RB family genes control T cell production and promote thymic involution through reducing Foxn1 expression in thymic epithelial cells.
Abstract
Thymic involution during aging is a major cause of decreased production of T cells and reduced immunity. Here we show that inactivation of Rb family genes in young mice prevents thymic involution and results in an enlarged thymus competent for increased production of naive T cells. This phenotype originates from the expansion of functional thymic epithelial cells (TECs). In RB family mutant TECs, increased activity of E2F transcription factors drives increased expression of Foxn1, a central regulator of the thymic epithelium. Increased Foxn1 expression is required for the thymic expansion observed in Rb family mutant mice. Thus, the RB family promotes thymic involution and controls T cell production via a bone marrow–independent mechanism, identifying a novel pathway to target to increase thymic function in patients.
The thymus plays a critical role in adaptive immunity as the site of mature T cell production (Miller, 2011). The thymus grows rapidly during early life, generating a large T cell repertoire. It then enters an involution process, leading to decreased production of naive T cells, resulting in impaired immune function in the elderly and preventing complete reconstitution of the immune system in various pathologies (Boehm, 2008; Rodewald, 2008; Carpenter and Bosselut, 2010). The mechanisms controlling thymic involution are poorly understood, hampering the development of therapeutic strategies to enhance immune function in a wide variety of patients (Napolitano et al., 2008; Sauce and Appay, 2011).
Although the thymus is composed mostly of T lymphocytes, T cell development requires a complex microenvironment including endothelial, dendritic, and thymic epithelial cells (TECs; Manley et al., 2011). TECs are highly proliferative during thymic expansion, and then their cell cycle slows considerably (Gray et al., 2007; Aw and Palmer, 2011; Manley et al., 2011). Recent data show that keratinocyte growth factor (KGF; or FGF-7) and growth hormone treatment in aged rodents and humans leads to transient thymic expansion and increased production of naive T cells (Min et al., 2007; Napolitano et al., 2008). In particular, increased TEC numbers were observed upon KGF treatment (Min et al., 2007; Rossi et al., 2007). IL-22 also can support thymic regeneration in mice after radiation treatment (Dudakov et al., 2012). These data suggest that increased numbers of TECs can enhance, at least temporarily, thymic function. Nevertheless, the molecular mechanisms regulating cell cycle activity in TECs are still poorly characterized, and no strategies have been devised yet for long-term thymic growth.
Through its ability to bind the E2F transcription factors, the RB family of proteins (RB, p107, and p130) plays a major role in the control of cell cycle progression. Growth factors and external signals activate Cyclin and Cyclin-dependent kinase (CDK) protein complexes. Upon activation, Cyclin–CDK complexes phosphorylate RB family proteins, resulting in their inactivation. Inactivation of RB family proteins by phosphorylation activates E2F, thereby promoting transcription of genes involved in the G1/S transition of the cell cycle (Iaquinta and Lees, 2007; Chinnam and Goodrich, 2011)
There is no reported thymic phenotype in mice in which any one of the Rb family gene is inactivated, possibly because of the strong functional overlap between the three proteins (Dannenberg and te Riele, 2006). Nevertheless, emerging evidence suggests that some members of the RB pathway may play a role in thymic biology, including E2F2, Cyclin D1 (CCND1), p18Ink4c, and p27Kip1 (Robles et al., 1996; Franklin et al., 1998; Pierce et al., 1998; Klug et al., 2000; Rodriguez-Puebla et al., 2000; Iglesias et al., 2004; Scheijen et al., 2004; Chien et al., 2006). However, the mechanisms underlying how the cell cycle machinery affects thymus development and involution are still unknown.
Here we report that deletion of Rb family genes in the thymus of mice leads to increased proliferation in TEC populations and prevents thymic involution. Furthermore, we found that the RB family regulates the transcription of Foxn1, an important regulator of TEC differentiation and function. Finally, we demonstrate that the increased expression of Foxn1 is required for the thymus expansion observed in Rb family mutant mice. These data identify a new RB-E2F-Foxn1 module as a critical regulator of thymic involution and function.
RESULTS AND DISCUSSION
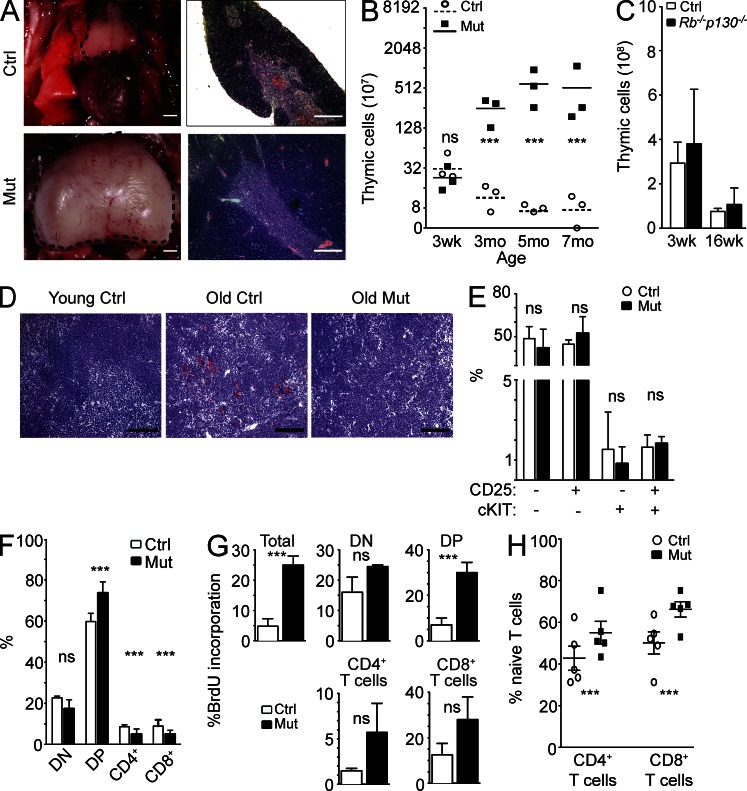
We previously reported that inactivation of the entire Rb gene family in young adult mice (3–6-wk-old Mx1-Cre Rblox/lox; p130lox/lox; p107−/− mice) results in rapid death as the result of hyperproliferation in multiple organs (Viatour et al., 2008; Chen et al., 2011). In contrast, reintroduction of one copy of p107 (Mx1-Cre Rblox/lox; p130lox/lox; p107+/− or Mx1-Cre p107-Single mice) rescues the lethality of the triple knockout mice and significantly extends their lifespan up to 9–12 mo of age (Viatour et al., 2008, 2011). At that age, although p107-Single mice that lack Cre-recombinase are still healthy, Mx1-Cre p107-Single mice exhibit weight loss and respiratory distress. Upon autopsy, we found that the thymus of Mx1-Cre p107-Single mice was significantly increased in size (Fig. 1 A, left), compressing the lungs. This thymic growth correlated with increased cellularity (Fig. 1 B). The plateau observed after several months of continuous growth in mutant mice may be the result of mechanical or vascular constraints, limits of the BM to generate T cell precursors, or a thymus-intrinsic effect.
Figure 1.
Thymic growth and increased T cells in Mx1-Cre p107-Single mice (Mx1-Cre Rblox/lox; p130lox/lox; p107+/−). (A, left) Representative control (Ctrl; top) and mutant (Mut; bottom) thymi (dashed lines). (right) H&E staining of representative control (top) and mutant (bottom) thymi. (B) Thymic cellularity (×107, log2 scale) of control and Mx1-Cre p107-Single mice after inactivation of the Rb family in young mice (n = 3 for each time point; asterisks indicate P < 0.05; ns indicates no significant difference; p-values: 3 wk = 0.35, 3 mo = 0.018, 5 mo = 0.04, and 7 mo = 0.03). Horizontal bars indicate the mean. (C) Comparison of thymic cellularity between control and Mx1-Cre Rb/p130 mutant mice at 3 wk (Ctrl: 29 ± 9.5 × 108 cells; Mx1-Cre Rb/p130: 38 ± 24.6 × 108 cells; P = 0.59; n = 3) and 16 wk (Ctrl: 7.6 ± 1.4 × 108 cells; Mx1-Cre Rb/p130: 10.8 ± 7.4 × 108 cells; P = 0.49; n = 3). (D) Oil Red O staining (red) on sections from 2-wk-old control (left), 9-mo-old control (middle), and 9-mo-old Mx1-Cre p107-Single mutant (right) thymi (performed on three mice each). Bars: (A) 200 µm; (D) 100 µm. (E) Quantification of intrathymic T cell progenitor populations between control and mutant mice (n = 3). Lineage-negative populations include CD25− cKit− (Ctrl: 48.65 ± 8.27%; Mut: 42.07 ± 12.95%; P = 0.54), CD25+ cKit− (Ctrl: 44.75 ± 2.90%; Mut: 53.37 ± 11.49%; P = 0.42), CD25− cKit+ (Ctrl: 3.08 ± 3.7%; Mut: 1.65 ± 1.64%; P = 0.45), and CD25+ cKit+ (Ctrl: 3.28 ± 1.28%; Mut: 2.65 ± 0.69%; P = 0.60). (F) Quantification of DN (Ctrl: 22.60 ± 0.82%; Mut: 17.33 ± 4.43%; P = 0.08), DP (Ctrl: 59.73 ± 3.98%; Mut: 73.61 ± 5.43%; P = 0.004), CD4+ T cell (Ctrl: 8.67 ± 0.83%; Mut: 5.00 ± 2.51%; P = 0.04), and CD8+ T cell populations (Ctrl: 8.93 ± 2.96%; Mut: 4.98 ± 1.91%; P = 0.03). (G) T cell BrdU incorporation 1 h after injection was quantified by FACS analysis (n = 3; p-values: Total = 0.002, DN = 0.14, DP = 0.05, CD4+ = 0.24, and CD8+ = 0.22). (H) Quantification of splenic naive T cells in mutants and control mice (n = 3 each; CD4+: Ctrl, 42.76 ± 5.81%; Mut, 55.06 ± 5.42% [P = 0.05]; CD8+: Ctrl, 50.14 ± 5.32%; Mut, 66.24 ± 3.64% [P = 0.03]). All error bars indicate standard error.
