Abstract
Small intestinal epithelial cells (sIECs) have a significant share in whole body metabolism as they perform enzymatic digestion and absorption of nutrients. Furthermore, the diet plays a key role in a number of complex diseases including obesity and diabetes. The impact of diet and altered genetic backgrounds on human metabolism may be studied by using computational modeling. A metabolic reconstruction of human sIECs was manually assembled using the literature. The resulting sIEC model was subjected to two different diets to obtain condition-specific metabolic models. Fifty defined metabolic tasks evaluated the functionalities of these models, along with the respective secretion profiles, which distinguished between impacts of different dietary regimes. Under the average American diet, the sIEC model resulted in higher secretion flux for metabolites implicated in metabolic syndrome. In addition, enzymopathies were analyzed in the context of the sIEC metabolism. Computed results were compared with reported gastrointestinal (GI) pathologies and biochemical defects as well as with biomarker patterns used in their diagnosis. Based on our simulations, we propose that (i) sIEC metabolism is perturbed by numerous enzymopathies, which can be used to study cellular adaptive mechanisms specific for such disorders, and in the identification of novel co-morbidities, (ii) porphyrias are associated with both heme synthesis and degradation and (iii) disturbed intestinal gamma-aminobutyric acid synthesis may be linked to neurological manifestations of various enzymopathies. Taken together, the sIEC model represents a comprehensive, biochemically accurate platform for studying the function of sIEC and their role in whole body metabolism.
INTRODUCTION
The major purpose of the human digestive system is to process food to provide the body with essential nutrients and energy (1). After being partially digested at the level of mouth and stomach, the food components (i.e. 60–70% of complex carbohydrates, 80–90% of dietary proteins and ∼85% fat) reach the duodenum, the proximal part of the small intestine, for complete enzymatic digestion and absorption (2). The small intestine is unique in its variety of cell types, including enterocytes [i.e. columnar small intestinal epithelial cells (sIECs) for digestion and absorption of nutrients], goblet cells (for mucus secretion), entero-endocrine cells (for hormone secretion) and paneth cells (i.e. stem cells that differentiate to form enterocytes). The large surface area, provided by the presence of hundreds of enterocytes, contributes to the maximal absorptive capacity of the small intestine.
Small intestinal enterocytes account for the majority of the enzymatic digestion and nutrient absorption (1). In fact, enterocytes are highly metabolically active cells and provide ∼25% of the endogenous glucose and cholesterol (3,4). As such, they contribute significantly to the metabolism of the whole body, acting as a gateway for nutrients. They channel essential nutrients to the liver (upon hormonal influence) as well as account for first pass drug metabolism (5). The liver and the small intestine are anatomically in close proximity, connected through the portal vein, and they have related physiological and metabolic functions. The small intestine provides ∼75% of blood flow to the liver via the hepatic portal system (6) and the liver, in turn, supplies biliary constituents through the common bile duct to the duodenum. The liver synthesizes bile acids from cholesterol, which are then delivered to the intestinal lumen to aid in the digestion and absorption of fat with their emulsifying properties. The bile acids are then actively absorbed by the enterocytes, but half of the bile acids can also diffuse through the lumen into the enterocytes. Bile acids are then sent back to the liver via the portal circulation, permitting their extensive recycling through this entero-hepatic circulation before they are finally excreted in feces (7).
Inborn errors of metabolism (IEMs) are the hereditary metabolic defects that are encountered in all major metabolic pathways occurring in man (8). IEMs have a myriad of pathological effects, affecting multiple organ systems that may lead to fatal phenotypes. IEMs can arise due to mutations in single genes (9) or multiple genes (10), which add another level of complexity to their diagnosis. Mass spectrometric analysis of whole blood samples from infants is the usual diagnostic method (e.g. looking for concentration changes of specific biomarkers). However, there exists a series of other tests, including molecular genetic testing for specific mutations, enzyme assays and further biochemical tests (e.g. urine tests and blood tests for blood gases and electrolytes), which together are employed to confirm the presence of an IEM (11). There exist multiple classification systems for these disorders depending on the clinical phenotypes, affected organs, mode of inheritance, occurrence in a specific metabolic pathway and other factors. While IEMs of the amino acid metabolism are usually treatable, IEMs involving biosynthesis of complex molecules (e.g. lysosomal storage or peroxisomal biogenesis disorders) have generally no specific treatment available (8). The use of special dietary formulations and medications usually alleviates clinical symptoms of the aminoacidopathies, and hence, is widely used as treatment strategy (8).
