Abstract
Probabilities of numbers of ligands proximal to an ion lead to simple, general formulae for the free energy of ion selectivity between different media. That free energy does not depend on the definition of an inner shell for ligand-counting, but other quantities of mechanistic interest do. If analysis is restricted to a specific coordination number, then two distinct probabilities are required to obtain the free energy in addition. The normalizations of those distributions produce partition function formulae for the free energy. Quasi-chemical theory introduces concepts of chemical equilibrium, then seeks the probability that is simplest to estimate, that of the most probable coordination number. Quasi-chemical theory establishes the utility of distributions of ligand-number, and sharpens our understanding of quasi-chemical calculations based on electronic structure methods. This development identifies contributions with clear physical interpretations, and shows that evaluation of those contributions can establish a mechanistic understanding of the selectivity in ion channels.
1. Introduction
Recent modeling and simulation aimed at understanding the K+/Na+ selectivity of potassium ion channels have focused on the statistical mechanics of ion solvation that leads to selectivity. The interactions of ions with their surrounding media are typically strong and complicated enough to present serious challenges to molecular theory and interpretation. Even the simplest cases – Li+ in water, for example – are not simple; a primitive aspect of hydration in that case, the average coordination number of Li+(aq), has yet to be entirely settled [1,2]. Excluded-volume interactions and thermal fluctuations contribute to the free energy of ion solvation, counterbalancing strong associative binding interactions, as well as weaker, longer-ranged interactions of electrostatic and van der Waals origin. Where one such effect does not overwhelm all others, comparisons among ions are not easy to predict. Rationalizations of measurements or exhaustive computations can be superficially appealing, but uncompelling. The accumulated results for potassium ion channels confirm the subtleties of simple rationalizations of these free energy comparisons [3–7].
Here we outline the modern statistical mechanical theory that is applicable to metal ions in solutions, quasi-chemical theory. A springboard for this discussion will be the current challenge of explaining K+/Na+ selectivity in the K-channel proteins [3–11]. Molecular simulation [3] has highlighted the flexibility of the structure at the heart of the selectivity questions, the selectivity filter (Fig. 1). The theory surveyed here, being statistical mechanics, is well suited for analysis of the statistical and structural variability inherent to this selectivity filter. Our discussion will not be exhaustive, but our goals will be to establish the utility of distributions of the number of ligands coordinating an ion, to sharpen our understanding of primitive quasi-chemical models that use electronic structure computational tools, and to clarify the implications of reduced models that have become popular for K+/Na+ selectivity in the K-channel proteins. Thus, our objective is to build a concise, physically transparent theoretical framework to support a unified interpretation of recent computations on the selectivity of K-channels.
Fig. 1.

Configuration from simulation [7] of potassium ion channels [12]. The inset shows a potassium ion (purple) in the S2 site, and eight (8) peptide backbone carbonyl groups that can ligate the ion.
1.1. Historical overview
Despite the strength and complexity of ion–water interactions noted above, there has been substantial progress over the past decade in designing molecular theory to treat chemically complicated ion–solvent interactions [1,13–20]. This theory has been called ‘quasi-chemical’ to emphasize the commonality with historical theories of phase transitions [20]. Kirkwood referred to those historical approaches as ‘the method of local configurations,’ [21] and noted that the method of local configurations ‘and the quasi-chemical method led to closed expressions for the partition function, but both are based upon approximations, the nature of which is not entirely clear.’ An enormous literature has grown up for analysis of quasi-chemical approximations to the statistical thermodynamics of lattice gases, and by now a clear understanding of quasi-chemical approximations is available in that context. The quasi-chemical theory discussed here avoids lattice-gas models and in doing so addresses additional complexities that deserve further clarification. One example is that electrostatic intermolecular interactions, long-ranged and strong beyond near-neighbor range, are an essential aspect of realistic models of aqueous solutions. Those interactions are not considered realistically in typical lattice-gas models of solutions.
The review of Friedman and Krishnan [22] also provides helpful perspective on the recent development of quasi-chemical theories. At the time of that review, the importance of near-neighbor water–ion configurations and chemical interactions was well recognized, as it is today. That recognition led to ‘hybrid’ or ‘chemical’ models, progenitors of the present molecular quasi-chemical theory, but a convincing formulation of the statistical mechanical problem remained elusive [22]: ‘unfortunately, it seems that there are important problems with calculations of this type which need to be overcome before they can provide a reliable interpretation of hydration phenomena…’ Since then, the theoretical formulation has become conclusive [14,19,20], and computations have matured to address particularly intractable steps. Indeed, direct numerical simulation has recently followed literally the ideas of quasi-chemical theory, and found them specifically useful [7,8,23,24]. Coincidentally, physical intuition about the selectivity of the KcsA potassium ion channel has returned to these models in the recent decade.