Histopathological analysis showed no gross alteration of the thymic architecture in mutant mice compared with controls (Fig. 1 A, right). As expected (Yang et al., 2009; Aw and Palmer, 2011), the thymus from 6–9-mo-old mice accumulated lipids, as indicated by Oil Red O staining. In contrast, the thymus of young control (2–3 mo of age) and old mutant mice (6–9 mo of age) showed little lipid accumulation (Fig. 1 D). The only defect we detected in mutant mice outside of the thymus was epidermal hyperplasia (not depicted; previously described in Ruiz et al., 2004). Thymic size in Mx1-Cre Rb/p130 double mutant mice was indistinguishable from that of control mice (Fig. 1 D). Overall, these data show that loss of RB family function in Mx1-Cre p107-Single mice results in the expansion of the thymus, while preserving overall thymic architecture.
Flow cytometry analysis of progenitor thymocyte populations (Fig. 1 E and Fig. S1 A) revealed no significant differences in the frequency of these cell populations in the thymus of control and mutant mice. There were no significant changes in the number of double-negative (DN) cells between control and mutant mice. Double-positive (DP) mature intrathymic T cells were slightly more numerous in the mutant mice, whereas CD4+ and CD8+ populations were smaller (Fig. 1 F and Fig. S1 B). These changes may reflect a higher percentage of DP thymocytes failing positive selection in the mutant animals (Morris and Allen, 2012). As expected (Pénit and Vasseur, 1997), BrdU/propidium iodide (PI) cell cycle analysis showed some BrdU incorporation in CD4+ and CD8+ populations in control mice, but proliferation rates were higher in mutant CD4+ and CD8+ cells (Fig. 1 G). These observations show that loss of RB family function results in a general expansion of thymocytes rather than expansion of a specific subpopulation as would be seen in a T cell lymphoma. Notably, this increase in the number of intrathymic T cells was accompanied by an increase in the number of peripheral CD62L+ CD44− naive T cells in Rb family mutant mice (Fig. 1 H and Fig. S1 C). This last finding is consistent with a recent observation that forced expression of thymic Foxn1 leads to a decrease in peripheral memory T cells (Zook et al., 2011). Thus, inactivation of the Rb gene family results in a thymic expansion that correlates with increased production of naive T cells in mice.
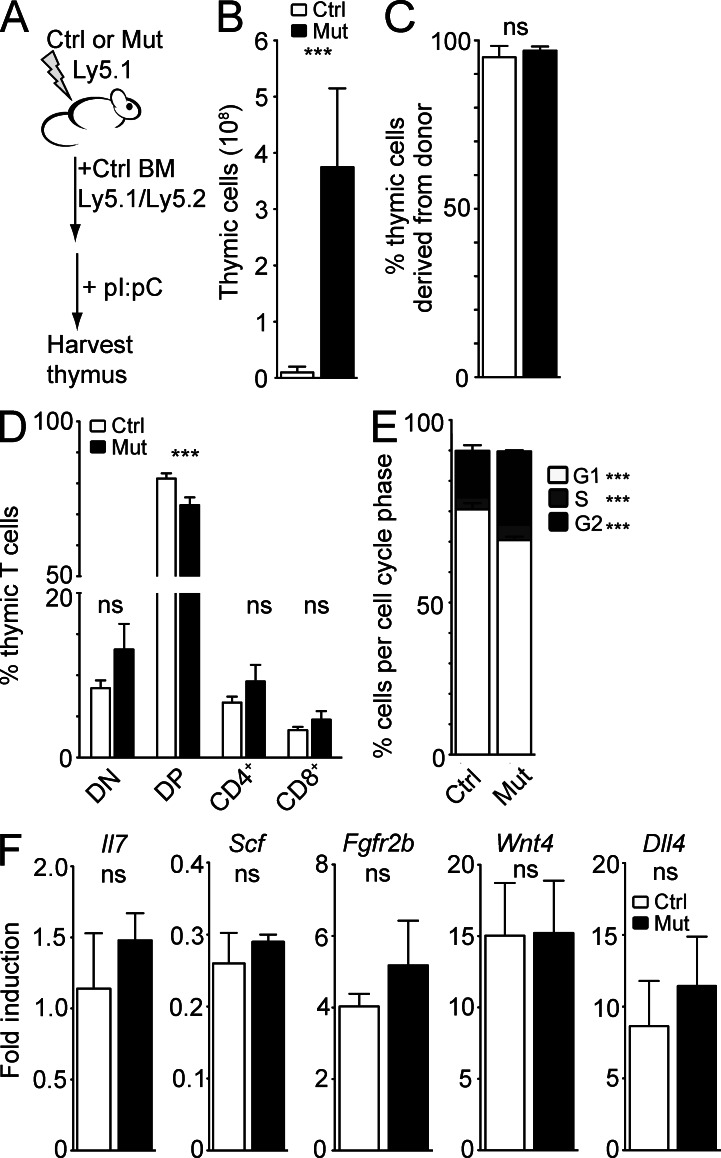
To investigate the origin of thymic expansion in Mx1-Cre p107-Single mice, we generated chimeric mice by transplanting Ly5.1+/ Ly5.2+ wild-type BM cells into 3-mo-old Ly5.1+ lethally irradiated mutant or control mice (Fig. 2 A). Upon reconstitution of hematopoiesis, chimeric mice were injected with polyinosine:polycytosine (pI:pC). 3 mo later, the thymi of Mx1-Cre p107-Single mice reconstituted with wild-type hematopoietic cells were enlarged significantly compared with the thymi from chimeric wild-type/control mice (Fig. 2 B). In both cases, as expected, BM cells and thymocytes from chimeric mutant and control mice expressed both Ly5.1 and Ly5.2 isoforms, confirming their wild-type origin (Fig. 2 C). Although all T cell populations were expanded in chimeric mutant mice (Fig. 2 D), there was, overall, no significant difference in the proportional representation of DN, DP, CD4+, and CD8+ populations between wild-type and mutant chimeric mice (Fig. 2 E). Nevertheless, the proliferative index of T cell populations in mutant mice was increased compared with controls (Fig. 2 E). Mutant T cells failed to exhibit increased proliferation upon transplantation of mutant BM cells in recipient mice (not depicted). Thus, although there may be a contribution of increased T cell proliferation to the thymic expansion observed in mutant mice, the thymus expansion observed in Rb family mutant mice does not depend on the deletion of Rb family genes in BM-derived cells.
Figure 2.
