. Despite such strong correlation, individuals who were discordant at both SNPs allowed us to explore which of the two SNPs is most strongly associated with the outcome and therefore more likely to be the functional variant.
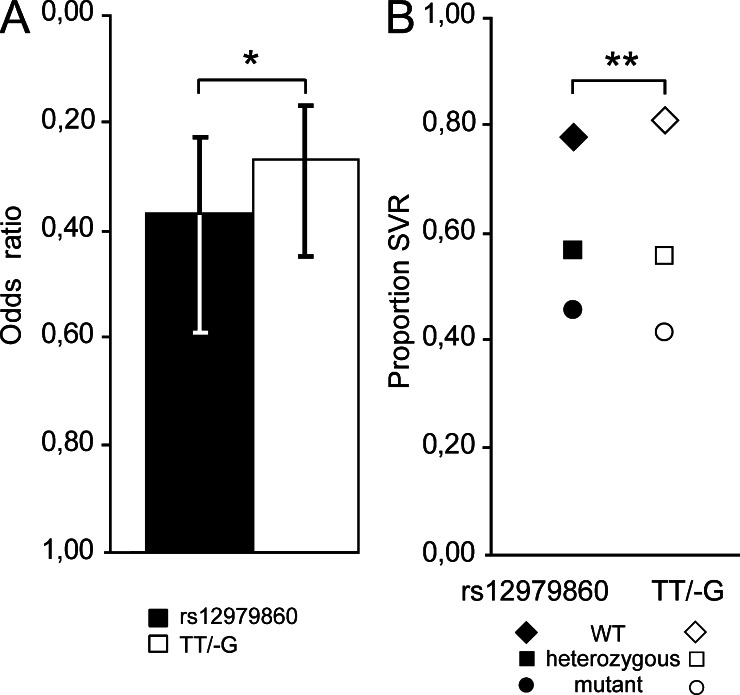
We assessed whether IL28B polymorphisms were associated with response to PEG-IFN-α/RBV therapy among chronically infected patients. When considering all viral genotypes together, both polymorphisms were associated with response to treatment (Fig. 1, Table 1, and Table S2). The strongest and most significant association was found for TT/-G (OR = 0.38, 95% CI 0.28–0.51, P = 2.51−10), compared with rs12979860 (0.46, 95% CI 0.35–0.62, P = 1.52−7). TT/-G still provided the strongest and most significant association in a multivariate model, after adjustment for age, sex, HCV RNA level, fibrosis stage, and viral genotype (OR = 0.27, 95% Cl 0.17–0.45, P = 2.70−7), compared with rs12979860 (OR = 0.37, 95% CI 0.23–0.59, P = 2.47−5). When both polymorphisms were included in the same model, the mutant -G allele of TT/-G was still associated with reduced clearance, whereas the mutant T allele of rs12979860 was associated with increased clearance (OR>1, Table 2). TT/-G had a better ability than rs12979860 to discriminate SVR rates (P = 0.04, Fig. 1 A). The SVR proportion was 0.80, 0.55, and 0.41 for patients carrying the TT/TT, TT/-G, and -G/-G genotypes, compared with 0.77, 0.56, and 0.45 for those carrying rs12979860 CC, CT, and TT genotypes (P = 0.02, Fig. 1 B). Altogether, this confirms that TT/-G is a better marker for HCV clearance than rs12979860 and that the mutant -G allele, but not the rs12979860 mutant T allele, is associated with reduced clearance. The rs12979860 mutant T allele likely represents a proxy for the effect of the mutant -G allele.
Figure 1.
TT/-G is a better predictor of response to treatment than rs12979860. (A) Association of the mutant -G allele of TT/-G and the mutant T allele of rs12979860 with HCV clearance. * , P = 0.04. Error bars represent 95% confidence level. (B) TT/-G versus rs12979860 in SVR to treatment in chronically HCV-infected patients. **, P = 0.02. Analyses were performed in 540 Caucasian patients from the Swiss Hepatitis C Cohort Study. Numbers of patients in each group are described in Table 1. All genotypes were validated by an independent laboratory as described in the materials and methods. Odds ratios are for an additive model, accounting for the effect of one or two copies of the mutant allele. Multivariate models are adjusted for age and sex, HCV RNA level, fibrosis stage, and viral genotype. P-values were calculated using the integrated discrimination improvement (IDI) test.
Table 1.
