Abstract
This chapter explores the structural responses of a massive, hetero-oligomeric protein complex to a single allosteric activator as probed by a wide range of chemical, biochemical, and biophysical approaches. Some of the approaches used are amenable only to large protein targets, whereas others push the limits of their utility. Some of the techniques focus on individual subunits, or portions thereof, while others examine the complex as a whole. Despite the absence of crystallographic data for the complex, the diverse techniques identify and implicate a small region of its catalytic subunit as the master allosteric activation switch for the entire complex.
Keywords: Protein structure, Hetero-oligomeric protein complexes, Chemical crosslinking, Proteolysis, Electron microscopy, Chemical footprinting, Small-angle X-ray scattering, Allosteric switch
1. Introduction
The large protein complex that is featured in this chapter as a model system for the detection of conformational changes caused by an allosteric activator is skeletal muscle phosphorylase kinase (PhK), the first protein kinase to be purified and characterized (1). It remains one of the most complex enzymes known, being composed of 16 subunits (αβγδ)4, and having a total mass of 1.3 × 106 Da (2). The γ subunit is catalytic, but accounts for only 13.7% of PhK’s mass. The remaining subunits are regulatory and impose quaternary constraint on the activity of the catalytic subunit in PhK’s nonactivated basal state. A wide variety of allosteric activators act upon the regulatory subunits of PhK, and these include ADP, GDP, glycogen, acidic phospholipids, certain gangliosides, and phosphorylation of the α and β subunits; however, the most fundamental allosteric activator is Ca2+, because the enhanced activity induced by all other activators is completely Ca2+-dependent. It appears that there is a fundamental tier of Ca2+-dependent conformational changes leading to activation, with additional conformational changes induced by the other activators being layered upon this fundamental tier to cause as much as a 100-fold cumulative activation at physiological pH. Consequently, we have sought to elucidate the Ca2+-dependent conformational changes in the PhK complex as the first step in understanding its overall allosteric activation. Although the large mass of the complex limits some of the physicochemical approaches that can be used, in contrast, it opens the door to a large number of informative approaches not practical with smaller enzymes.
The primary target for Ca2+ ions in the PhK complex is undoubtedly the δ subunit, which is calmodulin (CaM), although a recent sequence analysis suggests the presence of calcineurin B-like domains in the α and β subunits that may also be potentially capable of binding Ca2+ (3). The behavior of the CaM subunit of PhK has long been known to be unusual in two ways. First, it remains a tightly bound subunit of the complex even in the complete absence of Ca2+; and second, it has been reported to bind only three Ca2+ ions, despite its four EF-hands (4). Results from a variety of techniques are consistent with the catalytic γ subunit being a major interaction partner for the CaM (δ) subunit within the PhK complex: an isolated γδ dimer retains Ca2+-dependent kinase activity (5); the carboxy-terminal regulatory domain of γ (γCRD) beyond its catalytic domain contains two distinct regions with high affinity for CaM (6), and removal of these regions from the isolated γ subunit results in Ca2+-independent kinase activity (7); and a γδ conjugate can be formed by chemical crosslinking of the (αβγδ)4 complex (8, 9). A variety of chemical, biochemical, and biophysical approaches have been utilized to determine the effects of Ca2+ on PhK’s structure and properties.
2. Effects of Ca2+ on Individual Subunits in the PhK Complex
Many of the approaches used have sought to identify specific sub-units of PhK that are structurally affected, either directly or indirectly, by the binding of Ca2+. As will be described, each of its four different subunits has been shown to be affected. Yet, one of the most informative and specific techniques for detecting conformational changes, namely, hydrogen–deuterium exchange, has yet to be evaluated with PhK ± Ca2+, because of the very large size of the complex.
2.1. Single-Pair Fluorescence Resonance Energy Transfer
Fluorescence resonance energy transfer (FRET) is an extremely sensitive method for assessing conformational dynamics in proteins, and in the case of PhK, it was used to determine the conformations of its δ (CaM) subunit in the absence and presence of Ca2+ (10). A double mutant of CaM was first derivatized with a fluorescent donor–acceptor pair at opposite ends of the molecule. A small amount of this derivatized CaM was then exchanged for wild-type CaM in the (αβγδ)4 PhK complex by incubation with low concentrations of urea (0.5 M) in the absence of Ca2+, following which, the PhK was repurified by size-exclusion HPLC. Because of the theoretical possibility of different PhK complexes containing different numbers of derivatized CaM subunits (1–4), single molecule fluorescence spectroscopy was employed to assess the conformational substates of the PhKδ subunit, with the distribution of FRET distances representing compact to extended conformations. This approach revealed that the conformational substates of CaM bound as the δ subunit of the PhK complex compared to free CaM were greatly skewed toward a higher percentage of extended conformations in the complex (40% vs. 26%). The binding of Ca2+ to PhKδ further increased the percentage of extended conformers to 45%, which again was markedly different than the conformational substate distributions of free Ca2+ CaM. These results indicate that the other subunits of PhK exert a dramatic effect on the conformation of CaM bound as PhKδ.
Intrinsic to the above interpretation that populations of bound CaM molecules with the donor–acceptor pair at different apparent distances represent different conformational substates of the δ subunits is the assumption that the conformational substates of the δ subunit are in dynamic equilibrium, as is observed with free CaM. The large size and consequent slow diffusion rate of PhK allowed evidence to be gained in support of this assumption. Trajectory studies over time, some for up to 4 ms, followed single molecules of PhK having bound derivatized CaM that was observed to repeatedly jump from one conformational substate to another (10). Thus, the small δ subunits of PhK exercise considerable conformational plasticity, even when loaded with Ca2+ and interacting with the other subunits within the complex.