Genome-scale metabolic network reconstructions capture genomic, physiological and biochemical accurate data for target organisms (12). More than 100 organism-specific metabolic reconstructions have been published, including for human (13) and mouse (14). Moreover, cell-type specific reconstructions have been published for hepatocytes (15), kidney cells (16), alveolar macrophages (17), red blood cells (18) and cardiomyocytes (19). Metabolic reconstructions serve as a knowledge base for a target organism and target cell-type and can be converted into mathematical models (12). Besides describing the exact stoichiometry for metabolic reactions occurring in a cell, these reconstructions also contain information about the enzymes catalyzing the reactions and the corresponding encoding genes through gene–protein-reaction (GPR) associations. These GPR associations are expressed using the Boolean rules (‘and’ and ‘or’ relationships), where ‘and’ represents the requirement of the gene products for a reaction. Conversely, the ‘or’ relationship is employed to represent the relationship between genes encoding for isozymes, which catalyze the same reaction. These GPRs permit to connect the genotype to the phenotype and enable prediction of phenotypic changes associated with genetic alterations, e.g. enzyme deficiencies (14,20–22) or single-nucleotide polymorphisms (23). An increasing number of studies have become available, highlighting the value of computational modeling for understanding metabolism in health and disease. These studies include the analysis of comorbidity (24), the prediction of off-target drug effects (16), the prediction of specific drug targets (25,26), the assessment of metabolic changes occurring during diabetes and ischemia (27) and the prediction of the metabolic consequences of genetic and epigenetic properties, such as dosage effect (28).
The aim of the present study was to model the digestion and absorption of dietary components by the small intestine and to analyze changes in the intestinal metabolism under different genetic backgrounds. We therefore, reconstructed a metabolic network specific for sIECs and subjected it to two different diets to obtain condition-specific metabolic models. Subsequently, we analyzed the effect of various IEMs on the models' metabolic capabilities. For IEMs that highly affected the enterocyte's metabolism, we compared the computed results with known intestinal features, such as phenotypes exhibiting extensive intestinal pathology associated with these disorders as well as with biomarker patterns used in their diagnosis.
RESULTS
In this study, we assembled manually a comprehensive, predictive metabolic network for small intestinal enterocytes, which captures known, essential physiology and biochemistry of this cell type. We then employed the metabolic model to assess the impact of diet on the absorption and secretion capabilities of enterocytes. Finally, we investigated the consequences of enzymopathies on the enterocyte's metabolic functionalities.
Reconstruction of the sIEC-specific metabolic network
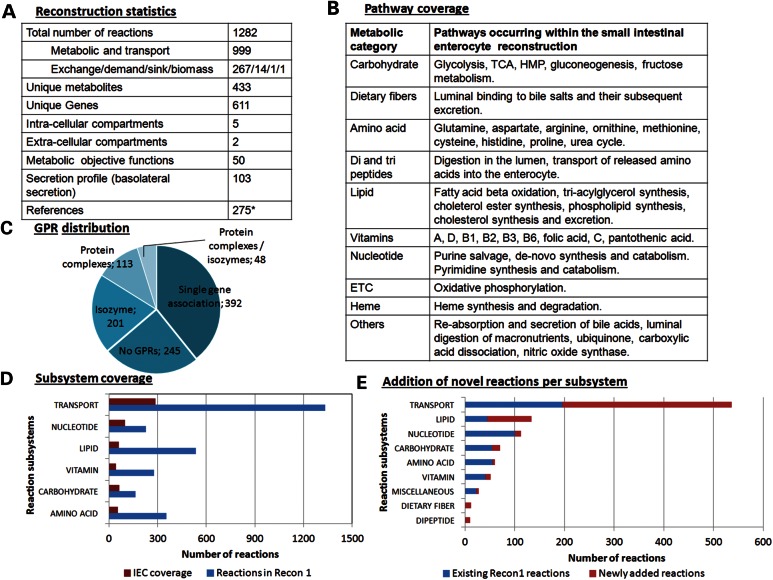
We reconstructed the first sIECs specific metabolic network in a bottom-up fashion through an extensive literature review. We identified metabolic pathways known to occur within these cells, including sIEC-specific pathways, such as citrulline and proline metabolism, the conversion of fructose to glucose and the re-synthesis of triglyceride and cholesterol-ester. We collected information on the various transport systems specific for sIECs (Fig. 1B, Supplementary Material, Table S1), resulting in the addition of 38% new enzymatic and transport reactions, which were not captured by a previously published global human metabolic reconstruction (13) (Fig. 1E). The final human small intestine-specific enterocyte metabolic reconstruction was deemed to be hs_sIEC611, where ‘hs’ stands for homosapiens, sIEC for sIECs, and 611 for the number of included genes. hs_sIEC611 accounts for 1282 reactions and 433 unique metabolites distributed over five intracellular (cytosol, mitochondria, nucleus, peroxisome and endoplasmic reticulum) and two extracellular compartments (extracellular space and lumen) (Fig. 1A). Information from more than 250 peer reviewed articles and books was reviewed (Supplementary Material, Table S2) and provides supporting evidence for the metabolic content of hs_sIEC611.