Association of IL28B polymorphisms with HCV clearance in chronic hepatitis C
| Response to treatment | Genotype | T | NR | SVR | Prop. SVRb | Univariate models | Multivariate modelsd | ||
| OR(95% CI) | P | OR (95% CI) | P | ||||||
| All viral genotypesa (n = 540) | |||||||||
| TT/-G | TT/TT | 37 | 152 | 0.80 | |||||
| TT/-G | 131 | 161 | 0.55 | ||||||
| -G/-G | 35 | 24 | 0.41 | 0.38 (0.28–0.51) | 2.51−10 | 0.27 (0.17–0.45) | 2.70−7 | ||
| rs12979860 | CC | 43 | 146 | 0.77 | |||||
| CT | 126 | 163 | 0.56 | ||||||
| TT | 34 | 28 | 0.45 | 0.46 (0.35–0.62) | 1.52−7 | 0.37 (0.23–0.59) | 2.47−5 | ||
| Viral genotype 1 and 4a (n = 288) | |||||||||
| TT/-G | TT/TT | 82 | 22 | 60 | 0.73 | ||||
| TT/-G | 174 | 112 | 62 | 0.36 | |||||
| -G/-G | 32 | 26 | 6 | 0.19 | 0.25 (0.16–0.40) | 4.61−9 | 0.16 (0.08–0.33) | 2.75−7 | |
| rs12979860 | CC | 80 | 24 | 56 | 0.70 | ||||
| CT | 173 | 109 | 64 | 0.37 | |||||
| TT | 35 | 27 | 8 | 0.23 | 0.31 (0.20–0.49) | 2.24−7 | 0.22 (0.11–0.41) | 3.46−6 | |
| Viral genotype 2 and 3c (n = 252) | |||||||||
| TT/-G | TT/TT | 15 | 92 | 0.86 | |||||
| TT/-G | 19 | 99 | 0.84 | ||||||
| -G/-G | 9 | 18 | 0.67 | 0.36 (0.15–0.86) | 2.13−2 | 1.33 (0.22–7.94) | 7.57−1 | ||
| rs12979860 | CC | 19 | 90 | 0.83 | |||||
| CT | 17 | 99 | 0.85 | ||||||
| TT | 7 | 20 | 0.74 | 0.54 (0.21–1.38) | 2.01−1 | 1.48 (0.26–8.53) | 6.59−1 | ||
| Spontaneous clearance (n = 633)a | |||||||||
| TT/-G | TT/TT | 189 | 63 | ||||||
| TT/-G | 292 | 29 | |||||||
| -G/-G | 59 | 1 | 0.28 (0.18–0.43) | 8.68−9 | 0.30 (0.19–0.47) | 1.54−7 | |||
| rs12979860 | CC | 189 | 62 | ||||||
| CT | 289 | 30 | |||||||
| TT | 62 | 1 | 0.29 (0.19–0.45) | 1.66−8 | 0.31 (0.20–0.49) | 3.64−7 | |||
CRI, chronic infection; SC, spontaneous clearance; NR, nonresponse to treatment; OR, odds ratio; CI, confidence interval; Prop, proportion.
Odd ratios and p-values are for an additive model, accounting for increased effect of one or two copies of the mutant allele.
Data indicate the proportion of patients with SVR for indicated host and viral genotypes.
Odd ratios and p-values are for a recessive model, comparing the presence of two copies versus zero or one copies of the mutant allele.
Multivariate models are adjusted for age and sex (spontaneous clearance), as well as HCV RNA level, fibrosis stage, and, whenever appropriate, viral genotype (response to treatment). Multivariate models include a smaller number of patients, due to missing covariates in some patients (n = 360 for all genotypes, n = 195 for genotypes 1 and 4, and n = 165 for genotypes 2 and 3). However, the results of univariate analyses restricted to the subset of individuals with all covariates were concordant with those of the whole group.
Table 2.
Independent contribution of rs12979860 and TT/-G to HCV clearance
| Response to treatment | Polymorphisms included in the same model | |
| OR (95% CI) | P | |
| All viral genotypes; n = 540 | ||
| Polymorphisms by pairs | ||
| TT/-G | 0.12 (0.03–0.40) | 5.94−4 |
| rs12979860 | 3.41 (1.01–11.4)a | 4.77−2 |
| Viral genotypes 1 and 4, n = 288 | ||
| Polymorphisms by pairs | ||
| TT/-G | 0.16 (0.05–0.55) | 3.60−3 |
| rs12979860 | 1.61 (0.48–5.40)a | 4.44−1 |
| Spontaneous clearanceb | ||
| Polymorphisms by pairs | ||
| TT/-G | 0.42 (0.14–1.30) | 1.31−1 |
| rs12979860 | 0.66 (0.22–1.99) | 4.59−1 |
Both TT/-G and rs12978960 were introduced in the same logistic regression model. OR, odds ratio; CI, confidence interval. OR represent the additive effect of the mutant allele (-G for TT/-G and T for rs12979860).