The thymic expansion in Rb family mutant mice does not originate from the hematopoietic compartment. (A) Control (Ctrl) and Mx1-Cre p107-Single (Mut) mice expressing the Ly5.1 surface marker were lethally irradiated and transplanted with BM from Ly5.1/Ly5.2 wild-type mice. (B) Thymic cellularity (×108) of control and Mx1-Cre p107-Single chimeric mice (n = 5; P = 0.02). (C) Quantified (Ctrl: 95.23 ± 2.2% donor-derived; Mut: 97 ± 1.01% donor-derived; P = 0.19; n = 3). (D) Quantification of T cells in control and mutant chimeric mice (DN: Ctrl, 8.54 ± 2.44%; Mut, 11.88 ± 7.33% [P = 0.42]; DP: Ctrl, 80.68 ± 3.75%; Mut, 71.98 ± 5.87% [P = 0.046]; CD4+: Ctrl, 7.2 ± 1.26%; Mut, 10.74 ± 3.5% [P = 0.11]; CD8+: Ctrl, 3.59 ± 0.76%; Mut, 5.35 ± 1.87% [P = 0.13%]; n = 3). (E) PI analysis of cell cycle activity in control and mutant chimeric mice (G1: Ctrl, 80.65 ± 2.02%; Mut, 70.53 ± 1.13% [P = 0.0001]; S: Ctrl, 3.93 ± 0.16%; Mut, 5.00 ± 0.66% [P = 0.02]; G2: Ctrl, 15.30 ± 1.85%; Mut, 24.5 ± 5.40% [P = 0.0001]; n = 4). (F) Cytokine and receptor expression in thymic extracts from control and mutant mice after lethal irradiation and rescue with wild-type BM as assayed by RT-qPCR (p-values: Il7 = 0.41, Scf = 0.37, Fgfr2b = 0.58, Wnt4 = 0.97, and Dll4 = 0.64; n = 3). Asterisks indicate P < 0.05; ns indicates no significant difference. All error bars indicate standard error.
Because thymic stroma-derived cytokines and growth factors are important for thymus growth and T cell proliferation and differentiation, we assessed mRNA expression levels for several cytokines and receptors produced in the thymus (Il7, Scf/kitl, Fgfr2b, Wnt4, and Dll4). After normalization to Eva1 levels (Guttinger et al., 1998; DeMonte et al., 2007; Min et al., 2007), we found no significant difference in per-cell levels of these mRNAs between chimeric mutant and chimeric control groups (Fig. 2 F). These data provide more evidence that the thymic milieu is essentially normal in the enlarged thymus of Mx1-Cre p107-Single mice, further suggesting that the thymic microenvironment is expanded and maintained in Rb family mutant mice.
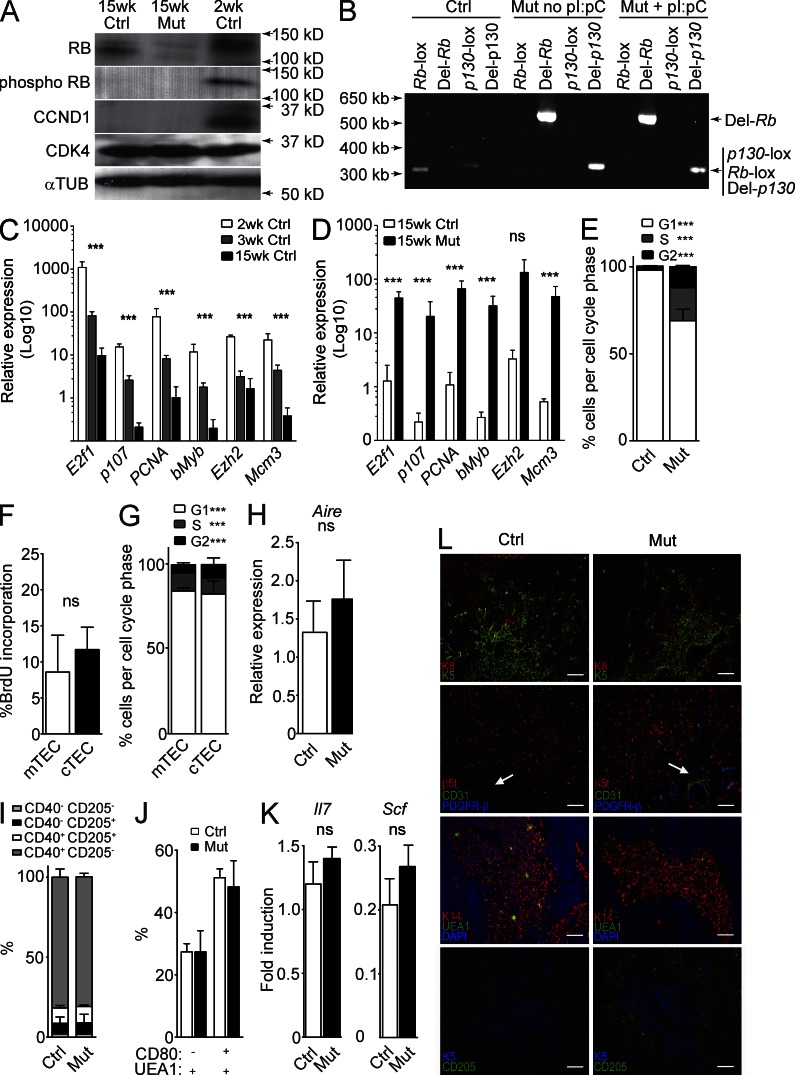
TECs are a critical component of the thymic microenvironment, providing many of the functions needed to control T cell differentiation and selection. Thus, we asked whether TECs may be responsible for the thymus phenotype observed in Mx1-Cre p107-Single mutant mice. We found that the pool of phosphorylated RB decreased with age, correlating with higher levels of CCND1 in TECs from younger mice; Cdk4 levels did not change with age (Fig. 3 A). Accordingly, transcription of canonical E2F targets in TECs significantly decreased with age (Fig. 3 C), further correlating thymic involution with increased RB activity. We also confirmed loss of RB by immunoblot analysis (Fig. 3 A) and deletion of Rb and p130 by PCR in injected and noninjected Mx1-Cre p107-Single TECs (Fig. 3 B). Deletion of Rb and p130 without pI:pC injections confirms that the Mx1 promoter can be induced by endogenous signals (Kühn et al., 1995); this deletion in noninduced mice also led to a significant increase in thymus size (thymic cellularity in control, 9.8 ± 0.2 × 107, versus noninjected Mx1-Cre p107-Single, 1.2 ± 0.7 × 109; P = 0.0001). The extent of deletion of the Rb family seen in the thymus of these noninduced mice suggests that the mutant cells may outcompete wild-type or partially deleted cells as the thymus becomes enlarged. Notably, inactivation of the Rb gene family in Mx1-Cre p107-Single mice prevented the down-regulation of E2F targets normally observed during aging (Fig. 3 D). Because CCND1 production is induced by mitogenic signals (Klein and Assoian, 2008), the lack of detectable CCND1 in 15-wk-old control and mutant mice argues that, at this age, TECs do not receive extracellular mitogenic signals, further suggesting that the thymic growth observed in Mx1-Cre p107-Single mice is caused by a TEC-intrinsic effect.
Figure 3.
Expansion of functional TECs in the thymus of Rb family mutant mice. (A) Immunoblot analysis (one of two replicates shown) of protein lysates from pooled CD45− EPCAM+ MHCII+ TECs from young (2 wk; n = 10), control (Ctrl; 15 wk injected; n = 10), and Mx1-Cre p107-single (15 wk; n = 3 injected, 2 noninjected) thymi against RB, phospho-RB, CCND1, CDK4, and α-Tubulin (loading control). (D) PCR analysis of genomic DNA isolated from CD45− EPCAM+ MHCII+ TECs for the lox-flanked or Cre-deleted exons of Rb and p130 in 15-wk-old control, non–pI:pC-injected Mx1-Cre p107-Single, and pI:pC-injected Mx1-Cre p107-Single mice (one of four shown). (C) RT-qPCR evaluation of expression of E2F target and S-phase genes in flow-purified TECs from 2-wk-old (n = 2), 3-wk-old (n = 5), and 15-wk-old control mice (n = 3), including E2f1 (p-values for all genes presented as 2–3 wk and 3–15 wk, P = 0.044 and 0.031, respectively), p107 (P = 0.001 and 0.04), PCNA (P = 0.026 and 0.018), bMyb (P = 0.026 and 0.041), Ezh2 (P = 0.0001 and 0.40), and Mcm3 (P = 0.018 and 0.073). (D) RT-qPCR evaluation of expression of E2F target and S-phase genes in flow-purified TECs from control and mutant TECs (15 wk; n = 3 each). Genes analyzed included E2f1 (P = 0.017), p107 (P = 0.004), PCNA (P = 0.033), bMyb (P = 0.026), Ezh2 (P = 0.059), and Mcm3 (P = 0.022). (E) PI analysis of cell cycle activity in TECs from 15-wk-old control and Mx1-Cre p107-Single mice (G1: Ctrl, 98.08 ± 1.18%; Mut, 69.17 ± 9.49% [P = 0.0003]; S: Ctrl, 1.21 ± 0.78%; Mut, 18.86 ± 6.32% [P = 0.0002]; G2: Ctrl, 1.24 ± 0.85%; Mut, 12.13 ± 3.68% [P = 0.0004]; n = 10 control and 4 mutant). (F) BrdU incorporation into mTECs and cTECs in mutant mice (mTEC: 8.95 ± 5.11%; cTEC: 11.7 ± 3.11%; P = 0.54; n = 2). (G) PI analysis of cell cycle activity in mTECs and cTECs from mutant mice (G1: mTEC, 84.17 ± 1.82%; cTEC, 82.4 ± 7.49% [P = 0.70]; S: mTEC, 10.63 ± 0.67%; cTEC, 9.30 ± 3.52% [P = 0.55]; G2: mTEC, 4.84 ± 1.25%; cTEC, 8.11 ± 3.88% [P = 0.02]; n = 2). (H) Expression levels of the mTEC-expressed gene, Aire, assessed by RT-qPCR in control and mutant thymic extracts, normalized for cell number by Eva1 levels (n = 4; P = 0.58). (I) Quantification of maturing cTEC subpopulations as identified by CD40 and CD205 expression (CD40− CD205−: Ctrl, 2.01 ± 0.44%; Mut, 1.89 ± 0.08% [P = 0.91]; CD40+ CD205−: Ctrl, 6.59 ± 4.00%; Mut, 7.23 ± 5.23% [P = 0.62]; CD40− CD205+: Ctrl, 9.61 ± 1.54%; Mut, 10.06 ± 0.96% [P = 0.79]; CD40+ CD205+: Ctrl, 81.77 ± 5.17%; Mut, 81.14 ± 2.11% [P = 0.65]; n = 3). (J) Quantification of maturing mTEC subpopulations as identified by CD80 and UEA1 staining (CD80− UEA1+: Ctrl, 27.34 ± 2.67%; Mut, 27.35 ± 6.82% [P = 0.87]; CD80+ UEA1+: Ctrl, 51.18 ± 2.84%; Mut, 48.24 ± 8.39% [P = 0.86]; n = 3). (K) Expression levels of Il7 and Scf assessed by RT-qPCR in control and mutant thymic extracts, normalized by Eva1 (n = 4; p-values: Il7 = 0.29 and Scf = 0.34). (L) Immunofluorescence staining of control (left) and mutant (right) thymi with antibodies for K5 and K8 (top row); β5t, CD31, and PDGFR-β (second row; white arrows indicate blood vessels); K14, UEA1, and DAPI (third row); or K5 and CD205 (bottom row; one of two shown). Bars, 100 µm. Asterisks indicate P < 0.05; ns indicates no significant difference. All error bars indicate standard error.