Figure 1.
Properties of the human metabolic reconstruction for small intestine epithelial cells. (A) Overview of the sIEC reconstruction. *peer-reviewed journal articles, primary literature and books. (B) Metabolic pathways that have been identified through a thorough literature review to be present in sIECs. (C) Overview of the GPR associations in the enterocyte reconstruction. (D) Coverage of metabolic and transport reactions in the enterocyte reconstruction with respect to Recon 1 (exchanges, luminal adjustments and novel metabolic and transport reactions existing in the enterocyte network have not been shown). (E) Addition of novel reactions to the enterocyte reconstruction, di-peptides, and dietary fibers were two subsystems that were absent in the previous human metabolic reconstruction, Recon 1 (13).
When comparing the number of reactions associated with the different metabolic pathways in sIECs (Fig. 1B), we found that the transport subsystem had the highest number of reactions associated (538; Fig. 1D and E), since a chief function of the enterocyte is the absorption of dietary nutrients from the lumen (apical uptake). Additionally, enterocytes take up nutrients from the arterial blood (basolateral uptake), depending on their own metabolic needs. Numerous metabolites, including bilirubin, nitric oxide and cholesterol, are also secreted into the lumen. Nutrient transporters are normally located at either the luminal or basolateral side, depending on the nature of the transporter. An exception is the GLUT-2 transporter, which is usually present at the basolateral side, but which can also migrate onto the luminal side to aid in glucose absorption under fully fed state (29). Moreover, the lipid metabolic pathway accounted for most metabolic reactions (134 reactions), followed by nucleotide metabolism (113 reactions) and carbohydrate metabolism (70 reactions). The supplementary text contains a detailed description of the covered subsystems.
To evaluate the predictive potential of a condition-specific model that can be derived from the sIEC-specific reconstruction, we tested for the model's capability to produce all defined biomass precursors under rich medium conditions (i.e. all metabolites, for which exchange reactions were defined, were allowed to be taken up) as well as under two defined dietary conditions, being an average American diet and a balanced diet. Moreover, we performed an extensive literature review to collect metabolic capabilities that can be accomplished by sIECs and formulated 50 different ‘metabolic tasks’, which sIECs, and thus our model, should be able to fulfill (Table 1, see Supplementary Material, Table S4 for associated references and flux values under different diets). We then tested the model's capability to carry a non-zero flux for each metabolic task under different simulation conditions using flux balance analysis (30). We also defined a sIEC-specific secretion profile based on a thorough literature review. A total of 267 exchange reactions were constrained accordingly for luminal uptake secretion and basolateral uptake secretion patterns (Supplementary Material, Table S5). We subsequently tested the model's capability to carry flux through the exchange reactions using flux variability analysis (FVA) (31) and compared the results with the defined secretion profile.
Table 1.