When TT/-G was entered into the model, the direction of the association of the T allele of rs12979860 was reversed (OR > 1).
93 individuals with spontaneous HCV clearance were compared to 540 patients with chronic infection
Similar observations were found after stratification by viral genotypes (Tables 1 and 2). In patients infected with HCV genotype 1/4, the strongest and most significant association was also found for TT/-G either in the univariate (OR = 0.25, 95% CI 0.16–0.40, P = 4.61−9 vs. OR = 0.31, 95% CI 0.20–0.49, P = 2.24−7 for rs12979860) or in the multivariate models (OR = 0.16, 95% CI 0.08–0.33, P = 2.75−7 vs. OR = 0.22, 95% CI 0.11–0.41, P = 3.46−6 for rs12979860). In patients infected with HCV genotype 2/3, TT/-G was the only SNP associated with response to treatment, but only in a recessive model (OR = 0.36, 95% CI 0.15–0.86, P = 2.13−2). Finally, TT/-G was also a better marker of spontaneous clearance (Tables 1 and 2, P = 8.68−9), compared with rs12979860 (P = 1.66−8).
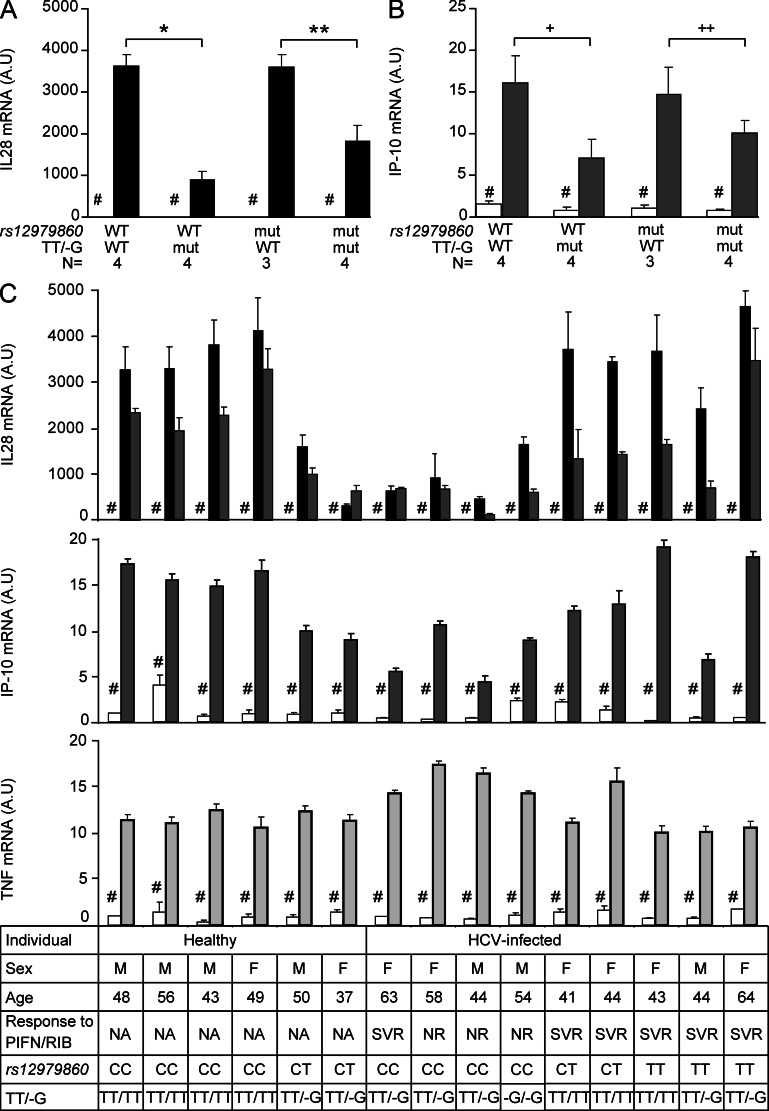
Because TT/-G is a better marker of HCV clearance, we speculated that it might correlate with IL28B mRNA expression. To address this issue, we measured IL28B mRNA expression in PBMCs from individuals carrying different allelic combinations of rs12979860 and TT/-G that were stimulated with the dsRNA analogue poly(I:C) (Fig. 2, A and C). Among individuals WT for rs12979860, PBMCs from those carrying one or two copies of the mutant allele -G of TT/-G had lower expression of IL28B mRNA compared with those from TT/-G WT individuals (P < 0.001). In contrast, among individuals WT for TT/-G, those carrying one or two copies of the mutant allele T of rs12979860 had similar expression of IL28B mRNA than those WT for rs12979860. Finally, among individuals carrying one or two copies of the mutant allele T of rs12979860, those carrying one or two copies of the mutant allele -G of TT/-G had lower expression of IL28B mRNA compared with those from TT/-G WT individuals (P = 0.007). The pattern of IL28B expression after stimulation was similar among patients with cured hepatitis C (SVR) and those with chronic infection (nonresponse) according to TT/-G, suggesting that the viremic state does not significantly influence acute response to stimulation in vitro (Fig. 2 C). Altogether, these data demonstrate that IL28B mRNA expression is driven by the presence of one or two mutant alleles of TT/-G but not by rs12979860. Because IL28B has a strong homology with IL28A due to ancestral gene duplication events, we sequenced previously cloned amplicons to unambiguously confirm that they were part of IL28B but not IL28A (unpublished data).
Figure 2.