Next, we sought to quantify the extent to which loss of RB family activity in TECs increased TEC numbers in the mutant thymus. CD45−, MHCII+ TECs increased 18-fold compared with CD45+ hematopoietic cells in the thymus of the mutant mice (control, 0.9 ± 0.49% TECs; mutant, 16.42 ± 6.9% TECs; n = 3 each; P = 0.018). Mutant TECs also displayed a higher proliferative index compared with their wild-type counterparts (Fig. 3 E). Additional separation of CD45− MHCII+ TECs from mutant thymus based on the Ly51 surface marker (Williams et al., 2009) showed that mutant cortical TECs (cTECs; Ly51+) and medullary TECs (mTECs; Ly51–) displayed a high proliferative activity (Fig. 3, F and G). Compared with Ly51− TECs, we noted an increase in the number of Ly51+ TECs in G2, which is sometimes seen in cells with loss of RB function (Stark and Taylor, 2006; Conklin et al., 2012).
To determine whether there was a change in proportional representation of particular TEC subsets, we quantified levels of the mature mTEC marker Aire (Gray et al., 2007; Dooley et al., 2008) in control and mutant thymi. We found no significant difference in Aire expression in Mx1-Cre p107-Single mice (Fig. 3 H), suggesting that there is no preferential expansion of either cTECs or mTECs in Rb family mutant mice. We also measured by flow cytometry the proportions of cTEC precursors as identified by CD40 and CD205 expression (Shakib et al., 2009; Nowell et al., 2011). There was no difference in CD40/CD205 subpopulations between control and mutant mice (Fig. 3 I and Fig. S1 D). Similarly, there were no differences in mTEC precursor populations as measured by CD80 and UEA1 expression (Fig. 3 J and Fig. S1 E; Akiyama et al., 2012). When we measured mRNA expression levels for two thymocyte growth cytokines that are expressed by differentiated TECs (Il7 and Scf), we found that the expression levels of these genes on a per-cell basis were not significantly different between control and mutant thymi (Fig. 3 K). These data support the idea that the major difference between control and Mx1-Cre p107-Single mice lies in the number of TECs and not in the differentiation state of these cells.
To examine further the differentiation state of RB family–deficient TECs, we performed immunofluorescence analysis of markers for various cell populations in the thymic microenvironment, including Keratin 5 (K5), Keratin 8 (K8), Keratin 14 (K14), the proteasomal β-5t subunit (β5t), and CD205 (Rodewald, 2008; Shrimpton et al., 2009; Takahama et al., 2012). We found no evidence for alteration of thymus structure in mutant mice (Fig. 3 L). We occasionally observed subcapsular cysts in very large mutant thymi (not depicted), perhaps reflecting the physical limitations placed by the thoracic cavity upon the enlarged thymus. This observation highlights the fact that we performed our analyses of TEC subpopulations at a late time point, when the cells analyzed may represent only the descendants of those cells that initially underwent complete recombination and may have been selected for because of their increased proliferation. Nevertheless, these data indicate that inactivation of the RB family in mice leads to increased TEC proliferation without significantly altering their differentiation.
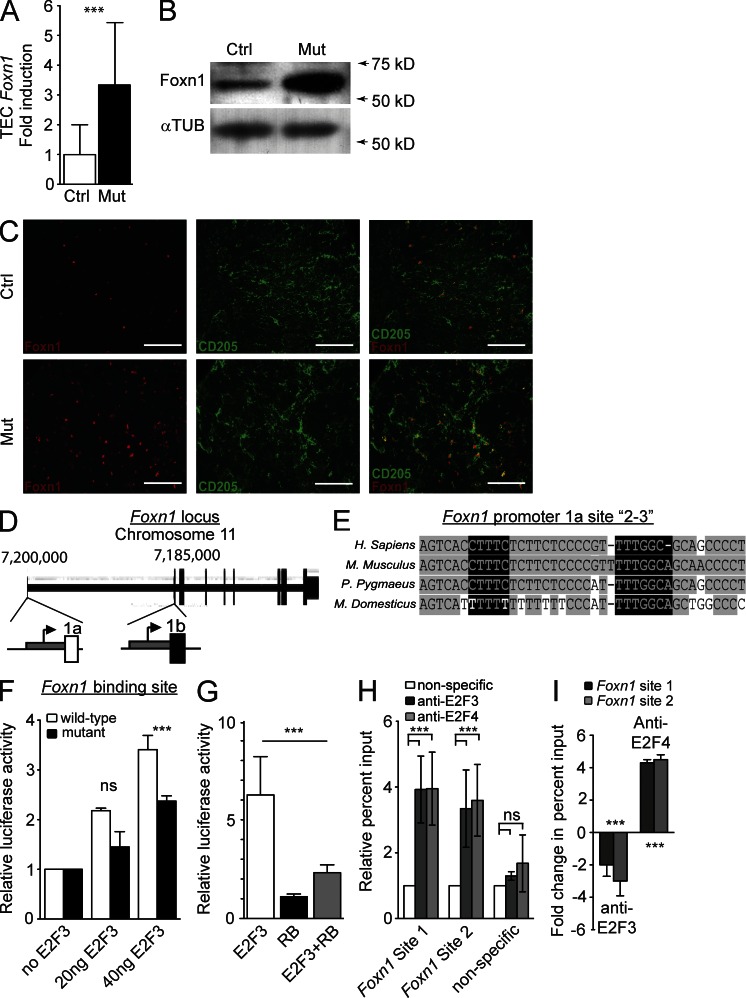
Thymus organogenesis is coordinated by the successive action of a network of transcription factors (Rodewald, 2008; Manley and Condie, 2010). We surmised that one mechanism by which loss of RB family function prevents thymic involution might be by maintaining the expression of such factors. However, we found that the expression of Eya1, Pax1, Pax9, Six1, Hoxa3, and Tbx1 was similar in control and mutant TECs at 3 mo of age (not depicted). In contrast, Foxn1 RNA and protein levels were significantly increased in mutant TECs compared with controls (Fig. 4, A and B). This increase in Foxn1 levels in the mutant thymi may come from increased levels per cell but may also be caused by increased numbers of Foxn1-expressing cells, as suggested by immunofluorescence analysis (Fig. 4 C). Forced overexpression of Foxn1 was shown to be insufficient to lead to an increase in thymic size (Zook et al., 2011). Thus, the thymic outgrowth in Rb family mutant mice is probably also dependent on the increased proliferation seen in mutant TECs, and this increased proliferation is likely to be greater than that with increased Foxn1 levels alone.
Figure 4.
