Summary
The life cycle of Myxococcus xanthus includes coordinated group movement and fruiting body formation, and requires directed motility and controlled cell reversals. Reversals are achieved by inverting cell polarity and re-organizing many motility proteins. The Frz chemosensory pathway regulates the frequency of cell reversals. While it has been established that phosphotransfer from the kinase FrzE to the response regulator FrzZ is required, it is unknown how phosphorylated FrzZ, the putative output of the pathway, targets the cell polarity axis. In this study, we used Phos-tag SDS-PAGE to determine the cellular level of phospho-FrzZ under different growth conditions and in Frz signaling mutants. We detected consistent FrzZ phosphorylation, albeit with a short half-life, in cells grown on plates, but not from liquid culture. The available pool of phospho-FrzZ correlated with reversal frequencies, with higher levels found in hyper-reversing mutants. Phosphorylation was not detected in hypo-reversing mutants. Fluorescence microscopy revealed that FrzZ is recruited to the leading cell pole upon phosphorylation and switches to the opposite pole during reversals. These results are consistent with the hypothesis that the Frz pathway modulates reversal frequency through a localized response regulator that targets cell polarity regulators at the leading cell pole.
Introduction
Myxococcus xanthus, a δ-proteobacterium, displays a spectrum of social behaviors that require cell-cell communication and the coordinated movement of cells (Pathak et al., 2012). Cell motility allows cells to move over solid surfaces to efficiently utilize nutrients (Zhang et al., 2012a). When nutrients become scarce, a developmental program is initiated, and cells aggregate into complex multicellular structures, called fruiting bodies, where they differentiate into resistant spores (Zusman et al., 2007).
M. xanthus employs two motility systems for movement: social motility or ‘S-motility, for when cells move in groups, and gliding motility (‘adventurous’ or ‘A’-motility), for when cells move as solitary individuals (Hodgkin & Kaiser, 1979). Social motility is similar to twitching motility and powered by Type IV pili that extend from the leading cell pole, attach to neighboring cells or extracellular polysaccharides, and then retract, pulling the cells forward (Li et al., 2003, Bulyha et al., 2009). However, in contrast to for example. Pseudomonas cells, M. xanthus cells move forward continuously without twitching.
Myxococcal gliding motility depends on force generated by distributed motor complexes in the cell envelope, and is energized by proton motive force (reviewed in (Nan & Zusman, 2011) (Mignot et al., 2007, Nan et al., 2011, Sun et al., 2011). M. xanthus uses both motility systems simultaneously for colony expansion, however the two mechanisms are differentially effective on different surfaces: social motility predominates on soft surfaces while gliding motility requires harder surfaces (Shi & Zusman, 1993). The two motility systems are encoded by separate sets of genes, and mutants are readily distinguished by their motility behavior on hard or soft agar plates (Hodgkin & Kaiser, 1979, Shi & Zusman, 1993).
In order to achieve directed motility and efficient colony expansion, M. xanthus cells must periodically change their direction of movement. Reversals in M. xanthus are accomplished by inverting the cell polarity axis so that periodically the lagging cell pole becomes the leading cell pole, and vice versa. In isolated cells, reversal of cell polarity occurs on average every 8–9 minutes, but is achieved within ~30 seconds (Bustamante et al., 2004, Blackhart & Zusman, 1985b, Zhang et al., 2010). During reversals, the proteins required for both motility machineries are simultaneously reoriented (Bulyha et al., 2009, Mignot et al., 2005, Mignot et al., 2007).
The polarity axis of cells is manifested in the asymmetric polar distribution of two proteins, MglA and MglB. MglA is a GTPase that localizes to the leading cell pole in its GTP-bound form, whereas MglB, its cognate GTPase activating protein (GAP), localizes to the lagging cell pole (Leonardy et al., 2010, Zhang et al., 2010). Recently, the response regulator protein RomR was found to be required for polar localization of MglA (Leonardy et al., 2007, Keilberg et al., 2012, Zhang et al., 2012b). During cell reversals, MglA, MglB and RomR swap their localization patterns, thereby inverting cell polarity to initiate the re-orientation of the motility machineries (Leonardy et al., 2007, Leonardy et al., 2010, Zhang et al., 2010). The cell polarity axis is essential for directing the motility machineries, since several motility proteins fail to localize correctly in the absence of MglA (Zhang et al., 2010, Leonardy et al., 2010). Indeed, ΔmglA mutants appear non-motile (Hartzell & Kaiser, 1991). However, it is still unknown how the coordinated rearrangement of the social and gliding motility systems is achieved, and how it might be mediated by the coordinated relocation of MglA/B and RomR proteins.
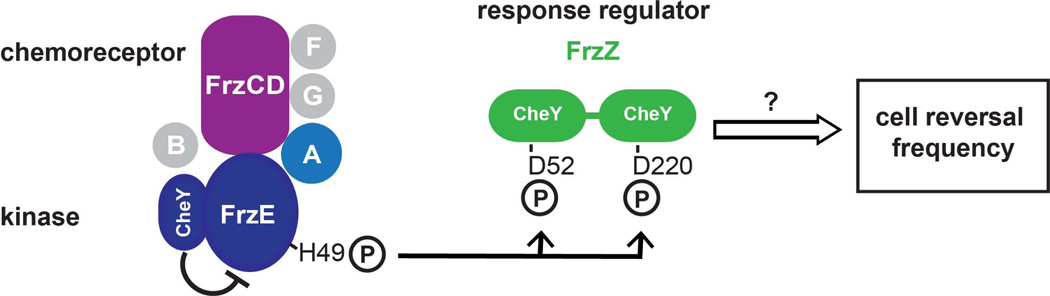
In order to control the frequency of cell reversals, M. xanthus uses a dedicated two-component chemosensory system, the Frz pathway, which is encoded by a divergent operon that encompasses chemotaxis signaling homologues (Zusman, 1982, Blackhart & Zusman, 1985b, Blackhart & Zusman, 1985a) (Fig 1). The core components are FrzCD, a methyl-accepting chemoreceptor (MCP), FrzA, a coupling protein, FrzE, a CheA-family protein kinase, and FrzZ, a dual response regulator (McCleary et al., 1990, Acuna et al., 1995, Trudeau et al., 1996). Additional components encoded within the frz operon modulate the methylation status and adaptation of the chemoreceptor (FrzG and FrzF), or have other functions (FrzB) (Bustamante et al., 2004, Scott et al., 2008). Deletion of the frz genes in most cases causes cells to reverse infrequently (> 60 min, hypo-reversing) (Bustamante et al., 2004). These cells maintain their polarity axis as well as social and gliding motility, but show reduced colony expansion, and, when starved, fail to aggregate into distinct fruiting bodies, but form ‘frizzy’ aggregates instead (Fig. 4A) (Zusman, 1982, Bustamante et al., 2004, Blackhart & Zusman, 1985b). In contrast, a partial N-terminal deletion of the chemoreceptor FrzCD, (Δ6–182, frzCDc) causes hyper-reversals (every 2 min), presumably by constitutive signaling through the pathway (Fig. 4A) (Bustamante et al., 2004). The response regulator FrzZ was identified as the putative output of the Frz pathway, since FrzZ acts downstream of FrzE, and deletion of frzZ in a frzCDc mutant relieves the hyper-reversing phenotype of frzCDc (Inclan et al., 2007). FrzZ contains two response regulator receiver domains that are connected by a short linker region and can each be phosphorylated at an aspartate residue (Inclan et al., 2007, Trudeau et al., 1996) (Fig. 1).
Figure 1. The Frz chemosensory signaling pathway regulates the frequency of cellular reversals in M. xanthus.
Schematic illustration of the Frz two-component system. The CheA-like kinase FrzE autophosphorylates at H49 in presence of ATP, the chemoreceptor FrzCD and the coupling protein FrzA. FrzF and FrzG modulate the methylation of the chemoreceptor, the function of FrzB is unknown. Autophosphorylation of the FrzE kinase domain is inhibited by an internal receiver domain (‘CheY’, residues 662–766). The phosphoryl group is transferred to D52 or D220 of the response regulator FrzZ, which comprises two CheY-like receiver domains. Phosphorylation of FrzZ controls the frequency of cell reversal in an unknown manner.