Expression of IL28B and IP-10 mRNA relies on TT/-G but not rs12979860 polymorphism. PBMCs from healthy and HCV-infected individuals were stimulated with poly(I:C) for 4 h (black) and 8 h (dark gray). The mRNA expression of IL28B and IP-10 was measured by RT-PCR. Results grouped according to different allelic combinations of the mutant T allele of rs12979860 and the mutant -G allele of TT/-G are shown in A (IL28B) and B (IP-10). Individual results are shown in C. As a control, TNF expression was measured after stimulation with LPS (C, bottom). AU, arbitrary units; mut, mutant allele; NA, not assessed; NR, nonresponse to treatment. For each individual, stimulation was performed in triplicate. P-values are calculated by linear regression. * , P < 0.001; **, P = 0.007; +, P < 0.001; ++, P = 0.006. N stands for the number of individuals. Error bars represent standard error. #, unstimulated cells.
Although the number of individuals carrying discrepant genotypes was low, the reduction in IL28B mRNA expression among samples from individuals carrying the mutant TT/-G allele was strongly significant. Recently, Prokunina-Olsson et al. (2013) also identified TT/-G as a better predictor of response to treatment than rs12979860 among African Americans, whereas it did not improve this prediction among Caucasians. This can be explained by the lower level of LD between the two polymorphisms among African Americans (R2 = 0.71) compared with Caucasian (R2 = 0.92). Thus, the high proportion of rs12979860 and TT/-G discrepant individuals in the African American population would allow further validation of the functional role of TT/-G on IL28B mRNA expression.
Infection with HCV activates the endogenous IFN system, which leads to the induction of ISGs and contributes to viral clearance. Pretreatment plasmatic levels of ISGs, such as IFN-γ–inducible protein 10 (IP-10), are predictors of HCV clearance. Because the induction of IP-10 is thought to rely on both type I and type III IFNs, we analyzed IP-10 mRNA expression in PBMCs stimulated with poly(I:C) for 8 h (Fig. 2, B and C). As for IL28B, IP-10 expression was lower among individuals carrying the mutant allele of TT/-G but not in those carrying the mutant allele of rs12979860 (Fig. 2 B), indicating that Il28B expression could be a determinant for the induction of some ISGs. These observations suggest a strong link between the mutant -G allele of TT/-G, reduced expression of IL28B, lower induction of ISGs, and HCV treatment failure. However, the mechanisms by which IL28B TT/-G genotypes are related to ISG induction and clinical phenotypes remain to be elucidated. As a control, TNF mRNA expression after LPS stimulation was not influenced by both polymorphisms (Fig. 2 C).