The RB family controls the expression of Foxn1. (A) Expression of Foxn1 in CD45– MHCII+ TECS from control (Ctrl) and mutant (Mut) mice relative to Eva1 expression (n = 4; P = 0.03). (B) Immunoblot analysis of Foxn1 levels in control and mutant TECs from 12 pooled control and 5 pooled mutant thymi. α-Tubulin is included as a loading control (n = 2). (C) Immunofluorescence staining of Foxn1 and CD205 individually and merged (right) in control (top) and mutant (bottom) thymi (one of two shown). Bars, 100 µm. (D) Schematic representation of the Foxn1 promoter. Region 1a is active in the skin and thymus, whereas region 1b is only active in the skin. (E) Sequence alignment on the UCSC genome browser (Fujita et al., 2011) of the promoter region 1a encompassing two E2F sites shows conservation across multiple mammalian species (Kent, 2002). Gray highlighting indicates conserved sequences, and black highlighting indicates putative E2F binding sites. (F) Transfection into 210R TECs of an expression plasmid coding for E2F3 together with a luciferase reporter plasmid for either the wild-type region 1a of the Foxn1 promoter or a region 1a where site 3 has been mutated (n = 3; P = 0.03). (G) Transfection into 210R cells of E2F3 and RB expression and the Foxn1 luciferase reporter plasmid (n = 3; P = 0.02). (H) ChIP analysis of E2F3 and E2F4 to sites 1 and 2–3 in TEC100 cells. To adjust for inter-experiment variability of scale of percent input, experiments were normalized to nonspecific antibodies (IgG and anti-p16INK4a). This ratio is represented here. In addition, binding to a nonspecific region of the fifth chromosome was included as a negative control (n = 3; for site 1 P = 0.02, for site 2–3 P = 0.04). (I) Fold change in percent input as assessed by ChIP analysis of E2F3 and E2F4 binding to the Foxn1 promoter in TEC100 cells after transient overexpression of RB (n = 3 compared with untransfected controls; p-values: E2F3 Foxn1 site 1 = 0.06 and site 2–3 = 0.04; E2F4 Foxn1 site 1 = 0.01 and site 2–3 = 0.02). Asterisks indicate P < 0.05; ns indicates no significant difference. All error bars indicate standard error.
These observations suggested that, in addition to its canonical role in regulating progression through the cell cycle, the RB family also controls TEC expression of Foxn1. Little is known about the transcriptional control of Foxn1 (Balciunaite et al., 2002). There are two alternate promoters and first exons in the Foxn1 gene (Schorpp et al., 1997; Cunliffe et al., 2002): the more distal promoter region (1a) is transcriptionally active in both the skin and the thymus; the second promoter region (1b) is active only in keratinocytes of the skin (Fig. 4 D). We found by computational analysis that region 1a harbors several conserved putative E2F binding sites. We labeled some of these putative sites “site 1” and “site 2–3”, with site 2–3 containing two independent but adjacent putative sites (Fig. 4 E).
To test the possibility that E2Fs might act at the Foxn1 promoter, we cotransfected an expression vector coding for E2F3, an activating member of the E2F family, with a reporter plasmid that contains the region 1a of the mouse Foxn1 promoter upstream of a cDNA coding for the luciferase reporter into the 210R TEC cell line (Friend et al., 1994). We found that E2F3 activated region 1a of the Foxn1 promoter in this context. A mutation abolishing E2F binding in one of the two sites in site 2–3 partially inhibited the ability of E2F3 to activate the Foxn1 promoter (Fig. 4 F). Coexpression of RB markedly inhibited E2F3-mediated activation of the Foxn1 promoter (Fig. 4 G). We further tested direct binding of E2Fs to the Foxn1 promoter by performing chromatin immunoprecipitation (ChIP) experiments in several TEC lines (TEC100, TEC71, and 201R; Friend et al., 1994; Wang et al., 1998). We found that E2F3 and E2F4, a repressor E2F, can bind to both site 1 and site 2–3 but do not bind significantly to regions of nonspecific DNA on the fifth chromosome of the mouse (Fig. 4 H for TEC100; TEC71 and 210R not depicted). We repeated similar experiments after the transient ectopic expression of RB in TEC100 cells and found a decrease in endogenous Foxn1 expression (fold change: Rb mRNA, 3.1 ± 0.11; Foxn1 mRNA, −1.8 ± 0.11; n = 3 each), as well as a switch between the E2F3 activator and the E2F4 repressor at the Foxn1 promoter (Fig. 4 I). Collectively, these data show that Foxn1 is a direct target of E2F regulation; E2F binds to and controls transcription at the Foxn1 promoter, and RB inhibits this activation.
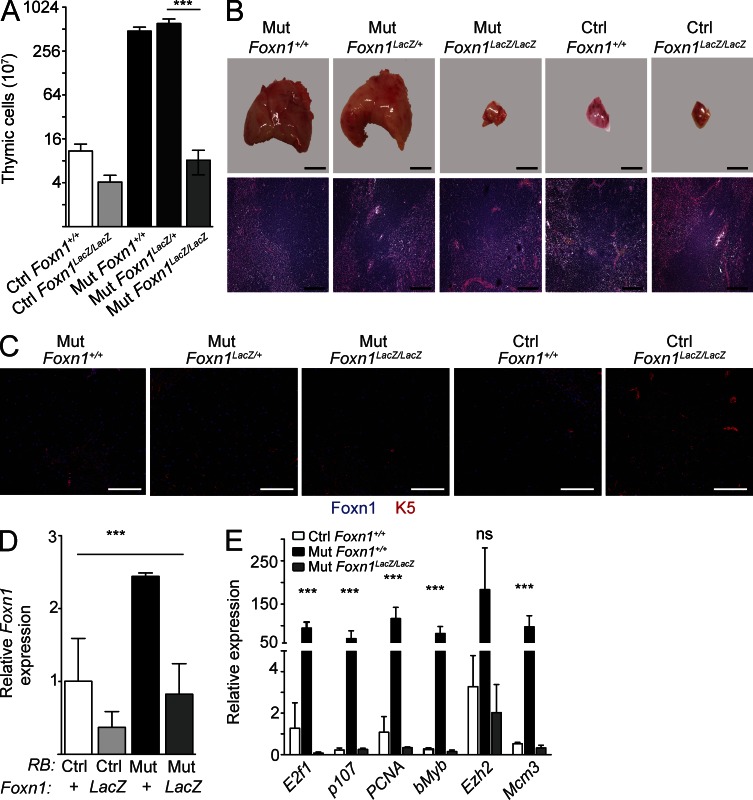
To assess the in vivo functional role of increased Foxn1 levels in the thymus expansion observed in Mx1-Cre p107-Single mice, we studied thymic growth in Mx1-Cre p107-Single; Foxn1LacZ mutant mice. Foxn1LacZ is a hypomorphic allele of Foxn1, which causes down-regulation of Foxn1 protein expression from soon after birth; of interest, Foxn1LacZ heterozygous mice have no appreciable thymic defects (Chen et al., 2009). Similar to Mx1-Cre p107-Single mice, Mx1-Cre p107-Single Foxn1LacZ/+ mice displayed significant thymic expansion. In contrast, Mx1-Cre p107-Single Foxn1LacZ/LacZ mice had a thymus equivalent in size to thymi from control (Rblox/lox; p130lox/lox; p107+/−; Foxn1+/+ without Cre) mice (Fig. 5, A and B). As expected, Foxn1 levels were reduced in the thymus of Mx1-Cre p107-Single Foxn1LacZ/LacZ mice compared with Mx1-Cre p107-Single Foxn1LacZ/+ or Mx1-Cre p107-Single mice (Fig. 5, C and D). We also observed decreased expression of E2F targets in the thymus of Mx1-Cre p107-Single Foxn1LacZ/LacZ mice (Fig. 5 E). This result suggests, as has been observed in zebrafish (Ma et al., 2012), that Foxn1 plays a role not only in differentiation of TECs but also in promoting proliferation through regulation of S phase genes. These observations also indicate that decreased expression of Foxn1 is sufficient to rescue the thymic phenotype observed in Rb family mutant mice, indicating that increased Foxn1 levels induced by unrestricted E2F activity upon inactivation of Rb family genes are a major cause of the expanding thymus in these mice.
Figure 5.
Reduction of Foxn1 levels reverses the enlarged thymus phenotype in Mx1-Cre p107-Single mice. (A) Thymic cellularity in control (Ctrl) and Rb family mutant (Mut) mice in the presence and in the absence of the Foxn1LacZ allele (n = 3; P = 0.02). (B) Representative images of whole thymus explants (top) and of H&E-stained sections (bottom; n = 5) from mice in A. (C) Representative immunofluorescence analysis of Foxn1 and K5 expression in sections prepared from control and Rb family mutant mice in the presence and absence of the Foxn1LacZ allele (n = 2). Bars: (B, top) 500 µm; (B, bottom) 200 µm; (C) 100 µm. (D) Analysis by RT-qPCR of Foxn1 message level in TECs from p107-single Foxn1 +/+, P107-single Foxn1Lacz/Lacz, Mx1-Cre p107-single Foxn1+/+, and Mx1-Cre p107-single Foxn1LacZ/LacZ (n = 3, none pI:pC injected; P = 0.023). (E) Evaluation of E2F target and S-phase genes in control, Mx1-Cre p107-single, and Mx1-Cre p107-single Foxn1LacZ/LacZ TECs (n = 3; 15 wk). Genes analyzed included E2f1 (P = 0.031), p107 (P = 0.008), PCNA (P = 0.02), bMyb (P = 0.01), Ezh2 (P = 0.12), and Mcm3 (P = 0.043). Asterisks indicate P < 0.05; ns indicates no significant difference. All error bars indicate standard error.
