Abstract
Abnormal metabolism of the tau protein is central to the pathogenesis of a number of dementias, including Alzheimer's disease. Aberrant alternative splicing of exon 10 in the tau pre-mRNA resulting in an imbalance of tau isoforms is one of the molecular causes of the inherited tauopathy, FTDP-17. We showed previously in heterologous systems that exon 10 inclusion in tau mRNA could be modulated by spliceosome-mediated RNA trans-splicing (SMaRT). Here, we evaluated the potential of trans-splicing RNA reprogramming to correct tau mis-splicing in differentiated neurons in a mouse model of tau mis-splicing, the htau transgenic mouse line, expressing the human MAPT gene in a null mouse Mapt background. Trans-splicing molecules designed to increase exon 10 inclusion were delivered to neurons using lentiviral vectors. We demonstrate reprogramming of tau transcripts at the RNA level after transduction of cultured neurons or after direct delivery and long-term expression of viral vectors into the brain of htau mice in vivo. Tau RNA trans-splicing resulted in an increase in exon 10 inclusion in the mature tau mRNA. Importantly, we also show that the trans-spliced product is translated into a full-length chimeric tau protein. These results validate the potential of SMaRT to correct tau mis-splicing and provide a framework for its therapeutic application to neurodegenerative conditions linked to aberrant RNA processing.
INTRODUCTION
Tau is a microtubule-associated protein that accumulates in characteristic filamentous inclusions in affected neurons in several related neurodegenerative diseases collectively termed tauopathies (1). Tauopathies include Alzheimer's disease (AD), a subgroup of frontotemporal lobar degeneration (FTLD-tau) and the inherited tauopathy, frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17), caused by dominant mutations in the MAPT gene, encoding tau. Tau regulates microtubule dynamics as well as a number of microtubule-dependent properties such as axonal transport, and might also have signaling functions (2). Six tau isoforms are expressed in the human brain by alternative splicing of exons 2, 3 and 10 in the tau pre-mRNA [reviewed in 3]. Of particular significance, exon 10 (E10) encodes the second of four microtubule-binding repeated motifs in the C-terminal part of the protein. Thus, exclusion or inclusion of E10 gives rise to tau isoforms with three (3R tau, E10−) or four (4R tau, E10+) microtubule-binding domains. 3R Tau and 4R tau are expressed in approximately equal amounts in the normal adult human brain, but only 3R tau is expressed during early development.
E10 splicing has an important significance in the pathogenesis of tauopathies as a number of FTDP-17 MAPT mutations affect cis-acting elements regulating E10 alternative splicing. FTDP-17 mutations in splicing regulatory elements result in a 2- to 3-fold increase in E10 inclusion and thus in an excess of 4R tau (4–6). Elevated levels of 4R tau have been reported in sporadic AD and the MAPT H1 haplotype, a risk factor for AD, promotes E10 inclusion in tau mRNA (7). Therefore, aberrant E10 splicing is not only causative of disease in FTDP-17, but might be also a contributing factor to AD.
Tau is a major target for therapeutic intervention in tauopathies but no effective treatment has been developed to date (8). In the specific case of FTDP-17 caused by tau mis-splicing, correction of isoform imbalance by direct intervention at the RNA level offers several advantages over conventional therapies (9). Not only does RNA therapy avoids side effects of drugs, but, most importantly, the repaired product is under endogenous transcriptional control and has the same expression pattern as the normal transcript. A powerful and versatile method to reprogram RNA is spliceosome-mediated RNA trans-splicing (SMaRT) (10, 11). SMaRT creates a hybrid mRNA by a trans-splicing reaction between the 5′ splice site of a target pre-mRNA and the 3′ splice site of a separate pre-trans-splicing RNA molecule (PTM). We showed previously in heterologous systems that tau E10 could be included or excluded by trans-splicing reprogramming of tau transcripts (12, 13). While this validated tau RNA as a target for trans-splicing reprogramming, development of SMaRT into therapeutic applications requires that RNA reprogramming is obtained with endogenous tau transcripts in differentiated neurons and that the trans-spliced product is translated into a chimeric protein. A suitable animal model of tau mis-splicing to evaluate SMaRT in neurons is the htau mouse. Htau mice are a transgenic line that expresses the entire human wild-type human MAPT gene from a PAC transgene in a mouse Mapt null background (14). In these animals, human tau is in a genomic context and both 4R and 3R tau isoforms are expressed in adult animals, with an excess of 3R tau over 4R tau (14, 15). Thus, compared with either adult human neurons, expressing both isoforms in an equimolar ratio, or wild-type adult mouse neurons, expressing 4R tau only, htau mice have an abnormal pattern of tau splicing.
Here, we show that the 4R/3R tau isoform ratio can be increased in neurons from htau mice in culture and in vivo by RNA trans-splicing after lentivirus-mediated delivery of tau-targeted PTMs. Furthermore, we show that the trans-spliced product is translated into a full-length tau protein. The demonstration of tau trans-splicing in post-mitotic neurons provides a platform to explore further the therapeutic potential of trans-splicing reprogramming for neurological disorders.