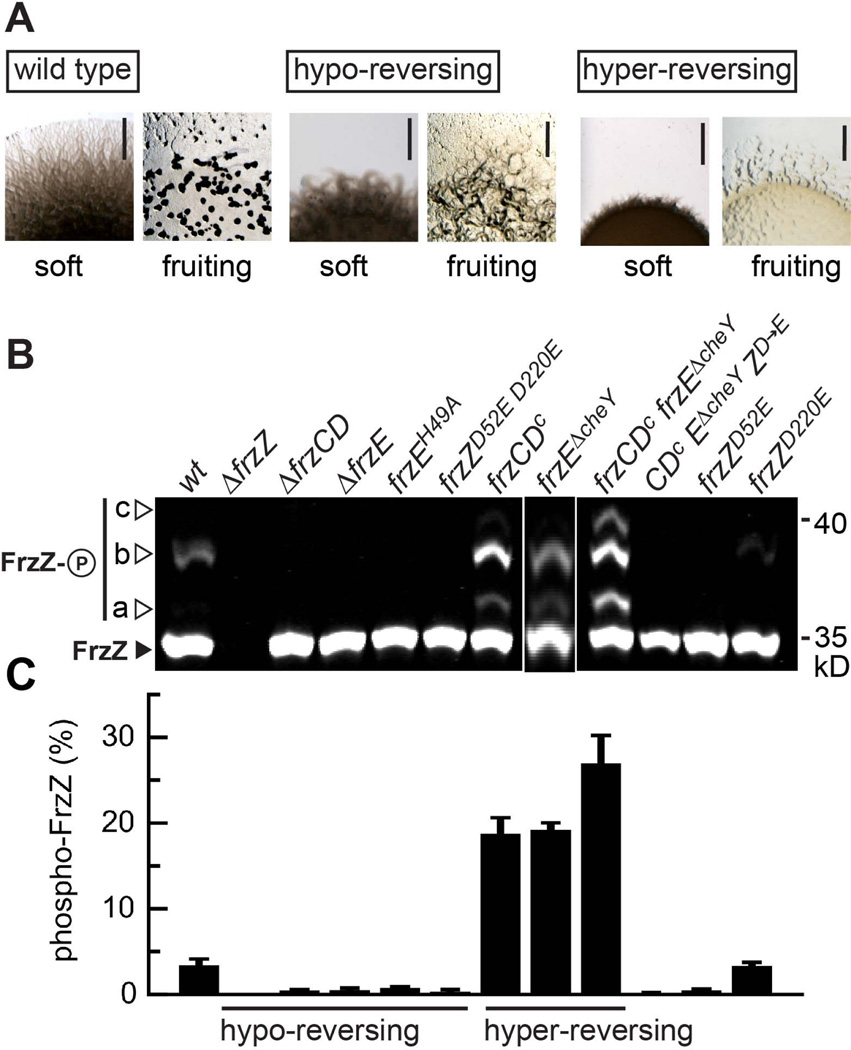
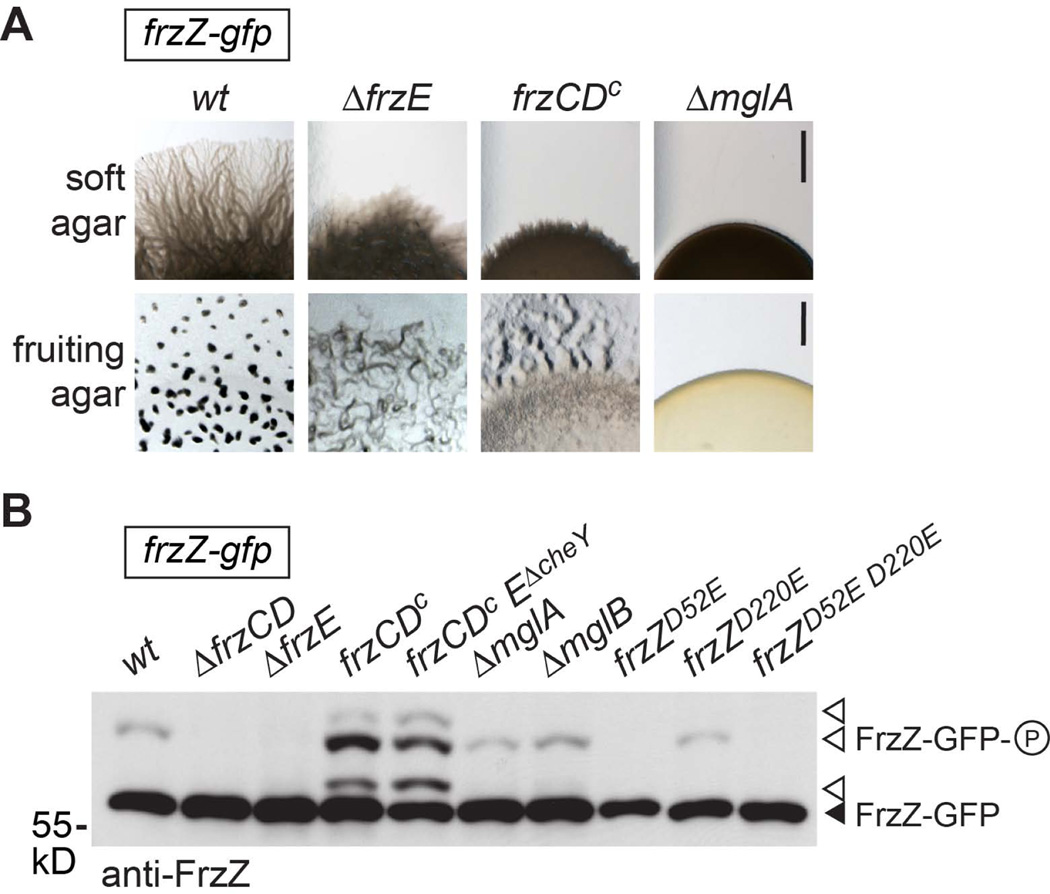
Figure 4. The cellular pool of phosphorylated FrzZ is linked to motility and developmental phenotypes.
A. Phenotype of M. xanthus wild type and representative mutant strains that display hypo-reversing (ΔfrzZ) or hyper-reversing (frzCDc) phenotypes as previously shown (Bustamante et al., 2004, Inclan et al., 2007). Images show the colony edge of cultures spotted to agar plates and incubated for 48 h (gliding motility on soft agar surface, top) or 96 h (fruiting body formation under starvation conditions, bottom). Scale bars represent 2 mm. B. Analysis of FrzZ phosphorylation in vivo. Cell extracts of different signaling mutants (as indicated) were analyzed by Phos-tag (50 µM) SDS-PAGE followed by Western blot using anti-FrzZ antibody. The image shows the signal detected after probing with fluorescent secondary antibody, and is representative of at least three independent experiments. The closed arrow-head marks the signal for unphosphorylated FrzZ (30.4 kD), open arrow-heads highlight different signals (a–c, please refer to Fig. 2B) that correspond to the different phosphorylated forms of FrzZ C. Quantitative analysis of phospho-FrzZ in M. xanthus strains as shown in panel B. The relative fluorescence intensity of phosphorylated FrzZ signals for at least three independent experiments is shown with error bars that represent the standard error of the mean. Compared to wild type, the level of phospho-FrzZ is elevated in hyper-reversing mutants and diminished in hypo-reversing mutants.
Phosphotransfer between the FrzE histidine kinase and the FrzZ receiver domains has been demonstrated in vitro using purified proteins: FrzE autophosphorylates at H49 in the presence of ATP, the chemoreceptor FrzCD and the coupling protein FrzA (Inclan et al., 2007) (Fig. 1). The phosphoryl group is then rapidly transferred to D52 or D220 of FrzZ. Indeed, FrzZ function is lost when both aspartate residues D52 and D220 are changed to glutamate residues, indicating that phosphorylation of FrzZ is required for the regulation of reversal frequency (Inclan et al., 2007, Inclan et al., 2008). However, the D220E single mutant shows normal colony expansion, whereas colony expansion in the D52E mutant is significantly reduced (Inclan et al., 2007).
It has been established that the MglA/MglB polarity axis is inverted to initiate cell reversal at a certain frequency that is dictated by the Frz signaling pathway (Leonardy et al., 2010, Zhang et al., 2010, Keilberg et al., 2012, Zhang et al., 2012b). However, it is unclear how signals are transmitted via the FrzZ response regulator, and how phosphorylation affects FrzZ function with respect to cell reversals. In this study, we investigated the phosphorylation of FrzZ in vivo under various growth conditions. We found the cellular pool of phosphorylated FrzZ to be directly correlated to the cellular reversal frequency. Furthermore, we show that FrzZ upon phosphorylation is associated with the leading cell pole, switching to the opposite cell pole when cells reverse. These results suggest that phosphorylated FrzZ targets proteins that establish cell polarity at the leading cell pole to modulate the frequency of cell reversals.
Results
Detection of phosphorylated FrzZ in cell extracts
We were interested in studying how phosphorylated FrzZ, the putative output of the Frz chemosensory pathway, impacts reversal frequency in M. xanthus. In order to track the level of phosphorylated FrzZ in vivo, we took advantage of the Phos-tag compound, which selectively forms a complex with phosphorylated amino acids in the presence of divalent cations (Kinoshita et al., 2009). In the presence of MnCl2, Phos-tag induces a mobility shift in phosphorylated proteins in SDS-PAGE, causing them to migrate slower than the non-phosphorylated form. This method, originally developed for the study of eukaryotic Ser/Thr kinases, has been successfully adapted for bacterial histidine kinases (e.g. E. coli PhoP), and validated against standard methods that require radioactive 32P-labeling (Barbieri & Stock, 2008). We used Phos-tag SDS-PAGE followed by Western immunoblotting with purified FrzZ antiserum to simultaneously detect the phosphorylated and the unphosphorylated form of FrzZ in whole cell extracts.
Figure 2A shows that extracts from wild type strain DZ2 or frzCDc hyper-reversing cells grown on agar plates displayed a strong signal for FrzZ corresponding to its molecular weight (closed arrowhead) and an additional fainter, slower migrating signal, which represents the phosphorylated form of FrzZ (open arrowhead). This second signal was never observed when cell extracts were probed by Western blot after SDS-PAGE without Phos-tag (data not shown).
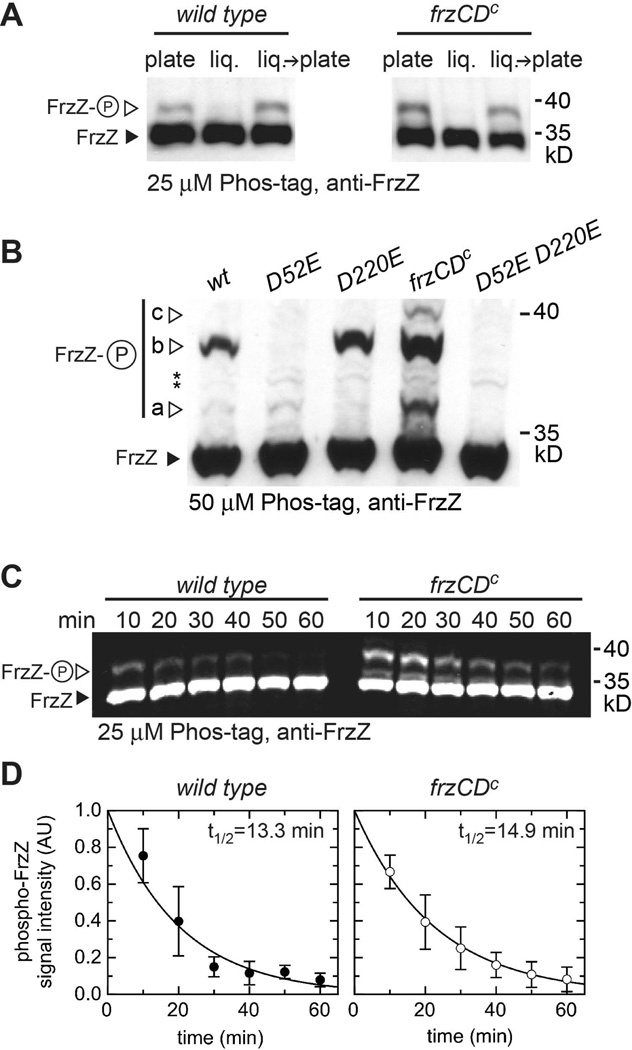
Figure 2. Detection of phosphorylated FrzZ in cell extracts.
