Abstract
Mutations in Parkin or PINK1 are the most common cause of recessively inherited parkinsonism. Parkin and PINK1 function in a conserved mitochondrial quality control pathway, in which PINK1, a putative mitochondrial kinase, directs Parkin, a cytosolic E3 ubiquitin ligase, selectively to dysfunctional mitochondria to promote their isolation, immobilization and degradation by macroautophagy (hereafter, mitophagy). As Parkin recruitment to mitochondria is robustly induced by PINK1 expression on the outer mitochondrial membrane, Parkin recruitment to mitochondria was used as an assay for PINK1 function. Unexpectedly, mutation of serine residues within the activation segment of PINK1 uncovered a temperature-sensitive variant of PINK1 (tsPINK1). tsPINK1 allowed for the first time the disassociation of PINK1 activity from its expression and localization. Additionally, extensive mutagenesis identified three disease-associated variants in the activation segment and one in an α-helix N-terminal to kinase domain (Q126P) that are similarly thermally labile, suggesting that their activity could be restored post-translationally (e.g. by reducing the temperature or by a chemical or pharmacologic chaperone). Together, these findings suggest that tsPINK1 may represent a valuable tool for the analysis of the PINK1/Parkin pathway in human cells; additionally, as the serine residue promoting thermal lability is conserved among Mus musculus, Danio rerio, Drosophila melanogaster and Caenorhabditis elegans, it may serve as the basis for developing other temperature-sensitive models for the study of recessive parkinsonism and mitophagy. Finally, these results suggest that PINK1 kinase function could be restored for a subset of patients with PINK1 mutations, and perhaps alter the course of their disease.
INTRODUCTION
Parkinson's disease, the second most common neurodegenerative disorder, is characterized by the cardinal motor signs of slowness of movement (bradykinesia), resting tremor, rigidity and postural instability (1). With a possible modest exception (2), no disease-modifying therapies are currently available for its treatment. Although most cases of Parkinson's disease are sporadic, roughly 10% of cases are inherited in a Mendelian fashion, and to date mutations in five genes have been definitively linked to familial forms of parkinsonism (3). Two of the five genes identified, encoding the proteins Parkin and PINK1 (for PTEN-induced putative kinase 1), are believed to form a common pathway important for mitochondrial quality control (4). Although Parkin and PINK1 account for only a small fraction of parkinsonism cases, the pathway they form may prove a valuable therapeutic target for the treatment of sporadic Parkinson's disease (3).
In the PINK1/Parkin pathway, PINK1, a mitochondrial kinase, accumulates selectively on the surface of severely impaired mitochondria (5–8). Parkin, an E3 ubiquitin ligase, which normally resides inert in the cytosol, is recruited to the surface of the impaired mitochondria (9), by PINK1, where the latent activity of Parkin is increased by a mechanism that is incompletely understood (6–9). Activated Parkin effects the isolation and degradation of the impaired mitochondria likely by ubiquitinating a number of targets on the outer mitochondrial membrane (9–12). Failure of the PINK1/Parkin pathway, due to mutation in PINK1 or Parkin, may allow impaired mitochondria to accumulate in long-lived cells subjected to high levels of oxidative stress—such as neurons in the substantia nigra pars compacta, which are preferentially affected in Parkinson's disease (reviewed in 4).
The robust activation of Parkin by PINK1 inspired a cell-based assay for carrying out a structure–function analysis of PINK1. PINK1 is a serine/threonine kinase by homology; however, it has been challenging to demonstrate human PINK1 functions as a kinase, likely due to difficulty expressing properly folded human PINK1 in recombinant systems (13). As PINK1 is likely better expressed within intact human cells, this study used a simple cell-based assay of PINK1 activity—Parkin recruitment to mitochondria—to carry out a structure–function analysis focused on the kinase domain of PINK1. Unexpectedly, several patient-associated and engineered mutations were found to render the kinase temperature sensitive, generating a potentially useful tool for studying the PINK1/Parkin pathway in mammalian cells and with implications for the potential treatment of a subset of patients, as is reported below.
RESULTS
PINK1 requires residues conserved among active protein kinases for activity
PINK1 is homologous to serine/threonine kinases and possesses the complement of conserved residues believed to be required for phosphyl transfer (14–16). These include the three small residues of the glycine-rich loop, the aspartic acids of the catalytic loop and the DFG motif, and the catalytic lysine in the third β-pleated sheet of the N-lobe. In vitro studies with human recombinant PINK1 to date have failed to demonstrate that these residues are individually required for the activity of PINK1, as they are in other active kinases, possibly due to difficulty purifying properly folded PINK1 for in vitro experiments (13,14).
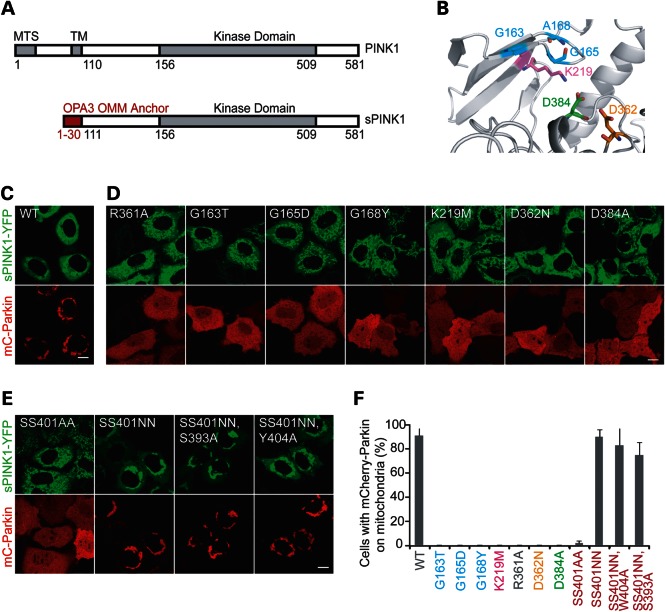
As active PINK1 is well expressed in intact human cells, a cell-based Parkin recruitment assay was used to assess the requirement of these residues for PINK1 activity in HeLa cells. In this assay, a stabilized form of PINK1 (sPINK1), resulting from the fusion of the cytosolic domain of PINK1 to the outer mitochondrial membrane anchor of OPA3, was used to recruit mCherry-Parkin to mitochondria (Fig. 1A). sPINK1-YFP was chosen because it has a dominant effect in cells with well-coupled mitochondria containing negligible levels of endogenous PINK1 on the outer mitochondrial membrane, and because it provides a strong fluorescent signal co-localizing with mitochondria when properly expressed, allowing for rapid assessment of PINK1 variant expression and localization (5).
Figure 1.
Structure–function analysis of PINK1 using Parkin recruitment as a reporter of PINK1 function. (A) A schematic representing the fusion of the outer mitochondrial membrane (OMM) anchor of OPA3 to the cytosolic domain of PINK1 to form a PINK1 fusion protein (sPINK1) that is stably expressed on the OMM. Amino acid residue numbers for full-length human PINK1 are black, whereas those for OPA3 are in red. (B) Homology model of the ATP-binding cleft of PINK1, with residues conserved among active kinases highlighted and labeled, including small residues contributing the glycine-rich loop (blue), the lysine of the β3 sheet (magenta), the aspartic acid of the catalytic loop (orange) and the aspartic acid of the DFG motif of the activation segment. (C–E) Representative images of HeLa cells co-transfected with sPINK1-YFP (green) with or without amino acid substitutions and mCherry-Parkin (red). Scale bars = 10 µm. (F) The graph represents the percent of cells with Parkin on mitochondria, following co-transfection with sPINK1. In total, at least 150 cells total were scored for each condition in three replicates. The experiment was performed on at least two separate occasions. The color scheme in the labels corresponds to that used in the homology model in (B). Substitutions of serine residues in the activation segment are depicted in red.
Residues in sPINK1 that are conserved among active kinases (G163, G165, A168, K219, D362 and D384) were substituted for residues found in pseudokinases (17), alanine or methonine (Fig. 1B–D and F). All sPINK1 variants exhibited a stable mitochondrial pattern of expression by confocal microscopy, consistent with the substitutions resulting in a properly folded and targeted protein (Fig. 1D). Wild-type sPINK1 recruited mCherry-Parkin to >90% of the cells ∼18 h after the transient co-transfection of sPINK1 and Parkin (Fig. 1C and F). In contrast, substitution of any of the residues conserved among active kinases eliminated Parkin recruitment to mitochondria (Fig. 1D and F). These findings supported the notion that PINK1 functions as an active kinase in intact cells.
PINK1 does not require activation loop phosphorylation for activity
Many kinases are synthesized in an inactive form and are activated post-translationally (reviewed in 18). For those kinases possessing an arginine in the −1 position relative to the catalytic aspartic acid (so called, RD kinases), the mechanism of activation is most often phosphorylation of a serine or threonine residue within the activation segment (the segment beginning with the DFG motif and ending with the APE motif). The resulting phospho-residue stabilizes the activation segment through an electrostatic interaction with a positively charged pocket formed by the arginine of the RD motif (19), often along with other positively charged residues (reviewed in 18). As PINK1 possesses the RD motif (R361, D362) and sPINK1 R361A failed to recruit Parkin similarly to the catalytic mutants (Fig. 1D and F), the possibility that PINK1 might require activation segment phosphorylation for full activity was explored.
As is discussed further below, human PINK1 possesses three serine residues (S393, S401 and S402) as well as one tyrosine residue within the activation segment. Only one of these, S402, is conserved among the Mus musculus, Danio rerio, Drosophila melanogaster and Caenorhabditis elegans PINK1 orthologs (see below). To test whether S402 phosphorylation is required for PINK1 activation experimentally, alanine residues were substituted for the S402 residue alone as well as for the S401, S402 diresidue. In cells expressing sPINK1 S402A or SS401/402AA and examined immediately after removal from the incubator (set at 37°C, 5% CO2), Parkin recruitment was observed in only a minority of cells (Fig. 1E and F; data not shown), demonstrating that S402 is required for full PINK1 activity.
As loss of activity with the S402A and SS401/402AA mutations could reflect a structural disruption rather than failure to form phosphoserine residue(s), the more conservative substitution of asparagines for the S401, S402 diresidue was also tested (Fig. 1E and F). The SS401/402NN variant was active for Parkin recruitment, demonstrating that S401 and S402 do not need to be phosphorylated for Parkin recruitment even though a polar residue at 402 or both 401 and 402 is required for activity (Fig. 1E and F). In addition, the triple variants (S393A, SS401/402NN) and (SS401/402NN, Y404A) were active for Parkin recruitment, suggesting that phosphorylation of S393 or Y404 is also dispensable for PINK1 activity (Fig. 1E and F). The quadruple variant (S393, SS401NN, Y404A) was not active for Parkin recruitment, which may reflect the substantial alteration to the activation segment induced by the four mutations (data not shown). Additionally, the activity of S402N and the potentially phosphomimetic S402D were directly compared and found to be similar both in the context of the OPA3-PINK1 chimera and in the context of full-length PINK1 (Supplementary Material, Fig. S1). Together these results strongly suggested that activation segment phosphorylation is not required for PINK1 activity in the PINK1/Parkin pathway.