RESULTS
Design of a lentiviral vector for delivery of tau-targeted PTMs
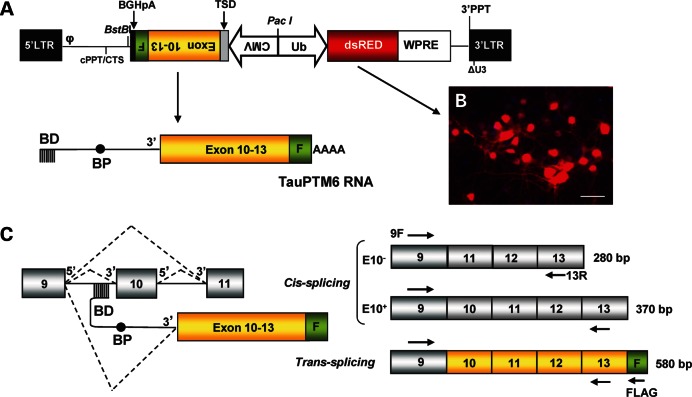
For efficient transduction and long-term expression of tau-targeted PTMs into differentiated neurons, we generated a lentiviral vector (LV) capable of delivering and integrating tau PTMs into the host genome (Fig. 1A). We used a self-inactivating LV carrying a PTM expression cassette and a DsRed reporter gene under the control of the human ubiquitin promoter. We previously generated two tau-targeted PTMs: TauPTM6 and TauPTM9 designed to introduce or exclude E10, respectively, in the mature trans-spliced tau mRNA (12, 13). TauPTM6 was selected for LV construction in order to reverse the excess of tau isoforms lacking E10 (i.e. 3R tau) in htau mouse brain. The trans-splicing domain of TauPTM6 comprises a 125 nt binding domain complementary to the 3′ end of tau intron 9, a branch point and the 3′AG splice acceptor site. The coding sequence of the PTM consists of tau exons 10 to 13, followed by the FLAG epitope sequence, to monitor trans-splicing at the RNA and protein levels. The bovine growth hormone polyadenylation signal was included at the 3′ end of the PTM. The PTM sequence was cloned under the CMV promoter and the resulting expression cassette was cloned in the 3′-5′ orientation in the LV backbone (Fig. 1A). The LV was designated as LV-TauPTM6. The transduction efficiency of LV-TauPTM6 was assessed by transducing cultured mouse cortical neurons and monitoring DsRed expression and was estimated to be 70–80% (Fig. 1B). Once integrated into the host genome LV-TauPTM6 drives the expression of TauPTM6 RNA that is designed to hybridize with endogenous tau pre-mRNA to produce a trans-spliced product containing PTM-encoded exons 10 to 13 and the FLAG sequence at the 3′ end (Fig. 1C). Tau pre-mRNA molecules not involved in a trans-splicing event would be cis-spliced to produce tau mRNA either including or excluding E10.
Figure 1.
Trans-splicing strategy for lentiviral delivery of TauPTM6. (A) Map of the LV designed for simultaneous delivery of TauPTM6 and DsRed. The viral backbone comprises: LTR, long-terminal repeat; φ, HIV encapsidation sequence; cPPT, central polypurine tract; CTS, central termination sequence; 3′PPT, 3′polypurine tract; ΔU3, deletion of the U3 portion of 3′LTR. The DsRed reporter cassette consists of DsRed under the human ubiquitin promoter (in the 5′–3′ direction) followed by the WPRE sequence. In the 3′–5′ direction, under the CMV promoter, the TauPTM6 sequence consists of a trans-splicing domain (TSD) comprising a 125 nt binding domain (BD) complementary to the 3′ end of tau intron 9, a branch point (BP) and the 3′AG splice acceptor site followed by tau exons 10–13, the FLAG epitope sequence (F) and the bovine growth hormone (BGH) polyadenylation signal. (B) DsRed expression in mouse cortical neurons transduced with LV-TauPTM6. Scale bar: 100 μm. (C) RT–PCR strategy to detect cis- and trans-splicing products. Schematic representation of cis- and trans-splicing events from tau pre-RNA (left) and the corresponding mRNA products (right). Arrows indicate the position of forward (9F) or reverse (13R, FLAGR) PCR primers. The expected size of PCR products is indicated for each splicing product.
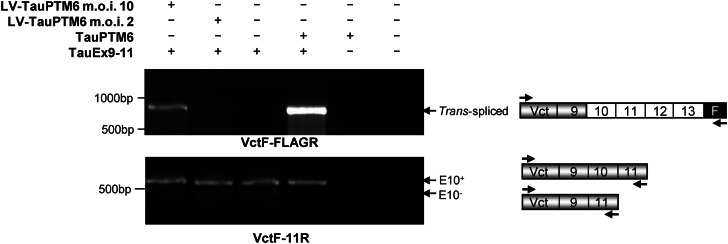
The potential of lentivirus-delivered TauPTM6 to promote trans-splicing at the level of E10 was assessed in HeLa cells first transduced with LV-TauPTM6 and then transfected with the TauEx9-11WT minigene consisting of MAPT exons 9, 10 and 11 and minimal intronic sequences flanking E10 (16). An 866 bp trans-spliced product containing minigene-derived exon 9 and TauPTM6-derived exons 10–13 and FLAG epitope sequences was detected by RT–PCR using a minigene-specific forward primer and the PTM-specific reverse primer, FLAGR, complementary to the FLAG sequence (Fig. 2, top panel). Although TauEx9-11WT produces mainly 4R tau RNA, RT–PCR detection of both cis- and trans-spliced products showed a multiplicity of infection (m.o.i.)-dependent increase in E10 inclusion in the mature RNA (Fig. 2, bottom panel).
Figure 2.
Trans-splicing modulation of E10 inclusion in tau transcripts. HeLa cells were transduced with LV-TauPTM6 and 2 to 3 days later were transfected with the TauEx9-11 minigene consisting of tau exons 9-10-11 and 498 bp 5′ and 264 bp 3′ of intronic sequences flanking E10. HeLa cells co-transfected with TauEx9-11 and TauPTM6 in pcDNA3 were used as a positive control. RT–PCR analysis with the forward primer VctF, specific for the target minigene and the PTM-specific reverse primer FLAGR, complementary to the FLAG epitope sequence, demonstrate trans-splicing between minigene transcripts and TauPTM6 (top panel). Both cis- and trans-spliced products were detected by RT–PCR with the forward primer VctF and the reverse primer 11R, corresponding to an exon 11 sequence; transduction with LV-TauPTM6 resulted in an increase in E10 inclusion (bottom panel).