2.2. Partial Proteolysis
Limited proteolysis followed by analysis on SDS–PAGE gels is a simple, yet effective, method for detecting ligand-induced conformational changes in proteins (see Note 1). It is especially useful for large, hetero-oligomeric complexes, because specific subunits affected by a ligand can usually be detected. Using a wide variety of 11 different proteases, the α subunit within the nonactivated (αβγδ)4 PhK complex was observed to generally be the most rapidly degraded, followed by the β subunit, with the γ and finally δ subunits being relatively resistant to hydrolysis (11). Utilizing partial proteolysis as a conformational probe successfully detected activated conformers of PhK produced by a number of ligands and phosphorylation. In the case of the allosteric activator Ca2+, the β subunit was protected from hydrolysis by trypsin (11), whereas the rate of hydrolysis of the α subunit by chymotrypsin was stimulated threefold (12). These results indicate that conformational changes induced in the δ subunit by the binding of Ca2+ are transmitted to both of PhK’s large regulatory α and β subunits.
2.3. Immunochemical Analysis
Conformation-sensitive antibodies represent another highly sensitive tool that is unaffected by the size of the protein target, and as above, it allows specific subunits in a complex to be targeted. Based upon their earlier work using overlapping synthetic peptides corresponding to the γCRD to identify two noncontiguous CaM-binding domains within that region of γ (6), Blumenthal and colleagues generated monospecific polyclonal antibodies against four synthetic peptides that covered the entire length of the γCRD. Using ELISA, these antibodies were then screened as conformational probes of the regulatory region of the γ subunit within the (αβγδ)4 complex (13). Ca2+ was reported to exert its greatest effect on the antibodies against regions nearest to the catalytic domain of γ, with their binding being enhanced, suggesting the activator causes increased exposure of the epitopes in question. Comparing the effects of these antibodies on Ca2+-dependent activity vs. their dependence on Ca2+ for binding allowed insightful inferences to be made regarding the functional roles of these regions of the γCRD on the allosteric activation by Ca2+.
Monoclonal antibodies specific for the individual subunits of PhK have also proved to be sensitive conformational probes for the effects of a variety of activators, but none of them detected structural changes caused by Ca2+ (14, 15). These antibodies were, however, valuable in mapping the relative locations of epitopes of PhK’s individual subunits by immunoelectron microscopy (immunoEM), which is shown for a single αβγδ protomer in Fig. 1. Note that the catalytic γ subunit is surrounded by portions of its three regulatory subunits (14–17).
Fig. 1.
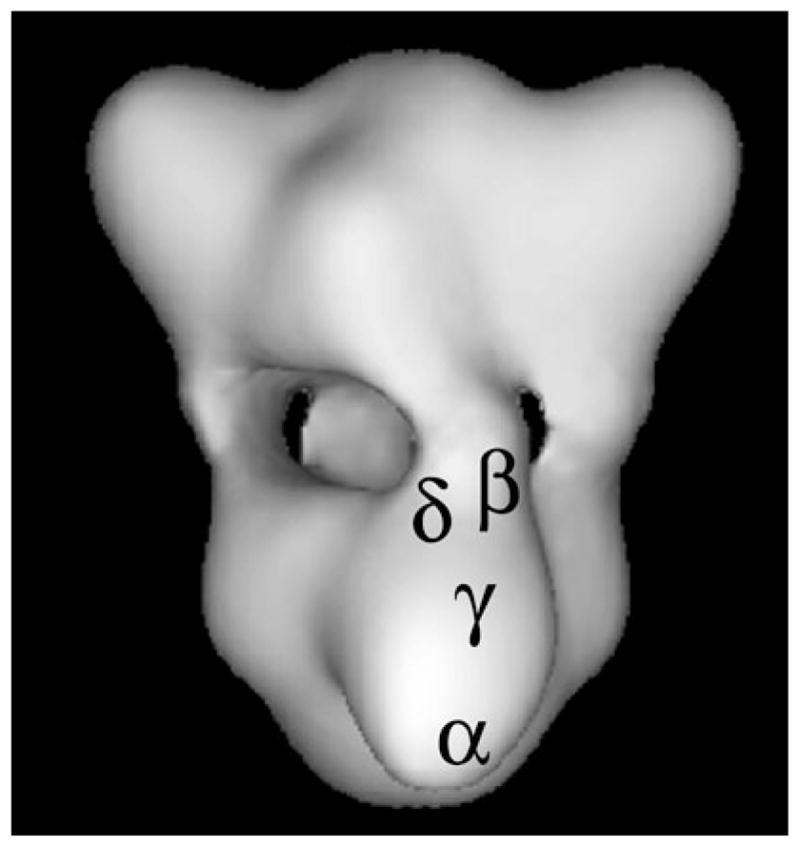
Three-dimensional structure of PhK at 25 Å resolution reconstructed from ~5,000 images of frozen hydrated particles in cryoEM (16). Each lobe is an (αβγδ)2 octamer and four bridges separate the two lobes. The positions of specific regions of the four subunits of a single αβγδ protomer are denoted. The regions of the α, β, and γ subunits represent epitopes recognized by monoclonal antibodies that were localized by immunoEM of negatively stained complexes (14, 15), with those locations then transferred to the cryoEM structure. The δ subunit was directly visualized by scanning transmission EM of PhK in which a fraction of its δ subunits had been exchanged with mutant calmodulin derivatized with Nanogold (17). Its location was also then transferred to the cryoEM model.
2.4. Chemical Footprinting
Chemical modification of proteins has been used for many years to assess the functional roles and solvent accessibility of specific amino acids. The latter, often referred to as chemical footprinting, is especially useful for detecting conformational changes in oligomeric complexes at the subunit level by measuring differences in the extent of their modification in the absence and presence of an allosteric ligand (see Note 2). The most useful chemical footprinting reagents contain functional groups that allow the reagent to be readily detected and that react preferentially with specific amino acid side chains (Table 1). Using 3H-iodoacetate (a reagent that preferentially targets protein thiols) and formaldehyde (a compound that bis-methylates lysine ε-amines in the presence of NaCNBH3), Ca2+ was found to increase labeling of the catalytic γ subunit in the PhK complex by both reagents (18), consistent with the immunochemical results described above.