S402A substitution in the activation loop renders PINK1 temperature sensitive
In evaluating the S402A and SS401/402AA variants, substantial variability in Parkin recruitment was noted between samples (data not shown). To test whether time out of the CO2 incubator prior to fixation might account for the variability, S402A was either fixed immediately or fixed 90 min following removal from the incubator. Unexpectedly, Parkin was recruited to mitochondria in <1% of cells fixed immediately and >90% of cells exposed to ambient air for 90 min prior to fixation (Supplementary Material, Fig. S2A and B), demonstrating that exposure to ambient air activates these variants.
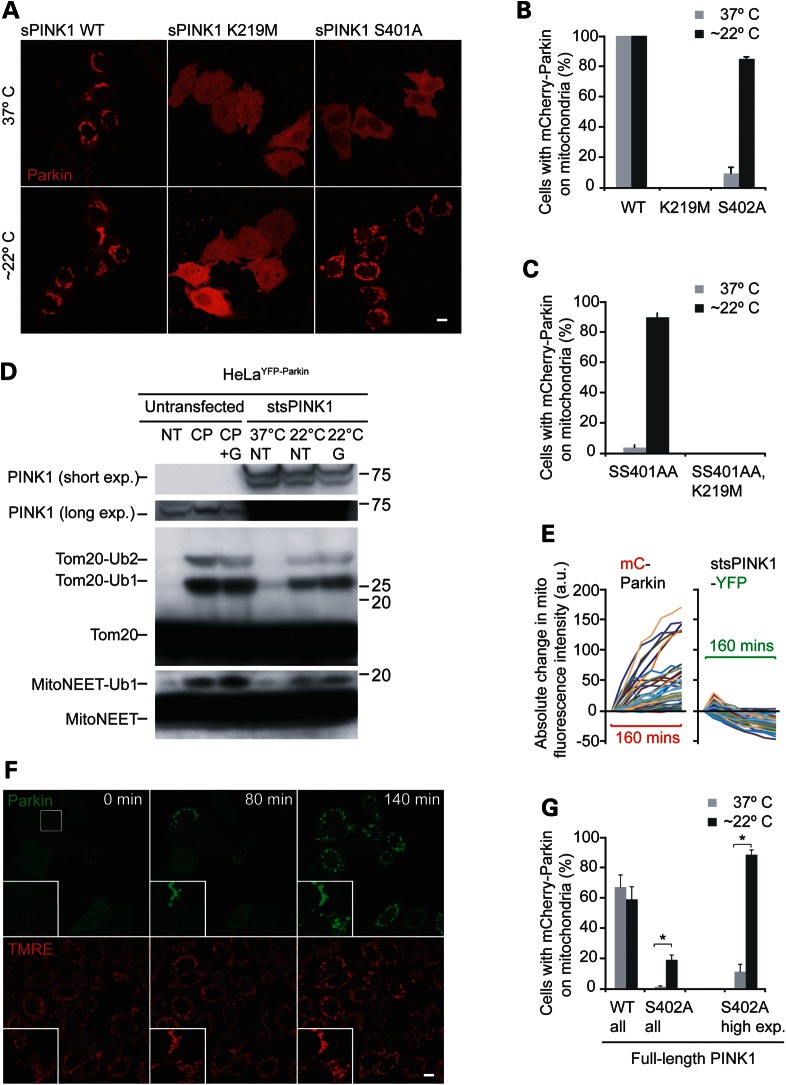
Transfer from the CO2 incubator to ambient air has a number of effects on the immediate environment of the cells, including a decrease in temperature and an increase in the pH of the cell culture media (DMEM, Dulbecco's modified Eagle's medium). To test whether the decrease in temperature is responsible for the activation of sPINK1 S402A following exposure to ambient air, cells expressing sPINK1 variants were transferred from the home incubator set at 37°C to one of two identical CO2 incubators set at 26 or 37°C for 2 h. In cells expressing sPINK1 S402A, Parkin recruitment to mitochondria increased from 9.6 ± 4.0% of cells incubated at 37°C to 85 ± 1.2% of cells incubated at 26°C (Fig. 2A and B). In contrast, temperature did not affect Parkin recruitment in cells expressing wild-type sPINK1 or the catalytically inactive variant sPINK1 K219M (Fig. 2A and B). Similar results were found for cells expressing sPINK1 S402A or sPINK1 SS401/402AA and bathed in CO2-independent media (buffered by a phosphate-based system), following transfer to an ambient air incubator set at 37°C or placed on the bench at room temperature (∼22°C) for 2 h (data not shown and see below). Together, these findings demonstrated that sPINK1 S402A and sPINK1 SS401/402AA are temperature-sensitive variants of sPINK1, at least with respect to Parkin recruitment.
Figure 2.
Evaluation of putative temperature-sensitive variants of PINK1, S402A and SS401/402AA. (A and B) Stabilized wild-type PINK1 (sPINK1), a catalytically inactive sPINK1 variant (sPINK1 K219M), or sPINK1 S402A was co-transfected with mCherry-Parkin (red). The cells were subsequently moved from the home CO2 incubator set at 37°C to one of two otherwise identical CO2 incubators set at 37 or 22°C for 2 h, after which the cells were fixed and evaluated by confocal microscopy. Representative confocal images are shown in (A). At least 150 cells per condition (in three replicates obtained on at least two occasions) were scored for the presence of mCherry-Parkin on mitochondria (B). (C) Cells co-transfected with sPINK1 SS401/402AA or PINK1 SS401/402AA, K219M and mCherry-Parkin were incubated at room temperature or 37°C in CO2-independent media for 2 h, fixed and scored as in (B). (D) HeLa cells stably expressing YFP-Parkin were left untransfected (left three lanes) or transfected with sPINK1 SS401/402AA (stsPINK1) (right three lanes). The untransfected cells were not treated (NT), treated with CCCP (CP) or treated with CCCP + geldanamycin (CP + G) for 2 h. Cells transfected with stsPINK1 and either not treated (NT) or treated with geldanamycin (G) were incubated at 37 or 22°C in CO2- independent media for 2 h. Lysates were run on SDS–PAGE gels, transferred to PDVF membranes and probed for PINK1, TOMM20 and MitoNEET immunoreactivity. The positions of predicted, ubiquitinated forms of TOMM20 and MitoNEET are labeled. (E) Cells co-transfected with mCherry-Parkin and stsPINK1-YFP were removed from the home incubator to a microscope stage at room temperature (∼22°C) and imaged live every 20 min for 160 min. Graphs represent the absolute change in mitochondrial fluorescence intensity of mCherry-Parkin (left graph) and stsPINK1-YFP (right) relative to time point 0 min for 37 individual cells from six fields. All cells were imaged with the same settings. (F) Cells co-transfected with stsPINK1-Flag (not depicted) and YFP-Parkin (green) and loaded with the potentiometric mitochondrial dye TMRE (red) were removed from the home incubator set at 37°C to a microscope stage at room temperature (∼22°C) and imaged live. A white square in the first image depicts the position of the highlighted region appearing in the lower right corner of all of the images. (G) HeLa cells transfected with mCherry-Parkin and wild-type or S402A full-length PINK1-YFP (in a 1:4 ratio) were scored for mCherry-Parkin in mitochondria-like puncta in all cells expressing mCherry-Parkin (left side of the graph) or in all cells with clear PINK1-YFP expression (right side of the graph). *P-value <0.01. All error bars in images = 10 µm.
As sPINK1 SS401/402AA (hereafter, stsPINK1) exhibited greater differential activity in Parkin recruitment between 37 and ∼22°C compared with sPINK1 S402A, it was chosen for further characterization (data not shown). To test whether recruitment of Parkin by stsPINK1 requires its kinase activity, the K219M mutation was introduced into stsPINK1. The K219M mutation failed to recruit Parkin at 37 or 22°C, demonstrating that stsPINK1, when active, likely recruits Parkin by a similar mechanism as wild-type PINK1 (Fig. 2C).
Mitochondrial uncouplers, such as CCCP, are believed to activate Parkin by causing endogenous PINK1 to accumulate on the outer mitochondrial membrane (5). To assess whether temperature activates Parkin by similarly causing an accumulation of stsPINK1 protein, stsPINK1 protein levels were evaluated by immunoblotting. stsPINK1-YFP appeared as two major bands on the gel, as was observed previously for other OPA3-PINK1-YFP fusion proteins (5). In contrast to endogenous PINK1 following treatment with CCCP, no substantial difference in the intensity of stsPINK1 was appreciated between the cells treated at 37°C and the cells treated at ∼22°C (Fig. 2D). This demonstrated that increased expression of stsPINK1 was not responsible for its increased activity at ∼22°C. This conclusion was supported by live cell analysis of stsPINK1-YFP in single cells, in which the fluorescent intensity of stsPINK1-YFP co-localizing with a mitochondria marker stayed the same or decreased as the intensity of mCherry-Parkin co-localizing with mitochondria increased (Fig. 2E). An increase in mCherry-Parkin fluorescence intensity was observed in all 37 cells over the course of 140 min, with 31 of 37 cells (83.8%) exhibiting a >50% increase in intensity; whereas all but one of the cells exhibited a decrease in stsPINK1-YFP intensity, with an average decrease of 24.2 ± 12.3%. Together these findings established that the temperature-dependent change in stsPINK1 activity was not solely due to a change in its abundance.
To test whether Parkin recruited by stsPINK1 promotes the ubiquitination of outer mitochondrial membrane proteins, the SDS gel migration of two small outer membrane proteins, TOMM20 and MitoNEET, was tested. Degradation of TOMM20 and MitoNEET has been shown to be promoted by exogenously expressed Parkin previously (20). In cells stably expressing YFP-Parkin and treated with CCCP for 2 h, higher molecular weight bands corresponding to ubiquitinated forms of TOMM20 and MitoNEET appeared, consistent with the activation of Parkin by endogenous PINK1 under these conditions (Fig. 2D). Incubation of stsPINK1 at 22°C similarly induced the appearance of TOMM20 and MitoNEET high molecular weight bands, suggesting that stsPINK1 also increases the activity of Parkin toward outer mitochondrial membrane proteins. Geldanamycin, an inhibitor of the chaperone Hsp90 of which PINK1 is a client (21), had a modest effect on PINK1 protein levels and the activity of Parkin under these conditions. Together, these findings demonstrate that temperature-activated stsPINK1 likely promotes Parkin E3 ligase activity in a manner similar to the activation of Parkin by endogenous PINK1 following mitochondrial uncoupling.