Trans-splicing reprogramming of endogenous tau RNA after lentivirus-mediated PTM delivery
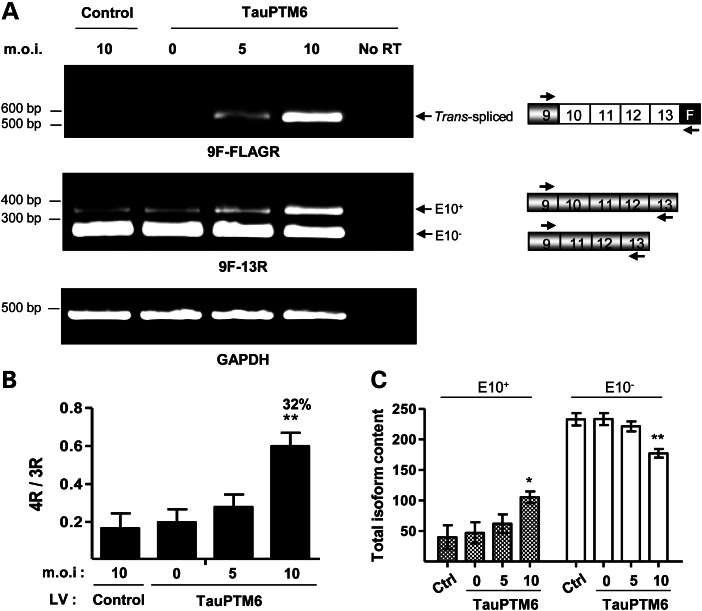
To determine if trans-splicing with an endogenous tau pre-mRNA target could be achieved through LV-TauPTM6 transduction, we used human neuroblastoma SH-SY5Y cells. SH-SY5Y cells were transduced with LV-TauPTM6 at different m.o.i. or with a control LV. RNA was extracted 1 week after transduction and analyzed by RT–PCR using a forward primer, 9F, complementary to an exon 9 sequence, and the PTM-specific reverse primer, FLAGR, to detect trans-spliced products. A 580 bp product corresponding to the size expected for a trans-spliced product was detected (Fig. 3A, top panel). Due to their fetal characteristics, proliferating SH-SY5Y cells mainly express 3R tau isoforms (17). Amplification of reverse-transcribed RNA using the 9F forward primer and a reverse primer, 13R, complementary to an exon 13 sequence, a combination that detects both cis- and trans-spliced products, showed an increase in 4R tau mRNA (Fig. 3A, middle panel). Both the level of the trans-spliced product and the increase in 4R tau RNA were m.o.i.-dependent, being particularly evident at the highest viral vector concentration (m.o.i. = 10). The 4R/3R ratio was lower than 0.1 in control cells but raised to 0.5 in cells transduced with LV-TauPTM6 at m.o.i. = 10 (Fig. 3B). The increase in the isoform ratio corresponds to a percentage of isoform conversion of ∼32% at m.o.i. = 10. The increase in the 4R/3R ratio was due to a concomitant increase in 4R tau RNA and a decrease in 3R tau RNA (Fig. 3C).
Figure 3.
Trans-splicing reprogramming of endogenous tau RNA in SH-SY5Y cells after lentivirus-mediated PTM delivery. Human neuroblastoma SH-SY5Y cells were transduced with LV-TauPTM6 at the indicated m.o.i., or with a control LV at m.o.i. = 10. Total RNA was extracted 7 days after transduction and reverse-transcribed. (A) PCR analysis of reverse-transcribed RNA from transduced cells. Top panel: PCR analysis with 9F-FLAGR primers detects a product of 580 bp only in cells transduced with LV-TauPTM6. Middle panel: 3R and 4R tau isoforms were detected with primers 9F and 13R. An increase in 4R tau RNA was observed in LV-TauPTM6-transduced cells. GAPDH mRNA was used for normalization (bottom panel). No RT: PCR with no RT using RNA from cells transduced with LV-TauPTM6 at m.o.i. = 10. (B) Quantitative analysis of 4R/3R tau RNA ratio. The 4R/3R ratios are expressed as mean ± SEM (*P < 0.05, **P < 0.01, n = 3). The percentage of isoform conversion is indicated above the respective bars. (C) Individual values of 4R and 3R tau RNA content for each condition. Values are expressed in arbitrary units as mean ± SEM (*P < 0.05, **P < 0.025, n = 3).
Correction of tau mis-splicing in htau mouse neurons in culture
To evaluate tau trans-splicing reprogramming in differentiated neurons, primary neurons from htau mice were transduced with LV-TauPTM6. Htau mice are a transgenic line that expresses the human wild-type human MAPT gene from a human PAC transgene in a mouse Mapt null background (14). Primary neurons from htau mice were transduced at 3 days in vitro (DIV3) at various viral vector m.o.i. and analyzed between 7 and 10 days after transduction. The number of viral particles integrated per neuron was estimated by qPCR analysis of DNA extracted from transduced neurons. The values obtained were consistent with the m.o.i. calculated from viral titers.