1.2. The inner shell and coordination-number distributions
The recent theoretical developments may be founded on the basic relation,
| (1) |
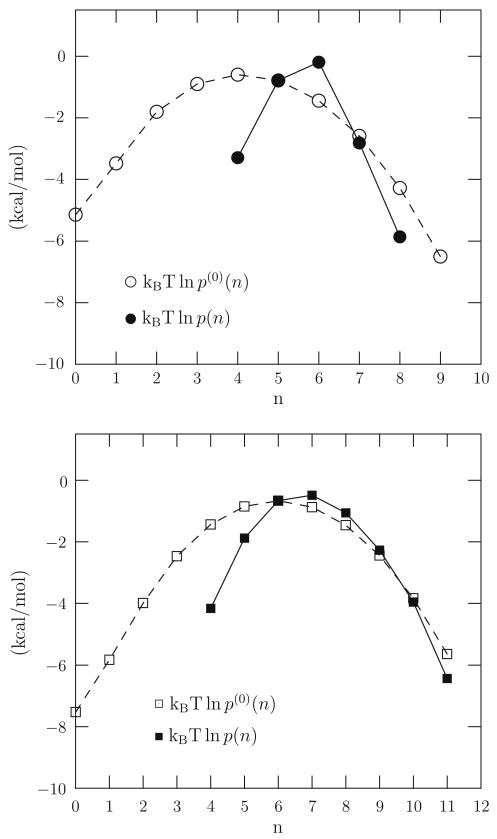
between traditional solution free energies and coordination-number distributions [27]. Here is the chemical potential in excess of the ideal gas result at the same density and temperature (T) of species X in solution. , defined below, is that free energy conditional on the specification that the number of ligands in an inner shell of a distinguished X be n, exactly. The inner shell need not be macroscopic, and in interesting cases it will be microscopic in size. The probability for observing n ligands in the inner shell is pX(n). The quantity is the probability that n ligands occupy the defined inner shell, but with solute-medium interactions turned-off. Alternatively, we can view as an intrinsic propensity of the ligands to occupy the inner-shell volume; this intrinsic propensity is then altered by the solute-ligand interactions to become pX(n). Fig. 2 gives examples of these distributions obtained by molecular dynamics simulation of aqueous solutions of Na+(aq) and K+(aq) with water molecules serving as the ligands.
Fig. 2.
Occupancy distributions obtained by molecular dynamics simulation for the cases of Na+(aq) (upper) and K+(aq) (lower). The calculations used the NAMD program [25] and the TIP3P [26] model of water. T = 298 K (Langevin thermostat) and the cubic simulation system contained 306 water molecules for the pure water simulation; the ion-water system had an additional ion. The total number density (counting water molecules and the ion, if present) is 33:33 nm−3. For Na+(aq) the inner shell radius was 3.1 Å, and for K+(aq) it was 3.5 Å. These are the radii of the first minimum in the ion-oxygen radial distribution in liquid water at infinite dilution.
Because of its simple, familiar structure, the relation Eq. (1) will be intuitive for many. Nevertheless, it was specifically written-out only recently and in the context of the present topic [27]. In addition, Eq. (1) should clarify arguments that have arisen in recent debates on the mechanism of K+/Na+ selectivity [3–7]. Because of the complexity of those arguments [28], a simple derivation of Eq. (1) is warranted, and we give such a derivation in the Appendix.
Access to the solution free energy
| (2) |
is a specific use for our fundamental Eq. (1). This point can be solidified by writing
| (3) |
This suggests the evaluation of the cross information entropy [29] between the two probabilities; the thermodynamic partial molar entropy should be obtained by the proper temperature derivative of . A numerical demonstration of Eq. (2) is given in Fig. 3.
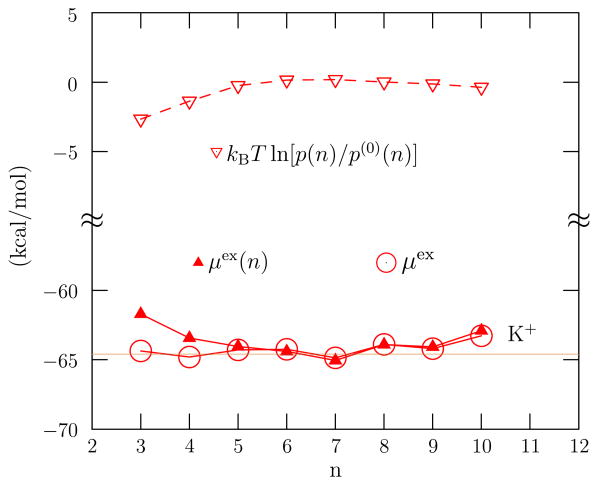
Fig. 3.