As PINK1 stably expressed on mitochondria can recruit Parkin in the absence of mitochondrial uncoupling, it was hypothesized that stsPINK1 should also be able to recruit Parkin to well-coupled mitochondria, which have negligible levels of endogenous PINK1 on the outer membrane (5). To test this hypothesis, HeLa cells expressing YFP-Parkin (green) and unlabeled stsPINK1 (not visualized) were loaded with the potentiometric mitochondrial dye TMRE (red). The double-labeled cells were imaged live at room temperature, immediately following removal from an incubator set to 37°C. YFP-Parkin failed to co-localize with TMRE-stained mitochondria at 0 min in all but one cell in the field, but by 160 min YFP-Parkin co-localized with TMRE-positive mitochondria in 9 of 11 cells expressing YFP-Parkin in the field (Fig. 2F). These findings demonstrated that Parkin is recruited to mitochondria by stsPINK1 in the absence of substantial mitochondrial depolarization.
To evaluate whether stsPINK1 activity can be ‘tuned’ by varying the temperature, mCherry-Parkin recruitment was assessed following incubation at a range of temperatures for 2 h. Consistent with the idea that stsPINK1 activity can be ‘tuned’, an inverse relationship between temperature and mCherry-Parkin recruitment was observed in cells expressing stsPINK1 (Supplementary Material, Fig. S2C).
As the temperature conferring S402A mutation may act differently in the context of the full-length PINK1 compared with the chimeric protein OPA3-PINK1, the S402A mutation was introduced into full-length PINK1-YFP. When co-expressed with mCherry-Parkin, PINK1-YFP S402A recruited Parkin to mitochondria in 0.700 ± 0.970% of all cells with visible mCherry-Parkin expression cells at 37°C (Fig. 2G and Supplementary Material, Fig. S2). In samples incubated at 22°C for 2 h, however, Parkin recruitment to mitochondria increased to 18.4 ± 3.58% in all cells with visible mCherry-Parkin expression (Fig. 2G and Supplementary Material, Fig. S2D). Similar results were obtained with PINK1-YFP SS401/402AA, indicating that the temperature-sensitive property of these variants is not altered by the protein context of full-length PINK1.
Notably, the activity of PINK1-YFP S402A at 22°C was substantially less than that of wild-type PINK1-YFP (18.4 ± 3.58 vs. 58.6 ± 8.61% of cells with Parkin recruitment), indicating that either S402A is less stable than wild-type in the context of full-length PINK1 or that S402A exhibits differential activity at 37 and 22°C but does not regain the full activity of wild-type at 22°C (Fig. 2G and Supplementary Material, Fig. S2D).
Many of the cells co-transfected with mCherry-Parkin and PINK1-YFP S402A had undetectable or barely detectable YFP signal but clearly visible mCherry signal, indicating very low expression of PINK1-YFP S402A. This is consistent with the constitutive degradation of PINK1 in cells with healthy mitochondria (4). When only cells with clear PINK1-YFP S402A expression were assessed for Parkin recruitment, Parkin recruitment was observed in 88.1 ± 3.57% of cells incubated at 22°C for 2 h compared with 10.7 ± 5.10% of cells incubated at 37°C (Fig. 2G and Supplementary Material, Fig. S2D). This finding suggests that the difference in activity between OPA3-PINK1 S402A and PINK1 S402A at 22°C may be due to their relative abundance in the cell, stemming from their relative stability.
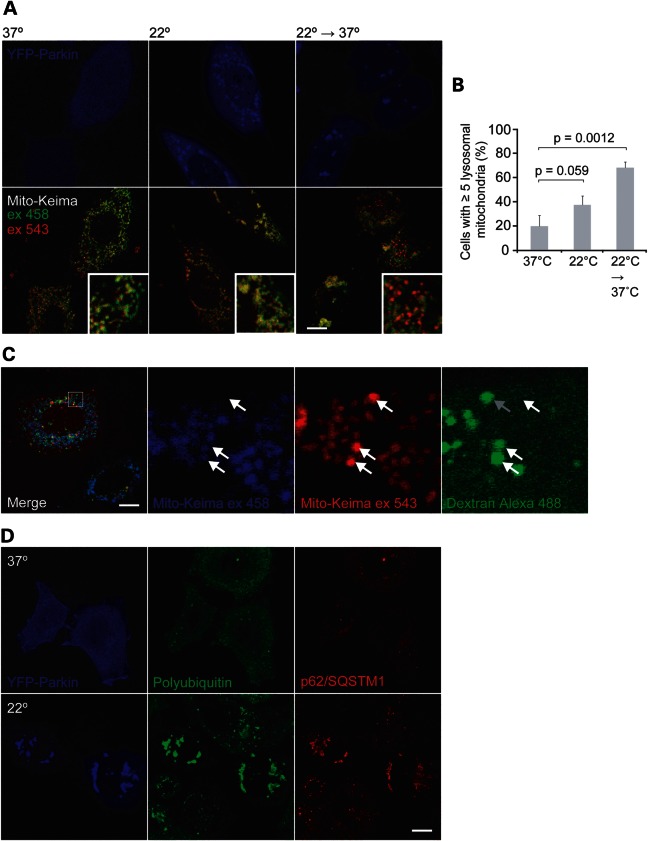
Exogenously expressed Parkin robustly increases selective mitochondrial turnover through macroautophagy (mitophagy) following its activation (9). To further validate the ability of stsPINK1 to activate YFP-Parkin in response to temperature, mitochondrial turnover was assessed using a genetically encoded pH-sensitive mitochondrial probe called mito-Keima. Mito-Keima, a DsRed variant that is targeted to the mitochondrial matrix, is excited preferentially by a 458 nm laser line at a pH of 7.4 and preferentially by the 543 nm laser line at a pH of 4.5 (the pH of lysosomes). By comparing the intensity of mito-Keima emissions following sequential excitation with 458 and 543 nm light, mitochondria that have been delivered to acidified lysosomes can be identified (22).
In cells transiently expressing stsPINK1, YFP-Parkin and mito-Keima and incubated at 37°C, YFP-Parkin seldom co-localized with mitochondria and few cells contained lysosomal mitochondria (Fig. 3A and B). When the cells were exposed to 22°C for 4 h, YFP-Parkin was recruited to mitochondria in the majority of cells, and a modest non-significant increase in lysosomal mitochondria was observed. As it was reasoned that the delivery of mitochondria to lysosomes via autophagy might be inhibited at one or more steps at the lower temperature, cells were incubated at 22°C for 2 h to allow activation of Parkin by stsPINK1 and then incubated at 37°C for an additional 2 h to allow delivery of mitochondria to lysosomes. Lysosomal mitochondria were observed in the majority of the cells sequentially incubated at 22 and 37°C. The lysosomal localization of mito-Keima was verified by labeling mitochondria with Dextran Alexa 488 (Fig. 3C). These findings were consistent with stsPINK1 promoting Parkin-dependent mitophagy in the absence of depolarization, similar to endogenous PINK1 promoting Parkin-dependent mitophagy following mitochondrial depolarization. Interestingly, they also suggested that step(s) in the delivery of Parkin-recruited mitochondria to lysosomes by autophagy are inhibited at 22°C. This system may prove useful in evaluating the Parkin-induced mitophagy pathway.
Figure 3.
Evaluation of Parkin activation by a temperature-sensitive variant of PINK1 (stsPINK1). (A and B) Cells transfected with YFP-Parkin (blue), stsPINK1-Flag (not labeled) and mito-Keima (green and red) were incubated at 37°C for 4 h, 22°C for 4 h or 22°C for 2 h and then 37°C for an additional 2 h. Representative confocal images obtained live (A). Mito-Keima was consecutively excited with a 458 nm laser line, which preferentially excites Keima at a neutral pH, and a 543 nm laser line, which preferentially excites Keima at an acidic pH such as that maintained within lysosomes. (B) Cells with ≥5 puncta preferentially excited by 543 nm relative to 458 nm were scored for at least 150 cells per condition (in three replicates on at least two occasions). (C) Cells transfected with untagged Parkin (not depicted), stsPINK1-Flag (not depicted) and mito-Keima (green and red). To label lysosomes, cells were loaded with 10 kDa of Dextran Alexa 488 overnight and then chased for 4 h. Cells were exposed to 22°C for 2 h and then returned to 37°C for 2 h prior to imaging live. Arrows identify mitochondria preferentially excited by 543 nm relative to 458 nm light (indicating exposure to acidic environment) co-localizing with Dextran-labeled lysosomes. The gray arrow in the last image identifies a Dextran/Keima co-localizing puncta that changed position in the tens of seconds between the acquisition of the Keima image and the Dextran Alexa 488 image excited at 458 nm. (D) Cells co-transfected with YFP-Parkin (blue) and stsPINK1-Flag (not depicted) were exposed to 37°C for 4 h or 22°C for 4 h, fixed and immunostained for polyubiquitin (green) and p62/SQSTM1 (red). All scale bars represent 10 µm.
Additional signs of Parkin activation by endogenous PINK1 following depolarization include the recruitment of the adaptor protein p62/SQSTM1 (6,23,24), which likely binds polyubiquitin chains attached to outer mitochondrial membrane proteins and causes mitochondrial clumping (23,24). stsPINK1 similarly induced the polyubiquitination of mitochondria and recruitment of p62/SQSTM1 in a temperature-dependent manner (Fig. 3D). Considered with the findings above, these data demonstrated that the PINK1 variant stsPINK1 activates Parkin in a temperature-dependent manner.
Structure–function analysis of the activation segment
Given that stsPINK1 protein abundance was unchanged between inactive and active conditions, it was reasoned that the most likely mechanism for the stsPINK1 activation is a conformational change. To explore a possible structural basis for this conformation change as well as to explore the mechanism of activation segment stabilization in the absence of phosphorylation, a structure–function analysis of the activation segment was conducted using a homology model based on other kinases in the active conformation and two series of mutagenesis experiments.