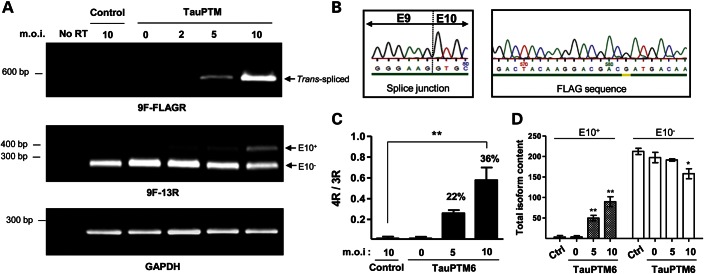
RNA extracted from transduced neurons was analyzed by RT–PCR using primers hybridizing in exon 9 and in the FLAG sequence (9F and FLAGR), as above. As in SH-SY5Y cells, a 580 bp trans-spliced product was generated from RNA from neurons transduced at m.o.i. = 5 and 10 (Fig. 4A, top panel). Transduction with lower amounts of LV-TauPTM6 (m.o.i. = 2) failed to produce detectable amounts of trans-spliced RNA. Sequencing of the 580 bp RT–PCR product confirmed that a correct exon 9–exon 10 junction had been created and that the full tau exon 10 to exon 13 sequence encoded by TauPTM6 as well as the FLAG sequence at the 3′ end was incorporated into the trans-spliced product (Fig. 4B). 3R and 4R tau isoforms in transduced htau neurons were detected by RT–PCR with primers hybridizing in exons 9 and 13 (9F–13R), as above. Htau neurons at early stages of culture express only 3R tau; trans-splicing of tau RNA was accompanied by the production and m.o.i.-dependent increase of 4R tau mRNA (Fig. 4A, middle panel). The 4R/3R ratio raised from 0.1 in control neurons to 0.4 and 0.6 in neurons transduced with LV-TauPTM6 at m.o.i. = 5 and 10, respectively (Fig. 4C). The change in the ratio corresponds to a percentage of isoform conversion of ∼36% at m.o.i. = 10. As for SH-SY5Y cells, the increase in the 4R/3R ratio was due to an increase in 4R tau RNA accompanied by a decrease in 3R tau RNA, the latter being clearly apparent at m.o.i. = 10 (Fig. 4D).
Figure 4.
Correction of tau mis-splicing in cultured cortical neurons from htau mouse after lentivirus-mediated PTM delivery. Cultured cortical neurons from htau mice were transduced at DIV3 with LV-TauPTM6 at the indicated m.o.i., or with a control LV at m.o.i. = 10. Total RNA was extracted 7 days after transduction and reverse-transcribed. (A) PCR analysis of reverse-transcribed RNA from transduced neurons. Top panel: a 580 bp PCR product was detected with 9F-FLAGR primers only in neurons transduced with LV-TauPTM6 at m.o.i. = 5 and 10. Middle panel: 3R and 4R tau isoforms were detected with primers 9F and 13R. GAPDH mRNA was used for normalization (bottom panel). No RT: PCR with no RT using RNA from cells transduced with LV-TauPTM6 at m.o.i. = 10. (B) Details of the sequence of the 9F-FLAGR PCR product showing a correct splice junction between exons 9 and 10 and the presence of the FLAG epitope sequence at the 3′ end of the trans-spliced product. (C) Quantitative analysis of 4R/3R tau RNA ratios. 4R/3R ratios are expressed as mean ± SEM (**P < 0.01, n = 3). The percentage of isoform conversion is indicated above the respective bars. (D) Individual values of 4R and 3R tau RNA content. Values are expressed in arbitrary units as mean ± SEM (*P < 0.05, **P < 0.025, n = 2).
Translation of trans-spliced tau RNA into a chimeric protein
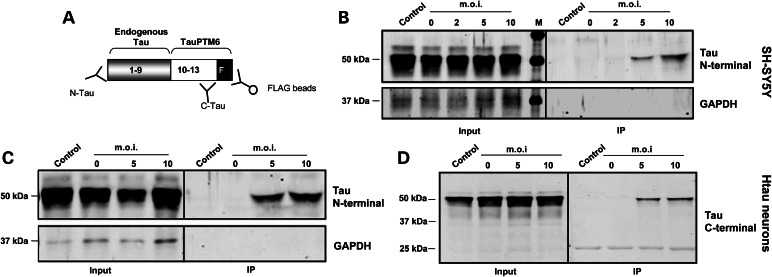
We next analyzed whether the trans-spliced product detected at the RNA level was translated into a bona fide chimeric tau protein. SH-SY5Y cells and primary neurons from htau mice were transduced with LV-TauPTM6 at different m.o.i. and 10 days later FLAG-tagged proteins were immunoprecipitated using an immobilized FLAG antibody (Fig. 5A). Immunoprecipitation products were analyzed by western blotting using an antibody to N-terminal amino acids 1–16 of human tau. Both in SH-SY5Y cells (Fig. 5B) and in htau neurons (Fig. 5C), a protein of ∼50 kDa, co-migrating with tau from whole cell lysates (input samples), was detected in cells transduced with LV-TauPTM6 at m.o.i. = 5 and 10 and not in cells transduced with a control LV. This demonstrates the translation of the trans-spliced product into a full-length tau protein, with an intact N-terminus and the FLAG epitope at the C-terminus. Similarly to the trans-spliced RNA, the amount of FLAG-immunoprecipitated tau increased with increasing m.o.i. (Fig. 5B and C).
Figure 5.