Effect of diet on the metabolic tasks defined for sIECs
| Metabolic task | Physiological function | D1 versus D2 |
|---|---|---|
| Secretion of lactate from glucose uptake | Glucose utilization for energy production. | ↑ |
| Glutamine to glucose conversion | Endogenous glucose production. | ↑ |
| Glutamine to proline conversion | Delivery to the liver. | ↑ |
| Glutamine to ornithine conversion | For spermine synthesis. | ↑ |
| Glutamine to citrulline conversion | For arginine synthesis. Biomarker of active enterocyte mass (85). This metabolic task is exclusive to enterocytes (86). | ↑ |
| Glutamine to lactate conversion | Utilization of glutamine, sparing glucose. | ✓ |
| Glutamine to aspartate conversion | For nucleotide synthesis. | ✓ |
| Glutamine to carbon dioxide conversion | Utilization of glutamine, sparing glucose. | ↑ |
| Glutamine to ammonia conversion | Glutaminase activity to generate glutamate. | ✓ |
| Putrescine to methionine conversion | Regeneration of methionine. | ↑ |
| Basolateral secretion of alanine | Delivery to the liver. | ↑ |
| Basolateral secretion of lactate | Delivery to the liver. | ↑ |
| Synthesis of arginine | Semi-essential amino acid. | ↑ |
| Synthesis of proline | To be sent to liver. The pyroline-5-carboxylate synthase is specific to the small intestine (87). | ✓ |
| Synthesis of alanine from glutamine | To be sent to the liver. | × |
| Basolateral secretion of proline | To be sent to the liver. | ↑ |
| Basolateral secretion of arginine | To be sent to the liver. | ↑ |
| Basolateral secretion of ornithine | To be sent to the liver. | ↑ |
| Synthesis of spermine | Protective effect in small intestine. | ↑ |
| Synthesis of spermidine | To be sent to the liver. | ↑ |
| Synthesis of nitric oxide | Aids in GI motility. | ↑ |
| Synthesis of cholesterol | Membrane constituent. | ↑ |
| De novo purine synthesis | Energy intermediate. | ↑ |
| Salvage of purine bases | Energy intermediate. | ↑ |
| Purine catabolism | Energy intermediate. | ↑ |
| Pyrimidine synthesis | Maintenance of the purine level and important byproduct release. | ↑ |
| Pyrimidine degradation (uracil) | Maintenance of purine and pyrimidine levels and important byproduct release. | ↑ |
| Fructose to glucose conversion | Fructose utilization. | ↑ |
| Uptake and secretion of cholic acid | Enterohepatic circulation. | ↑ |
| Uptake and secretion of glyco-cholic acid | Enterohepatic circulation. | ↑ |
| Uptake and secretion of tauro-cholic acid | Enterohepatic circulation. | ↑ |
| HMP shunt pathway | Generation of reducing equivalents and precursor for nucleotide synthesis. | ↑ |
| Malate to pyruvate conversion | Maintenance of cytosolic pyruvate pool. | ↑ |
| Synthesis of urea | Elimination of ammonia. | ↑ |
| Cysteine to pyruvate conversion | Maintenance of cytosolic pyruvate pool. | ↑ |
| Methionine to cysteine conversion | Synthesis of cysteine. | ↑ |
| Synthesis of triacylglycerol | To be transported in chylomicrons for body needs. | ↑ |
| Phosphatidylcholine synthesis in mitochondria | Membrane constituent. | ↑ |
| Binding of bile acids to dietary fibers | Cholesterol lowering effect of dietary fibers. | ✓ |
| Synthesis of FMN | Conversion of riboflavin to active form. | ↑ |
| Synthesis of FAD | Fatty acid oxidation. | ↑ |
| Synthesis of 5-methyl-tetrahydrofolate | Generated from folic acid, an important co-factor in one carbon metabolism. | ↓ |
| Putrescine to GABA conversion | Exerts protective effect in small intestine. | ↑ |
| Superoxide dismutase activity | Free radical scavenger. | ✓ |
| Carbonic anhydrase activity | For availability of bicarbonate. | ↑ |
| TCA cycle flux | Energy generation, anaplerosis, and amphibolic role. | ↑ |
| Histidine to form-imino-glutamic acid conversion | One-carbon metabolism. | ↑ |
| Heme synthesis | Prosthetic part of hemoglobin. | ↑ |
| Heme degradation | For availability of iron. | ↑ |
| Cell maintenance | Cell viability. | ↑ |
Please refer to Supplementary Material, Table S4 for the references for each metabolic task. Tick mark represents that corresponding task was performed and the flux value obtained for both the diets were similar, upper arrow represents higher flux values obtained in the American diet when compared with the balanced diet, and cross represents that task could not be performed by either diet. D1: Average American diet, D2: balanced diet.