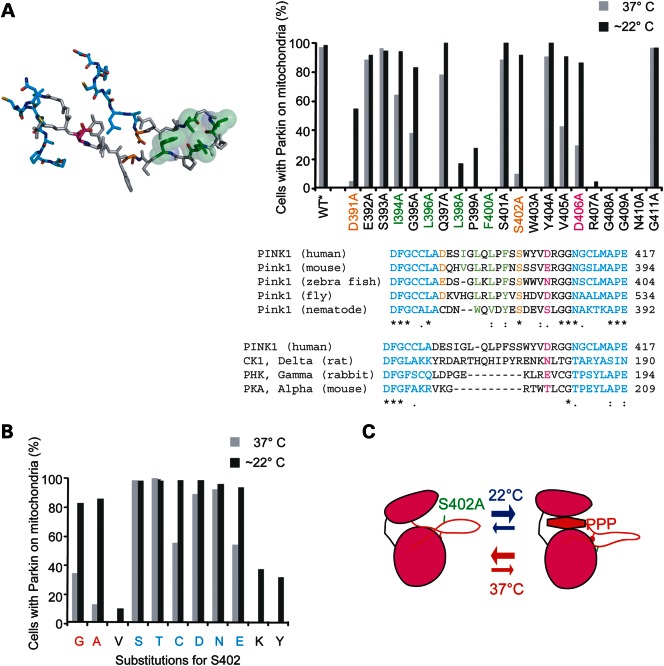
In the first series, alanine-scanning mutagenesis was used to determine whether other residues in the activation segment are required for its activity at physiological and sub-physiological temperatures. A homology model of the activation segment of PINK1 based on serine/threonine kinases in the active conformation suggested that the uncommonly long segment (34 amino acid residues in length compared with 25 residues in PKA) might be stabilized in part by a hairpin structure, although the activation loop in the model is not well templated and so the model is interpreted with caution. The hairpin suggested by the model is similar to the constitutively active casein kinase 1 (25), which also possesses an extended activation segment (35 residues in length) (Fig. 4A, left panel). A patch of bulky hydrophobic residues (L396, L398 and F400) that stabilized the tip on the hairpin in the model were required for activity at both 37 and ∼22°C, consistent with the activation segment forming a hairpin to stabilize the N- and C-anchors in the active conformation (colored green in Fig. 4A). These hydrophobic residues are evolutionarily conserved, supporting their structural importance to the activation segment. Attempts to rescue the activity of PINK1 following disruption of the proposed hydrophobic patch by introducing an electrostatic interaction or a disulfide bond across the proposed hairpin or reconfiguring the hydrophobic patch, however, were unsuccessful, suggesting either that these hydrophobic residues may be required for a reason other than stabilizing the proposed hairpin or that other residues (e.g. from the C-lobe) or distance between the targeted residues prevented the formation of the intended interactions (Supplementary Material, Fig. S3). Of note, the substitutions I394D, L398I and I394L/L398I were substantially more active at ∼22°C compared with 37°C, suggesting that, like SS401/402AA, these substitutions may have utility as temperature-sensitive alleles.
Figure 4.
Structure–function analysis of the activation segment of PINK1 to explore the mechanism of temperature sensitivity. (A) Residues of the activation segment between the N- and C-anchors (anchor residues colored blue) were singly substituted for alanine in human sPINK1. The location of these residues is shown in the homology model of that activation segment (left) as well as in the alignment of PINK1 orthologs. In addition, an alignment compares the activation segment of human PINK1 with that of other kinases with known structure. S402 is colored orange as is D391, which lies on the opposite S402 in the homology model. D406, which occupies the position of the primary phosphorylated residue of RD serine/threonine kinases, is colored magenta. A group of hydrophobic residues, which form a hydrophobic patch at the tip of the hairpin structure, are colored green. The sPINK1 variants were screened for activity at 22 and 37°C, using the Parkin recruitment assay as described in Figure 2C. (B) S402 was substituted for a series of residues. Small residues are colored red, whereas medium polar residues are colored blue in the x-axis label. The sPINK1 variants were screened for activity at 22 and 37°C, using the Parkin recruitment assay as described in (A). (C) Model depicting a possible mechanism for the activation of stsPINK1 by lowering temperature. The stability of the ATP-binding cleft is tied to the position of the N- and C-anchors of the activation segment, which in turn depends on positioning of the loop between the anchors. S402, which lies at the base of the hairpin, forms a hydrogen bond with the C-lobe or another residue in the hairpin, thereby stabilizing the hairpin in the face of high thermal motion at physiological temperatures. Lowering the temperature allows stabilization of the hairpin despite the absence of a stabilizing hydrogen bond by decreasing thermal motion within the activation segment.
Two glycine residues near the C-anchor were also required at both 37 and ∼22°C, suggesting that flexibility is needed in this region to allow positioning of the C-anchor in an active conformation.
In the model, the hydroxyl group of S402 (colored orange in Fig. 4A) lies at the base of the hairpin, where it might help anchor the activation segment by forming a hydrogen bond with a residue in the C-lobe. D391 (colored orange in Fig. 4A), which occupies a similar position as S402 but on the opposite side of the hairpin, also exhibited differential activity at 37 and ∼22°C upon substitution to alanine. It may similarly help anchor the hairpin to the C-lobe. The importance of S402 and D391 to the stability of the activation segment is suggested also by their conservation among PINK1 orthologs (Fig. 4A, right bottom).
The −9 or −10 position relative to the APE motif, which is occupied by a phosphoserine or phosphothreonine in many RD kinases (18), is occupied by an aspartic acid (D406) in PINK1 (Fig. 4A, right bottom). The aspartic acid could interact with catalytic loop arginine (R383) to help stabilize the activation segment, as is observed with the analogous residues in the structure of the constitutively active kinase Phk (26). The D406A substitution also displayed differential activity at 37 and ∼22°C, suggesting that like S402 and D391 it may help stabilize the position of the activation segment within the rest of the kinase. Notably, the C. elegans PINK1 ortholog contains a serine in this position, suggesting that unlike human PINK1, it may require phosphorylation at this site for activity, similar to the regulation of canonical RD kinases, such as PKA.
More limited temperature-dependent activation was observed for several residues in the activation segment. These include several positions in which non-synonymous mutations have been identified in cases of parkinsonism, as is explored further below.
In the second mutagenesis series, S402 was substituted for 1 of 10 amino acids with side chains that exhibit a range of chemical properties. High activity at both 37 and ∼22°C was observed for medium-sized polar residues such as serine, threonine, asparagine and cysteine, whereas low activity at 37°C and high activity at ∼22°C (i.e. temperature sensitivity) was observed for small residues like glycine and alanine. The non-polar residue isoleucine and large or bulky residues like tyrosine and lysine exhibited low activity at both 37 and ∼22°C. These findings were consistent with the idea that S402 (or another substituted medium-sized polar residue) may form a hydrogen bond that helps stabilize the activation loop in the active conformation in the face of thermal motion at physiological temperatures (Fig. 4C).
Stabilized tsPINK1 disassociates PINK1 activity from changes in PINK1 expression and localization for the analysis of PINK1/Parkin pathway
The activity of endogenous PINK1 toward Parkin in response to mitochondrial dysfunction correlates with a dramatic increase in the expression of endogenous PINK1 on the outer mitochondrial membrane (5). One potential advantage of stsPINK1 stems from its ability to disassociate PINK1 activity from acute changes in PINK1 protein levels; stsPINK1 allows the activity of PINK1 to be altered, whereas its expression on the outer mitochondrial membrane is held relatively constant.
Two sets of experiments were pursued to illustrate potential uses of stsPINK1. The first sought to demonstrate how parallel experiments comparing Parkin activation by stsPINK1 and by mitochondrial uncoupling might be used to help separate the effects of a drug or gene on PINK1 expression from the effects that a drug or gene may have on other steps in the PINK1-Parkin pathway. The second aimed to show how stsPINK1 might be used in experiments that require comparing the effects of active and inactive PINK1 at closely matched levels of expression.
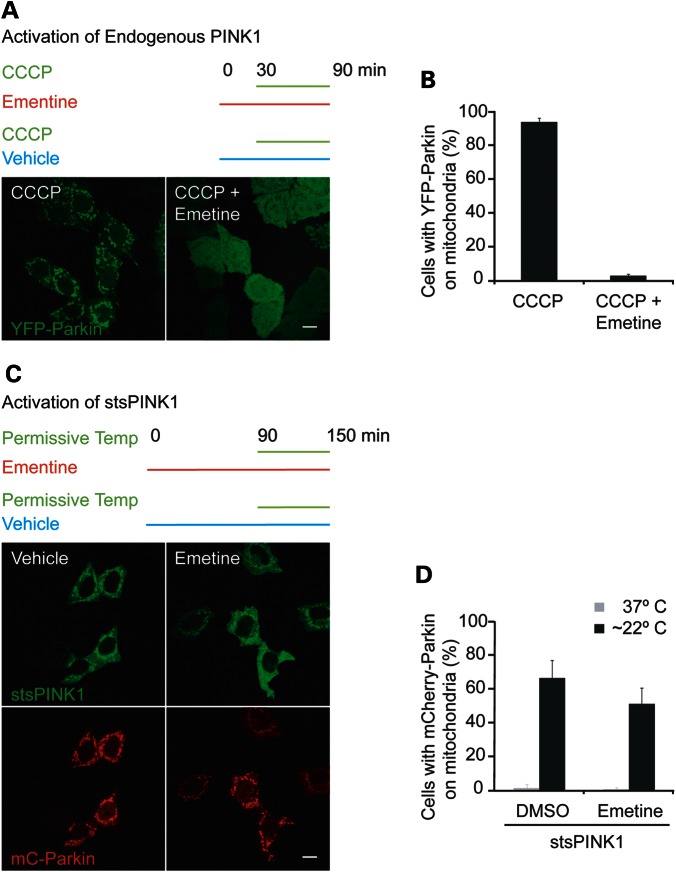
The first set of experiments evaluated a favored model of Parkin activation by PINK1, which suggests that PINK1 accumulation on the outer mitochondrial membrane triggers Parkin activation (5). In this model, PINK1 is proposed to accumulate by a mechanism that requires continued PINK1 translation in conjunction with a block in PINK1 degradation that follows loss of the inner mitochondrial membrane potential. Consistent with this model, it has been reported that inhibiting protein synthesis blocks the recruitment of Parkin by the mitochondrial uncoupler CCCP (5). One interpretation of these findings (hypothesis 1) is that new endogenous PINK1 synthesis is required for the activation of Parkin by depolarization; however, an alternative interpretation (hypothesis 2) is possible in which the synthesis of a protein other than PINK1 is required for the activation of Parkin by depolarization, as pharmacological inhibitors of translation (such as cycloheximide and emetine) prevent the translation of all proteins in the cell. It was reasoned that if hypothesis 2 is true and synthesis of an unidentified protein is required for Parkin activation by depolarization, then protein synthesis inhibition should also block Parkin activation by stsPINK1. Alternatively, if hypothesis 1 is true and the synthesis of endogenous PINK1 alone is required for Parkin activation by depolarization, then protein synthesis inhibition should not block Parkin activation by stsPINK1.
To evaluate these hypotheses, the ability of emetine to inhibit Parkin recruitment in response to CCCP and in response to temperature activation of stsPINK1 was compared. In HeLa cells stably expressing YFP-Parkin, pretreatment with emetine for half an hour prior to the addition of CCCP for an hour inhibited the translocation of Parkin to mitochondria (Fig. 5A and B), consistent with the previously reported findings (5). In contrast, in HeLa cells co-expressing mCherry-Parkin and stsPINK1-YFP, treatment with emetine for 90 min prior to incubation at ∼22°C for 1 h failed to substantially block mCherry-Parkin recruitment to mitochondria (Fig. 5C and D). Similar results were obtained when cycloheximide was used to block protein synthesis (data not shown). These results strongly support the hypothesis that new PINK1 protein synthesis is required for Parkin recruitment following loss of the inner mitochondrial membrane potential.