Translation of trans-spliced RNA into a chimeric protein. (A) Diagram of the strategy used for the detection of chimeric proteins by immunoprecipitation. An immobilized mouse monoclonal FLAG antibody was used to immunoprecipitate chimeric proteins. Immunoprecipitated proteins were analyzed by western blotting with N-terminal or C-terminal tau antibodies. The N-terminal part of the protein (grey box) corresponds to the part encoded by endogenous RNA (exons 1–9) and the C-terminal part (white box) corresponds to the part encoded by the PTM (exons 10–13 and FLAG epitope). (B–D) Immunoprecipitation analysis of FLAG-containing proteins from transduced cells. SH-SY5Y cells (B) or cortical neurons from htau mice at DIV3 (C and D) were transduced with LV-TauPTM6 at the indicated m.o.i. or with a control lentivirus (m.o.i. = 10). Transduced cells were lysed 14–16 days later and proteins immunoprecipitated with a FLAG antibody were analyzed by western blotting with an N-terminal tau antibody, a GAPDH antibody (B and C) or a C-terminal tau antibody (D) (input: 5 μl of cell lysate before immunoprecipitation). All samples had similar levels of tau before immunoprecipitation. No GAPDH immunoreactivity was detected in immunoprecipitates, ruling out non-specific precipitation by the FLAG antibody or by the affinity gel.
The N-terminal tau antibody detects only trans-splicing occurring with tau RNA. Immunoprecipitates from transduced htau neurons were also probed with an antibody to the C-terminus of tau, raised against amino acids 243–441 of human tau, a sequence encoded by TauPTM6. TauPTM6 includes the entire C-terminus of tau, from exons 10 to exon 13, as well as the FLAG epitope sequence. Thus, trans-splicing of TauPTM6 on targets other than tau would result in chimeric proteins containing the C-terminus of tau, detected by the C-terminal tau antibody used, as well as the FLAG epitope sequence. The C-terminal tau antibody detected a single ∼50 kDa band in FLAG immunoprecipitates, co-migrating with tau from whole cell lysates (Fig. 5D).
Detection of a single band with the C-terminal tau antibody in FLAG-immunoprecipitated proteins indicates that translated potential off-target trans-spliced products are minimal. Untranslated off-target products would be apparent at the RNA level only and would require methods such as rapid amplification of complementary DNA ends (RACE) to be detected. Two out of three off-target trans-spliced products will be out of frame and degraded by nonsense-mediated decay; only off-target products that are translated could represent a problem. Detection of a single band with a C-terminal tau antibody also rules out the possibility of direct translation from the PTM, which would have produced a smaller product immunoreactive for the tau C-terminus.
Correction of tau mis-splicing in vivo in adult htau mouse brain
Although tau trans-splicing can be obtained in cultured neurons from htau mice after lentivirus-mediated PTM delivery, culture conditions only partly reproduce in vivo conditions compatible with therapeutic applications. Therefore, we next conducted a small-scale pilot study in adult htau mice. For PTM delivery to the brain, we constructed second generation LVs, based on the original LV-TauPTM6 design. The CMV promoter was replaced by either the synapsin promoter for enhanced and neuron-specific expression or by the ubiquitin promoter. The bovine growth hormone polyadenylation signal was replaced by the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) for increased RNA stability. The DsRed reporter cassette was omitted to avoid promoter competition. The new LVs were designated as LVSyn-TauPTM6 and LVUb-TauPTM6, respectively (Fig. 6A).
Figure 6.
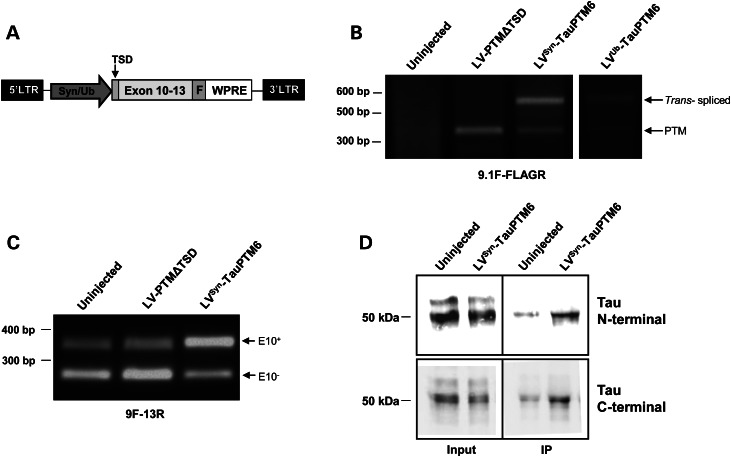
Correction of tau mis-splicing in vivo in adult htau mouse brain. (A) Map of LVSyn-TauPTM6/LVUb-TauPTM6 designed for TauPTM6 delivery in vivo. The viral backbone is the same as for LV-TauPTM6 (Fig. 1). TauPTM6 is under the control of the synapsin promoter or of the ubiquitin promoter and is followed by the WPRE sequence (TSD: trans-splicing domain; F: FLAG epitope sequence). LVSyn-TauPTM6 or LVUb-TauPTM6 was stereotaxically injected into the prefrontal cortex of 12–14-week-old htau mice; control mice were uninjected or injected with an LV expressing a PTM lacking the trans-splicing domain (LV-PTMΔTSD). RNA and protein were analyzed 8 months after injection. (B). RT–PCR analysis with the 9.1F-FLAGR primer combination. In addition to a 360 bp product, demonstrating PTM expression, a 550 bp trans-spliced product was detected after injection with LVSyn-TauPTM6 or LVUb-TauPTM6. (C) RT-PCR analysis with the 9F-13R primer combination, demonstrating an increase in 4R tau mRNA after injection with LVSyn-TauPTM6. (D) Immunoprecipitation analysis of FLAG-containing proteins using N- or C- terminal tau antibodies (Input: 5 μl of brain homogenate before immunoprecipitation). An increased signal was detected with both antibodies after injection with LVSyn-TauPTM6.