Table 1.
A partial list of chemical functional groups that are targeted preferentially by reactive amino acid side chains
| Functional group | Targeting nucleophile | Optimal pH | Amino acid target | |
|---|---|---|---|---|
| Name | Structure | |||
| N-Hydroxysuccinimide ester |
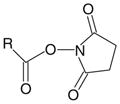
|
Amine | >7 | Lysine |
| Protein N-termini | ||||
| Imido ester |
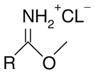
|
Amine | 8–9 | Lysine |
| Protein N-termini | ||||
| Maleimide |
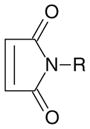
|
Sulfhydryl | 6.5–7.5 | Cysteine |
| Cyclohexanedione |
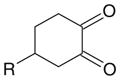
|
Guanidine | >7 | Arginine |
| Phenyl azide |
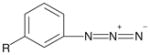
|
Amine | 7 | Cysteine |
| Phenol | Arginine | |||
| Double bond | Lysine | |||
| Histidine | ||||
| Tyrosine | ||||
| Phenylalanine | ||||
| Tryptophan | ||||
2.5. Chemical Crosslinking
Crosslinking is a technique used to covalently link two or more chemical functional groups, in this case amino acid side chains, and it can detect conformational changes in proteins and within protein complexes (19). As an extension of general protein chemical modification, chemical crosslinking is an empirical technique that is governed by the same variables described for chemical footprinting; however, because crosslinking utilizes reagents containing more than one reactive functional group (Table 1), the number of potential products formed is far greater than those generated using simple monofunctional reagents. As depicted in Fig. 2, crosslinking is an ongoing process in which coupling reactions between two proteins may progress from a single heteromeric conjugate to large polymeric arrays of indeterminable stoichiometry. The specificity of crosslinking is directly dependent on the selectivity of the crosslinking reagent used and stringent control of the crosslinking reaction, both of which are determined empirically by screening the protein target with a variety of cross-linkers under a range of conditions (19). For an oligomeric complex, the overall goal of such a screening is to limit the formation of crosslinked conjugates to a few readily identifiable species, so that each is formed in sufficient quantity for analysis of its subunit composition by apparent mass (SDS–PAGE), cross-reaction against subunit-specific antibodies (Western blots), or its primary structure (gas phase sequencing, MS). Although crosslinking can occur within a given subunit of a protein complex, we refer in this section only to crosslinking between subunits of that complex (see Note 3). For in-depth reviews on chemical crosslinking, see the books of Hermanson (20) and Wong (21).
Fig. 2.
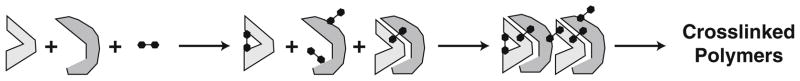
Protein crosslinking. Protein side chains react with bifunctional reagents to form mono-derivatized and cross-linked forms of interacting proteins. Subclasses within each form simultaneously occur through different combinations of mono-derivatization and crosslinking. In the continuous process of crosslinking, conjugates of increasing size may also be formed, progressing from dimers, tetramers, etc. to large extensively crosslinked polymers.
An allosteric ligand may alter the extent of formation of a given conjugate, or produce a different subunit crosslinking pattern, or both. In the case of PhK, Ca2+ was shown to increase the formation of αγγ and αδ conjugates that were initially observed after cross-linking the native complex with phenylene dimaleimide (PDM) (18). Similarly, a Ca2+-dependent increase in formation of a βδ conjugate by the crosslinker m-maleimidobenzoyl-N-hydroxysuccinimide ester was observed; however, Ca2+ promoted crosslinking of different regions of the δ subunit to β in that conjugate (18). A novel αγδδ conjugate was observed when PhK was crosslinked by PDM only in the presence of Ca2+ (18). Formation of a γδ conjugate by the zero-length crosslinker N-ethoxycarbonyl-2-ethoxy-1,2-dihydroquinoline (EEDQ) was inhibited by Ca2+ (9), and a Ca2+-dependent linkage between the γ and α subunits was revealed when PhK was crosslinked with N-5-azido-2-nitrobenzoyl-oxysuccinimide (22), 4-phenyl-1,2,4-triazoline-3,5-dione (23), or formaldehyde (12). Moreover, nearly all the other known allosteric activators of PhK besides Ca2+ increased crosslinking between the α and γ subunits in the PhK complex (24), suggesting structural similarities among the conformers induced by these activators and that enhanced αγ crosslinking is a marker for activated forms of PhK (24). Evidence for the latter came from showing that the complex remained partially activated after crosslinking in the presence of activators, even after they were subsequently removed (18, 22, 24), suggesting that structural features of the activated conformers were trapped by crosslinking.
Recent advances in MS and search engine technologies have considerably strengthened the utility of the crosslinking approach by enhancing the identification and characterization of crosslinked peptides from digests of nanomolar amounts of protein conjugates (ref. 25 and references therein). Although Fig. 2 indicates products from the crosslinking of proteins to already be complex, the number of potential products generated after digesting conjugates of large proteins is immense, and such products are essentially impossible to characterize in their entirety without computational assistance. We have recently employed search engine technologies developed by our laboratory (26) and by Yu et al. (27) in combination with FT-ICR MS to determine specific crosslinked residues in βγ (28) and γδ (9) conjugates formed in the PhK complex. In each case, the crosslink to γ was within its CRD, with the γδ EEDQ zero-length conjugate being of special interest, because the residue of δ (CaM) crosslinked to a γ-lysyl group was an aspartyl residue in its third EF hand, indicating that within the PhK complex that Asp, normally a Ca2+ ligand in CaM, is in a salt-bridge with the Lys, thus explaining the binding of only three Ca2+ ions by PhKδ (4). These crosslinking studies provided the first direct evidence for linkages in the PhK complex between functional domains of its regulatory α, β, and δ subunits and the regulatory domain of its catalytic γ subunit (Fig. 3).