Figure 5.
Evaluating the effect of protein synthesis inhibition on Parkin activation by mitochondrial depolarization and tsPINK1, respectively. (A and B) HeLa cells stably expressing YFP-Parkin (green) were pretreated with the protein synthesis inhibitor emetine for 30 min or not pretreated prior to treatment with CCCP. Representative confocal images are shown in (A) and quantification for at least 150 cells per condition (in three replicates on two occasions). (C and D) HeLa cells co-transfected with mCherry-Parkin and stsPINK1-YFP were pretreated with emetine for 90 min or not pretreated and then incubated at room temperature (∼22°C) for 1 h prior to fixation. Representative confocal images are shown in (C). (D) At least 150 cells per condition (in three replicates on two occasions) were scored for mCherry-Parkin on mitochondria. Scale bar in all images = 10 µm.
The finding that emetine blocks further PINK1 accumulation in response to CCCP but does not block Parkin recruitment by accumulated PINK1 was exploited to test the differential activity of full-length PINK1 S402A at 37 and 22°C in response to CCCP. To aid this experiment, a HeLa cell line lacking PINK1 expression was generated through targeted disruption of the genomic sequence of exon 2 of PINK1, using TALE nucleases recognizing that locus (Supplementary Material, Fig. S4A–C). In cells transiently expressing PINK1-YFP, Parkin recruitment to mitochondria is greatly accelerated compared with cells with only endogenous PINK1, possibly due to the several-fold increase in PINK1 message following transient transfection (5). In order to more carefully control the levels of PINK1 S402A that accumulate, the cells were treated with CCCP for 10 min at 37°C, then emetine, which blocks protein synthesis, was added to stop the accumulation of additional PINK1. The cells were then incubated at 37 or 22°C for an additional 1 h. In cells incubated at 22°C for 1 h, Parkin recruitment was observed in the majority of the cells (76.8 ± 4.96%), whereas significantly fewer cells incubated at 37°C displayed Parkin recruitment (32.0 ± 10.5%). This suggested that PINK1 S402A also behaves in a temperature-sensitive manner in response to loss of the mitochondrial inner-membrane potential. This experiment further suggested that although PINK1 S402A has decreased activity at 37°C, it retains some activity and its deficiency in Parkin recruitment might be overcome with higher expression. Likely, the expression level of full-length PINK1 S402A would need to be optimized for experiments requiring PINK1 accumulation in response to mitochondrial depolarization (e.g. by using a stable cell line with PINK1 S402A or SS401/402AA expression driven by a weak promoter).
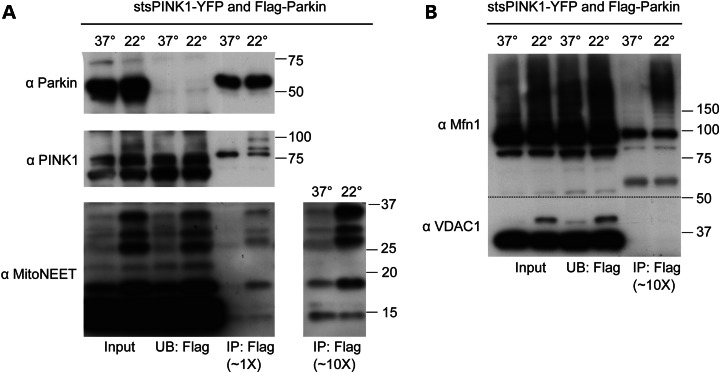
The second set of experiments demonstrating potential uses for stsPINK1 sought to illustrate how stsPINK1 might be used in experiments that require comparing the effects of active and inactive PINK1 at closely matched levels of expression. Specifically, it addressed the question of whether an increase in the association between Parkin and PINK1 by co-immunoprecipitation is evident under conditions in which Parkin is recruited to mitochondria by PINK1. It has been hypothesized that Parkin recruitment to mitochondria reflects primarily the formation of stable complex between Parkin and newly accumulated active PINK1 following mitochondrial depolarization (hereafter, the ‘stable complex’ model) (reviewed in 27). stsPINK1, which allows the disassociation of PINK1 activity and PINK1 expression and localization, offers a unique system for evaluating this model. Incubation of HeLa cells expressing stsPINK1-YFP and Flag-Parkin at 22°C for 2 h increased the appearance of high molecular weight bands of the outer mitochondrial membrane proteins MitoNEET, Mitofusin 1 and VDAC1 when compared with cells incubated at 37°C (Fig. 6A and B, left two lanes), demonstrating temperature-dependent activation of Parkin by stsPINK1. Flag-Parkin was successfully immunodepleted from the samples using anti-Flag M2 antibody-conjugated agarose (Fig. 6A, middle two lanes compared with left two lanes). In the bound fraction, however, no increase in total stsPINK1-YFP abundance was observed between 37 and 22°C. These findings suggest that a net increase in Parkin and PINK1 binding does not account for Parkin recruitment following temperature activation, in contrast with the predictions of the ‘stable complex’ model.
Figure 6.
tsPINK1 used to evaluate the hypothesis that Parkin and PINK1 directly bind. (A) Flag-Parkin and a temperature-sensitive variant of stsPINK1-YFP were transiently expressed in HeLa cells overnight. The media was replaced with CO2-independent media and the cells were exposed to an impermissive temperature (37°C) or a permissive temperature (22°C) for 2 h. Flag-Parkin was captured from cell lysates with an anti-Flag antibody conjugated to an agarose support. The input, the unbound (UB) fraction and the bound (IP) fraction were separated on two SDS–PAGE gels and transferred to PDVF membranes. The bound fraction was diluted to approximately 1 time or 10 times the concentration of Flag-Parkin in the input. (A) Membrane cut above the 37 kDa marker was probed with antibodies against PINK1 and MitoNEET, stripped and then reprobed with an antibody against Parkin. (B) Samples from (A) were separated on a separate gel, transferred to a PDVF membrane, which was cut above the 50 kDa marker (dotted line) and probed for Mitofusin 1 and VDAC1.
Interestingly, a substantial proportion of stsPINK1-YFP in the bound fraction migrated more slowly, consistent with its having been ubiquitinated following temperature activation. In a similar experiment, untagged stsPINK1 in the bound fraction was unmodified, suggesting that the modification is likely on the YFP moiety of stsPINK1-YFP (Supplementary Material, Fig. S5A). If Parkin is the enzyme modifying the YFP moiety of stsPINK1-YFP with ubiquitin, this suggests that in this system Parkin and stsPINK1-YFP may be in close proximity at least transiently to allow the modification of YFP to occur. However, it is also possible that stsPINK1-YFP is modified by a different ubiquitin ligase or is ubiquitinated by Parkin only as a bystander in this system, and, thus, it cannot be concluded from this experiment that Parkin and PINK1 transiently interact physiologically.
In contrast to the absence of an increased association between Parkin and stsPINK1-YFP, a substantial increase in the putative Parkin-substrate MitoNEET was observed in the bound fraction following Parkin activation (Fig. 6A). Unmodified MitoNEET did not increase in the bound fraction following Parkin activation, suggesting that either Parkin has an increased affinity only for ubiquitinated forms of MitoNEET or that, after binding Parkin, MitoNEET remains unmodified for only a short period of time. Similar to its increased affinity for ubiquitinated MitoNEET, activated Parkin also appeared to have an increased affinity for ubiquitinated forms of Mitofusin 1 relative to the unmodified form (Fig. 6B). In contrast, binding between Parkin and VDAC1 was not observed under the same conditions, suggesting that either Parkin does not directly bind VDAC1 or that the immunoprecipitation conditions disrupt Parkin and VDAC1 binding but not the binding between Parkin and other outer mitochondrial membrane proteins, such as MitoNEET (Fig. 6B).
To further examine whether a positive correlation exists between Parkin/PINK1 co-immunoprecipitation and recruitment of Parkin to mitochondria by PINK1, the abilities of wild-type sPINK1 and the catalytically dead sPINK1 K219M to co-immunoprecipitate Parkin were compared. Suggesting a lack of correlation between PINK1/Parkin co-immunoprecipitation and Parkin recruitment by PINK1, sPINK1 wild-type and sPINK1 K219M co-immunoprecipitated with Parkin to a similar extent (Supplementary Material, Fig. S5B), even though wild-type sPINK1 and sPINK1 K219M exhibit markedly different activities with respect to both Parkin recruitment (see Fig. 1) and activation of Parkin E3 ligase activity, as exhibited by the modification of outer mitochondrial membrane protein MitoNEET (Supplementary Material, Fig. S5B).
Taken together, these findings raise the possibility that Parkin recruitment to mitochondria may reflect a stable interaction between PINK1-activated Parkin and its outer mitochondrial membrane substrates rather than increased binding between Parkin and PINK1 directly, as has been also been suggested elsewhere (28).
Parkin recruitment assay for the analysis of disease-associated PINK1 variants
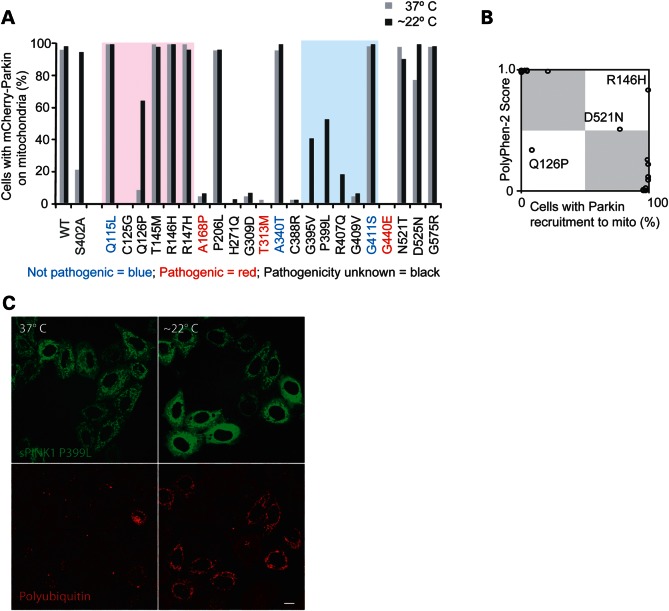
Several disease-associated PINK1 variants have been identified in the activation segment, including G395V, P399L, R407Q, G409V and G411A (reviewed in 29). That three of these residues (namely, G395, P399 and R407) exhibited thermal lability when mutated to alanine prompted us to screen a panel of 22 PD-associated variants for activity at 37°C and for rescue of activity following a reduction in temperature.