LVSyn-TauPTM6, LVUb-TauPTM6 or an LV expressing a PTM lacking the trans-splicing domain (PTMΔTSD) were stereotaxically injected into the prefrontal cortex of 12–14 week-old htau mice. Trans-splicing was evaluated 8 months after injection by RT-PCR using the FLAGR reverse primer and the forward primer, 9.1F, that hybridizes with exon 9 as well as with exon 11, included in the PTM coding sequence, in order to detect PTM expression as well as trans-splicing (12). In addition to a 360 bp product, demonstrating PTM expression, a 550 bp trans-spliced product was detected after injection with LVSyn-TauPTM6 or LVUb-TauPTM6 (Fig. 6B). Sequencing of the 550 bp product confirmed that a correct exon 9–exon 10 junction had been generated. This demonstrates that trans-splicing has occurred in vivo between endogenous tau RNA and TauPTM6. Unlike embryonic neurons cultured from htau mice, that express only 3R tau (Fig. 4A), adult htau mice express all human tau isoforms, with an excess of 3R tau over 4R tau [(14, 15) and Fig. 6C]. Amplification of reverse-transcribed RNA using the 9F-13R primer combination, detecting both isoforms, demonstrated an increase in 4R tau mRNA concomitant with a decrease in 3R tau after injection with LVSyn-TauPTM6 (Fig. 6C). FLAG-tagged proteins were immunoprecipitated from brain extracts and immunoprecipitation products were analyzed by western blotting using N- or C-terminal tau antibodies, as above. An increased signal was detected with both antibodies after injection with LVSyn-TauPTM6 (Fig. 6D). Thus, the tau trans-spliced product is translated into a full-length tau protein, with an intact N-terminus and the FLAG epitope at the C-terminus.
DISCUSSION
Tau mis-splicing is one of the molecular causes of inherited tauopathies. Using RNA trans-splicing in htau mice with a lentiviral delivery system to achieve long-term expression of trans-splicing molecules, we have demonstrated efficient correction of endogenous tau mis-splicing in neurons in vitro and in vivo in the brain of adult mice. Importantly, the trans-spliced RNA is translated into a full-length chimeric tau protein. Therefore, our results highlight the therapeutic potential of SMaRT to correct tau mis-splicing.
Another example of successful use of trans-splicing in neurons in vivo is SMN exon 7 inclusion in the SMN2 transcript in a mouse model of spinal muscular atrophy (SMA) (18–20). In this paradigm, production of a functional SMN protein after trans-splicing has been inferred through changes in total SMN protein (18–20), but not demonstrated directly. Successful translation of a chimeric protein after trans-splicing of endogenous transcripts has been reported for keratin 14, mutated in epidermolysis bullosa (21). Achieving trans-splicing in the brain necessitates an appropriate PTM delivery system. In the SMA example mentioned above, PTMs were delivered by intraventricular injection of plasmid DNA in neonatal animals. While this is suitable for experimental purposes in animals, this delivery method is not directly applicable in a therapeutic context. Intraventricular injection in human subjects or adult animals is effective for proteins and oligonucleotides, but not suitable for plasmids and has a number of other limitations as highlighted by Butchbach (22). On the other hand, we have shown that tau reprogramming could be achieved in vivo in adult animals by direct stereotaxic injection and long-term expression of a viral vector into the brain parenchyma, a delivery strategy compatible with therapeutic applications.
Htau mice represent an excellent in vivo system in which to evaluate strategies to correct tau mis-splicing. Htau mice develop a tau pathology with tau hyperphosphorylation and aggregation as well as extensive neuronal death during aging (14, 15). Htau mice express human tau at about four times the level of endogenous mouse tau and the tau pathology observed in these animals is likely due to a combination of tau overexpression and expression of both 3R and 4R tau (as opposed to the expression of 4R tau only in wild-type adult mice). Expression of 3R tau in adult mice is unlikely to be sufficient to induce tau pathology in htau mice. Indeed, other transgenic mice expressing the entire human MAPT gene in a Mapt−/− background have been described (23). These animals have a similar pattern of tau isoform expression as htau mice, but the overall level of tau is lower than in htau mice, and they do not develop a tau pathology (23). To date, there has been no animal model reproducing the pathological phenotype caused by splicing defects occurring in FTDP-17, in which the effect of splicing modulation can be evaluated.
For therapeutic applications, trans-splicing can elicit either increase or reduction of E10 inclusion from wild-type tau transcripts as well as from transcripts containing FTDP-17 mutations affecting E10 splicing (13). Trans-splicing modulation of E10 splicing is independent of the specific mutation affecting splicing and can therefore have a general application for the correction of the downstream consequences of mutations affecting splicing regulatory elements. Other methods of tau mis-splicing correction include the use of antisense oligonucleotides blocking splice junctions. Although antisense oligonucleotides would represent a more direct approach than trans-splicing and are promising for the treatment of brain diseases (24, 25), they require, in most cases, sustained delivery by intraventricular infusion. Modulation of tau splicing using antisense oligonucleotides has been obtained in vitro using minigenes or neuronal cell lines (26–28); however, there has been no report to date of their use in vivo.
In summary, the demonstration of successful in vivo trans-splicing correction of tau mis-splicing in adult brain provides a framework to be evaluated further for its therapeutic application for tauopathies and other neurological disorders, especially for neurodegenerative conditions caused by abnormal RNA processing (29, 30), including abnormal activity of RNA-binding proteins (31).
MATERIALS AND METHODS
Lentiviral vectors
The lentiviral backbone used is derived from the original pTrip vector (32) with several modifications, as described previously (33). To produce LVs expressing TauPTM6, the TauPTM6 cassette, including the CMV promoter and the BGH polyadenylation sequence (pA), was amplified from the TauPTM6 construct in pcDNA3.1 (designed and produced by VIRxSYS Corporation, Gaithersburg, MD, USA) described previously (12) and subcloned into the viral backbone vector. Details of cloning, lentivirus production and transduction of cell lines are described under Supplementary data.