Fig. 3.
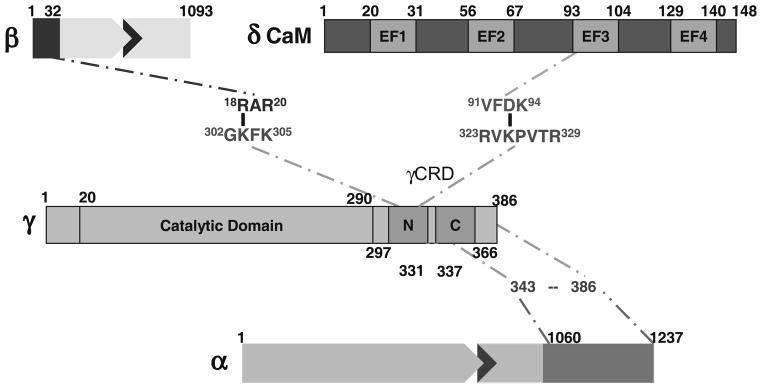
Linkage map of PhK’s regulatory subunit interactions with the γCRD determined by chemical crosslinking.
3. Effects of Ca2+ on the Overall Structure of the PhK Complex
The experimental approaches described above indicate that the structure of every individual subunit of the PhK complex is altered by the binding of Ca2+ to its δ subunit. A number of biophysical techniques corroborate the notion that Ca2+ does indeed induce widespread structural changes throughout the large PhK complex, involving alterations in its secondary, tertiary, and quaternary structure.
3.1. Conformational Changes Observed Through Spectroscopy
Tertiary structure was examined through second derivative UV absorption spectroscopy to monitor changes in the microenvironments of PhK’s 428 Phe, 460 Tyr, and 124 Trp residues upon the binding of Ca2+. The second derivative spectrum for the nonactivated enzyme showed six negative peaks having the following residue assignments: Phe (~245 and 251 nm), Tyr (~260 and 270 nm), overlapping Tyr/Trp signal (~276 nm), and Trp (~285 nm) (29). The only peak shifts observed to occur upon the binding of Ca2+ were for those assigned to Tyr residues, which were shifted to a longer wavelength, suggesting that those residues then occupied a more apolar environment (30).
The conclusions from second derivative UV absorption spectroscopy were consistent with those from intrinsic fluorescence spectroscopy. Although the intrinsic fluorescence emission peaks of Tyr and Trp residues in proteins overlap, these residues can be distinguished through the synchronous fluorescence technique (31, 32), which utilizes simultaneous scanning by the excitation and emission monochromators. Again, the binding of Ca2+, especially at pH values >7.0, showed burial of Tyr residues (29, 33). Given that the α and β subunits of PhK contain 82% of its Tyr residues, the spectral changes described most likely reflect structural changes in those subunits.
Circular dichroism was used to investigate changes in the secondary structure of PhK caused by the binding of Ca2+. The spectrum showed two minima near 208 and 220 nm, which is characteristic of proteins containing significant α-helical content. At pH values >7.0, the addition of Ca2+ shifted the second peak to 223 nm and increased its intensity, whereas the ellipticity at 208 nm was reduced without a change in peak position (29). Because the ratio of negative ellipticity at 222–208 nm is an index of the extent of interaction between α-helices and β-sheets (34, 35), the binding of Ca2+ can be considered to decrease the interactions between these structural elements in the PhK complex.
Examination of secondary structure by Fourier transform infrared (FTIR) spectroscopy also showed Ca2+-induced spectral changes, especially at 1,652 cm−1, a frequency near the band normally assigned to α-helices (36). The second derivative spectrum of the nonactivated conformer of PhK in this region suggested the presence of simple α-helices, whereas the Ca2+-activated conformer showed a new shoulder at 1,647 cm−1 (37). Second derivative minima near 1,645 ± 4 cm−1 are typically assigned to unordered structure (36) or distorted helices (38). Thus, the binding of Ca2+ by PhK may decrease the amount of highly ordered α-helices within the complex.
For all of the spectral techniques in this section but FTIR, it was possible to vary the temperature, and without exception, the spectral signals were maintained to a significantly higher temperature with the nonactivated, Ca2+-free conformer (37). Thus, the secondary and tertiary structures were maintained to higher temperatures. Upon binding Ca2+, PhK becomes increasingly labile to thermal perturbation, suggesting that activation by Ca2+ is associated with a less stable conformation of the complex.
3.2. Conformational Changes Detected Zeta Potential by Analysis
The zeta potential reflects the colloid shear surface to solvent interface environment (Stern Layer), a relatively ordered layer of solvent molecules that diffuses as part of the colloid particle. A change in electrostatic surface potential as a result of alterations in the interactions of solvent-accessible charged residues will alter the Stern Layer of a protein, and consequently, its zeta potential (39). Thus, zeta potential values provide an estimate of effective surface charge. At neutral pH, the nonactivated conformer of PhK has a zeta potential of −33 mV; however, upon binding Ca2+, its zeta potential becomes dramatically less negative, to between −22 (36) and −13 mV (29). That this change in zeta potential is associated with PhK’s activation, rather than just the binding of Ca2+ ions, is confirmed by the observation that increasing the pH to >7, which also activates the enzyme through deprotonation of residues on its regulatory α and/or β subunits, causes a similar change in zeta potential (29). Consequently, activation of PhK by either mechanism is associated with either the burial or neutralization of acidic surface side chains or the increased exposure of basic residues. Inasmuch, however, as the activity at elevated pH remains Ca2+-dependent, there must be additional Ca2+-dependent conformational changes besides those associated with just the change in zeta potential. These have, in fact, been detected as an increase in the ratio of ellipticity at 222 nm vs. 208 nm (29), as described in the previous section; however, the increase in this ratio occurs only in conformers of the PhK complex having less negative zeta potential values. The pH- and Ca2+-dependent conformers of PhK that have been observed, depicted in Fig. 4, are inter-dependent, and multiple, independent approaches, many described herein, corroborate the conformational transitions shown. The conformer with the greatest catalytic activity is the Ca2+-bound form at high pH.