Effects of different diets on the metabolic tasks of sIECs
We were interested in investigating the consequences of different diets on the sIEC metabolism. Therefore, we simulated an average American (32) and a balanced diet (1,33,34) and tested the model's capability to perform the 50 different metabolic tasks (Table 1, Supplementary Material, Table S4). When comparing the results for the maximal possible flux values of the different metabolic tasks, we found an increased flux for the synthesis of important products, such as glucose, ornithine and citrulline, from glutamine in the average American diet compared with the balanced diet (Table 1). Simultaneously, the glutamine oxidation flux to carbon dioxide was higher under the average American dietary regime, reflecting the higher glutamine availability in this diet. The conversion of fructose to glucose was not possible in the balanced diet due to the absence of fructose in this diet (1). The synthesis flux of 5-methyltetrahydrofolate (5-methyl-THF) was higher in the balanced diet compared with the average American diet due to the influx of its precursor folate in the simulation of the balanced diet. 5-Methyl-THF is an important one carbon substituted folate and participates in amino acid and purine metabolism. Further, when compared with a balanced diet, the average American diet caused a higher flux of luminal inputs, which was particularly the case for fat (i.e. triglycerides and cholesterol) and for carbohydrates. This increased input flux led to a 45% higher flux through the biomass reaction in the average American diet. Another noteworthy observation was the failure to synthesize alanine from glutamine in the absence of glucose, which we observed for both diets (when glucose was removed under this particular simulation condition). The literature review revealed that the major source of pyruvate, which serves as the precursor for lactate and alanine production, is glucose rather than glutamine in sIECs. This conversion route has been demonstrated by 13C-nuclear magnetic resonance studies (3).
To evaluate potential effects of intestinal nutrient absorption on whole body metabolism, we analyzed secretion patterns obtained from the two diets. We performed FVA for the models corresponding to the two diets and compared the computed maximum flux values of each exchange reaction on the basolateral side. A total of 80 metabolites showed differences in their secretion profiles between the two diets (Supplementary Material, Table S5). For instance, higher flux through the secretion reactions for vitamin E (2-fold), choline (1.4-fold) and retinol (2-fold) was observed when the balanced diet was compared with the average American diet. The balanced diet is designed to provide essential vitamins (both water- and fat-soluble ones) and minerals, which should be principally also provided in an average American diet. However, the model failed to secrete components, such as biotin, pantothenic acid and chloride, since these components were missing in the average American diet chart (32). Interestingly, we observed a 2-fold elevation in maximal possible flux values for the secretion of cholesterol esters, and triglycerides when the model was provided with the average American diet compared to the balanced diet. Additionally, the maximal possible secretion flux values for cholesterol and glucose were 4- and 3-fold higher, respectively, in the average American diet than in the balanced diet. When we compared the essential fatty acids secretion pattern, linoleic acid (C18:2) secretion was 2-fold higher in the average American diet, whereas linolenic acid (C18:3) secretion was 2-fold higher in the balanced diet.
Impact of enzymopathies on the enterocyte metabolism
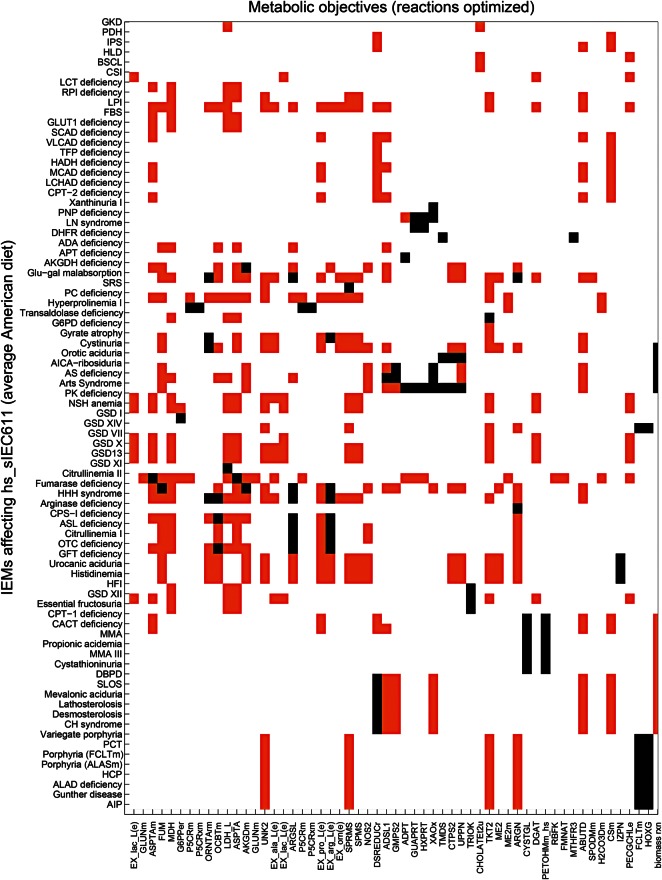