In order to validate the Parkin recruitment assay for assessing the functional relevance of the PINK1 sequence variants, three PINK1 variants of proven pathogenicity and three variants of proven non-pathogenicity, according to their annotations in the PD mutation database (29), were evaluated for activity at 37°C. All sPINK1 variants exhibited stable protein expression on mitochondria, and cells with similar sPINK1 expression levels were scored for Parkin recruitment. Consistent with the validity of the assay for assessing PINK1 function, all three non-pathogenic variants recruited Parkin to mitochondria in the majority of cells, similar to wild-type (Fig. 7A, blue text in the x-axis label), whereas all three pathogenic variants disrupted Parkin recruitment to mitochondria (Fig. 7A, red text in the x-axis label), similar to the catalytically inactive variants of PINK1.
Figure 7.
PINK1 variants linked to parkinsonism evaluated for rescue at lower temperatures. (A) A series of PINK1 variants were introduced into sPINK1. Some are annotated as ‘pathogenic’ (red), some as ‘not pathogenic’ (blue) and most as ‘pathogenic nature unclear’ (black) in the PD mutation database. Wild-type sPINK1 and sPINK1 S402A in gray serve as positive controls for variants with activity at 37°C and variants exhibiting temperature sensitivity, respectively. The sPINK1 variants were screened for activity at 22 and 37°C, using the Parkin recruitment assay as described in Figure 2C. The pink shading on the graph represents those residues in the region N-terminal to the kinase domain, and the blue shading represents those residues in the activation segment of the kinase domain. (B) Scatter plot representation of Parkin recruitment by sPINK1 variants in (A) at 37°C along the x-axis and the PolyPhen-2 score of the PINK1 variants along the y-axis, where 0.0 indicates substitutions predicted to be most benign and 1.0 indicates substitutions predicted to most damaging. Boxes shaded gray indicate areas of plot where both methods would predict the substitution to be either benign or damaging. (C) cells co-transfected with sPINK1 P399L (green) and mCherry-Parkin (not pictured) and exposed to either 22 or 37°C for 2 h were immunostained for polyubiquitin and imaged by confocal microscopy. Scale bar = 10 μm.
The majority of non-synonymous sequence variations in PINK1 (including 16 of the 22 variants examined here) are currently annotated in the PD mutation database as being of unknown clinical significance (29). This is due to insufficient genetic evidence from other patients and healthy controls to demonstrate that they are a cause of, and not merely associated with, Parkinson's disease. In all, nine variants of unknown clinical significance disrupted Parkin recruitment at 37°C, whereas seven variants recruited Parkin at 37°C. Of the variants in the activation segment, one (G411S) is known to be non-pathogenic, whereas the other four were found to substantially disrupt Parkin recruitment at 37°C. Also of note, a cluster of residues N-terminal to PINK1 kinase domain, which are conserved among metazoan PINK1 orthologs, disrupted Parkin recruitment without affecting the global stability of sPINK1, as judged by fluorescence intensity. This suggests that residues N-terminal to the conserved kinase domain are important for PINK1 function likely independently of their effect on kinase maturation or localization.
To further evaluate the Parkin recruitment assay for assessing the disruptiveness of PINK1 disease-associated variants, results from the recruitment assay were compared with predictions of the PolyPhen-2 algorithm, a widely used bioinformatic approach to assess the likely effects of non-synonymous variations (30). Predictions were said to be discordant if the HumDIV-trained PolyPhen-2 score was in the benign range and Parkin was recruited to mitochondria by sPINK1 in fewer than 50% of cells or the HumDIV-trained PolyPhen-2 score was in the probably damaging range and Parkin was recruited to mitochondria by sPINK1 in >50% of cells. Otherwise, the predictions were said to be concordant. In total, concordant predictions were made by the two methods for 19 of the 22 variants (86.3%). Of the three discordant predictions, one variant was predicted to be ‘benign’ by PolyPhen-2 but had low activity in the Parkin recruitment assay, and two variants predicted to be ‘possibly damaging’ by PolyPhen-2 were active in the Parkin recruitment assay. Together, these results suggest that the Parkin recruitment assay may provide non-redundant information to help determine whether or not a mutation is likely to be pathogenic.
The disease-associated PINK1 variants were assessed next for the rescue of activity following incubation at 22°C for 2 h. Substantial rescue of activity was observed for three of the four disease-associated mutations in the activation segment that exhibited little or no activity at 37°C (G395V, P399L and R407Q), both in the screening experiment and in a subsequent replication experiment (Fig. 7A–C and Supplementary Material, Fig. S6A and B). This was consistent with the findings from alanine-scanning mutagenesis of this region and with the notion that the activation segment of PINK1 is a ‘hot spot’ for thermally labile activity (see Fig. 4B). Less expectedly, the activity of a variant Q126P was also partially restored following incubation at room temperature. Q126P, although not strictly conserved among PINK1 orthologs, is nested near conserved residues in a region that is predicted to form an α-helix. The Q126P mutation would be predicted to disrupt the α-helix, leading to mispositioning of the adjacent conserved residues.
Together, these findings suggest that the activity of a subset of PINK1 disease-associated variants might be rescued following post-translational stabilization, e.g. by temperature.
DISCUSSION
This study provides a structure–function analysis of PINK1, using a simple cell-based reporter: Parkin recruitment to mitochondria. Consistent with PINK1 acting as a serine–threonine kinase, individual substitution of seven of the residues conserved among active kinases disrupted the recruitment of Parkin to mitochondria. Although it is formally possible that these mutations prevented Parkin recruitment by a mechanism other than blocking phosphyl transfer, these findings strongly suggest that PINK1 is an active kinase in the PINK1/Parkin pathway. These results are also consistent with a recent report using recombinant PINK1 orthologs from several insect species to demonstrate that the aspartic acid of the DFG motif (equivalent of D384 in human PINK1) is required for the phosphorylation of an optimized substrate peptide (13). The study also found that recombinant human PINK1 exhibits low activity, offering a possible explanation for a previous failure to show that D384 is required for activity (14).
Activation segment phosphorylation, including at residue S402, was found to be inessential for the activity of PINK1 in the Parkin recruitment assay. This is in contrast to most RD kinases including PKA, which are initially translated in an inactive form and are phosphorylated in a conserved position within the activation segment, which stabilizes the segment in an active conformation (reviewed in 18).
While this manuscript was in preparation, Okatsu et al. (31) reported that the S402A substitution in PINK1 disrupted Parkin recruitment and activation at physiological temperatures, as is reported here. Okatsu et al. also found that the PINK1 S402A substitution (and other substitutions disrupting the catalytic activity of PINK1) blocked PINK1 autophosphorylation. From these data, Okatsu et al. (31) concluded that PINK1 is phosphorylated at S402 and that formation of the phosphoserine (or a phosphomimetic) at this site is required for the recruitment and activation of Parkin by PINK1. Although all of the data presented in Okatsu et al. (31) are consistent with the data reported above, in this study the substitutions S402N and S402C (which cannot be phosphorylated and are not phosphomimetic) were also found to retain function at physiological temperatures. These findings strongly suggest that the S402A mutation disrupts activity not by preventing the formation of a phosphoserine but rather through loss of a polar residue that is required for full activity at physiological temperatures (but not at subphysiological temperatures). Loss of autophosphorylation of PINK1 by the S402A mutant, as reported by Okatsu et al. (31), may reflect its decreased catalytic activity at physiological temperatures rather than the loss of a phosphorylation site. Alternatively, S402A may be a site of autophosphorylation. Nonetheless, the retained activity of S402C and S402N (which are neutral in charge and cannot be phosphorylated) suggests that phosphorylation at S402 is likely not required for Parkin recruitment.
Further analysis of the activation segment points to two features as possibly stabilizing the activation segment of PINK1 in the absence of activation segment phosphorylation. First, similar to the constitutively active RD kinase casein kinase I, PINK1 has a relatively long activation segment (34 amino acid residues in PINK1 compared with 25 residues in PKA), which may allow for extensive contacts with the C-lobe, as was observed in the casein kinase I structure (25). Additionally, in PINK1, an aspartic acid occupies the position of the primary phosphorylation site in a manner that is analogous to the constitutively active kinase Phk (26). The C-terminal group of the aspartic acid may form an electrostatic interaction with the conserved arginine of the catalytic loop, as is observed in the Phk structure (26), and as is observed between the phosphoserine and the catalytic loop arginine in the structure of PKA (32).
In the homology model, using active kinases as the template, the activation loop of PINK1 appeared to form a hairpin structure and in this respect was reminiscent of the activation loop of CK1. In the case of PINK1, this structure appeared to be stabilized by a conserved hydrophobic patch near its tip. However, whether PINK1 actually forms a hairpin structure remains uncertain, as this portion of the homology model is poorly templated and attempts to demonstrate the importance of the hydrophobic patch through reciprocal mutations in pairs of residues near or within the patch were unsuccessful.
Although PINK1 did not require activation segment phosphorylation for activity, it is still possible that PINK1 is translated in an inactive form and is activated by a mechanism other than activation segment phosphorylation. Supporting this view, PINK1 appears to promote Parkin activity more robustly when PINK1 is membrane bound, at least in indirect assays of PINK1 activity (28). Increased local concentration of PINK1 as a consequence of membrane association may activate PINK1 through PINK1–PINK1 allosteric interactions, as has been observed for some members of the epidermal growth factor kinase family (33). PINK1 autophosphorylation, which has been directly observed (31,34), could also be promoted by the increased local concentration and activate PINK1, although PINK1 autophosphorylation has not as of yet been shown to promote its activity [see reference (34) and discussion above]. Alternatively, PINK1 may be primarily regulated through its expression and localization, both of which change dramatically in response to loss of the mitochondrial inner-membrane potential (5).
Unexpectedly, the S402A substitution in PINK1 rendered the kinase temperature sensitive. Although several temperature-sensitive kinases have been described, generally they have been identified in kinases that are required for cellular growth or survival or have been engineered based on homology to previously identified temperature-sensitive kinases (see, for instance, reference 35). This has limited the isolation of temperature-sensitive kinases largely to essential kinases or kinases highly homologous to essential kinases. To our knowledge, this is the first description of a temperature-sensitive residue identified with a cellular assay of kinase function that does not depend on cell survival or cell growth. Although the identification of temperature-sensitive variants of PINK1 was fortuitous in this case, it suggests a systematic mutagenesis approach for identifying temperature-sensitive variants in inessential kinases.
Additional mutagenesis of the activation segment suggested that the segment is a ‘hot spot’ for thermally labile residues. In this context, it is notable that a temperature-sensitive allele in the Drosophila protein kinase CK2α was identified in the activation segment of that protein, raising the possibility that the activation segment of kinases may be a good target for the generation of temperature-sensitive kinases more generally (35).