Fig. 4.
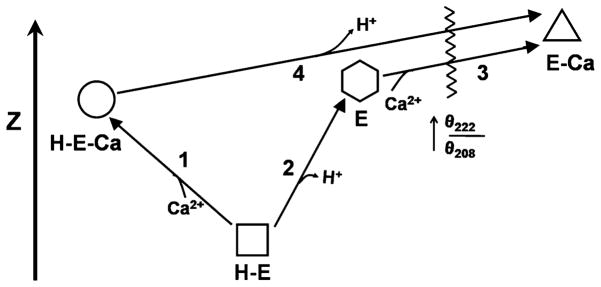
Schematic depicting the effects of Ca2+ and pH on the structure of PhK as defined through circular dichroism and zeta potential. The four conformers, two at pH 6.8 (H-E and H-E-Ca) and two at pH 8.2 (E and E-Ca), are vertically arranged according to the values of their zeta potential, with the Ca2+-free form at pH 6.8 having the most negative zeta potential and negligible catalytic activity. The numbers represent conformational transitions between forms, and the waved line intersecting conformational transitions 3 and 4 represents the occurrence of a greatly increased θ222/θ208 ratio. The Ca2+-bound conformer at pH 8.2 has by far the greatest catalytic activity. Copyright 2008 Wiley: used with permission from Liu, Weiya; Priddy, Timothy S.; Carlson, Gerald M., Physiochemical changes in phosphorylase kinase associated with its activation, Protein Science, John Wiley and Sons.
3.3. Conformational Changes Detected by Electron Microscopy
Electron microscopy with single-particle reconstruction provides a means for determining low-to-medium resolution three-dimensional structures of large protein complexes that are refractory to crystallization, such as PhK. In the single molecule approach, images of proteins are obtained from either molecules stained on carbon grids (negative staining) or frozen in vitreous ice (cryoEM), generating fields of the particles, which are considered as rigid body copies in different orientations (40). Each image is assigned directional (Euler) angles on the basis of an initial model, which may be deduced (41) or determined experimentally by methods such as the random conical tilt (42). Fourier transforms of the images are then aligned, averaged, and combined using best fit Euler angles from the model in three-dimensional Fourier space, followed by back transformation to generate a real-space reconstruction. The process is then repeated using the new reconstruction as a starting model and continued iteratively to further refine the structure. Reconstructions of the nonactivated and Ca2+-activated conformers of PhK were carried out with images of negatively stained molecules of the kinase using a simple spheres starting model (41), based on a previously deduced structure of the kinase (43). To account for potential differences in negative staining and enzyme preparations, reconstructions from three different PhK preparations were carried out ±Ca2+. Because the differences among the reconstructions for each conformer proved to be minor, all the particles from each kinase preparation were combined and averaged for each of the conformers. Comparison of the two averaged structures showed that Ca2+ promoted significant redistribution of density in both the lobe and bridge regions of the complex without significantly altering its maximum dimension (260 Å). Major structural changes in the lobes occurred in regions corresponding to the approximate positions mapped for the α, γ, and δ subunits in the complex (Fig. 1), corroborating Ca2+-dependent structural changes detected for these subunits by crosslinking (12, 18, 22, 23). Ca2+ also perturbed the structure of the bridges, which is consistent with the position of the δ subunits proximal to the bridges and with the Ca2+-dependent changes in crosslinking observed between the δ and β subunits, which are thought to compose the bridges (44).
3.4. Conformational Changes Detected by Small-Angle X-Ray Scattering
The overall shape and dimensions of proteins can be determined from their small-angle X-ray scattering (SAXS) patterns. A useful conformational parameter derived from SAXS measurements is P(r), which is the probable distribution of distances between scattering centers in a molecule and is obtained by calculating the inverse Fourier transform of the scattering data. P(r) reflects the overall asymmetry and domain structure of the scattering particle (45). SAXS complements the EM approach, in that it provides molecular weight, volume, radius of gyration (Rg: root mean square distance of the molecule’s parts from its center of gravity), and the maximum dimension of a molecule. In turn, EM provides moderate resolution structural information that can be used to model SAXS data and augment structural details for potential shape changes between conformers of allosteric proteins. Utilizing this modeling approach, simple geometric shapes were used to simulate the bridges and lobes of PhK in analyzing its SAXS data collected ± Ca2+ (46). As was observed by EM, the binding of Ca2+ caused a redistribution of density in the lobe and bridge regions of PhK without significantly changing its Rg or maximum dimension (270 Å).