The transcriptional activation of Foxn1 in RB family mutant cells is reminiscent of the interactions between RB, E2F, and PPARγ (peroxisome proliferator-activated receptor gamma subunit): increased levels of PPARγ upon loss of RB function in mesenchymal cells change the fate of these cells toward the adipocytic lineage and may affect the spectrum of tumors in mice (Calo et al., 2010). However, a unique aspect of the RB-E2F-Foxn1 module is that it keeps in check the protumorigenic consequences of losing RB family function, at least in part by activating prodifferentiation functions of Foxn1.
Our data demonstrate that the RB pathway plays a key cell-autonomous role in TECs to control maximal thymic size and aging. This result is striking when one considers that thymic size likely is controlled by complex feedback interactions between stromal cells and maturing T cells. Perhaps surprising, given the well-characterized tumor suppressor role of RB family members, loss of RB family function in the thymus does not perturb the differentiation of TECs or T cells and simply increases the proliferation of some of these cells. These observations raise the possibility that loss of RB family function could be used to restore or promote thymic expansion and T cell production in patients (van den Brink et al., 2004; Wils and Cornelissen, 2005). Clearly, however, constant inactivation of RB family function in the thymus, although initially beneficial for the immune system of mice, eventually becomes detrimental to the animals when the mutant thymus has reached a large size in the chest. Thus, an important future step will be to identify novel means to transiently inactivate the RB family in a controlled fashion to minimize the potential negative effects of this inactivation while optimizing the benefits to the immune system of patients.
MATERIALS AND METHODS
Animals.
All mice where housed in the Stanford University School of Medicine Research Animal Facility in accordance with institutional and National Institutes of Health guidelines. All animal care and experiments were approved by the Stanford University Administrative Panel on Laboratory Animal Care and followed the guidelines of this animal use committee. Mice were of a mixed C57/129 background. Mx1-Cre Rblox/lox p130lox/lox p107−/− mice were previously described (Viatour et al., 2008). Mx1-Cre p107-Single mice were generated by breeding Mx1-Cre Rblox/lox p130lox/lox p107−/− mice with Mx1-Cre Rblox/lox p130lox/lox mice. The Foxn1-IRES-LacZ transgene was described previously (Chen et al., 2009). These mice were bred through successive generations with Mx1-Cre Rblox/lox p130lox/lox p107−/− to generate Rblox/lox p130lox/lox p107−/+ Foxn1LacZ/+ and Rblox/lox p130lox/lox p107−/+ Foxn1LacZ/+ with and without Mx1-Cre. As appropriate, mice were injected intraperitoneally with pI:pC at 3–8 wk of age. Thymi were removed from euthanized mice and processed as appropriate for the particular experiment.
BM transplantation.
Chimeric mice were generated by lethal irradiation (9 Gr) of 3-mo-old Mx1-Cre p107-Single and control mice expressing only the Ly5.1 antigen. Irradiated mice were rescued by retroorbital transplantation of 2 × 106 wild-type BM cells expressing both the Ly5.1 and Ly5.2 antigens. pI:pC was injected 12 wk after transplantation. Tissues were harvested 12–16 wk later.
Immunofluorescence.
Immunofluorescence was performed on frozen sections or sections from paraffin-embedded tissues as described previously (Chen et al., 2009; Gordon et al., 2010; Bryson et al., 2011; Park et al., 2011). Images were obtained by confocal microscopy using a confocal LSM 510 Meta (Carl Zeiss) with a Plan-Apochromat 20×/0.8 objective and analyzed with AxioVision 4.8 software (Carl Zeiss). Primary antibodies were directed against Foxn1 (goat anti–mouse, G-20; Santa Cruz Biotechnology, Inc.), K5 (rabbit anti–mouse; Covance), K8 (Troma-1; Developmental Studies Hybridoma Bank), K14 (rabbit anti–mouse; Covance), β5t (rabbit anti–mouse; MBL International Corporation), CD205 (rat biotinylated; Abcam), UEA-1–biotin (Vector Laboratories), CD31 (goat anti–mouse; BD), and PDGFR-β (goat anti–mouse; R&D Systems). Secondary antibodies included donkey anti–goat Cy5, donkey anti–rabbit 594, donkey anti–rat FITC, donkey anti–rabbit Texas Red, and streptavidin-FITC (Jackson ImmunoResearch Laboratories, Inc.) and donkey anti–goat Alexa Fluor 594 (Invitrogen).
Histology.
Hematoxylin and eosin (H&E) staining was performed on paraffin-embedded sections. For H&E staining, thymi were fixed with 4% paraformaldehyde overnight, dehydrated in gradient ethanol solutions, and embedded in paraffin. 5–10-µm sections were cut and stained with H&E. Oil Red O staining was performed on frozen sections as follows: sections were fixed in 4% formalin for 10 min, washed with running water, rinsed in 60% isopropanol, stained for 15 min in fresh Oil Red O solution, washed in 60% isopropanol, stained briefly with hematoxylin, and washed with running water for 10 min. Microscopy was performed on a DM2000 microscope (Leica) at room temperature using a DFC500 camera (Leica). Images were analyzed using LASV3.8 software.
Cytometry.
FACS analysis sorting was performed on an LSRI, Vantage, or FACs ARIA LSRII (BD). Data were acquired on CellQuest or FACSDiva (BD) and analyzed with FlowJo software (Tree Star). Antibodies for Kit, CD25, CD3ε, CD4, CD8, MHCII, CD40, CD205, CD80, CD45, Ly5.1, Ly5.2, Ly51, Mac1, Ter119, B220, Gr1, and Sca1 were purchased from eBioscience. FITC-conjugated UEA1 was purchased from Vector Laboratories. Cell cycle analysis was performed by intraperitoneal injection of BrdU, followed by euthanasia 1–4 h after injection. Cells were FACS sorted and analyzed for BrdU content as described previously (Viatour et al., 2008). TECs were isolated by enzymatic digestion. Thymi were cut into ∼1-mm3 pieces, and fragments were digested by a mixture of DNase I, and collagenase A/D:Dispase for 60 min at 37°C. Cells were passed through a 45-µm filter and washed in PBS/2% serum. Red cells were lysed using ammonium lysis buffer on ice. After three PBS/serum washes, cells were blocked in PBS/serum for 15 min. Suspensions were enriched for cells of interest by incubating with anti-MHCII PE or anti-EPCAM PE or biotin-conjugated antibodies, followed by incubation with anti-PE or antibiotin magnetic beads (Miltenyi Biotec) and passing over a MACS MS or LS column (Miltenyi Biotec). Cells were incubated for 30 min on ice with appropriate antibodies, washed, and resuspended for analysis in PBS/serum.
Plasmids and sequence analysis.
Promoter sequence identification and alignment are as described on the University of California, Santa Cruz (UCSC) genome browser. The Foxn1 promoter (from −1550 to 64) was cloned in pGL3 basic. Point mutations were introduced by subcloning a 129-bp fragment that spans from an Xma1 site at −212 to the ECORV site at −83 and includes point mutations of the 2–3 E2F binding sites. E2F3- and RB-expressing plasmids have been described previously (Angus et al., 2003; Burkhart and Sage, 2008). Primer sequences are available upon request.
ChIP.
ChIP analysis in TEC100, TEC71, and 210R cell lines (a gift from A. Farr, University of Washington, Seattle, WA) and RT–quantitative PCR (RT-qPCR) were performed as described in Burkhart et al. (2010). Chromatin was sonicated using a fisher probe sonicator 30% output power for eight cycles of 30 s. The chromatin was precleared before being diluted and bound by 4 µg of the primary antibody overnight at 4°C. Each ChIP was then incubated with 8 µg rabbit anti–mouse IgG (MP Biomedicals) or anti–mouse p16 antibody (N-20; Santa Cruz Biotechnology, Inc.) as a secondary for 1 h. Nucleoprotein complexes were pulled down using Pansorbin cells (EMD Millipore). Complexes were digested with Proteinase K and RNase A and purified by a QIAquick PCR Purification kit (QIAGEN). Analysis was performed using qPCR with primers directed against Foxn1, Cyclin A, and GAPDH promoters (primer sequence available upon request) using SYBR-green reagents (Applied Biosystems). PCR was run on a C1000 thermal cycler (Bio-Rad Laboratories).