Estimates for the excess chemical potential, the open circles are the right-side of Eq. (2). The thin solid line is the result of direct numerical simulation. Notice that the variations of the upper results with n are of the same order-of-magnitude as the transfer free energies discussed below (Fig. 5), and that the circles have diminished variation with n, which follows from Eq. (2). Notice also the wide separation in energy range of these different pieces. Simulation results are from Ref. [27]; the SPC/E water model and the ion parameters of Ref. [30] were used. Coordination-number constraints were explicitly imposed within a Monte Carlo simulation.
2. Quasi-chemical theory
The n = 0 instance of Eq. (2) has been used to evaluate when quasi-chemical theory [1,13–15,20,19,31] has been viewed from the perspective of processing simulation data [32]. That choice is motivated by the points that: (a) in favorable cases n = 0 events can be directly observed, and thus kT ln p(n = 0) directly evaluated; (b) −kT ln p(0) (n = 0) has the physical interpretation of a packing contribution [33] so a non-trivial body of available theory, techniques, and results [34] can be exploited; (c) close contacts having been eliminated, is simpler than ; a Gaussian statistical model of the distribution of binding energies might suffice. As an example, recent studies [33,32,35] sought an inner-shell definition that permitted so that the free energy could be approximated by
| (4) |
The countervailing advantage of the partition function formulae of the Appendix is that they should be insensitive to infrequent coordination-number cases. If p(n = 0) is impractical to evaluate because it is small, then Eq. (16) might be satisfactory without the n = 0 contribution. Indeed, it is natural to consider all values of n for which data are available and Eq. (16) does that [7].
2.1. Chemical detail in quasi-chemical theories
Concepts of chemical equilibrium among the possible occupancy states (n) gives further perspective on the preceding results, a perspective suggested also by detailed derivations [20]. This additional perspective helps for treating metal ions in chemical detail in liquid water [1,16,18-20], or other bulk solution settings. Introduce the chemical equilibrium ratio defined by Knρn = pX(n)/pX(n = 0) where ρ is the bulk density of ligands [33]. The Kn so defined characterizes the equilibrium
| (5) |
where L denotes the ligand species [14,19,20]. Note that the combination Knρn is dimensionless, and this specification adopts the natural absolute standard state [20]. Of all the probabilities appearing in Eq. (3), the probability of the most probable coordination number n = n̄ is the easiest to estimate. In fact, pX(n̄) ≈ 1 is often a benign a priori estimate. With this idea in mind, write the n = 0 instance of Eq. (2)
| (6) |
The chemical equilibrium ratio of Eq. (6) can be expressed as the combination
| (7) |
of contributions naturally implied by Eq. (5). All quantities in this combination are well-defined computational targets [14,19,20]. is the chemical equilibrium ratio describing the reaction Eq (5) considered ideally, i.e., without interactions with a medium external to the combining molecules [20,36]. The rightmost term of Eq. (7) is
| (8) |
the excess chemical potential of the ion under the constraint that no inner-shell binding is possible, i.e., with pX(n = 0) = 1 in Eq. (2). On the basis of Eq. (8), we observe an exact cancellation of between Eqs. (6) and (7) producing
| (9) |
Since can be calculated without consideration of the solution medium, the computational methods of atomic and molecular physics can be brought to bear directly [37]. Provided the structures of isolated clusters do not violate the specification of n̄ with the inner-shell definition, it is irrelevant whether they are the same as probable solution structures. No approximation has been introduced as this stage.
The effects of the medium are present in the remaining contributions of Eq. (9). Considering , calculations would initially consider structures that are probable or easily available. It is natural and permitted to sample structures and then to treat the sampled structures as rigid [14,19,20]. Several or many structures might be considered, and the results then combined in the thermo-dynamically consistent fashion [19,20]. For example, such structures might be drawn from simulation calculations, and it is known [19,20] how to process free energy results for structures sampled in that way.