To study the difference in the sIEC metabolism during healthy and diseased states, we analyzed the effect of IEMs on the enterocyte metabolism in the context of the average American diet. hs_sIEC611 captured 109 IEMs out of the 235 IEMs that we have recently mapped onto the global human metabolic reconstruction (20) (Supplementary Material, Table S6). Of these, we considered for further analysis of 103 IEMs, which are caused by mutations in a single gene, meaning that they had one gene-one reaction (66 IEMs) or one gene-many reaction (37 IEMs) associations. Additionally, we analyzed the in silico effect of cystinuria (OMIM: 220 100), which is caused by mutation in either solute carrier family 3 (SLC3A1, GeneID: 6519) or solute carrier family 7 (SLC7A9, GeneID: 11 136). Cystinuria was included in the analysis as the intestinal dibasic amino acid transport system is defective in these patients (35), and thus evidence of the expression of both the genes in human sIECs exists. For each IEM, we deleted the corresponding metabolic reaction(s) and computed the maximal possible flux value for each metabolic task. We found that 76% (79 of 104) IEMs affected at least one of the defined metabolic tasks in hs_sIEC611, when simulated on an average American diet (Fig. 3). Thirty-six of these IEMs (45%) resulted in intestinal pathology or biochemical defect and correspond to reported true positive cases (Table 2). When compared with the reported biomarker patterns used for diagnosis of the simulated IEMs, the model failed to predict the elevated biomarker patterns for 10 IEMs, including hyperprolinemia and glycogen storage disease XI (Supplementary Material, Table S7). We also investigated the basolateral secretion profiles in the different IEM conditions (Fig. 4). The model corresponding to carbamoyl phosphate synthetase I deficiency (OMIM: 237 300) showed a reduced secretion of citrulline, which is consistent with the known reduced citrulline level in these patients. The methylmalonic acidemia model predicted reduced secretion flux for cysteine, which is in agreement with reduced cysteine levels seen in methylmalonic acidemia patients (36). Analysis of the model's metabolic capability revealed that this reduced cysteine secretion flux is due to the model's inability to convert methionine to cysteine. The computational analysis thus provides a mechanistic explanation for the observed biomarker, which represents a novel testable hypothesis.
Figure 3.
Metabolic tasks affected by the different IEMs, under average American diet. Black bars where the metabolic task could not carry any non-zero flux (flux block). Orange bars represent reduced flux through the metabolic task. White bars represent no change. For full description of the IEMs and the corresponding references, please refer to Supplementary Material, Table S7.
Table 2.
IEMs that showed intestinal metabolic dysfunction during in silico simulation and their relation to small intestinal pathology and biochemical defects as reported in the literature
| IEMs | Deficient enzyme and specific gene mutation | Symptoms/organs affected | Relation to small intestine |
|---|---|---|---|
| Porphyrias | |||
| Porphyria (‘FCLTm’)/ erythropoietic protoporphyria | Ferrochelatase (FECH, Gene ID: 2235, E.C. 4.99.1.1). | Acute photosensitivity and liver failure, polyneuropathy (88). | One retrospective case study revealed black blood stained fluid in small intestine (89). Mouse model showed massive bile duct proliferation and biliary fibrosis (90). |
| Porphyria (‘ALASm’)/ sideroblastic anemia | 5-aminolevulinate synthase (ALAS2, Gene ID: 212, E.C. 2.3.1.37). | Fatigue, dizziness, a rapid heartbeat, pale skin, and an enlarged liver and spleen (OMIM: 300 751). | No direct link between small intestine malfunction and sideroblastic anemia could be found. However, erythropoietic protoporphyria patients have been reported to have small intestinal signs and have also been reported to develop eventually sideroblastic anemia (91). |