4. Evidence for the γCRD Being the Master Allosteric Activation Switch for the PhK Complex
Utilization of the various techniques reviewed in this chapter showed that the binding of Ca2+ ions to the δ subunit of the (αβγδ)4 PhK complex causes conformational changes in all four of its subunits. Specific regions affected include the regulatory domain of the γ subunit, γCRD (shown by immunochemistry), and the C-terminal region of the α subunit (shown by partial proteolysis at Phe-1014), itself a regulatory region of that subunit having multiple phosphorylation sites (2). In addition to perturbations by Ca2+ of the secondary and tertiary structures of PhK’s subunits, it also altered the quaternary interactions of all four subunits as probed by chemical crosslinking. Of particular note was the doubling by Ca2+ of αγ conjugate formation by the very short crosslinker formaldehyde, with the crosslinked region of α shown biochemically to be within its C-terminal regulatory domain (12). The interaction of this region of α with γ is further supported by two-hybrid screening results, which show an interaction between the C-terminal portion of γCRD and the C terminus of α (12). Zero-length crosslinking to form a γδ conjugate between the third EF hand of δ and the γCRD was also influenced by Ca2+; but unlike αγ formation by formaldehyde, γδ formation was halved (9), instead of doubled. That the α and δ subunits both interact with regions within the γCRD suggests a Ca2+-sensitive network of quaternary interactions involving these three subunits that is mediated by the γCRD. In this network, the Ca2+-induced conformational changes in δ observed by single-pair FRET (spFRET) would perturb γ–δ interactions and concomitantly the γ–α interactions. The locations of the subunits (δ near the bridges and the C-terminal region of α near the lobe tips) considered together with the Ca2+-induced structural changes observed in these regions by EM provide additional support for the existence of this Ca2+-sensitive α ↔ γ ↔ δ communication network, which stretches from near the center of the PhK complex to the tips of its lobes, a distance of ~84 Å. A high sequence similarity between regions of γCRD and the inhibitory region of troponin I containing the Ca2+-dependent allosteric activation switch for the actin ↔ troponin I ↔ troponin C system has been discussed (6, 9, 47), and of course, troponin C is a homolog of CaM, PhK’s δ subunit. These similarities, coupled with the direct evidence described herein, support the hypothesis that the γCRD is likewise the Ca2+-dependent allosteric activation switch for the PhK complex. Not only its proximal interactions with the δ subunit and the regulatory C-terminal region of the α subunit are Ca2+-dependent, but chemical crosslinking has also shown it to be situated relatively near the N-terminal regulatory region of the β subunit that is phosphorylated by cAMP-dependent protein kinase (2, 28). The interactions of γCRD with PhK’s three regulatory subunits, depicted in Fig. 3, would seem to require that it be conformationally dynamic in order to function as an allosteric activation switch, which is consistent with the experimental data. It might also be noted that the γCRD is very basic with a pI of 10.0 (9); thus, increased exposure of this domain upon activation could explain PhK’s significantly less negative zeta potential upon its binding of Ca2+ ions.
Footnotes
When evaluating partial proteolysis as a conformational probe, a wide variety of proteases should be screened, as in ref. 11, and of course, this should be done under the conditions that the enzyme in question demonstrates its allosteric response. For analysis of a response to an allosteric effector, the proteolysis should still be in its linear phase, so that either inhibition or stimulation can be readily observed. Also, it is important to test the same protease against several other protein substrates to ensure that any observed effect of a ligand is due to its action on the protein of interest, and not on the protease being used. Finally, with the widespread availability of mass spectrometry to readily analyze protein fragments, this technique is becoming even more useful.
Chemical footprinting and all other protein chemical modification methods are empirical techniques that are influenced by a number of variables, including the concentrations of reactants, time and temperature of the reaction, and the pH value (19). The variability inherent in these techniques derives primarily from the protein itself, specifically the numerous potentially reactive amino acid side chains that may be targeted selectively by different classes of reagents. With only a few exceptions, the amino acids that typically undergo chemical modification (tyrosine, lysine, cysteine, histidine, aspartic acid, and glutamic acid) contain ionizable nucleophilic side chains. The microenvironment (location in the protein) of a side chain functional group directly influences its accessibility, polarity, and noncovalent interactions with other residues or solvent. These and other factors ultimately lead to differences in reactivity for a given functional group that can occur even on the surface of a protein, thus causing difficulty in selectively targeting a particular side chain with a single reagent. To optimize selectivity and extent of modification of a protein, it is important to screen multiple reagents having different reactive functional groups (Table 1), water solubility, reporter groups, etc., while varying the time, pH, and concentration of the reactants. As with partial proteolysis, screens should be performed under conditions in which the target protein responds to its cognate allosteric ligand and undergoes chemical modification linearly with time.
A necessary precaution in determining whether a given crosslinking reagent is a suitable conformational probe is to verify that a covalent conjugate of interest is derived from crosslinking within a complex as opposed to between complexes. This verification can usually be achieved by size-exclusion chromatography (SEC) over a matrix capable of separating the native complex from its dimers and higher order multimers (22). Following crosslinking, the reaction is quenched (using an excess of a general nucleophilic reagent such as mercaptoethanol or lysine) and applied to SEC. The protein fractions that coelute with the native complex are collected and analyzed by SDS–PAGE. Any conjugates detected in these fractions represent intersubunit crosslinking within the complex.