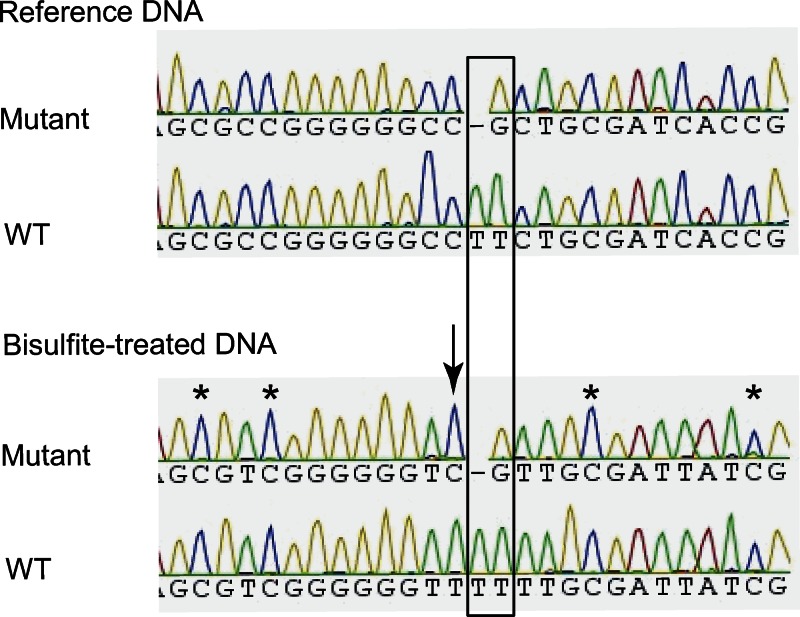
Genetic polymorphisms can influence gene function through different mechanisms. For instance, they can disrupt or create DNA regulatory elements affecting gene expression. In addition, SNPs can influence the ability of DNA sequences to undergo methylation, thereby influencing gene expression. Both rs12979860 and TT/-G are located within a CpG island which has been identified on the UCSC human genome draft by the Miklem and Hillier method. As assessed by bisulfite sequencing, TT/-G polymorphism was found to promote the methylation of a cytosine residue which is unmethylated in WT DNA sequences (Fig. 3). Further studies are needed to determine the relationship between TT/-G and surrounding CpG methylation and IL28B and/or ISG expression.
Figure 3.
The TT/-G creates a methylation site in the CpG region. The TT/-G substitution is associated with the methylation of the adjacent cytosine residue (indicated by arrow). As opposed to methylated cytosine residues (indicated by asterisks), unmethylated cytosine residues, C, are replaced by thymine, T, after bisulfite treatment and subsequent PCR amplification.
Prokunina-Olsson et al. (2013) showed that TT/-G introduces a frame shift in the DNA sequence, which transiently induces mRNA expression of an IFN analogue (IFNL4) in human hepatocytes stimulated with poly(I:C). Only patients carrying the mutant allele -G express IFNL4. The authors suggest that this protein could be a direct marker of HCV treatment failure and a new target for therapeutic intervention, raising major interest in the medical community. Nevertheless, issues regarding the molecular functions of IFNL4 remain to be clarified, such as the inability of recombinant IFNL4 to directly induce the Jak–STAT pathway in HepG2 cells, unless by transfection of an IFNL4 construct. Furthermore, the low secretion level of IFNL4 together with the lack of demonstration of its effective binding to the IFNL receptor and/or another specific receptor raises questions about its physiological function.
Using a large European cohort, we identified a new TT/-G polymorphism nearby IL28B that influences both IL28B and IP-10 mRNA expression and improves HCV clearance prediction in patients infected with HCV viral genotype 1/4 or 2/3. Because IL28B has antiviral properties, reduced IL28B expression may impair individual ability to clear HCV by itself. Whether this phenomenon is further influenced by another protein remains to be demonstrated. TT/-G genotyping may have an important impact on the management of both African American and Caucasian patients with chronic hepatitis C. Altogether, the identification of this TT/-G functional variant provides a new step in understanding the role of IL28B polymorphisms in the prediction of the response to chronic hepatitis C treatment.
MATERIALS AND METHODS
Study patients.
Patients were included from the Swiss Hepatitis C Cohort Study (SCCS), a multicenter study of >3,700 HCV-infected patients enrolled at eight major Swiss hospitals and their local affiliated centers since 2001 (Prasad et al., 2007; Bochud et al., 2009). Caucasian patients enrolled in the SCCS before August 1, 2010, with available DNA and written consent for genetic studies were selected. The study included patients with spontaneous HCV clearance (defined as presence of anti-HCV but undetectable HCV RNA without previous antiviral treatment) and those who had chronic infection with HCV genotypes 1, 2, 3, or 4 and were assessable for response to therapy with pegylated-IFN-α and ribavirin, i.e., who received >80% of the recommended dose of each drug. Demographic characteristics including age, sex, HCV risk factors, HCV genotypes, alcohol consumption, markers of chronic infection with the hepatitis B virus and HIV, HCV viral load, liver biopsy data, and HCV treatment were extracted from clinical databases. SVR was defined as an undetectable HCV RNA in serum more than 24 wk after treatment termination. Severe fibrosis was considered in patients with a METAVIR score F3 or higher.