A. Wild type or hyper-reversing frzCDc mutant strains were grown on CYE agar (‘plate’) or in CYE liquid culture (‘liq.’) for 72 h prior to analysis. ‘liq.→ plate’: cells from liquid culture were transferred to an agar plate for 10 min before cell lysis. Cells grown for 72 h in liquid medium were diluted several times to keep the culture growing exponentially. Cell lysates were rapidly subjected to SDS-polyacrylamide gel electrophoresis in the presence of 25 µM Phos-tag and 50 µM MnCl2, followed by Western immunoblotting with purified anti-FrzZ antiserum. The closed arrow-head marks the signal corresponding to unphosphorylated FrzZ (30.4 kD), open arrow-heads highlight signals for phospho-FrzZ. B. Different signals detected after Phos-tag SDS-PAGE correspond to phosphorylation at different aspartate residues. Signal a: FrzZ phosphorylated at D220, b: at D52, c: at both phosphorylation sites. Whole cell extract of wild type, FrzZ point mutants and the hyper-reversing strain frzCDc were subjected to SDS-PAGE (50 µM Phos-tag, 100 µM MnCl2), followed by Western blotting. Asterisks depict signals due to unspecific binding of anti-FrzZ antibody. C. Estimation of the half-life of phospho-FrzZ in wild type and a hyper-reversing mutant (frzCDc). Cells were lysed in 10 min intervals prior to electrophoresis in Phos-tag SDS-PAGE. Western blots were probed with fluorescent secondary antibody. D. Quantitative analysis of fluorescence signal intensity from panel C. Data and standard deviations summarize at least three independent experiments.
Interestingly, phospho-FrzZ was not detected when extracts from cells growing for 72 h in liquid CYE medium (‘liq.’) were analyzed, while cells grown on CYE agar (‘plate’) for the same period showed small but detectable phosphorylation (Fig. 2A). Moreover, the phosphorylation of FrzZ was rapidly restored when liquid grown cells were spotted on CYE agar for 10 min prior to cell lysis (Fig. 2A, ‘liq.’→‘plate’). The results were similar for the analysis of wild type and the frzCDc mutant strain, in which Frz signaling presumably is ‘constitutively active.’ Based on these observations, we performed all further phosphorylation analyses with cells grown on CYE agar plates, where Frz phosphorylation appears to be consistent.
To confirm that the observed slower migrating signals correspond to phosphorylated FrzZ, we analyzed cell extracts of mutants that carry single or double phosphorylation site substitutions, frzZD52E and frzZD220E (Fig. 2B). In these experiments, we increased the amount of Phos-tag (to 50 µM) and MnCl2 (to 100 µM) in the SDS-PAGE, which increased the resolution of phosphorylated proteins. Three different slowly migrating phospho-FrzZ bands were detected (labeled a, b, and c in Fig. 2B). We deduced that the three phospho-FrzZ signals correspond to FrzZ phosphorylated at D220 (signal a), D52 (signal b) or at both sites (signal c): only signal (b) was detected in frzZD220E, while signal (a) was picked up in the frzD52E mutant. Both signals were absent in a frzZD52E D220E double mutant, but more pronounced in a hyper-reversing mutant (frzCDc). Since Western blot membranes were overexposed to detect the faint signal for phosphorylation of D220, we also observed several additional bands due to unspecific binding of anti-FrzZ antibody (asterisks in Fig. 2B). Our observations are consistent with previous studies, which also reported that Phos-tag binding shifts the mobility of phosphorylated proteins specific to different phosphorylation sites (Hosokawa et al., 2010).
Phospho-FrzZ has a limited half-life
Phosphorylated aspartate in bacterial response regulators typically has a short half-life, which, although important for its regulatory function, can compromise quantitative analyses (Lukat et al., 1992, Scharf, 2010). To determine the half-life of phospho-FrzZ in our experiments, we incubated wild type and frzCDc cell extracts for 10–60 min on ice before stabilizing phospho-FrzZ by the addition of SDS-sample buffer. After Phos-tag SDS-PAGE, Western blot membranes were probed with anti-FrzZ and fluorescent secondary antibody, and fluorescence signal intensity was quantified. Figure 2C shows that the signal for unphosphorylated FrzZ (closed arrowheads) remained stable during the experiment, but the signal for phospho-FrzZ (open arrowheads) diminished over time similarly in both strains. The half-life of phospho-FrzZ in extracts incubated on ice was calculated from the exponential decay of signal intensity to be t1/2=13.3 min for wild type and t1/2=14.9 min for the hyper-reversing frzCDc mutant strain (Fig. 2C and D). The frzCDc strain reproducibly showed higher levels of phospho-FrzZ than wild type, consistent with its hyper-signaling phenotype (see below).
The experimental procedures used here required about 10 min from cell lysis until stabilization of phospho-FrzZ. Therefore, the fraction of phospho-FrzZ that was present in vivo before cell lysis can be estimated to be 1.4–1.6 fold higher at t0 (Fig. 2C and D). This suggests that in vivo only a fraction of FrzZ is available in phosphorylated form. The experimentally determined half-life of phospho-FrzZ in cell extracts kept at low temperature in the presence of phosphatase inhibitors likely does not accurately reflect the in vivo situation, since phosphatases or other interacting proteins are likely to increase or decrease phospho-FrzZ stability at 32°C. However, the half-life of phospho-FrzZ could play a direct role in setting cellular reversal frequencies, which range within 2–30 minutes in M. xanthus.
FrzZ phosphorylation is independent of cell movement
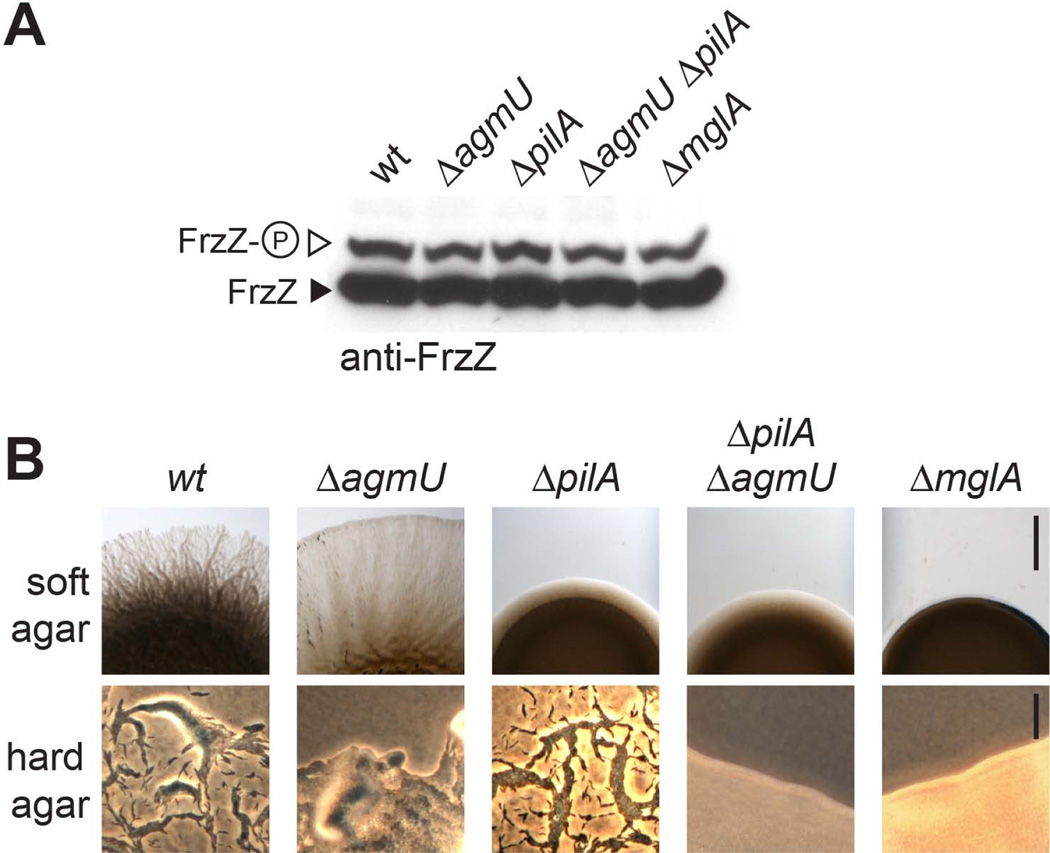
Since M. xanthus cells require a solid surface for their motility and FrzZ is not phosphorylated in cells growing in liquid culture (Fig. 2A), we were interested in determining whether cell motility per se impacts FrzZ phosphorylation. Our experiments suggest that this is not the case (Fig. 3): Frz signaling reflected by the level of phospho-FrzZ was similar to wild type in mutants defective in social motility (ΔpilA, (Wu & Kaiser, 1995)) or gliding motility (ΔagmU, (Nan et al., 2010)), as well as mutants defective in both motility systems (ΔpilAΔagmU double mutants or ΔmglA) (Fig. 3A). The different motility phenotypes on soft and hard agar surfaces are shown in Figure 3B. The observation that FrzZ is phosphorylated to wild type levels in a ΔmglA mutant also confirms that phosphorylation is independent of cell reversals, since cells cannot invert their polarity without MglA.