One distinct use of the stabilized tsPINK1 chimera (stsPINK1) is that it allows the isolation of PINK1 activity from changes in protein expression and localization. This permits the differentiation of pharmacological or genetic modifiers of PINK1 that primarily affect PINK1 expression and targeting from those that also affect PINK1 activity following its maturation.
As an illustration of this use of stsPINK1, the requirement of new protein synthesis for Parkin activation was re-evaluated. A global inhibitor of protein translation was previously reported to block Parkin recruitment in response to mitochondrial depolarization (5). This block is predicted by a proposed model of the PINK1/Parkin pathway, in which PINK1 accumulates on the outer mitochondrial membrane following depolarization due to a block in its degradation coupled with continued PINK1 translation and targeting. In turn, the accumulation of PINK1 on the outer mitochondrial membrane drives Parkin activation. That a short treatment with the protein synthesis inhibitor emetine did not block Parkin recruitment by stsPINK1 demonstrated that apart from PINK1 no other new protein synthesis is required for Parkin activation following mitochondrial uncoupling. Thus, these findings strongly suggest that PINK1 accumulation, in isolation, drives Parkin activation in response to mitochondrial uncoupling.
stsPINK1 also allows for experiments with closely matched expression of active and inactive PINK1. As an illustration of this kind of experiment, co-immunoprecipitation of stsPINK1 and Parkin at temperatures that were either permissive or impermissive for stsPINK1 activity was compared. In these experiments, a small fraction of Parkin appeared to be in complex with both inactive and active forms of stsPINK1. However, the amount of Parkin in complex with stsPINK1 did not increase between activating and inactivating temperatures and, thus, did not correlate with the increase in Parkin activity and Parkin recruitment to mitochondria observed between these conditions. These observations argue that Parkin recruitment to mitochondria does not reflect the formation of new stable Parkin/PINK1 complexes and is consistent with the previous observation that Parkin and PINK1 are largely associated with stable protein complexes of different sizes by gel filtration and blue native gel electrophoresis (28,36). In a marked contrast to stsPINK1, a complex between Parkin and MitoNEET, a likely substrate of exogenously expressed Parkin (20), was markedly increased following the temperature-dependent activation of PINK1, suggesting a correlation between Parkin recruitment and Parkin co-immunoprecipitation with at least some of its outer mitochondrial membrane substrates. In addition to the experiments described, stsPINK1, which allows the conditional activation of Parkin in the absence of mitochondrial uncoupling agents, may be useful for experiments in which the use of mitochondrial uncoupling agents would be detrimental (e.g. in experiments with neurons, which are more dependent on oxidative phosphorylation and are less tolerant of treatment with CCCP compared with many established cell lines).
In addition to its use for studies in human cells, that S402 is conserved among PINK1 orthologs in M. musculus, D. rerio, D. melanogaster and C. elegans suggests that the S402A mutation identified in human PINK1 may serve as a basis for developing temperature-sensitive models for the in vivo study of the PINK1/Parkin pathway.
The success of the Parkin recruitment assay in evaluating PINK1 residues of structural interest suggested that the assay might also be effective in evaluating PINK1 variants that have been associated with disease. All variants of proven pathogenicity and proven non-pathogenicity disrupted or supported Parkin recruitment, respectively, further validating the assay. For those variants annotated in the PD mutation database as of ‘unknown clinical significance’ (29), there was generally good agreement between results from the Parkin recruitment assay and predictions of variant disruptiveness by the widely used Polyphen-2 algorithm (30). Notably, one variant, Q126P, that was predicted to be ‘benign’ using the Polyphen-2 algorithm was found to substantially disrupt Parkin recruitment, suggesting that the Parkin recruitment assay may provide non-redundant information to support the likely pathogenicity of PINK1 variants.
That the Q126P substitution is truly pathogenic as the Parkin recruitment assay would predict is supported by its pattern of segregation in a German family described previously (37). In this family, two affected siblings (developing parkinsonism at 36 and 40) were found to be homozygous for the Q126P mutation, whereas three unaffected siblings in their 60s or 70s at the time of the study were found be heterozygous or homozygous for the wild-type allele (37). Consistent with the likely pathogenicity of Q126P, the variant does not appear in the dbSNP database (BUILD 136) or among called variants from the 1000 Genomes Project (1000 Genomes release 12) (38). This suggests that the variant is rare in the general population, as might be expected for a highly penetrate cause of recessive parkinsonism. Finally, although Q126 is not strictly conserved among PINK1 orthologs (which may account for a Polyphen-2 score in the ‘benign’ range), it lies within a predicted α-helix that is N-terminal to the core kinase domain. The change from glutamine to proline would likely break the α-helix and disrupt the position of neighboring conserved residues. These considerations support the view that the Q126P mutation is pathogenic as the Parkin recruitment assay predicted.
Considered together, these findings suggest that Parkin recruitment by sPINK1 variants may offer a general method for assessing variant function and, by extension, the likelihood that a variant is pathogenic. This assay has the advantages of being simple to perform with standard equipment and cell lines available at many institutions. It requires only transient co-transfection of two plasmids (encoding mCherry-Parkin and sPINK1-YFP) into established cell lines such as HeLa and assessment for mCherry/YFP co-localization 18–36 h later with a fluorescent microscope. The contrast between functional variants and non-functional variants is stark with mCherry/YFP co-localization in the vast number of cells for functional variants and very little or no co-localization for non-functional variants. This assay is limited, however, to those variants in the cytosolic portion of the kinase (residues 111–581), as they are the only ones present in the sPINK1 fusion protein.
The activities of several PINK1 variants were rescued at subphysiological temperatures. Three of these reside in the activation loop, whereas Q126P, mentioned above, occupies a position within a predicted α-helix N-terminal to the core kinase domain. That a subset of PINK1 variants can be rescued by a reduction in temperature suggests that they might also be stabilized post-translationally by chemical and pharmacological chaperones, as has been observed previously for other temperature-sensitive proteins. A mutation in the chloride channel responsible for cystic fibrosis (CFRT ΔF508), for instance, generates a protein that becomes misfolded and accumulates in the ER. Either a reduction in temperature or treatment with chemical or pharmacological chaperones favors local folding and partially restores targeting of the chloride channel to the plasma membrane (39). The rescue of activity following a reduction in temperature for a subset of disease-associated mutations in PINK1 suggests that chemical or pharmacological chaperones may similarly restore PINK1 function in these patients and perhaps alter the course of their disease.
The identification of temperature-sensitive variants in PINK1 offers a tool with potentially broad use for the study of the PINK1/Parkin pathway and mitophagy in human cells as well as in model organisms from roundworms to mice. Additionally, that some PINK1 variants are temperature sensitive has treatment implications for families whose disease is likely a result of those variants.
MATERIALS AND METHODS
Cell culture
The HeLaYFP-Parkin cell line, which stably expresses YFP-Parkin, and the parent control HeLa cell line have been described previously. The cells were grown in DMEM with high glucose and 10% FBS with or without phenol red and pencillin/streptomycin added (Invitrogen). HeLaYFP-Parkin cells were grown with hygromycin 200 µg/ml (Invivogen) supplemented to the media to maintain selection for YFP-Parkin. The PINK1−/− HeLa cell line was generated by TALEN technology with targeting site as follows: GTTGCTTCCAGGGAGAGGCCCAAGGTACCAGTGCACCAGGAGAAGGGCAGGAG, in which the binding site for the left and right TALEs are underlined; a KpnI restriction site lies in between. 16-mer left and right TALEs were assembled according to Huang et al. (40) and cloned into a final TALEN vector modified from Miller et al. (41). 0.8 µg of left and right TALEN constructs were co-transfected with 0.4 µg of pEYFP-C1 vector (Clontech) in HeLa cells, and GFP-positive cells were FACS-sorted and plated into 96-well plates 2 days after transfection. Single colonies were picked and screened by PCR with KpnI digestion. Positive clones were confirmed by immunoblotting.
Immunocytochemistry
For immunocytochemistry, the following primary antibodies were obtained from commercial or non-commercial sources as indicated: rabbit polyclonal TOMM20 (Santa Cruz, USA), anti-total polyubiquitinated protein (Clone FK1) and anti-p62 polyclonal guinea pig C-terminus (Progen, DE, USA). All primary antibodies were used at a 1:500–1:1000 dilution for immunocytochemistry. The following secondary antibodies were also used: anti-mouse IgG, anti-rabbit IgG and anti-guinea pig IgG antibodies preconjugated to Alexa 488, 594, 633 and 647 fluorescent dyes (Invitrogen). All secondary antibodies were used at a 1:1000 dilution.
Immunocytochemistry was performed as follows: cultured cells seeded on borosilicate chamber sildes (Nunc) were fixed in 4% paraformaldehyde in 1× phosphate buffered saline (PBS) for 15–30 min, washed with 1× PBS and then permeabilized with 0.5% Triton X-100 (Sigma). Permeabilized cells were blocked with 10% BSA in 1× PBS (PBS-B) for 15–60 min and then incubated in primary antibody diluted in PBS-B for 1 h at room temperature or overnight at 4°C. Cells were subsequently washed with PBS-B three times and then incubated with a secondary antibody for 45 min at room temperature. Finally, cells were washed with 1× PBS.
Plasmid vectors
The plasmids encoding mito-Keima, YFP-Parkin and mCherry-Parkin have been reported previously. A plasmid coding for the fusion protein comprising OPA3 (1–30)-PINK1 (111–581)-YFP (called sPINK1-YFP in text) was created by subcloning a PCR product containing PINK1 (111–581) into the BamHI site of the EYFP-N1/OPA3-YFP vector [from Dr SW Ryu (NINDS)], which contains OPA3 (1–30) between the HindIII and BamHI sites of EYFP-N1 (Clontech), as described previously. OPA3-PINK1-mCherry was constructed by subcloning mCherry from the mCherry-N1 vector (Clontech) into the OPA3-PINK1-YFP vector. Flag-Parkin and OPA3-PINK1-Flag were produced by subcloning Parkin and OPA3-PINK1 PCR products into the pcDNA5.1 vector (Invitrogen) containing a streptavidin and Flag tag, using a sequence and ligation-independent cloning method.
Amino acid substitutions in sPINK1 were introduced by site-directed mutagenesis, using the Stratagene Quikchange protocol. In brief, primers for the Quikchange protocol were designed using the PrimerX webtool (http://www.bioinformatics.org/primerx/; accessed on 10 April 2011), with the protocol set to ‘QuikChange Site-Directed Mutagenesis Kit by Stratagene’. PCR products containing the desired point mutation were amplified using the Pfu Turbo polymerase (Stratagene, USA) for 12–15 cycles. The plasmid template was digested by adding the DpnI restriction enzyme (1 µl, NEB) to 50 µl of PCR reaction mixture and incubating at 37°C for 60 min. A portion of the reaction mix was added directly to 10–50 µl of NEB 5-α high-competency E. coli cells in a 1:10 ratio, and the E. coli were transformed following a standard heat shock protocol provided by the manufacturer. Single colonies were isolated from LB plates containing the appropriate antibiotics, usually kanamycin or ampicillin (Sigma). Mutagenesis was verified by Sanger DNA sequencing of appropriate regions of the vectors and was performed by the DNA sequencing core facility at NINDS in the USA or by Geneservice in the UK.