| Acute intermittent porphyria | Hydroxymethylbilane synthase (HMBS, Gene ID: 3145, E.C. 2.5.1.61) | GI system, nervous system (OMIM: 176 000). | In one case, trans-mural infarction of distal ileum and the existence of sickle cell anemia caused death (49). |
| Variegate porphyria | Protoporphyrinogen oxidase (PPOX, Gene ID: 5498, E.C. 1.3.3.4) | Skin, peripheral nervous system, liver (OMIM: 176 200). | Two cases of co-existence of variegate porphyria and celiac disease have been reported. Coincidental existence of porphyrias and celiac disease also exists, villous atrophy has been seen (50). |
| Delta-amino-levulinate dehydratase deficiency | Delta-aminolevulinate dehydratase (ALAD, Gene ID: 210, E.C. 4.2.1.24) | Described as one form of acute hepatic porphyria (OMIM: 125 270) | Six cases have been reported. Severe abdominal pain may cause nervous symptoms (48). |
| Hereditary coproporphyria | Coproporphyrinogen-III oxidase (CPOX, Gene ID: 1371, E.C. 1.3.3.3) | Nervous system, skin (OMIM: 121 300) | Delay in transit time in small intestine has been observed (92). |
| Nucleotide disorders | |||
| Adenosine deaminase (ADA) deficiency | Adenosine deaminase (ADA, Gene ID: 100, E.C. 3.5.4.4) | Immune systems (OMIM: 102 700) | ADA deficient mice died at the third day of birth and has been reported to show pathological signs of small intestinal cell death (93). |
| Orotic aciduria | Uridine 5′-monophosphate synthase (UMPS, Gene ID: 7372, E.C. 2.4.2.10 and E.C. 4.1.1.23) | Heart, muscle, nervous system (OMIM: 258 900) | Orotic aciduria has been reported in short bowel syndrome (94). |
| Carbohydrate disorders | |||
| Glycogen storage disease XI | l-lactate dehydrogenase (LDHA, Gene ID: 3939, E.C. 1.1.1.27) | Kidney, muscle, skin (OMIM: 612 933) | Patients exhibit intestinal malabsorption and diarrhea (95). |
| Hereditary fructose intolerance | Fructose-bisphosphate aldolase B (ALDOB, Gene ID: 229, E.C. 4.1.2.13) | Liver (OMIM: 229 600) | GI symptoms include nausea, vomiting, abdominal pain, and meteorism (i.e. excessive gas accumulation) (96). |
| Glucose–galactose malabsorption | Sodium/glucose cotransporter 1 (SLC5A1, Gene ID: 6523) | GI system (OMIM: 606 824) | The transport protein, located at the apical surface of enterocytes, actively absorbs glucose and galactose into the cells. Defective transporter leads to primary GI disturbances, i.e. chronic diarrhea as seen in this disorder (97). |
| Pyruvate kinase deficiency | Pyruvate kinase 1 (PKLR, Gene ID: 5313, E.C. 2.7.1.40) | Liver, spleen, gall bladder (OMIM: 609 712 and OMIM: 266 200) | No direct link reported but an increased abundance of the pyruvate kinase M2 form has been used as an inflammatory marker in patients with IBD (98). |
| Congenital lactase deficiency/congenital alactasia | Lactase-phlorizin hydrolase (LCT, GeneID: 3938, E.C. E.C. 3.2.1.108 and 3.2.1.62) | Digestive system (OMIM:223 000) | Congenital lactase deficiency is a severe GI disorder characterized by watery diarrhea in infants fed with breast milk or other lactose-containing formulas. Other clinical signs include vomiting, failure to thrive, dehydration, disacchariduria including lactosuria, renal tubular acidosis and amino aciduria. In a clinical study of 16 patients, it was identifed that the mean height of the epithelial cells was reduced in all cases (99,100). |
| Glycogen storage disease I (GSD I) | Glucose-6-phosphatase and Glucose-6-phosphate translocase (G6PC or SLC37A4, GeneID: 2538, 2542, E.C. 3.1.3.9). GSD type 1 a is casued due to mutations in G6PC and type 1 b due to defect in SLC37A. | Liver, kidney, digestive system, respiratory system, skeletal system (OMIM:232 200) | In both of these disorders, there is frequently occurring chronic diarrhea. Additionally, in GSD Ib, loss of mucosal barrier function due to inflammation seems to be the main cause of diarrhea (101). There is disordered intestinal function in both (102), while untreated type 1b can lead to intestinal mucosal ulcers (103). |