Figure 3. FrzZ phosphorylation remains unchanged in different motility system mutants.
A. in vivo phosphorylation shown by Western blot of whole cell extract probed with anti-FrzZ antibody after Phos-tag (25 µM) SDS-PAGE. B. Motility phenotypes of the strains used in panel A on soft and hard agar surface. Scale bars represent 2 mm (soft agar) and 40 µm (hard agar), respectively. ΔpilA (pilin): defective in social motility. ΔagmU: defective in gliding motility; ΔpilA ΔagmU and ΔmglA: non-motile.
FrzZ phosphorylation levels in vivo are correlated with the Frz signaling phenotypes
We analyzed FrzZ phosphorylation levels in M. xanthus wild type and a set of strains that carry mutations in components of the Frz pathway and show various phenotypes in motility and development (Bustamante et al., 2004, Li et al., 2005, Inclan et al., 2007). Based on previous genetic studies and biochemical analysis in vitro, we predict that hypo-reversing mutants should abolish FrzZ phosphorylation whereas hyper-reversing mutants should increase phosphorylation (Fig. 1). Indeed, we did not detect phosphorylated FrzZ in mutants carrying in-frame deletions of the chemoreceptor FrzCD (ΔfrzCD), the CheA-like kinase FrzE (ΔfrzE), as well as a point mutation of the phospho-accepting residue H49 of FrzE (frzEH49A) (Fig. 4B and 4C). Furthermore, strain frzZD52E D220E with both phosphorylation sites mutated showed only the signal for unphosphorylated FrzZ (Fig. 4B). In contrast, we detected low but consistent levels of phospho-FrzZ (about 3% of total FrzZ) in wild type extracts (Fig. 4C).
Hyper-reversing mutants showed higher levels of phospho-FrzZ. For example, phospho-FrzZ increases to 18% of total FrzZ in the frzCDc constitutively active mutant (Fig. 4B and 4C). A FrzE mutant strain that carries a deletion of an inhibitory receiver domain (frzEΔcheY) showed higher FrzE kinase activity in vitro and hyper-reversals (Inclan et al., 2008, Li et al., 2005). In this strain, we found phospho-FrzZ levels to be increased to 21% (Fig. 4B and 4C). Furthermore, the double mutant frzCDc frzEΔcheY showed an additive increase in FrzZ phosphorylation, with phospho-FrzZ levels of about 28% of total FrzZ. In contrast, FrzZD52E D220E failed to be phosphorylated in the frzCDc frzEΔcheY background (abbreviated as “CDc EΔcheY ZD→E” in Fig. 4B), confirming that D52 and D220 are the only residues in FrzZ that accept phosphoryl groups via the Frz pathway (Fig. 4B and 4C). Frz signaling affected FrzZ phosphorylation, but not the expression or stability of FrzZ, since the total amount of FrzZ protein was similar in all tested mutants.
The phosphorylation experiments reported in Fig. 4 show a direct correspondence between the reversal phenotypes observed for Frz pathway mutants and the available cellular pool of phosphorylated FrzZ protein. These results indicate that the FrzZ response regulator serves as the main signaling output with respect to the initiation of the cell polarity switch.
FrzZ is recruited to the leading cell pole upon phosphorylation
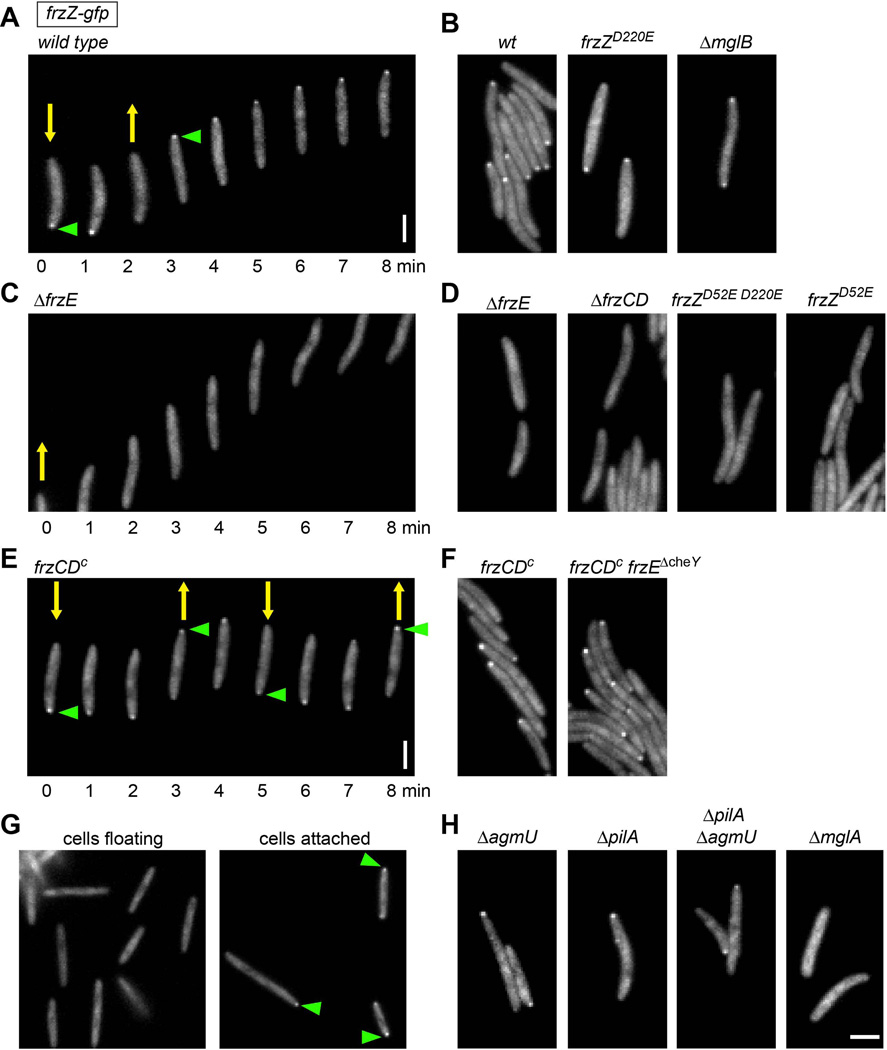
To address the localization of the FrzZ response regulator in live cells, we utilized a strain (DZ4833) that encodes green fluorescent protein (GFP) fused to the C-terminus of FrzZ. This strain expresses FrzZ-GFP as the only copy of FrzZ from its native promoter and shows wild type reversal frequency, motility, fruiting body formation, and phosphorylation (Fig. 5, suppl. Fig. S1 and data not shown). When analyzed by fluorescence microscopy, motile cells expressing FrzZ-GFP reproducibly displayed diffuse fluorescence over the length of cell bodies but also showed a distinct albeit faint cluster at the leading cell pole (Fig. 6A and 6B). Time-lapse analysis showed that the FrzZ-GFP cluster localized to the opposite cell pole during cell reversals (green arrowhead in Fig. 6A).
Figure 5.
A. M. xanthus strains in which FrzZ was replaced by fluorescently tagged FrzZ-GFP show phenotypes similar to strains expressing untagged FrzZ. Images show motility behavior on soft agar surface after 48 h incubation (top), as well as fruiting body formation under starvation conditions after 96 h (bottom). Shown is the colony edge of wild type and selected hypo-reversing (ΔfrzE), hyper-reversing (frzCDc) and non-motile (ΔmglA) strains that express FrzZ-GFP. Scale bars represent 2 mm. See supplementary Fig. S1 for phenotypic analysis of all fluorescent strains used in this study. B. Phosphorylation analysis of selected M. xanthus strains expressing FrzZ-GFP (57.3 kD). Cell extracts were analyzed by Phos-tag (50 µM) SDS-PAGE and Western Blot using anti-FrzZ antibody.
Figure 6.
Fluorescence microscopy of FrzZ-GFP in live M. xanthus cells reveals distinct and phosphorylation-dependent localization at the leading cell pole. Yellow arrows indicate the direction of cell movement. Green arrowheads indicate FrzZ-GFP accumulation at the leading cell pole. A, B. strains that show wild type levels of FrzZ phosphorylation. C, D. strains that shown no FrzZ phosphorylation. E, F. strains with elevated FrzZ phosphorylation levels. G. hyper-reversing strain frzZ-gfp frzCDc was grown in liquid culture fro 72 h and imaged after transferring cells to a glass slide. Left panel: floating cells; right panel: cells attached to surface. H. strains affected in social or gliding motility. Images were taken every 1 min for time-lapse acquisition (A, C, E). Scale bars represent 2 µm.
To test the possibility that the polar clusters of FrzZ-GFP are dependent on phosphorylation by the Frz chemosensory pathway, we introduced the frzZ-gfp fusion into mutants that affect reversal frequency, FrzZ phosphorylation, and motility. In the hypo-reversing frzZ-gfp ΔfrzE and frzZ-gfp ΔfrzCD strains, which are defective in Frz signaling, we observed diffuse fluorescence, but never defined clusters at the cell poles (Fig. 6C and D). We also imaged the fluorescence of a strain expressing FrzZD52E D220E-GFP, which has both phosphorylation sites mutated. In this strain, FrzZ displayed diffused localization as observed in the ΔfrzE and ΔfrzCD strains (Fig. 6D). Cells expressing FrzZD52E-GFP, also failed to show polar localization (Fig. 6D). However, FrzZD52E-GFP at the cell pole might be below the detection limit, since we observed only a minuscule amount of FrzZ phosphorylated at D220 in our phosphorylation analysis (Fig. 2B). In contrast, polar clusters similar to those observed in wild type were present in the strain expressing FrzZD220E-GFP, which exhibits wild type motility and phosphorylation (Fig. 6B). The hyper-reversing frzZ-gfp frzCDc and frzZ-gfp frzCDc frzEΔcheY strains, which display constitutive Frz signaling, showed a prominent bright FrzZ cluster at the leading cell pole; these polar clusters also switched their position at each cell reversal (Fig. 6E and F).