SNP genotyping.
TT/-G and rs12979860 were extracted from a GWAS-generated dataset (Rauch et al., 2010) or genotyped by TaqMan (Applied Biosystems), using the ABI 7500 Fast real time thermocycler, according to manufacturer’s protocols. For TT/-G and rs12979860, TT and C were defined as WT and -G and T as mutant alleles, respectively. TaqMan probes and primers were designed and synthesized using Applied Biosystems software (Table S3). Automated allele calling was performed using SDS software (Applied Biosystems). For quality control, all samples were also genotyped in an independent laboratory (KBioscience, Unit 7, Maple Park, Hoddesdon, Herts, UK) using KASP SNP genotyping system, a competitive allele-specific PCR incorporating a FRET quencher cassette. Patients with at least one missing genotype and/or discordant results regarding one polymorphism were excluded from the analyses.
Statistical analysis.
Statistical analyses were performed using Stata (version 11.1, StataCorp LP). LD and Hardy-Weinberg equilibrium test were assessed using the programs pwld and genhw, respectively, both implemented in Stata. Strong LD was defined as an R2 > 0.7. The association of IL28B polymorphisms with response to treatment and spontaneous clearance was performed by univariate and multivariate logistic regression. Age, duration of infection, and body mass index (BMI) were treated as continuous variables. Multiple logistic regression models were adjusted for age, sex, HCV RNA level, fibrosis stage, and, whenever appropriate, viral genotype. For IL28B SNPs, comparisons were made using an additive model (considering a similar effect for each additional copy of the minor allele), a neutral model (comparing separately patients with heterozygous and homozygous mutant genotypes to patients with WT genotypes), and, when appropriate, a recessive model (comparing patients with homozygous mutant genotypes to the others). Discrimination ability between two different logistic regression models was compared using the integrated discrimination improvement test (IDI) implemented in Stata (Pencina et al., 2008).
PBMCs isolation and RT-PCR analysis.
PBMCs were prepared from 1.6 mg/ml of fresh EDTA blood from nine patients and six healthy donors with written consent and approval of Ethics committee. In brief, whole blood diluted in PBS was overlaid above Ficoll-Paque Plus (GE Healthcare) and mononuclear cells extracted by gradient density centrifugation. Viability, determined by trypan blue exclusion, was >90%. PBMCs were stimulated with 50 µg/ml poly(I:C) and 0.1 µg/ml LPS for 4 or 8 h. Total RNA was isolated from PBMCs using an RNeasy Mini kit and the automated QIAcube (QIAGEN). 250 ng of total RNA was reverse transcribed using the QuantiTect Reverse Transcription kit (QIAGEN). The relative levels of IL28B, IP-10, and TNF transcripts were determined by RT-PCR, with a 7500 Fast real-time PCR system (Applied Biosystems), using the Power SYBR green PCR master mix (Applied Biosystems) with primers described in Table S3. Primers were designed using the Primer 3 software and validated by BLAST analysis. The relative levels of mRNA expression to HPRT were determined by the 2(−ΔΔCt) method and expressed in arbitrary units. HPRT expression was not influenced by cell stimulation. The PCR products were run on a 2% agarose gel, purified (QIAquick Gel extraction kit; QIAGEN), cloned into a pGEM-T Easy vector (Promega), and sequenced on an ABI3130XL Sequencer (Applied Biosystems).
Methylation analysis by bisulfite sequencing.
The TT/-G–containing region was amplified by PCR using primers detailed in Table S3. Regular sequencing was performed in eight patients (four homozygous for the WT C allele and four homozygous for the mutant T rs12979860 allele). Polymorphic region was also sequenced after bisulfite treatment of DNA, with a subsequent cloning step, in 26 additional patients (13 homozygous for the WT C allele and 13 homozygous for the mutant T rs12979860 allele). Specifically, 250 ng of genomic DNA were treated with sodium bisulfite using the EpiTect Bisulfite kit (QIAGEN) per manufacturer’s recommendations and the polymorphic TT/-G region was amplified using bisulfite sequencing primers designed using MethPrimer (http://www.urogene.org/methprimer/index1.html). PCR amplifications from 10 ng of DNA consisted of an initial activation of HotStartTaq DNA polymerase (QIAGEN) at 95°C for 15 min, followed by 40 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and 1 cycle at 72°C for 10 min. Amplicons were gel purified with the QIAquick Gel Extraction kit (QIAGEN) and cloned into a pGEM-T Easy vector (Promega). Five clones from each sample were sequenced using an ABI BigDye Terminator v.3.0 Cycle sequencing kit (Applied Biosystems) and an ABI3130XL Sequencer (Applied Biosystems).