3. Present status
A take-it-or-leave-it model of solvation free energies is not the goal of the quasi-chemical development arriving at Eq. (9). Instead the goal is a statistical thermodynamic structure that identifies a natural role for chemical computation. The explicit factor of ρn̄ in Eq. (9) helps to clarify this distinction. This factor describes an effect of variation of the density of the solution medium, a necessity for statistical thermodynamic theory. This stands in contrast to computational models that treat addition of an ion X to a microcluster of solvent as in a chemical equilibrium
| (10) |
which is different from Eq. (5). To the extent that the model Eq. (10) does not treat variations of the solution density, it is incomplete as statistical thermodynamic theory. The possibility of continuous variation of the composition of mixtures is also a necessity for statistical theory. For example, the theory must permit investigation of water-methanol mixtures [38]. For mixtures, the solvent micro-cluster of Eq. (10) would presumably reflect the bulk solvent composition, and that is generally different than the composition of the inner shell of the ion.
Nevertheless, chemical models based intuitively on concepts such as Eq. (10) have a record of producing reasonable hydration free energies of ions [39–43]. The suggested conclusion is that chemical interactions are a big part of those free energies; getting those chemical interactions right, even modelistically, is an important step toward getting the free energies right. For those models and for the quasi-chemical theory, there has been much less investigation of partial molar volumes and entropies [14,20,42]. Therefore those properties are not expected to work out so well as do the free energies. Simple estimation of the partial molar volumes of alkali metals ions in water on the basis of these ideas [14,20] concludes that those properties require an assessment of packing contributions which were exactly cancelled in the theory above. Also the partial molar volumes directly involve the mean coordination number 〈n〉, which should be close to the most probable coordination number n̄ appearing above.
3.1. Some practical points
There are several pragmatic points to bear in mind in considering the calculations based on the theory Eq. (9). The first point is that calculations of the two contributions to should be physically consistent because that permits some cancellation of approximation errors between the individual terms [1,13–15,19,20,31]. Because of this balance, the approach Eq. (9) is typically insensitive to parameters of the computational procedures. A second point is that partial molar volumes of metal ions in water are often small, even negative. Sophistication in evaluating volumetric effects is often unnecessary [14,20]. A third point is that for metal ions in water, the structures to be considered are often not particularly novel. The sampling of structures often need not be exhaustive. Finally, kT ln pX(n̄) can be estimated from routine simulation calculations, though the discrepancy of that estimate from the even more primitive pX(n̄) ≈ 1 has not been significant in applications to metal ions in water so far. These points can make calculations more convenient.
4. Discussion of ion selectivity
Recent analyses of the mechanism of K+/Na+ selectivity have typically followed the reasonable assumption that this selectivity is thermodynamically controlled [3–7]. That selectivity is governed then by the chemical potentials of the ions. The latter point is not always explicitly displayed in simulation calculations, which employ a variety of macroscopic conditions and a variety of computational processes. But we seek free energy changes for displacements of a single ion [44], or the differences of such quantities between ions of different types, and those free energy changes are differences of chemical potentials.
When free energies for two ions, e.g., Na+ and K+, are compared, the inner shells are usually defined for mechanistic insight to reflect characteristics specific to the ions. Then
| (11) |
The rightmost contribution to Eq. (11) is typically significant, as is documented in Fig. 4 for n = 8. The value n = 8 has been of special interest in the debates on the mechanism of K+/Na+ selectivity [3– 7] because 〈n〉 = 8 is a feature of the solved crystal structure. Nevertheless, Eq. (11) is valid for any coordination number, and other choices for n may be more useful here.
Fig. 4.
For the simulated aqueous solution specified in Fig. 2, contributions from coordination-number probabilities of Eq. (11) for n = 8, a coordination number of interest for the KcsA channel. Note that these contributions have similar sizes, comparable to the sizes of the transfer free energies discussed below (Fig. 5), but differing signs. The upward-pointing arrows with solid points indicate −kT ln p(n = 8) for each case. The downward-point arrows indicate for each case. Comparison utilizing just p(n = 8)'s would lead to a free energy balance here of 5 kcal/mol instead of 2 kcal/mol [7].