Since the level of FrzZ-GFP expressed in our experiments was similar in all of these mutants (Fig. 5B and data not shown), changes in fluorescence localization were not due to changes in FrzZ expression, but rather in its phosphorylation. Our observations in different Frz signaling mutants indicate that phosphorylation is required to recruit FrzZ to the leading cell pole.
The phosphorylation analysis indicated that FrzZ phosphorylation is low or non-existent when cells are grown in liquid culture (Fig. 2A). Indeed, when we imaged FrzZ-GFP fluorescence in floating cells immediately after they were transferred from liquid culture to a glass slide, we rarely observed polar clusters (< 5% of cells), even in hyper-reversing mutants (frzZ-gfp frzCDc, Fig. 6G, left panel). However, when cells were allowed to attach to the surface, polar FrzZ-GFP clusters were detected in most cells within minutes (>50% of cells, Fig. 6G, right panel). FrzZ-GFP clusters were also observed in individual cells attached to the glass surface where cells were unable to move. These observations imply that cells require a surface rather than cell-cell contact to initiate Frz signaling.
We also imaged FrzZ-GFP localization in mutants defective in social motility (ΔpilA), gliding motility (ΔagmU) and both motility systems (ΔagmU ΔpilA). All of these motility mutants showed polar fluorescence clusters (Fig. 6H), indicating that recruitment of phospho-FrzZ to the leading cell pole is indeed independent of cell movement. In contrast, non-motile cells that lack MglA only showed diffuse distribution of FrzZ-GFP (Fig. 6H), although FrzZ (and FrzZ-GFP) is phosphorylated to wild type levels in a ΔmglA strain (Fig. 3 and 5B). This suggests that MglA, which also localizes to the leading pole in moving cells and effects the correct distribution of several other proteins (Leonardy et al., 2010, Zhang et al., 2010), is directly or indirectly required to recruit phospho-FrzZ to the cell pole. In contrast, FrzZ-GFP localized to the leading cell pole (not shown) or to both cell poles (Fig. 6B) in a ΔmglB mutant, which is in agreement with the previous observation that MglA shows unipolar or bipolar distribution in the absence of MglB (Zhang et al., 2010, Leonardy et al., 2010).
FrzZ-GFP localization follows the MglA/MglB polarity axis
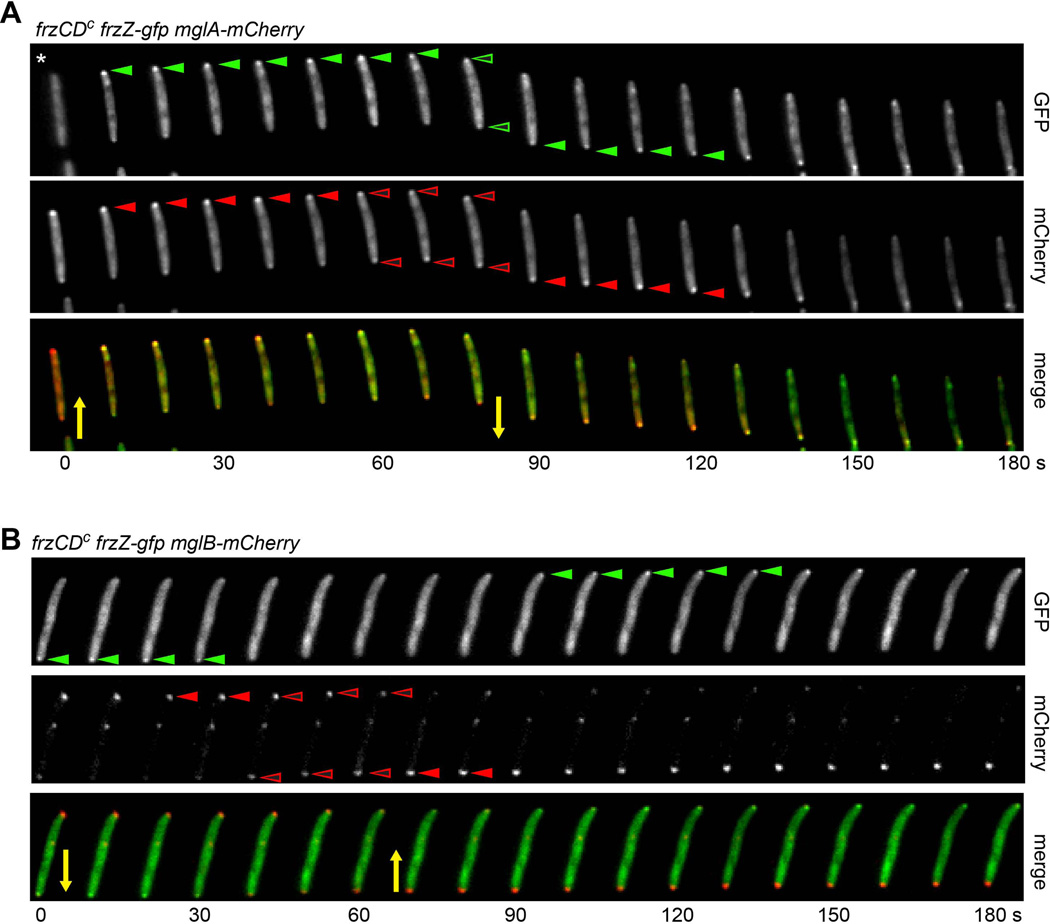
To further study the FrzZ-GFP localization pattern with respect to the cell polarity axis, we imaged FrzZ-GFP in hyper-reversing strains that simultaneously express MglA-mCherry or MglB-mCherry, respectively (Fig. 7A and B). We followed GFP and mCherry fluorescence in moving cells during reversals at 10 sec intervals. To observe FrzZ and MglA localizations simultaneously, the MglA-mCherry fusion was expressed in a frzZ-gfp frzCDc hyper-reversing mutant (strain DZ4847) as an additional copy of MglA from a vanillate-inducible promoter (Iniesta et al., 2012). Experiments were performed in the presence of 50 µM vanillate. MglA had previously been shown to occupy the leading cell pole in moving cells (Leonardy et al., 2010, Zhang et al., 2010), and, as expected, we observed co-localization of FrzZ-GFP and MglA-mCherry at the leading cell pole in >90% of cells that displayed both fluorescent foci (closed green and red arrowheads in Fig. 7A). When cells reversed, MglA-mCherry started to accumulate at the opposite cell pole, after a transient bipolar distribution lasting about 30 s (open red arrowheads in Fig. 7A). FrzZ-GFP changed its localization to the opposite cell pole simultaneously with MglA-Cherry, but a bipolar distribution at cell reversal was less pronounced and appeared to be delayed with respect to MglA-mCherry (open green arrowheads).
Figure 7. FrzZ-GFP localization follows the MglA/MglB polarity axis.
A. FrzZ and MglA co-localize at the leading cell pole and switch to the opposite cell pole at cell reversals. Time-lapse analysis of a reversing cell that expresses simultaneously FrzZ-GFP (green) and MglA-mCherry (red). Both proteins accumulate at the leading cell pole (closed arrowheads), and undergo a transient symmetric distribution at cell reversal (open arrowheads), which is led by MglA. Expression of MglA-mCherry was induced by addition of 50 µM vanillate. Note that in the first frame of the GFP channel (asterisk) the cell is out of focus. B. FrzZ and MglB switch to the opposite cell pole at cell reversals. Time-lapse analysis of a reversing cell that expresses simultaneously FrzZ-GFP (green) and MglB-mCherry (red). FrzZ-GFP accumulates at the leading cell pole (green arrowheads). MglB is asymmetrically distributed with a larger cluster at the lagging cell pole (red arrowheads), and shows a transient symmetric distribution at cell reversals (open red arrowheads). Yellow arrows indicate the direction of cell movement. Images were taken every 10 s.
MglB has been shown previously to display asymmetric bipolar localization, with a large cluster at the lagging cell pole and a smaller cluster at the leading cell pole (Fig. 7A) (Leonardy et al., 2010, Zhang et al., 2010). When cells reversed, the distribution of MglB clusters became transiently symmetric for about 30 s (open red arrowheads), before re-establishing an asymmetric distribution (red arrowheads) with inverted polarity (Fig. 7A). After about 60 s FrzZ-GFP visibly re-assembled at the new leading cell pole (green arrowheads), while MglB was now largely clustered at the lagging cell pole (red arrowheads, Fig. 7A). These observations suggest that FrzZ-GFP localization is tightly coordinated with the MglA/MglB cell polarity switch. In contrast, previous work has shown that the asymmetric polar localization of MglA or MglB do not depend on a functional Frz pathway (Zhang et al., 2010, Leonardy et al., 2010).
Discussion
Social behavior patterns in bacteria require coordinated cell motility, which in the gliding myxobacteria involves the control of cell reversals. The frequency of cell reversals in M. xanthus is regulated by a dedicated chemosensory two-component system, the Frz pathway. In this study, we addressed the function of the response regulator FrzZ, which serves as an output of the Frz signaling pathway and likely conveys the signal to set the reversal frequency to the polarity module that initiates cell reversals.