Immunoblot analysis.
Protein lysates were prepared by Triton X-100–based lysis from TECs pooled according to age and genotype from 5–12 animals each. Equal amounts of protein were loaded onto either manually prepared 10% Bis-Tris gels or Novex NuPAGE 4–12% gradient gels (Invitrogen) and run until appropriate separation was achieved. Gels were transferred to polyvinylidene difluoride membrane (GE Healthcare). Blots were probed with primary antibodies anti-Tubulin (T9026; Sigma-Aldrich); anti-Foxn1 H-270, anti-Foxn1 G-20, anti-RB C-15, and anti-CCND1 H-295 (Santa Cruz Biotechnology, Inc.); pooled anti–phospho-RB S807/811 and S608 (Cell Signaling Technology); and anti-CDK4 (DCS-31; Invitrogen). HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. Signal was detected using either ECL Western blotting reagent (Thermo Fisher Scientific) or ECL-Prime Western blot detection reagent (GE Healthcare).
Cell culture, transfections, and luciferase analysis.
TEC lines (TEC100, TEC71, and 210R) were grown in RPMI 1640 or DMEM supplemented with 10% serum, glutamine, penicillin, and streptomycin. Transfections were performed using either Lipofectamine 2000 (Invitrogen) or Fugene (Promega) and included experimental plasmids with TK-Renilla (Promega) and pcDNA plasmid to ensure transfection of equivalent amounts of DNA. Luciferase assays were performed at least three times in triplicates using the Dual Luciferase Assay System (Promega) and analyzed using the Hybrid Synergy Reader and Gen5 V2.00 software (BioTek).
Statistical analysis.
For analysis of two populations, statistical analyses were performed using a two-tailed paired Student’s t test. A p-value of <0.05 was required for significance (actual p-values are listed in figure legends). For analyses for three or more populations, significance was determined using ANOVA analysis.
Online supplemental material.
Fig. S1 describes complex gating strategies used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121716/DC1.
Supplementary Material
Acknowledgments
We thank Ms. Anne-Flore Zmoos for technical assistance and Dr. Andrew Farr for the generous gift of TEC lines.
This work was supported by the Lucile Packard Foundation for Children’s Health and the Leukemia and Lymphoma Society (to J. Sage), as well as fellowships from Human Frontier Science Program, the European Molecular Biology Organization, the Fonds de la Recherche Scientifique, the Leon Fredericq Foundation (to P. Viatour), and the California Institute of Regenerative Medicine grant number TG2-01159 (to P.M. Garfin). This work was also supported by the Alex’s Lemonade Stand Foundation for Childhood Cancer and the Paul F. Glenn Laboratories for the Biology of Aging at Stanford University.
There are no competing financial interests to disclose.
Author contributions: P.M. Garfin and P. Viatour conceived and performed most of the experiments, analyzed data, and wrote the manuscript. D. Min, J.L. Bryson, and B. Edris performed experiments and contributed to the manuscript. T. Serwold, C.C. Blackburn, E.R. Richie, N.R. Manley, and K.I. Weinberg contributed to the design of the project and writing of the manuscript. J. Sage conceived the project, contributed to data analysis and writing of the manuscript, and supervised and managed the project.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- cTEC
- cortical TEC
- DN
- double negative
- DP
- double positive
- mTEC
- medullary TEC
- PI
- propidium iodide
- pI:pC
- polyinosine:polycytosine
- qPCR
- quantitative PCR
- TEC
- thymic epithelial cell
References
- Akiyama T., Shinzawa M., Akiyama N. 2012. TNF receptor family signaling in the development and functions of medullary thymic epithelial cells. Front Immunol. 3:278 10.3389/fimmu.2012.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus S.P., Solomon D.A., Kuschel L., Hennigan R.F., Knudsen E.S. 2003. Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation. Mol. Cell. Biol. 23:8172–8188 10.1128/MCB.23.22.8172-8188.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw D., Palmer D.B. 2011. The origin and implication of thymic involution. Aging Dis. 2:437–443 [PMC free article] [PubMed] [Google Scholar]
- Balciunaite G., Keller M.P., Balciunaite E., Piali L., Zuklys S., Mathieu Y.D., Gill J., Boyd R., Sussman D.J., Holländer G.A. 2002. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat. Immunol. 3:1102–1108 10.1038/ni850 [DOI] [PubMed] [Google Scholar]
- Boehm T. 2008. Thymus development and function. Curr. Opin. Immunol. 20:178–184 10.1016/j.coi.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Bryson J.L., Coles M.C., Manley N.R. 2011. A method for labeling vasculature in embryonic mice. J. Vis. Exp. (56):e3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D.L., Sage J. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 8:671–682 10.1038/nrc2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D.L., Ngai L.K., Roake C.M., Viatour P., Thangavel C., Ho V.M., Knudsen E.S., Sage J. 2010. Regulation of RB transcription in vivo by RB family members. Mol. Cell. Biol. 30:1729–1745 10.1128/MCB.00952-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J.A., Danielian P.S., Nedelcu S., Berman S.D., Lees J.A. 2010. Rb regulates fate choice and lineage commitment in vivo. Nature. 466:1110–1114 10.1038/nature09264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.C., Bosselut R. 2010. Decision checkpoints in the thymus. Nat. Immunol. 11:666–673 10.1038/ni.1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Gu X., Liu Y., Wang J., Wirt S.E., Bottino R., Schorle H., Sage J., Kim S.K. 2011. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 478:349–355 10.1038/nature10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiao S., Manley N.R. 2009. Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood. 113:567–574 10.1182/blood-2008-05-156265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien W.M., Rabin S., Macias E., Miliani de Marval P.L., Garrison K., Orthel J., Rodriguez-Puebla M., Fero M.L. 2006. Genetic mosaics reveal both cell-autonomous and cell-nonautonomous function of murine p27Kip1. Proc. Natl. Acad. Sci. USA. 103:4122–4127 10.1073/pnas.0509514103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnam M., Goodrich D.W. 2011. RB1, development, and cancer. Curr. Top. Dev. Biol. 94:129–169 10.1016/B978-0-12-380916-2.00005-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Baker J., Sage J. 2012. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun. 3:1244 10.1038/ncomms2254 [DOI] [PubMed] [Google Scholar]
- Cunliffe V.T., Furley A.J., Keenan D. 2002. Complete rescue of the nude mutant phenotype by a wild-type Foxn1 transgene. Mamm. Genome. 13:245–252 10.1007/s00335-001-3079-6 [DOI] [PubMed] [Google Scholar]
- Dannenberg J.H., te Riele H.P. 2006. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl. Cell Differ. 42:183–225 10.1007/400_002 [DOI] [PubMed] [Google Scholar]
- DeMonte L., Porcellini S., Tafi E., Sheridan J., Gordon J., Depreter M., Blair N., Panigada M., Sanvito F., Merati B., et al. 2007. EVA regulates thymic stromal organisation and early thymocyte development. Biochem. Biophys. Res. Commun. 356:334–340 10.1016/j.bbrc.2007.02.131 [DOI] [PubMed] [Google Scholar]
- Dooley J., Erickson M., Farr A.G. 2008. Alterations of the medullary epithelial compartment in the Aire-deficient thymus: implications for programs of thymic epithelial differentiation. J. Immunol. 181:5225–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakov J.A., Hanash A.M., Jenq R.R., Young L.F., Ghosh A., Singer N.V., West M.L., Smith O.M., Holland A.M., Tsai J.J., et al. 2012. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 336:91–95 10.1126/science.1218004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin D.S., Godfrey V.L., Lee H., Kovalev G.I., Schoonhoven R., Chen-Kiang S., Su L., Xiong Y. 1998. CDK inhibitors p18(INK4c) and p27(Kip1) mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 12:2899–2911 10.1101/gad.12.18.2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend S.L., Hosier S., Nelson A., Foxworthe D., Williams D.E., Farr A. 1994. A thymic stromal cell line supports in vitro development of surface IgM+ B cells and produces a novel growth factor affecting B and T lineage cells. Exp. Hematol. 22:321–328 [PubMed] [Google Scholar]
- Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A., et al. 2011. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39:D876–D882 10.1093/nar/gkq963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J., Patel S.R., Mishina Y., Manley N.R. 2010. Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis. Dev. Biol. 339:141–154 10.1016/j.ydbio.2009.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D., Abramson J., Benoist C., Mathis D. 2007. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204:2521–2528 10.1084/jem.20070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger M., Sutti F., Panigada M., Porcellini S., Merati B., Mariani M., Teesalu T., Consalez G.G., Grassi F. 1998. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. J. Cell Biol. 141:1061–1071 10.1083/jcb.141.4.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaquinta P.J., Lees J.A. 2007. Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19:649–657 10.1016/j.ceb.2007.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias A., Murga M., Laresgoiti U., Skoudy A., Bernales I., Fullaondo A., Moreno B., Lloreta J., Field S.J., Real F.X., Zubiaga A.M. 2004. Diabetes and exocrine pancreatic insufficiency in E2F1/E2F2 double-mutant mice. J. Clin. Invest. 113:1398–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E.A., Assoian R.K. 2008. Transcriptional regulation of the cyclin D1 gene at a glance. J. Cell Sci. 121:3853–3857 10.1242/jcs.039131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug D.B., Crouch E., Carter C., Coghlan L., Conti C.J., Richie E.R. 2000. Transgenic expression of cyclin D1 in thymic epithelial precursors promotes epithelial and T cell development. J. Immunol. 164:1881–1888 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Ma D., Wang L., Wang S., Gao Y., Wei Y., Liu F. 2012. Foxn1 maintains thymic epithelial cells to support T-cell development via mcm2 in zebrafish. Proc. Natl. Acad. Sci. USA. 109:21040–21045 10.1073/pnas.1217021110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N.R., Condie B.G. 2010. Transcriptional regulation of thymus organogenesis and thymic epithelial cell differentiation. Prog Mol Biol Transl Sci. 92:103–120 10.1016/S1877-1173(10)92005-X [DOI] [PubMed] [Google Scholar]
- Manley N.R., Richie E.R., Blackburn C.C., Condie B.G., Sage J. 2011. Structure and function of the thymic microenvironment. Front. Biosci. 16:2461–2477 10.2741/3866 [DOI] [PubMed] [Google Scholar]
- Miller J.F. 2011. The golden anniversary of the thymus. Nat. Rev. Immunol. 11:489–495 10.1038/nri2993 [DOI] [PubMed] [Google Scholar]
- Min D., Panoskaltsis-Mortari A., Kuro-O M., Holländer G.A., Blazar B.R., Weinberg K.I. 2007. Sustained thymopoiesis and improvement in functional immunity induced by exogenous KGF administration in murine models of aging. Blood. 109:2529–2537 10.1182/blood-2006-08-043794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.P., Allen P.M. 2012. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat. Immunol. 13:121–128 10.1038/ni.2190 [DOI] [PubMed] [Google Scholar]
- Napolitano L.A., Schmidt D., Gotway M.B., Ameli N., Filbert E.L., Ng M.M., Clor J.L., Epling L., Sinclair E., Baum P.D., et al. 2008. Growth hormone enhances thymic function in HIV-1-infected adults. J. Clin. Invest. 118:1085–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell C.S., Bredenkamp N., Tetélin S., Jin X., Tischner C., Vaidya H., Sheridan J.M., Stenhouse F.H., Heussen R., Smith A.J., Blackburn C.C. 2011. Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 7:e1002348 10.1371/journal.pgen.1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.S., Martelotto L.G., Peifer M., Sos M.L., Karnezis A.N., Mahjoub M.R., Bernard K., Conklin J.F., Szczepny A., Yuan J., et al. 2011. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat. Med. 17:1504–1508 10.1038/nm.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pénit C., Vasseur F. 1997. Expansion of mature thymocyte subsets before emigration to the periphery. J. Immunol. 159:4848–4856 [PubMed] [Google Scholar]
- Pierce A.M., Fisher S.M., Conti C.J., Johnson D.G. 1998. Deregulated expression of E2F1 induces hyperplasia and cooperates with ras in skin tumor development. Oncogene. 16:1267–1276 10.1038/sj.onc.1201666 [DOI] [PubMed] [Google Scholar]
- Robles A.I., Larcher F., Whalin R.B., Murillas R., Richie E., Gimenez-Conti I.B., Jorcano J.L., Conti C.J. 1996. Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proc. Natl. Acad. Sci. USA. 93:7634–7638 10.1073/pnas.93.15.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald H.R. 2008. Thymus organogenesis. Annu. Rev. Immunol. 26:355–388 10.1146/annurev.immunol.26.021607.090408 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Puebla M.L., LaCava M., Miliani De Marval P.L., Jorcano J.L., Richie E.R., Conti C.J. 2000. Cyclin D2 overexpression in transgenic mice induces thymic and epidermal hyperplasia whereas cyclin D3 expression results only in epidermal hyperplasia. Am. J. Pathol. 157:1039–1050 10.1016/S0002-9440(10)64616-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S.W., Jeker L.T., Ueno T., Kuse S., Keller M.P., Zuklys S., Gudkov A.V., Takahama Y., Krenger W., Blazar B.R., Holländer G.A. 2007. Keratinocyte growth factor (KGF) enhances postnatal T-cell development via enhancements in proliferation and function of thymic epithelial cells. Blood. 109:3803–3811 10.1182/blood-2006-10-049767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Santos M., Segrelles C., Leis H., Jorcano J.L., Berns A., Paramio J.M., Vooijs M. 2004. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 131:2737–2748 10.1242/dev.01148 [DOI] [PubMed] [Google Scholar]
- Sauce D., Appay V. 2011. Altered thymic activity in early life: how does it affect the immune system in young adults? Curr. Opin. Immunol. 23:543–548 10.1016/j.coi.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Scheijen B., Bronk M., van der Meer T., De Jong D., Bernards R. 2004. High incidence of thymic epithelial tumors in E2F2 transgenic mice. J. Biol. Chem. 279:10476–10483 10.1074/jbc.M313682200 [DOI] [PubMed] [Google Scholar]
- Schorpp M., Hofmann M., Dear T.N., Boehm T. 1997. Characterization of mouse and human nude genes. Immunogenetics. 46:509–515 10.1007/s002510050312 [DOI] [PubMed] [Google Scholar]
- Shakib S., Desanti G.E., Jenkinson W.E., Parnell S.M., Jenkinson E.J., Anderson G. 2009. Checkpoints in the development of thymic cortical epithelial cells. J. Immunol. 182:130–137 10.4049/jimmunol.0990018 [DOI] [PubMed] [Google Scholar]
- Shrimpton R.E., Butler M., Morel A.S., Eren E., Hue S.S., Ritter M.A. 2009. CD205 (DEC-205): a recognition receptor for apoptotic and necrotic self. Mol. Immunol. 46:1229–1239 10.1016/j.molimm.2008.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Taylor W.R. 2006. Control of the G2/M transition. Mol. Biotechnol. 32:227–248 10.1385/MB:32:3:227 [DOI] [PubMed] [Google Scholar]
- Takahama Y., Takada K., Murata S., Tanaka K. 2012. β5t-containing thymoproteasome: specific expression in thymic cortical epithelial cells and role in positive selection of CD8+ T cells. Curr. Opin. Immunol. 24:92–98 10.1016/j.coi.2012.01.006 [DOI] [PubMed] [Google Scholar]
- van den Brink M.R., Alpdogan O., Boyd R.L. 2004. Strategies to enhance T-cell reconstitution in immunocompromised patients. Nat. Rev. Immunol. 4:856–867 10.1038/nri1484 [DOI] [PubMed] [Google Scholar]
- Viatour P., Somervaille T.C., Venkatasubrahmanyam S., Kogan S., McLaughlin M.E., Weissman I.L., Butte A.J., Passegué E., Sage J. 2008. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 3:416–428 10.1016/j.stem.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P., Ehmer U., Saddic L.A., Dorrell C., Andersen J.B., Lin C., Zmoos A.F., Mazur P.K., Schaffer B.E., Ostermeier A., et al. 2011. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J. Exp. Med. 208:1963–1976 10.1084/jem.20110198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Nelson A., Kimachi K., Grey H.M., Farr A.G. 1998. The role of peptides in thymic positive selection of class II major histocompatibility complex-restricted T cells. Proc. Natl. Acad. Sci. USA. 95:3804–3809 10.1073/pnas.95.7.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.M., Mella H., Lucas P.J., Williams J.A., Telford W., Gress R.E. 2009. Single cell analysis of complex thymus stromal cell populations: rapid thymic epithelia preparation characterizes radiation injury. Clin. Transl. Sci. 2:279–285 10.1111/j.1752-8062.2009.00128.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils E.J., Cornelissen J.J. 2005. Thymopoiesis following allogeneic stem cell transplantation: new possibilities for improvement. Blood Rev. 19:89–98 10.1016/j.blre.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Yang H., Youm Y.H., Dixit V.D. 2009. Inhibition of thymic adipogenesis by caloric restriction is coupled with reduction in age-related thymic involution. J. Immunol. 183:3040–3052 10.4049/jimmunol.0900562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook E.C., Krishack P.A., Zhang S., Zeleznik-Le N.J., Firulli A.B., Witte P.L., Le P.T. 2011. Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood. 118:5723–5731 10.1182/blood-2011-03-342097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.