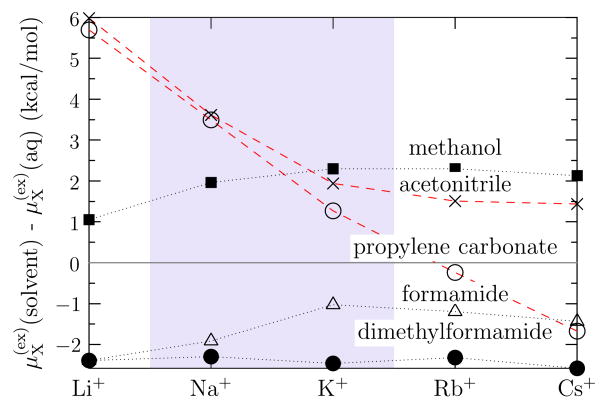
The present theoretical framework is not limited to cases where an ion binding site is defined by an experimental crystal structure. We emphasize this by noting ion selectivity in phase equilibria (Fig. 5) [45]. Those data illuminate many of the mechanistic issues that are currently debated for the K+/Na+ selectivity of K-channels [3–7]. Firstly, the adjective ‘liquid-like’ is unobjectionable here, and if structural fluctuations were key contributions to selectivity, then these data would be the more interesting for that reason. Secondly, some of these data (particularly for propylene carbonate and acetonitrile) exhibit obvious ion-size effects, an issue that has been debated for K+/Na+ selectivity of K-channels [3–7]. Propylene carbonate and acetonitrile are not prominent solvents in biophysical problems, and they are not proposed here to be models for K+/Na+ selectivity of K-channels. Nevertheless, these data can clarify concepts that are applied to K+/Na+ selectivity of K-channels. Thirdly, these systems are models for the case that no biomolecular structural specificity is involved. Finally, these data demonstrate that the selectivity free energies sought are typically less than 6 kcal/mol, though absolute solvation free energies are an order-of-magnitude larger. The several ingredients to the statistical thermodynamic formulation discussed here (Eq. (4)) are larger than the net selectivity free energy that is sought, and have different signs.
Fig. 5.
Standard free energies of transfer from aqueous solution to dilute solution of the solvent indicated at infinite electrolyte dilution, T = 298.15 K, and standard pressure [45]. Absolute values for these transfer free energies involve extrather-modynamical assumptions [45,46]. Differences of these transfer free energies between different ions do not depend on those extrathermodynamical assumptions. These data give a perspective on the possibility of selectivity that is broader than just comparison of Na+ and K+ (shaded), but do not address biomolecular features that might arise with ion channels.
On the fine scale of these transfer free energies, the data (Fig. 5) exhibit fascinating variety. The phenomena underlying recent debates on the mechanism of K+/Na+ selectivity of K-channels support some of that variety also, and that makes the discussion more complex. The theoretical framework developed here is complete, and isolates contributions with clear physical interpretations. Indeed, it has been argued [44] that Eq. (4) provides a basis for testing statistical thermodynamic concepts used to rationalize numerical results because it sharply distinguishes those concepts. Thus, careful assessment of each of the contributions of Eq. (4) would be a natural path for establishing a mechanistic understanding of the selectivity of ion channels. The multi-state analysis expressed by Eq. (2) pursues that goal of concept-testing further [13,27,44,47,48]. The more refined theory Eq. (9) gives additional insight [49] permitted by the physical organization of the problem based on Eq. (5).
5. Future, challenges, and directions
Theory and computations that analyze the experimental data shown in Fig. 5 should be a specific near-term target for understanding K+/Na+ selectivity in the K-channel proteins. A longer-range challenge is the direct numerical calculation of fully on the basis of validated electronic structure methods so that chemical effects can be investigated non-heuristically; indeed initial steps in that direction have already been taken [1,6,49]. This is clearly a large-scale computational challenge, but also clearly feasible [37].
6. Conclusions
This method of local configurations evaluates chemical potentials by defining an inner shell for detailed consideration. This produces intermediate results, particularly the probabilities of the number of inner-shell ligands, that suggest mechanistic interpretations. Thermodynamic results of the calculations are not affected by the inner-shell definition, but results of mechanistic interest are affected.
Two coordination-number distributions arise in a full discussion of the statistical thermodynamic problem, not only the naturally observed coordination-number distribution pX(n), but also the occupancy distribution of the same inner-shell volume without the ion. The latter distribution describes an intrinsic propensity for ligands to occupy the inner-shell volume.
Each of these distributions may be eliminated individually to obtain a partition function formula, e.g., Eq. (16), for the free energy that is sought. Since just one distribution need be evaluated, that can be an advantage. A corresponding disadvantage is that a full range of coordination numbers must be considered instead of just one coordination number that might offer special advantages.
The primitive quasi-chemical approximation finds another way to address these two probabilities. That approach introduces concepts of chemical equilibrium, then seeks the probability that is simplest to estimate, namely the probability of the most probable coordination number n̄. Quasi-chemical approximations include anharmonic packing effects in the combination Eq. (8) by acknowledging an exact cancellation and computing the remainder approximately. Simple neglect of multiple coordinate states is not an approximation of that quasi-chemical approach, as Eq. (9) shows.