Animals
Htau transgenic mice, in a C57BL/6 background, were obtained from Jackson Laboratories (Bar Harbour, Maine, USA; B6.Cg-Mapttm1 (EGFP) Klt Tg(MAPT)8cPdav/J. Stock number: 005 491). Mice were genotyped by PCR to confirm the presence of the human MAPT transgene and the mouse Mapt null background using the primers listed in Supplementary data. All animal procedures were conducted in accordance with the UK Home Office, Animals Scientific Procedures Act 1986.
Primary neuronal cultures and viral transduction
Cortical neurons were obtained from embryonic day 16 (E16) mouse embryos and dissociated with the papain dissociation system (Worthington Biochemical Corp., NJ, USA) following the manufacturer's instructions. Cells were plated onto 6-well plates coated with poly-d-lysine (10 μg/ml) at a density of 106 cells per well and cultured in Neurobasal medium (Invitrogen) supplemented with a 2% (v/v) B-27 serum-free supplement, 0.5 mm l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were incubated at 37°C in a 5% CO2/95% air atmosphere. For fluorescence microscopy, the cells were plated onto poly-d-lysine-coated glass coverslips in 12-well plates at a density of 2 × 105 cells per well and cultured under the same conditions. Viral transductions were performed at 3 days in vitro (DIV3), at an m.o.i. of 2, 5 or 10 by adding 5–30 μl of concentrated viral suspension (in phosphate buffered saline, PBS) in a final volume of 1 ml of Neurobasal medium per well. Twenty-four hours after transduction, the medium was replaced with fresh medium and the cells were cultured for 7–10 further days before RNA or protein extraction. To detect DsRed fluorescence, transduced neurons were fixed in 4% (w/v) paraformaldehyde for 10 min, then rinsed three times in PBS, with the last wash containing DAPI and mounted in DakoCytomation fluorescent mounting medium. Slides were viewed under a Zeiss Axioskop fluorescence microscope with the appropriate filter sets and imaged using Metamorph software. The number of viral particles integrated into the host genome was estimated by qPCR using DNA from transduced neurons. Quantification was performed with SYBR Green with two separate sets of primers, to detect either the LTR or the WPRE sequence.
Stereotaxic injections
Mice aged 12–14 weeks (weight 25–30g) were anesthetized using ketamine-HCl (100 mg/kg; Ketaset) and xylazine-HCl (10 mg/kg, Rompun 2%, Bayer), i.p. 2 µl of viral suspension (0.5–1 × 106 TU) were stereotaxically injected into the brain at the following coordinates (34): anteroposterior: +2.5 mm, lateral: ±0.5 mm, dorso-ventral: −2.0 mm, measured from the Bregma, at an injection rate of 0.2 µl/min. After surgery, the skin was sutured and the animals kept at 37°C until complete recovery. Carprofen (5 mg/kg, s.c.) was administered immediately after surgery and 24 h later. Mice were sacrificed by cervical dislocation 8 months after injection, the brain dissected and ∼1 mm3 of tissue around the injected area was dissected, snap frozen and stored at −80°C until RNA or protein extraction.
RT–PCR analysis of cis- and trans-splicing
Total RNA and DNA were isolated from transduced cells using the AllPrep DNA/RNA Mini kit (Qiagen). RNA from LV-injected brains was extracted using a RNeasy Lipid Tissue kit (Qiagen). Reverse transcription was performed using a TaqMan RT kit (Applied Biosystems) with oligo(dT) or an equimolar ratio of oligo(dT) and random hexamers. To detect cis- and trans-splicing products, reverse-transcribed RNA was amplified by PCR using Go Taq polymerase (Promega) using conditions and primers detailed in Supplementary data.
Immunoprecipitation and western blotting
Ten days after transduction, the cells were washed in PBS at 4°C and lysed in lysis buffer [50 mm Tris–HCl, pH 7.4; 150 mm NaCl; 1 mm EDTA; 1% (v/v) Triton X-100] containing a protease inhibitor cocktail (Complete Mini, Roche). Brain samples were homogenized in lysis buffer. FLAG-tagged proteins were immunoprecipitated using EZ-view anti-FLAG M2 Affinity gel (Sigma). Immunoprecipitates were analyzed by western blotting using antibodies to the N- or C-terminus of tau.
Additional details of reagents and protocols are described in Supplementary data.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Wellcome Trust, the Medical Research Council, Alzheimer's Research UK (formerly the Alzheimer's Research Trust) and the Psychiatry Research Trust. Funding to pay the Open Access publication charge for this article was provided by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Gary Mansfield and Dr Madaiah Puttaraju (VIRxSYS Corporation, Gaithersburg, MD, USA) for PTMs, Dr Uwe Maskos (Pasteur Institute, Paris) for viral packaging plasmids, Dr Jane Wu (Northwestern University Feinberg School of Medicine, Chicago) for the TauEx9-11WT tau minigene, Dr Sylvane Desrivieres for the use of stereotaxic injection equipment and Dr Wendy Noble (King's College London) for helpful discussions.
Conflict of Interest statement: none declared.