Cellular transfection
For confocal imaging experiments, cells seeded on borosilicate chamber slides were transfected with plasmids using 6 µl of the Fugene6 (Roche) or X-tremeGene9 transfection (Roche) reagent for every 1 µg of plasmid, using the manufacturer's instructions. For immunoblotting experiments, cells plated in dishes were transfected using Lipofectamine LTX reagent (Invitrogen) in a 1 µg plasmid:3 µl LTX ratio, using the manufacturer's instructions.
Confocal microscopy
Fixed cells and live cells were imaged using an inverted microscope (Zeiss LSM510 Meta) with a 63×/1.4 Oil DIC Plan Apo objective at 22°C. For live cell imaging experiments, HeLa cells were incubated with 30 nm TMRE or MitoTracker Deep Red (Molecular Probes) for 15 min prior to imaging. To quantify the fluorescence intensity of YFP and mCherry signals co-localizing with mitochondria, a mask for mitochondria was created from the Mitotracker Deep Red channel in ImageJ64 1.43u (NIH) using the ‘Convert to Mask’ function. The mask was inverted and then subtracted from the YFP and mCherry channels to produce images with only the YFP or mCherry signals, respectively, overlapping with the Mitotracker Deep Red signal. Cells were manually selected from these masked images using the Region of Interest tool. The same regions of interest were applied to all channels and all time points in the image sets. For each image, the average intensity was measured for all pixels within the region of interest with intensities between 1 and 255.
Immunoblotting/immunoprecipitation
Cells were lysed in 1% SDS lysate buffer (10 mm Tris/HCl pH 7.4, 150 mm NaCl). DNA was sheared by 5–10 passages through 26 and 30 gauge needles and lysates were obtained after centrifugation (16 000g). 1× LDS loading buffer (Invitrogen) containing 25 mm DTT was added to lysates to a final 1× concentration (42), and the protein content was resolved on 12–22% polyacrylamide gradient SDS–PAGE gels (Invitrogen) and transferred to PVDF membranes (Millipore). Proteins were detected by immunoreaction with the antibodies listed below. To block non-specific antibody-binding sites, the membrane was incubated in a 5% solution of skim milk dissolved in 1× PBS containing 0.1% Tween (PBS-T) for 30–60 min. The membrane was then incubated with primary antibody diluted in skim milk for 1 h at room temperature or overnight at 4°C. The membrane was washed three times in PBS-T and then incubated for 1 h with a secondary antibody conjugated to horseradish peroxidase (HRP) diluted in skim milk. The membrane was washed three times in PBS-T, and HRP bound to protein on the membrane was reacted with a chemiluminescent agent. Resulting radiation was detected with an X-ray film [Kodak (USA) or Fuji (Japan)] or directly with a Bio-Rad imaging system.
The following commercial primary antibodies were used for immunoblotting: anti-Porin HL Ab3 (also referred to as Ab6 in the literature) (Calbiochem), anti-Parkin (PRK8) (Santa Cruz), anti-TOMM20 polyclonal (Santa Cruz Biotechnology), anti-PINK1 polyclonal (Novus Biologicals) and anti-mitoNEET mouse monoclonal (Proteintech).
For immunoprecipitation of Flag-Parkin HeLa cells co-transfected with Flag-Parkin and stsPINK1-YFP, sPINK1-YFP K219M, sPINK1-YFP wild-type or untagged stsPINK1 were solubilized in an NP40 buffer containing 0.5% NP-40, 10 mm Tris, pH 7.5, 150 mm NaCl, plus one complete protease tablet (Roche). Lysates were incubated at 4°C overnight with 25–50 µl of anti-Flag M2 agarose gel (Sigma). The beads were washed three times in dilution buffer containing 10 mm Tris, pH 7.5, and 150 mm NaCl. Complexes were eluted by boiling the beads in 1× LDS buffer with 25 mm DTT for 5–10 min. Immunoprecipitation of YFP-Parkin from HeLa cells transiently co-transfected with YFP-Parkin and stsPINK1-mCherry in an analogous manner excepting the use of GFP-Trap agarose beads in place of anti-Flag M2 agarose gel.
AUTHORs' CONTRIBUTIONS
D.P.N., R.J.Y and J.E.W. conceived the project and designed the experiments. D.P.N. carried out the experiments. C.W. generated the PINK1−/− HeLa cell line. D.P.N, R.J.Y. and J.E.W. wrote the manuscript.
SUPPLEMENTARY MATERIAL
FUNDING
This work received funding support from the Medical Research Council (J.E.W. and D.P.N.), the intramural program at the National Institute of Neurological Disorders and Stroke (R.J.Y. and D.P.N.) and NIH training grant 1F30AG039185-01 (D.P.N.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the proteomics group at the MRC Mitochondrial Biology Unit, especially Joe Carroll, Ian Fearnley and Byron Andrews, as well as members of the Youle laboratory for helpful discussions. We also thank Dragan Maric and the NINDS flow cytometry facility as well as Jim Nagle and the NINDS sequencing facility for their research support. Finally, we thank Dr Jehan El-Bayoumi, Dr Alan Wasserman and the George Washington University Department of Medicine for their support of trainee research (D.P.N.).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ropper A.H., Adams R.D., Victor M., Samuels M.A. Adams and Victor's Principles of Neurology. 9th edn. New York, NY: McGraw-Hill Medical; 2009. [Google Scholar]
- 2.Olanow C.W., Rascol O., Hauser R., Feigin P.D., Jankovic J., Lang A., Langston W., Melamed E., Poewe W., Stocchi F., et al. A double-blind, delayed-start trial of rasagiline in Parkinson's disease. N. Engl. J. Med. 2009;361:1268–1278. doi: 10.1056/NEJMoa0809335. doi:10.1056/NEJMoa0809335. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron. 2010;68:201–206. doi: 10.1016/j.neuron.2010.10.014. doi:10.1016/j.neuron.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. doi:10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. doi:10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 7.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. doi:10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. doi:10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. doi:10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. doi:10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziviani E., Tao R.N., Whitworth A.J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. doi:10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole A.C., Thomas R.E., Yu S., Vincow E.S., Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. doi:10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodroof H.I., Pogson J.H., Begley M., Cantley L.C., Deak M., Campbell D.G., van Aalten D.M., Whitworth A.J., Alessi D.R., Muqit M.M. Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 2011;1:110012. doi: 10.1098/rsob.110012. doi:10.1098/rsob.110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D.W., Petsko G.A., Cookson M.R. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl Acad. Sci. USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. doi:10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E.M., Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. doi:10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 16.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. doi:10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 17.Scheeff E.D., Eswaran J., Bunkoczi G., Knapp S., Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. doi:10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. doi:10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 19.Johnson L.N., Noble M.E., Owen D.J. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. doi:10.1016/S0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 20.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., Hess S., Chan D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. doi:10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weihofen A., Ostaszewski B., Minami Y., Selkoe D.J. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum. Mol. Genet. 2008;17:602–616. doi: 10.1093/hmg/ddm334. doi:10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 22.Katayama H., Kogure T., Mizushima N., Yoshimori T., Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. doi:10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. doi:10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.S., Kimura M., Sato S., Hattori N., Komatsu M., et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longenecker K.L., Roach P.J., Hurley T.D. Three-dimensional structure of mammalian casein kinase I: molecular basis for phosphate recognition. J. Mol. Biol. 1996;257:618–631. doi: 10.1006/jmbi.1996.0189. doi:10.1006/jmbi.1996.0189. [DOI] [PubMed] [Google Scholar]
- 26.Lowe E.D., Noble M.E., Skamnaki V.T., Oikonomakos N.G., Owen D.J., Johnson L.N. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 1997;16:6646–6658. doi: 10.1093/emboj/16.22.6646. doi:10.1093/emboj/16.22.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra D.P., Youle R.J. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. doi:10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazarou M., Jin S.M., Kane L.A., Youle R.J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuytemans K., Theuns J., Cruts M., Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum. Mutat. 2010;31:763–780. doi: 10.1002/humu.21277. doi:10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. doi:10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M., et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 2012;3:1016. doi: 10.1038/ncomms2016. doi:10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J., Trafny E.A., Knighton D.R., Xuong N.H., Taylor S.S., Ten Eyck L.F., Sowadski J.M. 2.2 Å refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D Biol. Crystallogr. 1993;49:362–365. doi: 10.1107/S0907444993000423. doi:10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Gureasko J., Shen K., Cole P.A., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. doi:10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., Burchell L., Walden H., Macartney T.J., Deak M., et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. doi:10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuntamalla P.P., Kunttas-Tatli E., Karandikar U., Bishop C.P., Bidwai A.P. Drosophila protein kinase CK2 is rendered temperature-sensitive by mutations of highly conserved residues flanking the activation segment. Mol. Cell Biochem. 2009;323:49–60. doi: 10.1007/s11010-008-9963-6. doi:10.1007/s11010-008-9963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas K.J., McCoy M.K., Blackinton J., Beilina A., van der Brug M., Sandebring A., Miller D., Maric D., Cedazo-Minguez A., Cookson M.R. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. doi:10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prestel J., Gempel K., Hauser T.K., Schweitzer K., Prokisch H., Ahting U., Freudenstein D., Bueltmann E., Naegele T., Berg D., et al. Clinical and molecular characterisation of a Parkinson family with a novel PINK1 mutation. J. Neurol. 2008;255:643–648. doi: 10.1007/s00415-008-0763-4. doi:10.1007/s00415-008-0763-4. [DOI] [PubMed] [Google Scholar]
- 38.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown C.R., Hong-Brown L.Q., Biwersi J., Verkman A.S., Welch W.J. Chemical chaperones correct the mutant phenotype of the delta F508 cystic fibrosis transmembrane conductance regulator protein. Cell Stress Chaperones. 1996;1:117–125. doi: 10.1379/1466-1268(1996)001<0117:ccctmp>2.3.co;2. doi:10.1379/1466-1268(1996)00<0117:CCCTMP>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P., Xiao A., Zhou M., Zhu Z., Lin S., Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotech. 2011;29:699–700. doi: 10.1038/nbt.1939. doi:10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 41.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., et al. A TALE nuclease archietecture for efficient genome editing. Nat. Biotech. 2011;29:143–150. doi: 10.1038/nbt.1755. doi:10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. doi:10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.