These theories are not limited to crystallographically defined ion-binding sites, but apply to solutions generally. Available standard free energies of transfer of alkali metal ions from aqueous solution to alternative solvents offer examples of mechanisms suggested to explain K+/Na+ selectivity of K-channels [3-7], e.g., liquid-like structural fluctuations and ion-size effects. The role of specific biomolecular structures in the mechanism of K+/Na+ selectivity of K-channels, or other cases [11], may also be illuminated by comparison with those transfer free energies, because those solution data show fascinating variety without involvement of specific protein structures.
Acknowledgments
We thank T.L. Beck for a helpful reading of a preliminary version of this Letter. DA acknowledges the donors of the American Chemical Society Petroleum Research Fund for partial support of this research. P.D. and D.A gratefully acknowledge support from the National Science Foundation. S.V. and S.B.R. gratefully acknowledge support from Sandia's LDRD program and NIH through their Road Map for Medical Research. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the US Department of Energys National Nuclear Security Administration under contract DE-AC04-94AL8500.
Biographies

Dilip Asthagiri is an Assistant Professor in the Department of Chemical and Biomolecular Engineering at Johns Hopkins University. He has a PhD (1999) in Chemical Engineering from the University of Delaware, Newark, DE. He was a postdoctoral fellow (2002) at the Scripps Research Institute. Prior to his appointment at JHU in 2006, he was a postdoctoral fellow (2002–2005) and later a technical staff member at the Los Alamos National Laboratory.

Purushottam Dixit is a graduate student in the Department of Chemical and Biomolecular Engineering at Johns Hopkins University. He has a BS (2006) in Chemical Engineering from the Indian Institute of Technology, Bombay.

Safir Merchant is a graduate student in the Department of Chemical and Biomolecular Engineering at Johns Hopkins University. He has a BS/MS (2006) in Chemical Engineering from the Indian Institute of Technology, Bombay.

Michael Paulaitis is an Ohio Eminent Scholar in Chemical & Biomolecular Engineering at Ohio State University. He holds Chemical Engineering degrees from Princeton University (BS 1968), Stanford University (MS 1970), and the University of Illinois (PhD 1976). Prior to Ohio State, he spent 19 years on the Chemical Engineering faculty at the University of Delaware and 9 years on the Chemical Engineering faculty at Johns Hopkins University.

Lawrence Pratt is the Herman and George R. Brown Chair of Chemical Engineering at Tulane University. He holds Chemistry degrees from Michigan State University (BS 1972), and the University of Illinois (MS 1974 and PhD 1977). Prior to Tulane, he spent 23 years as a Technical Staff Member at Los Alamos National Laboratory.

Susan Rempe is a Principal Member of the Technical Staff at Sandia National Laboratories. She holds BA degrees from Columbia University and the University of Montana, and Chemistry degrees from the University of Washington (MS 1995 and PhD 1998). Prior to joining Sandia in 2001, she spent 3 years as a postdoctoral fellow at Los Alamos National Laboratory.

Sameer Varma is a Research Assistant Professor at the Illinois Institute of Technology. He holds BS and MS degrees in physics from the Indian Institute of Technology and a PhD in biophysics from the University of Illinois at Urbana-Champaign (2005). Prior to joining the Illinois Institute of Technology, he spent four years as a Postdoctoral Fellow at Sandia National Laboratories.
Appendix A. A simple derivation of the basic distribution formula Eq. (1)
Having defined an inner shell, introduce an indicator function
| (12) |
Then
| (13) |
This notation is customary, e.g., ε = UN+1 −UN − U1 is the binding energy of the ion for a given configuration. 〈〈…〉〉0 indicates averaging with respect to the configurations of the solution and a distinguished X solute without interactions between those two systems. The middle step of Eq. (13) uses the rule of averages [19,20] in which ion–ligand interactions in both the numerator and normalizing denominator are separated from the probability weights, and then explicitly displayed within the brackets that now indicate uncoupled averaging. Integration variables and ranges are the same in all cases here. The final step of Eq. (13) is a definition of the conditional expectation. The denominator of Eq. (13) is provided by the potential distribution theorem [20] . Together with the definition
| (14) |
Eq. (13) is the desired result, Eq. (1).
The definition of the inner shell and the introduction of the occupancy n is a stratification [50], i.e., a divide-and-conquer strategy. Chemical intuition can be utilized in defining the inner shell, important cases can get detailed attention, and in fact a small number of cases are involved in any consideration. Specialization of this generally valid maneuver led previously to a multi-gaussian model for hydration free energies [7,47,48]. Because of the character of the required averaging, the marginal probabilities do not involve ion–ligand interactions, but do depend on the characteristics of the defined inner shell. Conditional expectations might be evaluated by selecting a subensemble corresponding to n from a long simulation record. Alternatively, n can be directly constrained to a particular value, as was done for the results of Fig. 3 [27].