Previous in vitro experiments demonstrated that signal flow through the Frz pathway is represented by phosphotransfer from the CheA-like kinase FrzE to the response regulator FrzZ, but it is unknown how FrzZ phosphorylation impacts downstream regulator components and cell reversal frequency. Here, we investigated the relation between FrzZ phosphorylation and reversal frequency in vivo by tracking the phosphorylated and unphosphorylated form of the protein in different mutant backgrounds, and under different growth conditions. Our experiments indicate the following:
We found that FrzZ is phosphorylated when cells are grown on agar plates, although the level of phospho-FrzZ present in extracts is low, only about 3%, probably due to instability and turnover. In contrast, phosphorylation was not detected in extracts from liquid culture, even though the FrzCD receptor is reversibly methylated/demethylated in response to nutrients/repellents in liquid broth (McBride et al., 1992, McCleary et al., 1990). Indeed, cells phosphorylate FrzZ within 10 min of being spotted on plates (Fig. 2A). Phosphorylation of FrzZ probably does not require cell-cell contact per se since isolated cells that were well separated on a glass slide showed phosphorylation-dependent polar localization (Fig. 6G). This apparent requirement for a solid surface for Frz signaling is consistent with the requirement for a solid surface for motility, as M. xanthus is non-motile in a liquid medium. It is not known how cells detect and monitor their contact with surfaces and how this information is relayed to the Frz pathway, as non-motile mutants show normal FrzZ phosphorylation and polar localization (Fig. 6H). We detected wild type levels of phospho-FrzZ in cells capable of only social or gliding motility, as well as in non-motile mutants (Fig. 3). Moreover, we did not detect significant differences when wild type cells were grown on soft or hard agar surfaces, or on a membrane where cells cannot move at all (data not shown). FrzZ phosphorylation also appears to be independent of cell polarity, since wild type levels were observed in ΔmglA (Fig. 3A) and ΔmglB mutants (data not shown).
-
Our results indicate that reversal frequency is directly linked to the amount of phosphorylated FrzZ that is present in the cell, with a higher pool of phospho-FrzZ resulting in a higher reversal frequency (Fig. 4). In wild type cells, a phospho-FrzZ pool of approximately 3% of total FrzZ sets the reversal period to 8–9 min. The amount of phospho-FrzZ is probably kept at an equilibrium level by phospho-transfer input from the kinase FrzE, and by dephosphorylation from exposure to cellular phosphatases and/or by chemical instability. We estimate the half-life of phospho-FrzZ in the presence of phosphatase inhibitors to be about 13 min at ~2°C, and to be similar in wild type and hyper-signaling mutants (Fig. 2C and D). In hyper-reversing mutants, the phospho-FrzZ level increases to 20–30% of total FrzZ, at which level the reversal period is ~2 min (Bustamante et al., 2004, Li et al., 2005). In the absence of phospho-FrzZ, as observed in ΔfrzCD, ΔfrzE, ΔfrzZ or frzZD52E D220E mutant strains, reversals are very infrequent, usually less than once per hour (Bustamante et al., 2004, Inclan et al., 2007).
The FrzZ protein is small, consisting of only two weakly homologous receiver domains joined by a linker (Inclan et al., 2007, Trudeau et al., 1996). Phosphorylation experiments in vitro showed that FrzZ can accept a phosphoryl group from FrzE at two aspartate residues, D52 and D220, one in each of the two receiver domains (Inclan et al., 2007). Our analysis of the D52E and D220E mutants suggests a differential use of these phosphorylation sites in vivo (Fig. 2B). Specifically, we detected wild type levels of phospho-FrzZ when D220 was mutated, but only very little phospho-FrzZ in a D52E mutant. Moreover, the phosphorylation pattern in wild type and several hyper-reversing mutants showed a stronger signal for phospho-D52 than for phospho-D220. This indicates that the D52 site is preferentially targeted for phosphorylation in vivo. However, we cannot rule out that both sites are phosphorylated at the same rate, but phospho-D220 degrades at a higher rate and escapes detection by the methods used here.
Perhaps the most significant finding from this study was that FrzZ is localized diffusely in the cytoplasm but is recruited to the leading cell pole upon phosphorylation (Fig. 6). We constructed FrzZ-GFP fusions in wild type and Frz pathway mutants of M. xanthus to follow the localization of FrzZ and phospho-FrzZ in moving cells by fluorescence microscopy. Part of the cytoplasmic fluorescence we observed might be the result of protein cleavage, since a small amount of free GFP was detected in Western blots using anti-GFP antibody (data not shown). However, the accumulation of FrzZ-GFP at the leading cell pole was unexpected and striking: this localization was strictly dependent on FrzZ being phosphorylated by the Frz pathway, since non-phosphorylated FrzZ showed dispersed localization in several Frz mutants. We suggest that this localization pattern represents the activity of the Frz chemosensory system in modulating cell polarity coordinators like MglA/MglB or RomR in order to set a certain cell reversal frequency. Indeed, the ΔmglA mutant that produces normal levels of phospho-FrzZ, was unable to target phospho-FrzZ to the leading cell pole, suggesting that there may be a direct interaction between MglA and FrzZ. Moreover, MglA and FrzZ colocalize at the cell pole (Fig. 7A). However, MglA is a major determinant of cell polarity and has been shown to affect the localization of a number of downstream proteins that could equally serve as a target for phospho-FrzZ recruitment to the leading cell pole.
FrzZ-GFP switches from the old leading cell pole to the new one, along with MglA, during cell reversals (Fig. 7), and is maintained at that pole until the next reversal. We have no evidence for a gradual accumulation of FrzZ at the leading cell pole, although we cannot rule out this possibility. Our observations are consistent with phospho-FrzZ regulating the retention times of cell polarity components at the leading cell pole, thereby setting the reversal frequency period (Fig. 8). However, cell polarity is based on carefully balanced interactions and biochemical activities of at least three proteins (MglA, MglB and RomR) in different stoichiometries and at different sites in the cell (Leonardy et al., 2010, Zhang et al., 2010, Keilberg et al., 2012, Zhang et al., 2012b). The detailed mechanism by which phospho-FrzZ impacts cell polarity remains to be investigated. For example, phospho-FrzZ might interact with MglA directly, and alter its GTPase activity. Alternatively, sufficient phospho-FrzZ could recruit the GAP protein MglB (or another unidentified GAP protein) to the leading pole, which would indirectly result in MglA displacement. Another possibility that has been suggested is that phosphotransfer occurs from FrzZ to the response regulator RomR (Keilberg et al., 2012, Zhang et al., 2012b), which might act between the Frz pathway and the MglA/B polarity axis and serves as a polar determinant for MglA localization. An adaptor protein would most likely be required to mediate phosphotransfer between aspartate residues in the response regulators (Keilberg et al., 2012).
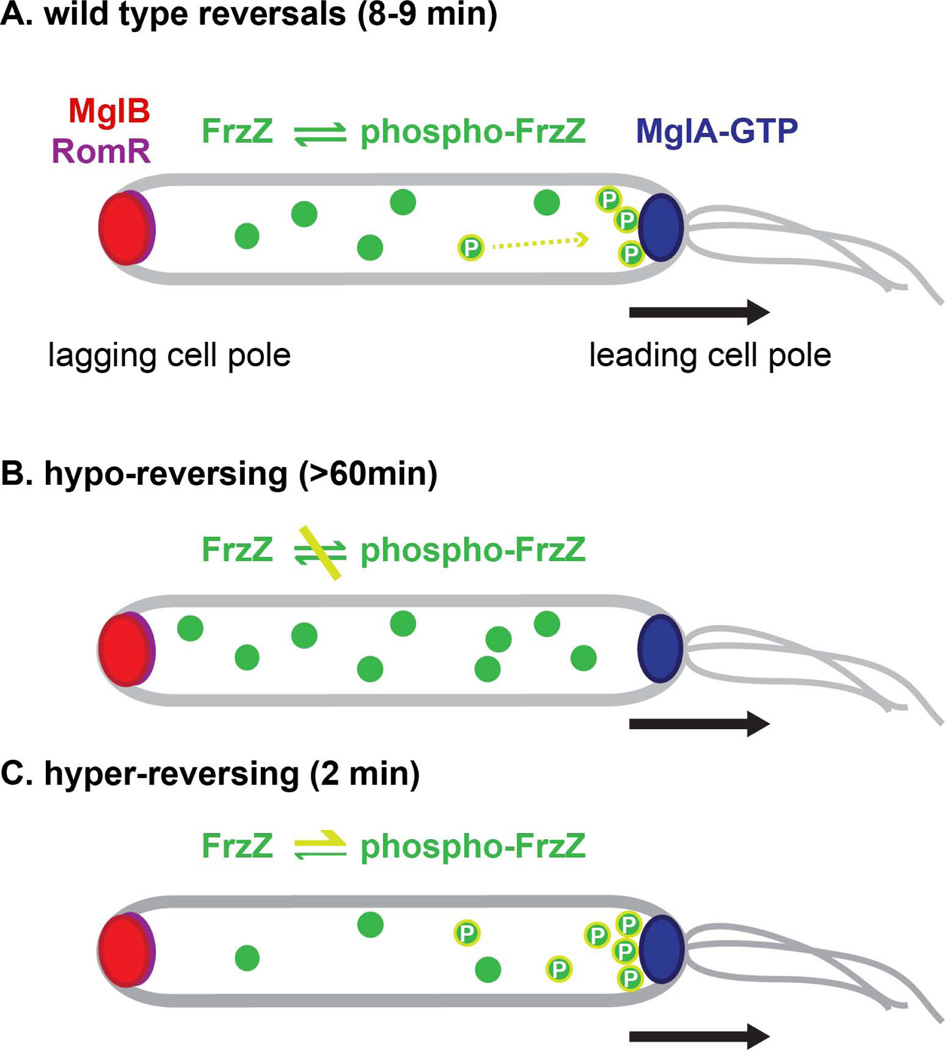
Figure 8. Model for the regulation of cell reversal frequency in M. xanthus.