References
- 1.Krebs EG, Love DS, Bratvold GE, Trayser KA, Meyer WL, Fischer EH. Purification and properties of rabbit skeletal muscle phosphorylase b kinase. Biochemistry. 1964;3:1022–1033. doi: 10.1021/bi00896a003. [DOI] [PubMed] [Google Scholar]
- 2.Brushia RJ, Walsh DA. Phosphorylase kinase: The complexity of its regulation is reflected in the complexity of its structure. Front Biosci. 1999;4:D618–641. doi: 10.2741/brushia. [DOI] [PubMed] [Google Scholar]
- 3.Carriere C, Mornon JP, Venien-Bryan C, Boisset N, Callebaut I. Calcineurin b-like domains in the large regulatory alpha/beta subunits of phosphorylase kinase. Proteins. 2008;71:1597–1606. doi: 10.1002/prot.22006. [DOI] [PubMed] [Google Scholar]
- 4.Burger D, Cox JA, Fischer EH, Stein EA. The activation of rabbit skeletal muscle phosphorylase kinase requires the binding of 3 Ca2+ per delta subunit. Biochem Biophys Res Commun. 1982;105:632–638. doi: 10.1016/0006-291x(82)91481-4. [DOI] [PubMed] [Google Scholar]
- 5.Chan KF, Graves DJ. Rabbit skeletal muscle phosphorylase kinase. Catalytic and regulatory properties of the active αγδ and γδ complexes. J Biol Chem. 1982;257:5948–5955. [PubMed] [Google Scholar]
- 6.Dasgupta M, Honeycutt T, Blumenthal DK. The gamma-subunit of skeletal muscle phosphorylase kinase contains two noncontiguous domains that act in concert to bind calmodulin. J Biol Chem. 1989;264:17156–17163. [PubMed] [Google Scholar]
- 7.Harris WR, Malencik DA, Johnson CM, Carr SA, Roberts GD, Byles CA, Anderson SR, Heilmeyer LM, Jr, Fischer EH, Crabb JW. Purification and characterization of catalytic fragments of phosphorylase kinase gamma subunit missing a calmodulin-binding domain. J Biol Chem. 1990;265:11740–11745. [PubMed] [Google Scholar]
- 8.Picton C, Klee CB, Cohen P. Phosphorylase kinase from rabbit skeletal muscle: Identification of the calmodulin-binding subunits. Eur J Biochem. 1980;111:553–561. doi: 10.1111/j.1432-1033.1980.tb04971.x. [DOI] [PubMed] [Google Scholar]
- 9.Jeyasingham MD, Artigues A, Nadeau OW, Carlson GM. Structural evidence for co-evolution of the regulation of contraction and energy production in skeletal muscle. J Mol Biol. 2008;377:623–629. doi: 10.1016/j.jmb.2007.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priddy TS, Price ES, Johnson CK, Carlson GM. Single molecule analyses of the conformational substates of calmodulin bound to the phosphorylase kinase complex. Protein Sci. 2007;16:1017–1023. doi: 10.1110/ps.062747407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trempe MR, Carlson GM. Phosphorylase kinase conformers. Detection by proteases. J Biol Chem. 1987;262:4333–4340. [PubMed] [Google Scholar]
- 12.Rice NA, Nadeau OW, Yang Q, Carlson GM. The calmodulin-binding domain of the catalytic γ subunit of phosphorylase kinase interacts with its inhibitory α subunit: Evidence for a Ca2+ sensitive network of quaternary interactions. J Biol Chem. 2002;277:14681–14687. doi: 10.1074/jbc.M201229200. [DOI] [PubMed] [Google Scholar]
- 13.Wangsgard WP, Dasgupta M, Blumenthal DK. Antipeptide antibodies as probes of subunit-dependent structural changes in the regulatory domain of the gamma-subunit of phosphorylase kinase. Biochem Biophys Res Commun. 1997;230:179–183. doi: 10.1006/bbrc.1996.5927. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson DA, Marion TN, Tillman DM, Norcum MT, Hainfeld JF, Seyer JM, Carlson GM. An epitope proximal to the carboxyl terminus of the α-subunit is located near the lobe tips of the phosphorylase kinase hexadecamer. J Mol Biol. 1994;235:974–982. doi: 10.1006/jmbi.1994.1051. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson DA, Norcum MT, Fizgerald TJ, Marion TN, Tillman DM, Carlson GM. Proximal regions of the catalytic γ and regulatory β subunits on the interior lobe face of phosphorylase kinase are structurally coupled to each other and with enzyme activation. J Mol Biol. 1997;265:319–329. doi: 10.1006/jmbi.1996.0739. [DOI] [PubMed] [Google Scholar]
- 16.Nadeau OW, Gogol EP, Carlson GM. Cryoelectron microscopy reveals new features in the three-dimensional structure of phosphorylase kinase. Protein Sci. 2005;14:914–920. doi: 10.1110/ps.041123905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Traxler KW, Norcum MT, Hainfeld JF, Carlson GM. Direct visualization of the calmodulin subunit of phosphorylase kinase via electron microscopy following subunit exchange. J Struct Biol. 2001;135:231–238. doi: 10.1006/jsbi.2001.4411. [DOI] [PubMed] [Google Scholar]
- 18.Nadeau OW, Sacks DB, Carlson GM. The structural effects of endogenous and exogenous Ca2+/calmodulin on phosphorylase kinase. J Biol Chem. 1997;272:26202–26209. doi: 10.1074/jbc.272.42.26202. [DOI] [PubMed] [Google Scholar]
- 19.Nadeau OW, Carlson GM. Protein interactions captured by chemical cross-linking. In: Golemis E, Adams PD, editors. Protein–protein interactions: A molecular cloning manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2005. pp. 105–127. [Google Scholar]
- 20.Hermanson GT. Bioconjugate techniques. 2. Elsevier Academic Press; Amsterdam; Boston: 2008. [Google Scholar]
- 21.Wong SS. Chemistry of protein conjugation and cross-linking. CRC Press; Boca Raton, FL: 1993. [Google Scholar]
- 22.Nadeau OW, Traxler KW, Fee LR, Baldwin BA, Carlson GM. Activators of phosphorylase kinase alter the cross-linking of its catalytic subunit to the C-terminal one-sixth of its regulatory α subunit. Biochemistry. 1999;38:2551–2559. doi: 10.1021/bi982060b. [DOI] [PubMed] [Google Scholar]