Noting the normalization of pX(n), Eq. (1) establishes a partition function,
| (15) |
for evaluating the solution free energy . Compared to a canonical ensemble partition function, for example, appears in a Boltzmann factor, as would the energy at level n. Then appears in the position of the degeneracy of level n. Note that both, and depend on the thermodynamic state including the temperature [51]. The quantities on the right-side of Eq. (15) depend also on the definition of the inner shell, though the thermodynamic property on the left does not. Although the defined inner shell is an open system, Eq. (15) is not a grand canonical ensemble formula and the thermodynamic property obtained is different from the grand canonical thermodynamic potential [14,20]. Nevertheless, the thermodynamic significance of is fully understood.
Of more specific interest, the normalization of establishes the inverse formula,
| (16) |
which requires only quantities that might be obtained by simulation of the physical system.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit:
Contributor Information
D. Asthagiri, Email: dilipa@jhu.edu.
P.D. Dixit, Email: dixitpd@gmail.com.
S. Merchant, Email: safir.merchant@gmail.com.
M.E. Paulaitis, Email: paulaitis.1@osu.edu.
L.R. Pratt, Email: lpratt@tulane.edu.
S.B. Rempe, Email: slrempe@sandia.gov.
S. Varma, Email: varma.sameer@gmail.com.
References
- 1.Rempe SB, Pratt LR, Hummer G, Kress JD, Martin RL, Redondo A. J Am Chem Soc. 2000;122:966. [Google Scholar]
- 2.Mason P, Ansell S, Neilson G. J Phys: Condens Matter. 2006;18:8437. doi: 10.1088/0953-8984/18/37/004. [DOI] [PubMed] [Google Scholar]
- 3.Noskov SY, Bernèche S, Roux B. Nature. 2004;431:830. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 4.Asthagiri D, Pratt LR, Paulaitis ME. J Chem Phys. 2006;125:024701. doi: 10.1063/1.2205853. [DOI] [PubMed] [Google Scholar]
- 5.Bostick DL, Brooks CL. Proc Natl Acad Sci USA. 2007;104:9260. doi: 10.1073/pnas.0700554104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varma S, Rempe SB. Biophys J. 2007;93:1093. doi: 10.1529/biophysj.107.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit PD, Merchant S, Asthagiri D. Biophys J. 2009;96:2138. doi: 10.1016/j.bpj.2008.12.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Noskov SY, Roux B. J Phys Chem B. 2009;113:8725. doi: 10.1021/jp901233v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noskov SY, Roux B. J Gen Physiol. 2007;129:135. doi: 10.1085/jgp.200609633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bostick DL, Arora K, Brooks CL. Biophys J. 2009;96:3887. doi: 10.1016/j.bpj.2008.12.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varma S, Sabo D, Rempe SB. J Mol Biol. 2007;376:13. doi: 10.1016/j.jmb.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 13.Pratt LR, LaViolette RA. Mol Phys. 1998;94:909. [Google Scholar]
- 14.Pratt LR, Rempe SB. In: Simulation and Theory of Electrostatic Interactions in Solution, AIP Conference Proceedings of the Computational Chemistry, Biophysics, and Aqueous Solutions. Pratt LR, Hummer G, editors. Vol. 492. American Institute of Physics; Melville, NY: 1999. p. 172. [Google Scholar]
- 15.Paulaitis ME, Pratt LR. Adv Prot Chem. 2002;62:283. doi: 10.1016/s0065-3233(02)62011-x. [DOI] [PubMed] [Google Scholar]
- 16.Asthagiri D, Pratt LR, Ashbaugh HS. J Chem Phys. 2003;119:2702. doi: 10.1063/1.2202350. [DOI] [PubMed] [Google Scholar]
- 17.Paliwal A, Asthagiri D, Pratt LR, Ashbaugh HS, Paulaitis ME. J Chem Phys. 2006;124:224502. doi: 10.1063/1.2202350. [DOI] [PubMed] [Google Scholar]
- 18.Asthagiri D, Pratt LR, Paulaitis ME, Rempe SB. J Am Chem Soc. 2004;126:1285. doi: 10.1021/ja0382967. [DOI] [PubMed] [Google Scholar]