Online supplemental material.
Table S1 shows patient characteristics. Table S2 shows genotypic association of IL28B polymorphisms with response to pegylated IFN-α and ribavirin in chronically infected HCV patients. Table S3 shows primers. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130012/DC1.
Supplementary Material
Acknowledgments
The authors thank all patients from the Swiss Hepatitis C Cohort Study, as well as collaborators from the clinical, laboratory, and data centers and all study nurses, with a special attention to Tanja Grandinetti, Dorothea Brosi, Tina Horschick, Marianne Ibe, Adeline Amador, and Maribelle Herranz Garcia. The authors thank Florence Fellmann for critical review of the manuscript.
P.-Y. Bochud is supported by the Swiss National Foundation (32003B-127613 and 324730-144054), the Leenaards Foundation, the Santos-Suarez Foundation, and the Loterie Romande. This work is part of the HepaCute network (Collaborative Project) supported by the European Commission under the Health Cooperation Work Program of the seventh Framework Program (Grant agreement no. 260844). The Swiss Hepatitis C Cohort Study is supported by grants from the Swiss National Science Foundation (3347C0-108782/1), the Swiss Federal Office for Education and Sciences 03.0599), and the European Commission (LSHM-CT-2004-503359; VIRGIL Network of Excellence on Antiviral Drug Resistance).
S. Bibert and P.-Y. Bochud are inventors on a patent application filed by the Centre Hospitalier Universitaire Vaudois on the basis of these genetic findings but they have no additional financial interests. The other authors have no conflicting financial interests.
Members of the Swiss Hepatitis C Cohort Study Group: Francesco Negro (Geneva, Chairman), Antoine Hadengue (Geneva, Chairman of Scientific Committee), Laurent Kaiser, and Laura Rubbia-Brandt (Geneva); Darius Moradpour, Giuseppe Pantaleo, and Patrick Francioli (Lausanne); Martin Rickenbach (Lausanne Data Center); Gladys Martinetti and Andreas Cerny (Lugano); Virginie Masserey Spicher, Meri Gorgievski, and Jean-François Dufour (Berne); Hans Hirsch and Markus Heim (Basel); Beat Helbling, Beat Müllhaupt, and Stephan Regenass (Zurich); Raffaele Malinverni (Neuchatel); Christa Meyenberger, Tilman Gerlach, and Guenter Dollenmaier (St Gallen); and Gieri Cathomas (Liestal).
Footnotes
Abbreviations used:
- GWAS
- genome-wide association studies
- HCV
- hepatitis C virus
- IP-10
- IFN-γ–inducible protein 10
- ISG
- IFN-stimulated gene
- LD
- linkage disequilibrium
- PBMC
- peripheral blood mononuclear cell
- SVR
- sustained virological response
References
- Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80:4501–4509 10.1128/JVI.80.9.4501-4509.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon B.R., Gordon S.C., Lawitz E., Marcellin P., Vierling J.M., Zeuzem S., Poordad F., Goodman Z.D., Sings H.L., Boparai N., et al. ; HCV RESPOND-2 Investigators 2011. Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1207–1217 10.1056/NEJMoa1009482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochud P.Y., Cai T., Overbeck K., Bochud M., Dufour J.F., Müllhaupt B., Borovicka J., Heim M., Moradpour D., Cerny A., et al. ; Swiss Hepatitis C Cohort Study Group 2009. Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J. Hepatol. 51:655–666 10.1016/j.jhep.2009.05.016 [DOI] [PubMed] [Google Scholar]
- di Iulio J., Ciuffi A., Fitzmaurice K., Kelleher D., Rotger M., Fellay J., Martinez R., Pulit S., Furrer H., Günthard H.F., et al. ; Swiss HIV Cohort Study 2011. Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology. 53:1446–1454 10.1002/hep.24263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill M.T., Duong F.H., Vogt J.E., Bibert S., Bochud P.Y., Terracciano L., Papassotiropoulos A., Roth V., Heim M.H. 2011. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 140:1021–1031 10.1053/j.gastro.2010.11.039 [DOI] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 461:399–401 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- Honda M., Sakai A., Yamashita T., Nakamoto Y., Mizukoshi E., Sakai Y., Yamashita T., Nakamura M., Shirasaki T., Horimoto K., et al. ; Hokuriku Liver Study Group 2010. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 139:499–509 10.1053/j.gastro.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Hou W., Wang X., Ye L., Zhou L., Yang Z.Q., Riedel E., Ho W.Z. 2009. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J. Virol. 83:3834–3842 10.1128/JVI.01773-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77 10.1038/ni875 [DOI] [PubMed] [Google Scholar]