- 23.Ayers NA, Nadeau OW, Read MW, Ray P, Carlson GM. Effector-sensitive cross-linking of phosphorylase b kinase by the novel cross-linker 4-phenyl-1,2,4-triazoline-3,5-dione. Biochem J. 1998;331(Pt 1):137–141. doi: 10.1042/bj3310137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadeau OW, Sacks DB, Carlson GM. Differential affinity cross-linking of phosphorylase kinase conformers by the geometric isomers of phenylenedimaleimide. J Biol Chem. 1997;272:26196–26201. doi: 10.1074/jbc.272.42.26196. [DOI] [PubMed] [Google Scholar]
- 25.Chu F, Baker PR, Burlingame AL, Chalkley RJ. Finding chimeras: A bioinformatics strategy for identification of cross-linked peptides. Mol Cell Proteomics. 2010;9:25–31. doi: 10.1074/mcp.M800555-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadeau OW, Wyckoff GJ, Paschall JE, Artigues A, Sage J, Villar MT, Carlson GM. Crosssearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins. Mol Cell Proteomics. 2008;7:739–749. doi: 10.1074/mcp.M800020-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu ET, Hawkins A, Kuntz ID, Rahn LA, Rothfuss A, Sale K, Young MM, Yang CL, Pancerella CM, Fabris D. The collaboratory for MS3D: A new cyberinfrastructure for the structural elucidation of biological macromolecules and their assemblies using mass spectrometry-based approaches. Journal of Proteome Research. 2008;7:4848–4857. doi: 10.1021/pr800443f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadeau OW, Anderson DW, Yang Q, Artigues A, Paschall JE, Wyckoff GJ, McClintock JL, Carlson GM. Evidence for the location of the allosteric activation switch in the multisubunit phosphorylase kinase complex from mass spectrometric identification of chemically crosslinked peptides. J Mol Biol. 2007;365:1429–1445. doi: 10.1016/j.jmb.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Priddy TS, Carlson GM. Physiochemical changes in phosphorylase kinase associated with its activation. Protein Sci. 2008;17:2111–2119. doi: 10.1110/ps.037895.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demchenko AP. Ultraviolet spectroscopy of proteins. Springer-Verlag; Berlin; New York: 1986. Derivative spectroscopy of aromatic amino acids and proteins; pp. 121–135. Rev. and enl. translation of the Russian ed. [Google Scholar]
- 31.Lloyd JBF. Synchronised excitation of fluorescence emission spectra. Nature physical sciences. 1971;231:64–65. [Google Scholar]
- 32.Miller JN. Recent advances in molecular luminescence analysis. Proceedings of the Analytical Division of the Chemical Society. 1979;16:203–208. [Google Scholar]
- 33.Cui FL, Wang JL, Cui YR, Li JP. Fluorescent investigation of the interactions between n-(p-chlorophenyl)-n′-(1-naphthyl) thiourea and serum albumin: Synchronous fluorescence determination of serum albumin. Anal Chim Acta. 2006;571:175–183. doi: 10.1016/j.aca.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Chothia C, Levitt M, Richardson D. Structure of proteins: Packing of alpha-helices and pleated sheets. Proc Natl Acad Sci USA. 1977;74:4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manavalan P, Johnson WC. Sensitivity of circular dichroism to protein tertiary structure class. Nature. 1983;305:831–832. [Google Scholar]
- 36.Susi H, Byler DM. Resolution-enhanced fourier transform infrared spectroscopy of enzymes. Methods Enzymol. 1986;130:290–311. doi: 10.1016/0076-6879(86)30015-6. [DOI] [PubMed] [Google Scholar]
- 37.Priddy TS, Middaugh CR, Carlson GM. Electrostatic changes in phosphorylase kinase induced by its obligatory allosteric activator Ca2+ Protein Sci. 2007;16:517–527. doi: 10.1110/ps.062577507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trewhella J, Liddle WK, Heidorn DB, Strynadka N. Calmodulin and troponin c structures studied by fourier transform infrared spectroscopy: Effects of Ca2+ and Mg2 + binding. Biochemistry. 1989;28:1294–1301. doi: 10.1021/bi00429a052. [DOI] [PubMed] [Google Scholar]
- 39.McNeil-Watson F, Tscharnuter W, Miller J. A new instrument for the measurement of very small electrophoretic mobilities using phase analysis light scattering (pals) Colloids and Surfaces A: Physicochemical and Engineering Aspects. 1998;140:53–57. [Google Scholar]
- 40.Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- 41.Nadeau OW, Carlson GM, Gogol EP. A Ca2+-dependent global conformational change in the 3D structure of phosphorylase kinase obtained from electron microscopy. Structure. 2002;10:23–32. doi: 10.1016/s0969-2126(01)00678-5. [DOI] [PubMed] [Google Scholar]
- 42.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50s ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 43.Norcum MT, Wilkinson DA, Carlson MC, Hainfeld JF, Carlson GM. Structure of phosphorylase kinase. A three-dimensional model derived from stained and unstained electron micrographs. J Mol Biol. 1994;241:94–102. doi: 10.1006/jmbi.1994.1476. [DOI] [PubMed] [Google Scholar]
- 44.Trempe MR, Carlson GM, Hainfeld JF, Furcinitti PS, Wall JS. Analyses of phosphorylase kinase by transmission and scanning transmission electron microscopy. J Biol Chem. 1986;261:2882–2889. [PubMed] [Google Scholar]
- 45.Trewhella J. Insights into biomolecular function from small-angle scattering. Curr Opin Struct Biol. 1997;7:702–708. doi: 10.1016/s0959-440x(97)80081-4. [DOI] [PubMed] [Google Scholar]
- 46.Priddy TS, MacDonald BA, Heller WT, Nadeau OW, Trewhella J, Carlson GM. Ca2+-induced structural changes in phosphorylase kinase detected by small-angle x-ray scattering. Protein Sci. 2005;14:1039–1048. doi: 10.1110/ps.041124705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paudel HK, Carlson GM. Functional and structural similarities between the inhibitory region of troponin I coded by exon VII and the calmodulin-binding regulatory region of the catalytic subunit of phosphorylase kinase. Proc Natl Acad Sci USA. 1990;87:7285–7289. doi: 10.1073/pnas.87.18.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]