Cell polarity is established by the dynamic interplay of the RomR, MglB and MglA proteins, and is based on in their asymmetric distribution to the lagging and leading cell pole. According to this model, cell reversals are initiated by inversion of cell polarity and the relocation of MglA, and MglB/RomR to opposite cell poles. (A) In wild type cells, FrzZ becomes phosphorylated via Frz pathway components and is recruited to the leading cell pole. Under these conditions, the retention time of MglA at the leading cell pole is limited to 8–9 min after which cells reverse. (B) Hypo-reversing mutants reverse very infrequently (less than once per hour). These reversals occur in the absence of a functional Frz pathway and have an unknown etiology. (C) Hyper-reversals are the result of elevated phospho-FrzZ levels. Under these conditions, MglA is retained at the leading cell pole for about 2 min before cells reverse. The mechanism by which FrzZ modulates the balance of RomR, MglB and MglA proteins to regulate the retention time of the cell polarity axis is unknown. A black arrow marks the direction of cell movement.
The activation-dependent localization pattern of FrzZ in M. xanthus is an example for the functional impact of the spatio-temporal distribution of regulatory proteins in the bacterial cell. FrzZ in M. xanthus relates in several aspects to the PleD response regulator in Caulobacter crescentus, in that both proteins exhibit phosphorylation dependent localization to a cell pole. PleD is recruited to the stalked pole upon phosphorylation to regulate cell cycle progression by the locally restricted production of cyclic-di-GMP (Paul et al., 2004). Interestingly, it was shown that phosphorylation of PleD promotes dimerization of the protein, which in turn is required for polar localization and enhanced calatytic activity (Paul et al., 2007). A similar dimerization-dependent mechanism might be feasible for FrzZ recruitment, since FrzZ and PleD share their organization in two tandem receiver domains. The two receiver domains of PleD, like D52 and D220 in FrzZ, seem to be used differentially for phosphorylation as well, with D53 of PleD being the main phosphoryl acceptor that induces structural rearrangements with respect to the second receiver domain to support dimerization, and eventually catalytic activity (Aldridge et al., 2003, Chan et al., 2004, Paul et al., 2007). However, in contrast to the diguanylate cyclase PleD, M. xanthus FrzZ lacks any domains that might suggest additional biochemical functions.
FrzZ localization in M. xanthus is also in some ways analogous to the E. coli chemotaxis response regulator, CheY, which is also recruited to its site of action, the flagellar motor, upon phosphorylation by the Che chemosensory pathway (Welch et al., 1993). The interaction of E. coli CheY with the motor component FliM results in conformational changes that lead to a switch in flagellum rotation. However, further investigations are required to reveal the molecular mechanism that results in cell reversal in M. xanthus upon FrzZ phosphorylation, and how it might help to coordinate single cells in multi-cellular behaviors.
Experimental procedures
Bacterial strains and growth conditions
M. xanthus strains were grown in CYE medium (10 mM MOPS, pH 7.6, 1% Casitone, 0.5% yeast extract and 4 mM MgSO4; (Campos et al., 1978)) at 32°C and 200 rpm. 1.5% agar was added for the preparation of agar plates. Antibiotics were used in the following concentrations: ampicillin 100 µg ml−1, kanamycin 100 µg ml−1, tetracycline 12.5 µg ml−1. E. coli strain DH5α was used for plasmid construction, and cultured in Luria-Bertani medium at 37°C.
Strain construction
Plasmids, oligonucleotides and bacterial strains that were used during this study are listed in Table 1. For the construction of pEMC68 and pCK121, a fragment encompassing base pairs 267 to 869 of the frzZ gene was amplified from chromosomal DNA of wild type strain DZ2 and strain DZ4706, respectively, using primers EMC37 and EMC35. The coding sequence of green fluorescent protein (gfp) was amplified from pEGFP-C1 with primers EMC32 and EMC36. Both DNA fragments were fused by PCR using primers EMC32 and EMC37. The DNA fragment and vector pBJ114 were digested with restriction enzymes EcoRI and BamHI, and ligation, transformation to E. coli DH5α and identification of correct constructs were performed using standard methods. M. xanthus wild type and different mutant strains were transformed with pEMC68 or pCK121 by electroporation. Transformants that incorporated the plasmid via a single recombination event at the frzZ locus were selected for their kanamycin resistance. Successful integration of frzZ-gfp was confirmed by Western blot, sequencing and microscopy.
Table 1.
| plasmids | relevant characteristics | Reference |
|---|---|---|
| pBJ114 | galS kanR, for the construction of insertion and deletion mutations in M. xanthus | (Julien et al., 2000) |
| pSWU30 | tetR, integrates at Mx8 phage attachment site attB | (Wu & Kaiser, 1995) |
| pMR3562 | tetR, integrates at 1.38 kb site into the M. xanthus chromosome | (Iniesta et al., 2012) |
| pEGFP-C1 | contains coding sequence for green fluorescent protein | Clontech |
| pSWU19mglBY | mglB-yfp; kanR, integrates at Mx8 attachment site | (Zhang et al., 2010) |
| pEMC68 | pBJ114 with frzZ bp 267-end to fused to GFP | this study |
| pCK121 | pBJ114 with frzZD52E D220E bp 267- end fused to GFP | this study |
| pCK126 | pSWU30 mglB-mCherry | this study |
| pCK133 | pMR3562 mglA-mCherry | this study |
| oligonucleotides | ||
| EMC32 | 5’-ATGGTGAGCAAGGGCGAGGAG-3’ | |
| EMC35 | 5’-CCTCGCCCTTGCTCACCATCTCGTTACCGGTGGGCATC-3’ | |
| EMC36 | 5’-CGGGATCCCTACTTGTACAGCTCGTCCATG-3’ | |
| EMC37 | 5’-CGGAATTCCGGAAAGACGAAGGCGTCG-3’ | |
| C221 | 5’-GCAAGCTTTTACTTGTACAGCTCGTCCATG-3’ | |
| C243 | 5‘-GCCTCGAGCTCTGGCGCGCCGGGCATGGTGAGCAAGGGCGAG-3‘ | |
| C259 | 5’-GCGAATTCGTGGGAAGGGCTCTTTCAG-3’ | |
| C272 | 5’-GCCATATG TCCTTCATCAATTACTCATC-3’ | |
| C273 | 5’-GCGAATTCTCACCACCCTTCTTGAGCTCGG-3’ | |
| C269 | GCGAATTCCTCTGGCGCGCCGGGCATGGTGAGCAAGGGCGAG-3’ | |
| C270 | 5’-GCGCTAGCTTACTTGTACAGCTCGTCCATG-3’ | |
| E. coli DH5α | host for plasmid construction | Invitrogen |
| M. xanthus strains | ||
| DZ2 | wild type | (Campos et al., 1978) |
| DZ4484 | ΔfrzZ | (Bustamante et al., 2004) |
| DZ4706 | frzZD52E D220E | (Inclan et al., 2007) |
| DZ4704 | frzZD52E | (Inclan et al., 2007) |
| DZ4705 | frzZD220E | (Inclan et al., 2007) |
| DZ4481 | ΔfrzE | (Bustamante et al., 2004) |
| DZ4480 | ΔfrzCD | (Bustamante et al., 2004) |
| DZ4487 | frzCDc | (Bustamante et al., 2004) |
| DZ4547 | frzEΔcheY | (Li et al., 2005) |
| DZ4629 | frzEH49A | (Inclan et al., 2008) |
| DZ4558 | frzCDc frzEΔcheY | laboratory collection |
| DZ4469 | ΔpilA | (Vlamakis et al., 2004) |
| DZ4846 | ΔagmU | Beiyan Nan |
| DZ4771 | ΔpilAΔagmU | (Nan et al., 2010) |
| TM12 | ΔmglA | (Mauriello et al., 2010) |
| DZ4832 | frzCDc frzEΔcheYΔfrzZD52E D220E | this study |
| DZ4833 | frzZ-gfp kanR | this study |
| DZ4834 | frzCDc frzZ-gfp kanR | this study |
| DZ4835 | ΔfrzE frzZ-gfp kanR | this study |
| DZ4836 | ΔfrzCD frzZ-gfp kanR | this study |
| DZ4837 | ΔmglA frzZ-gfp kanR | this study |
| DZ4838 | ΔmglB frzZ-gfp kanR | this study |
| DZ4839 | frzZD52E-gfp kanR | this study |
| DZ4840 | frzZD220E-gfp kanR | this study |
| DZ4841 | frzZD52E D220E-gfp kanR | this study |
| DZ4842 | ΔagmU frzZ-gfp kanR | this study |
| DZ4843 | ΔpilA frzZ-gfp kanR | this study |
| DZ4844 | ΔpilA ΔagmU frzZ-gfp kanR | this study |
| DZ4845 | frzZ-gfp attB::mglB-mCherry kanR tetR | this study |
| DZ4847 | frzZ-gfp 1.38kB::mglA-mCherry kanR tetR | this study |
To construct a strain that simultaneously expresses FrzZ-GFP and MglA-mCherry, plasmid pCK133 was introduced to strain DZ4833 and transformants were selected for tetracycline and kanamycin resistance. pCK133 is a derivate of pMR3562 (tetR), which integrates at a 1.38 kB site in the M. xanthus chromosome and supports expression from a vanillate-inducible promoter (Iniesta et al., 2012). First, the coding sequence of mCherry was introduced to the EcoRI and NheI sites of pMR3562 to replace EGFP (oligonucleotides C269 and C270). The mglA gene was subsequently cloned in frame with mCherry to the NdeI and EcoRI sites (C272 and C273) to create pCK133. Plasmid pMR3562 was a kind gift of Montserrat Elías-Arnanz (Universidad de Murcia).