REFERENCES
- 1.Ballatore C., Lee V.M., Trojanowski J.Q. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 2.Morris M., Maeda S., Vossel K., Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallo J.-M., Noble W., Rodriguez Martin T. RNA and protein-dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cell. Mol. Life Sci. 2007;64:1701–1714. doi: 10.1007/s00018-007-6513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H.H., Pickering Brown S., Chakraverty S., Isaacs A., Grover A., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–704. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini M.G., Murrell J.R., Goedert M., Farlow M.R., Klug A., Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl Acad. Sci. USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell J.W., Rodriguez-Martin T., Gibb G.M., Khan N.M., Grierson A.J., Hanger D.P., Revesz T., Anderton B.H., Gallo J.-M. Quantitative analysis of tau isoform transcripts in sporadic tauopathies. Mol. Brain Res. 2005;137:104–109. doi: 10.1016/j.molbrainres.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Niblock M., Gallo J.-M. Tau alternative splicing in familial and sporadic tauopathies. Biochem. Soc. Trans. 2012;40:677–680. doi: 10.1042/BST20120091. [DOI] [PubMed] [Google Scholar]
- 8.Noble W., Pooler A.M., Hanger D.P. Advances in tau-based drug discovery. Expert Opin. Drug Discov. 2011;6:797–810. doi: 10.1517/17460441.2011.586690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond S.M., Wood M.J. Genetic therapies for RNA mis-splicing diseases. Trends Genet. 2011;27:196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Puttaraju M., Jamison S.F., Mansfield S.G., Garcia-Blanco M.A., Mitchell L.G. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 11.Wally V., Murauer E.M., Bauer J.W. Spliceosome-mediated trans-splicing: the therapeutic cut and paste. J. Invest. Dermatol. 2012;132:1959–1966. doi: 10.1038/jid.2012.101. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Martin T., Garcia-Blanco M.A., Mansfield S.G., Grover A.C., Hutton M., Yu Q., Zhou J., Anderton B.H., Gallo J.-M. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: implications for tauopathies. Proc. Natl Acad. Sci. USA. 2005;102:15659–15664. doi: 10.1073/pnas.0503150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Martin T., Anthony K., Garcia-Blanco M.A., Mansfield S.G., Anderton B.H., Gallo J.-M. Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing. Hum. Mol. Genet. 2009;18:3266–3273. doi: 10.1093/hmg/ddp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andorfer C., Kress Y., Espinoza M., de Silva R., Tucker K.L., Barde Y.A., Duff K., Davies P. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J. Neurochem. 2003;86:582–590. doi: 10.1046/j.1471-4159.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- 15.Andorfer C., Acker C.M., Kress Y., Hof P.R., Duff K., Davies P. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. J. Neurosci. 2005;25:5446–5454. doi: 10.1523/JNEUROSCI.4637-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Z., Cote J., Kwon J.M., Goate A.M., Wu J.Y. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell Biol. 2000;20:4036–4048. doi: 10.1128/mcb.20.11.4036-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith C.J., Anderton B.H., Davis D.R., Gallo J.-M. Tau isoform expression and phosphorylation state during differentiation of cultured neuronal cells. FEBS Lett. 1995;375:243–248. doi: 10.1016/0014-5793(95)01221-y. [DOI] [PubMed] [Google Scholar]
- 18.Coady T.H., Shababi M., Tullis G.E., Lorson C.L. Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol. Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- 19.Coady T.H., Baughan T.D., Shababi M., Passini M.A., Lorson C.L. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS One. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shababi M., Glascock J., Lorson C.L. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum. Gene Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- 21.Wally V., Brunner M., Lettner T., Wagner M., Koller U., Trost A., Murauer E.M., Hainzl S., Hintner H., Bauer J.W. K14 mRNA reprogramming for dominant epidermolysis bullosa simplex. Hum. Mol. Genet. 2010;19:4715–4725. doi: 10.1093/hmg/ddq405. [DOI] [PubMed] [Google Scholar]
- 22.Butchbach M.E. Trans-splicing, more than meets the eye: multifaceted therapeutics for spinal muscular atrophy. Hum. Gene Ther. 2011;22:121–125. doi: 10.1089/hum.2011.903. [DOI] [PubMed] [Google Scholar]
- 23.McMillan P., Korvatska E., Poorkaj P., Evstafjeva Z., Robinson L., Greenup L., Leverenz J., Schellenberg G.D., D'Souza I. Tau isoform regulation is region- and cell-specific in mouse brain. J. Comp. Neurol. 2008;511:788–803. doi: 10.1002/cne.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southwell A.L., Skotte N.H., Bennett C.F., Hayden M.R. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S., et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalbfuss B., Mabon S.A., Misteli T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J. Biol. Chem. 2001;276:42986–42993. doi: 10.1074/jbc.M105113200. [DOI] [PubMed] [Google Scholar]
- 27.Donahue C.P., Muratore C., Wu J.Y., Kosik K.S., Wolfe M.S. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J. Biol. Chem. 2006;281:23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peacey E., Rodriguez L., Liu Y., Wolfe M.S. Targeting a pre-mRNA structure with bipartite antisense molecules modulates tau alternative splicing. Nucleic Acids Res. 2012;40:9836–9849. doi: 10.1093/nar/gks710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallo J.-M., Jin P., Thornton C.A., Lin H., Robertson J., D'Souza I., Schlaepfer W.W. The role of RNA and RNA processing in neurodegeneration. J. Neurosci. 2005;25:10372–10375. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony K., Gallo J.-M. Aberrant RNA processing events in neurological disorders. Brain Res. 2010;1338:67–77. doi: 10.1016/j.brainres.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Da Cruz S., Cleveland D.W. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 33.Avale M.E., Faure P., Pons S., Robledo P., Deltheil T., David D.J., Gardier A.M., Maldonado R., Granon S., Changeux J.P., et al. Interplay of β2* nicotinic receptors and dopamine pathways in the control of spontaneous locomotion. Proc. Natl Acad. Sci. USA. 2008;105:15991–15996. doi: 10.1073/pnas.0807635105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paxinos G., Franklin K.B. The mouse brain in stereotaxic coordinates. San Diego, CA, USA: Academic Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.