- Lange C.M., Zeuzem S. 2011. IL28B single nucleotide polymorphisms in the treatment of hepatitis C. J. Hepatol. 55:692–701 10.1016/j.jhep.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Lauer G.M., Walker B.D. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52 10.1056/NEJM200107053450107 [DOI] [PubMed] [Google Scholar]
- Mangia A., Thompson A.J., Santoro R., Piazzolla V., Tillmann H.L., Patel K., Shianna K.V., Mottola L., Petruzzellis D., Bacca D., et al. 2010. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 139:821–827: 827: e1 10.1053/j.gastro.2010.05.079 [DOI] [PubMed] [Google Scholar]
- McCarthy J.J., Li J.H., Thompson A., Suchindran S., Lao X.Q., Patel K., Tillmann H.L., Muir A.J., McHutchison J.G. 2010. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 138:2307–2314 10.1053/j.gastro.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meager A., Visvalingam K., Dilger P., Bryan D., Wadhwa M. 2005. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine. 31:109–118 10.1016/j.cyto.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Pencina M.J., D’Agostino R.B., Sr, D’Agostino R.B., Jr, Vasan R.S. 2008. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat. Med. 27:157–172, discussion :207–212 10.1002/sim.2929 [DOI] [PubMed] [Google Scholar]
- Poordad F., McCone J., Jr, Bacon B.R., Bruno S., Manns M.P., Sulkowski M.S., Jacobson I.M., Reddy K.R., Goodman Z.D., Boparai N., et al. ; SPRINT-2 Investigators 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 10.1056/NEJMoa1010494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad L., Spicher V.M., Zwahlen M., Rickenbach M., Helbling B., Negro F., and C.C.S.G. Swiss Hepatitis 2007. Cohort Profile: the Swiss Hepatitis C Cohort Study (SCCS). Int. J. Epidemiol. 36:731–737 10.1093/ije/dym096 [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., et al. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45:164–171 10.1038/ng.2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallón N.I., Naggie S., Benito J.M., Medrano J., Restrepo C., Goldstein D., Shianna K.V., Vispo E., Thompson A., McHutchison J., Soriano V. 2010. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 24:F23–F29 10.1097/QAD.0b013e3283391d6d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Kutalik Z., Descombes P., Cai T., Di Iulio J., Mueller T., Bochud M., Battegay M., Bernasconi E., Borovicka J., et al. ; Swiss Hepatitis C Cohort Study; Swiss HIV Cohort Study 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 138:1338–1345: 1345: e1–e7 10.1053/j.gastro.2009.12.056 [DOI] [PubMed] [Google Scholar]
- Robek M.D., Boyd B.S., Chisari F.V. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79:3851–3854 10.1128/JVI.79.6.3851-3854.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 10.1038/ni873 [DOI] [PubMed] [Google Scholar]
- Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 10.1038/ng.447 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 10.1038/ng.449 [DOI] [PubMed] [Google Scholar]
- Urban T.J., Thompson A.J., Bradrick S.S., Fellay J., Schuppan D., Cronin K.D., Hong L., McKenzie A., Patel K., Shianna K.V., et al. 2010. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 52:1888–1896 10.1002/hep.23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S., Berg T., Moeller B., Hinrichsen H., Mauss S., Wedemeyer H., Sarrazin C., Hueppe D., Zehnter E., Manns M.P. 2009. Expert opinion on the treatment of patients with chronic hepatitis C. J. Viral Hepat. 16:75–90 10.1111/j.1365-2893.2008.01012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S., Andreone P., Pol S., Lawitz E., Diago M., Roberts S., Focaccia R., Younossi Z., Foster G.R., Horban A., et al. ; REALIZE Study Team 2011. Telaprevir for retreatment of HCV infection. N. Engl. J. Med. 364:2417–2428 10.1056/NEJMoa1013086 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.