For colocalization analysis of FrzZ-GFP and MglB-mCherry, YFP was first exchanged for mCherry in pSWU19mglBY (Zhang et al., 2010), using primers C243 and C221. mglB-mCherry, including the promoter sequence, was subsequently amplified using primers C221 and C259. The resulting fragment was ligated into pSWU30 (tetR) using EcoRI and HindIII restriction sites, and plasmid pCK126 was used to transform DZ4833. The resulting strain DZ4845 carries mglB-mCherry as an additional copy of the gene at the phage attachment site attB, where it is expressed from its native promoter (Zhang et al., 2010). Plasmids pSWU30 and pSWU19mglBY were kindly provided by Tâm Mignot (CNRS Marseille).
Determination of motility and developmental phenotypes
M. xanthus strains were cultured to exponential phase in liquid CYE medium, harvested by centrifugation at 8000 × g for 10 min, washed once with MMC buffer (10 mM MOPS pH 7.6, 4 mM MgSO4, 2 mM CaCl2) and diluted to 4 × 109 cells ml−1 in MMC buffer. 10 µl of the suspension was spotted to CYE plates containing 1.5% (hard) or 0.5% (soft) agar to monitor motility behavior. To determine fruiting body formation upon starvation, 10 µl cell suspension was spotted to CF agar plates (10 mM MOPS pH 7.6, 8 mM MgSO4, 1 mM KH2PO4, 0.015% Casitone, 0.2% sodium citrate, 0.1% sodium pyruvate, 0.02% (NH4)2SO4, 1.5% agar; (Hagen et al., 1978). Agar plates were incubated at 32°C in the dark and phenotypes were documented after 48–96 h using a Nikon SMZ500 stereo microscope.
Phos-tag experiments
Unless otherwise indicated, M. xanthus strains were cultured to exponential phase in liquid CYE medium, harvested, and concentrated to 4 × 109 cells ml−1. 10 µl of the cell suspension were spotted on CYE agar plates and incubated for 48 h. Cells were scraped off the plate and resuspended in 200 µl ice-cold lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM MgCl2, 0.5 mM PMSF, 0.1% Triton X-100, 15 µg ml−1 DNase, 1× phosphatase inhibitor cocktail (Phosstop, Roche Diagnostics)). Cell debris was removed by centrifugation at 13,000 × g, 4°C for 1 min, and the protein concentration of the cleared lysate was determined by the Bradford method. Samples were prepared in reducing SDS-loading buffer, and an equivalent of 5–10 µg protein per lane was loaded to a 7.5% SDS-polyacrylamide gel containing MnCl2 and Phos-tag acrylamide (Wäko Chemicals, Japan). As indicated in the figures, gels either contained 25 µM Phos-tag and 50 µM MnCl2, or the double amount of 50 µM Phos-tag and 100 µM MnCl2.
Electrophoresis was performed at 4°C in Tris-glycine SDS-PAGE running buffer at 100–150 V for 1.5 h. Due to the limited half-life of phosphorylated FrzZ, experiments were performed on ice or at 2–4°C without interruption, and protein samples were not heated prior to loading to the gel. After electrophoresis, gels were equilibrated for 10 min in Western transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) with 1 mM EDTA to remove MnCl2, and another 10 min in Western transfer buffer. Proteins were transferred to nitrocellulose membrane, and Western immunoblots were performed using standard methods. Polyclonal, purified anti-FrzZ antibody was diluted 1:10,000 and secondary anti-rabbit antibody (HRP-conjugate, Biorad) was used 1:5,000. For experiments that required the quantification of Western blot signals, membranes were probed with fluorescent secondary antibody in 1:5,000 dilution (IRDye 800CW, Li-Cor), and analyzed using a Odyssey infrared fluorescence scanner and Image Studio software (Li-Cor).
Microscopy
Exponentially growing M. xanthus cells were diluted to 4 × 108 cells ml−1, and 5 µl of the suspension were spotted on thin agarose pads (1.5% agarose in CF salts) on slides, and covered with a cover glass. Slides were incubated for 30–60 min at 32°C in the dark prior to image acquisition. For fluorescence microscopy, image acquisition was performed on a DV Elite microscope setup (Applied Precision) equipped with a CCD camera (CoolSnap HQ, Photometrics), and using solid state illumination at 461/489 nm (GFP) and 529/556 nm (mCherry). Time-lapses were performed for 5–10 min at 10–60 s intervals. Cell viability was critical to observe FrzZ-GFP polar clusters. To minimize phototoxocity and photobleaching of the faint FrzZ-GFP signal a minimal exposure time of 0.08 s at 50% light transmission was chosen, and viability was judged from intact cell movement. All statements regarding the localization of fluorescent protein fusions are based on three or more independent microscopy experiments where > 200 viable cells were observed.
Supplementary Material
Acknowledgements
We want to thank Eva M. Campodonico, who shared valuable experimental experience and unpublished data, and constructed strains DZ4832 and DZ4833. We further thank Beiyan Nan and James E. Berleman for helpful discussion and comments on the manuscript, and Tâm Mignot and Montserrat Elías-Arnanz for the gift of plasmids. This work was supported by a fellowship by the Deutsche Forschungsgemeinschaft (DFG Ka 3361/1-1 to C.K.), and the National Institutes of Health (GM 020509 to D.R.Z).
Abbreviations
- GFP
green fluorescent protein
- CYE
Casitone/yeast extract growth medium.
Phosphorylation (-PO42−) of amino acids aspartate (D) or histidine (H) is abbreviated ‘phospho-’ in the text and represented by a circled letter ‘P’ in figures.
References
- Acuna G, Shi W, Trudeau K, Zusman DR. The 'CheA' and 'CheY' domains of Myxococcus xanthus FrzE function independently in vitro as an autokinase and a phosphate acceptor, respectively. FEBS Lett. 1995;358:31–33. doi: 10.1016/0014-5793(94)01389-i. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart BD, Zusman DR. Cloning and complementation analysis of the "Frizzy" genes of Myxococcus xanthus. Mol Gen Genet. 1985a;198:243–254. doi: 10.1007/BF00383002. [DOI] [PubMed] [Google Scholar]
- Blackhart BD, Zusman DR. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985b;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Hone A, Maier B, Hoppert M, Sogaard-Andersen L. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol. 2009;74:691–706. doi: 10.1111/j.1365-2958.2009.06891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante VH, Martinez-Flores I, Vlamakis HC, Zusman DR. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol. 2004;53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- Campos JM, Geisselsoder J, Zusman DR. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen DC, Bretscher AP, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hartzell P, Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J Bacteriol. 1991;173:7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- Hosokawa T, Saito T, Asada A, Fukunaga K, Hisanaga S. Quantitative measurement of in vivo phosphorylation states of Cdk5 activator p35 by Phos-tag SDS-PAGE. Mol Cell Proteomics. 2010;9:1133–1143. doi: 10.1074/mcp.M900578-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inclan YF, Laurent S, Zusman DR. The receiver domain of FrzE, a CheA-CheY fusion protein, regulates the CheA histidine kinase activity and downstream signalling to the A- and S-motility systems of Myxococcus xanthus. Mol Microbiol. 2008;68:1328–1339. doi: 10.1111/j.1365-2958.2008.06238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inclan YF, Vlamakis HC, Zusman DR. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol Microbiol. 2007;65:90–102. doi: 10.1111/j.1365-2958.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, Garcia-Heras F, Abellon-Ruiz J, Gallego-Garcia A, Elias-Arnanz M. Two systems for conditional gene expression in Myxococcus xanthus inducible by isopropyl-beta-D-thiogalactopyranoside or vanillate. J Bacteriol. 2012;194:5875–5885. doi: 10.1128/JB.01110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien B, Kaiser AD, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilberg D, Wuichet K, Drescher F, Sogaard-Andersen L. A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in Myxococcus xanthus. PLoS Genet. 2012;8:e1002951. doi: 10.1371/journal.pgen.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4:1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 2010;29:2276–2289. doi: 10.1038/emboj.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bustamante VH, Lux R, Zusman D, Shi W. Divergent regulatory pathways control A and S motility in Myxococcus xanthus through FrzE, a CheA-CheY fusion protein. J Bacteriol. 2005;187:1716–1723. doi: 10.1128/JB.187.5.1716-1723.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci U S A. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Kohler T, Zusman DR. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary WR, McBride MJ, Zusman DR. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Merlie JP, Jr, Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci U S A. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Mauriello EM, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak DT, Wei X, Wall D. Myxobacterial tools for social interactions. Res Microbiol. 2012;163:579–591. doi: 10.1016/j.resmic.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf BE. Summary of useful methods for two-component system research. Curr Opin Microbiol. 2010;13:246–252. doi: 10.1016/j.mib.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Scott AE, Simon E, Park SK, Andrews P, Zusman DR. Site-specific receptor methylation of FrzCD in Myxococcus xanthus is controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase. Mol Microbiol. 2008;69:724–735. doi: 10.1111/j.1365-2958.2008.06323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zusman DR. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci U S A. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci U S A. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau KG, Ward MJ, Zusman DR. Identification and characterization of FrzZ, a novel response regulator necessary for swarming and fruiting-body formation in Myxococcus xanthus. Mol Microbiol. 1996;20:645–655. doi: 10.1046/j.1365-2958.1996.5521075.x. [DOI] [PubMed] [Google Scholar]
- Vlamakis HC, Kirby JR, Zusman DR. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol Microbiol. 2004;52:1799–1811. doi: 10.1111/j.1365-2958.2004.04098.x. [DOI] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SS, Kaiser D. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ducret A, Shaevitz J, Mignot T. From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol Rev. 2012a;36:149–164. doi: 10.1111/j.1574-6976.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Franco M, Ducret A, Mignot T. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 2010;8:e1000430. doi: 10.1371/journal.pbio.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guzzo M, Ducret A, Li YZ, Mignot T. A Dynamic Response Regulator Protein Modulates G-Protein-Dependent Polarity in the Bacterium Myxococcus xanthus. PLoS Genet. 2012b;8:e1002872. doi: 10.1371/journal.pgen.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR. "Frizzy" mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982;150:1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.