- 19.Pratt LR, Asthagiri D. In: Free Energy Calculations: Theory and Applications in Chemistry and Biology, Springer series in Chemical Physics. Chipot C, Pohorille A, editors. Vol. 86. Springer; 2007. p. 323. Chapter 9. [Google Scholar]
- 20.Beck TL, Paulaitis ME, Pratt LR. The Potential Distribution Theorem and Models of Molecular Solutions. Cambridge University Press; 2006. [Google Scholar]
- 21.Kirkwood JG. J Chem Phys. 1940;8:623. [Google Scholar]
- 22.Friedman HL, Krishnan CV. In: Water A Comprehensive Treatise. Franks F, editor. Vol. 3. Plenum; New York: 1973. p. 1. [Google Scholar]
- 23.Rogers DM, Beck TL. J Chem Phys. 2008;129:134505. doi: 10.1063/1.2985613. [DOI] [PubMed] [Google Scholar]
- 24.Deng Y, Roux B. J Chem Phys. 2008;128:115103. doi: 10.1063/1.2842080. [DOI] [PubMed] [Google Scholar]
- 25.Kale L, et al. J Comp Phys. 1999;151:283. [Google Scholar]
- 26.Jorgensen W, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- 27.Merchant S, Asthagiri D. J Chem Phys. 2009;130:195102. doi: 10.1063/1.3132709. [DOI] [PubMed] [Google Scholar]
- 28.Bostick DL, Brooks CL., III Biophys J. 2009;96:4470. doi: 10.1016/j.bpj.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shore JE, Johnson RW. IEEE Trans Inform Theory IT/26. 1980:26. [Google Scholar]
- 30.Hummer G, Pratt LR, García AE. J Phys Chem. 1996:1206. [Google Scholar]
- 31.Varma S, Rempe SB. J Am Chem Soc. 2008;130:15405. doi: 10.1021/ja803575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah JK, Asthagiri D, Pratt LR, Paulaitis ME. J Chem Phys. 2007;127:144508. doi: 10.1063/1.2766940. [DOI] [PubMed] [Google Scholar]
- 33.Chempath S, Pratt LR, Paulaitis ME. J Chem Phys. 2009;130:054113. doi: 10.1063/1.3072666. [DOI] [PubMed] [Google Scholar]
- 34.Ashbaugh HS, Pratt LR. Rev Mod Phys. 2006;78:159. [Google Scholar]
- 35.Asthagiri D, Ashbaugh HS, Piryatinski A, Paulaitis ME, Pratt LR. J Am Chem Soc. 2007;129:10133. doi: 10.1021/ja071037n. [DOI] [PubMed] [Google Scholar]
- 36.McQuarrie DA. Statistical Mechanics. second. University Science Books; 2000. [Google Scholar]
- 37.Ishikawa Y, Sugita Y, Nishikawa T, Okamoto Y. Chem Phys Letts. 2001;333:199. [Google Scholar]
- 38.Owczarek E, Rybicki M, Hawlicka E. J Phys Chem B. 2007;111:14271. doi: 10.1021/jp076233v. [DOI] [PubMed] [Google Scholar]
- 39.Silva EFD, Svendsen HF, Merz KM. J Phys Chem A. 2009;113:6404. doi: 10.1021/jp809712y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryantsev VS, Diallo MS, Goddard WA., III J Phys Chem B. 2008;112:9709. doi: 10.1021/jp802665d. [DOI] [PubMed] [Google Scholar]
- 41.Rais J, Okada T. J Phys Chem B. 2008;112:5393. doi: 10.1021/jp711292c. [DOI] [PubMed] [Google Scholar]
- 42.Khaliullin RZ, Head-Gordon M, Bell AT. J Phys Chem B. 2007;111:10992. doi: 10.1021/jp073557a. [DOI] [PubMed] [Google Scholar]
- 43.Westphal EE, Pliego JR., Jr J Chem Phys. 2005;123:074508. doi: 10.1063/1.2001632. [DOI] [PubMed] [Google Scholar]
- 44.Chempath S, Pratt LR. J Phys Chem B. 2009;113:4147. doi: 10.1021/jp806858z. [DOI] [PubMed] [Google Scholar]
- 45.Marcus Y. Pure & Appl Chem. 1983;55:977. [Google Scholar]
- 46.Marcus Y. Pure & Appl Chem. 1986;58:1721. [Google Scholar]
- 47.Hummer G, Pratt LR, García AE. J Am Chem Soc. 1997;119:8523. [Google Scholar]
- 48.Hummer G, Pratt LR, García AE. J Phys Chem A. 1998;102:7885. [Google Scholar]
- 49.Varma S, Rempe SB. Biophys Chem. 2006;124:192. doi: 10.1016/j.bpc.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Hammersley JM, Handscomb DC. Monte Carlo Methods. Chapman and Hall; London: 1964. [Google Scholar]
- 51.Rushbrooke G. Trans Faraday Soc. 1940;36:1055. [Google Scholar]