| Fanconi-Bickel syndrome (FBS) | GLUT2 (SLC2A2, GeneID: 6514) | Nervous system, skeletal system, liver, kidney, pancreas (OMIM:227 810) | FBS patients exhibit intestinal malabsoprtion and diarrhea (104). Microvillus inclusion disease (MVID) is a rare congenital enteropathy associated with brush border atrophy and reduced expression of enzymes at the enterocyte's apical surface. MVID is associated with mutations in the MYO5B gene, which is expressed in all epithelial tissues. Patients with MVID were seen to develop FBS while receiving total parenteral nutrition (105). |
| Congenital sucrase–isomaltase deficiency (CSI) | Sucrase–isomaltase (SI, Gene ID:6476, E.C. 3.2.1.48 and 3.2.1.10) | Digestive system | CSI is caused by a deficiency in sucrase–isomaltase, an integral brush border membrane protein of the small intestine (OMIM:222 900). It is charaterized by fermentative diarrhea, abdominal pain and cramps (106). |
| Aminoacidopathies | |||
| Methylmalonic acidemia | Methylmalonyl-CoA mutase (MUT, Gene ID: 4594, E.C. 5.4.99.2) | Brain, kidney, pancreas | Co-occurrence of methylmalonic and propionic acidemia has been reported in patients, who also had gastroenteritis (65). |
| Methylmalonic aciduria III | Methylmalonyl-CoA epimerase (MCEE, Gene ID: 84 693, E.C. 5.1.99.1) | Nervous system (OMIM: 251 120) | Co-occurrence of methylmalonic and propionic acidemia has been reported in patients, who also had gastroenteritis (65). |
| Propionic acidemia | Propionyl-CoA carboxylase (PCCA or PCCB, Gene ID: 5095, Gene ID: 5096, E.C. 6.4.1.3) | Nervous system, muscle, heart (OMIM: 606 054) | Co-occurrence of methylmalonic and propionic acidemia has been reported in patients, who also had gastroenteritis (65). |
| Arginase deficiency | Arginase (ARG1, Gene ID: 383, E.C. 3.5.3.1) | Nervous system, RBC (OMIM: 207 800) | No small intestine disorder reported but a mouse model of arginase deficiency exhibited urea cycle perturbations in liver (107). |
| Cystinuria | b0, +AT transporter (SLC3A1 or SLC7A9, Gene ID: 6519, Gene ID: 11 136) | Kidney, urinary bladder (OMIM.220 100) | Cystinuria is caused by a defect in the intestinal dibasic amino acid transporter. However, no associated GI pathology has been reported (108). |
| Histidinemia | Histidine ammonia-lyase (HAL, Gene ID: 3034, E.C. 4.3.1.3) | Nervous system (OMIM: 235 800) | One case reported with GI disorder (109). |
| Ornithine transcarbamylase deficiency | Ornithine carbamoyltransferase, mitochondrial (OTC, Gene ID: 5009, E.C. 2.1.3.3) | Nervous system, liver, skin, hair (OMIM: 311 250) | Intestinal ornithine transcarbamylase /lactase ratio indicates degree of mucosal damage and atrophy (110). |
| Ornithine translocase deficiency/HHH (hyperornithinemia, hyperammonemia, homocitrullinuria) syndrome | Mitochondrial ornithine transporter 1 (SLC25A15, Gene ID: 10 166) | Nervous system, liver (OMIM: 238 970) | GI illness is a secondary complication in urea cycle disorders (58). |
| Snyder–Robinson syndrome | Spermine synthase (SMS, Gene ID: 6611, E.C. 2.5.1.22) | Nervous system, skeletal system (OMIM: 309 583) | Targeted deletion of the gene in mouse showed no pathology. However, deletion of the downstream enzyme to spermine synthase in polyamine metabolism, i.e. diamine acetyltransferase 1 (SAT1, Gene ID: 6303, E.C.2.3.1.57), in mouse showed GI carcinogenesis (111). |
| Citrullinemia | Argininosuccinate synthase or Citrin (ASS1 or SLC25A13, Gene ID: 445, Gene ID: 10 165, E.C. 6.3.4.5) | Nervous system, liver (OMIM: 215 700, OMIM: 603 471) | Contrastingly low levels of plasma citrulline is an indicator for villous atrophy and short bowel syndrome (85). |
| Lysinuric protein intolerance (LPI) | Y + L amino acid transporter 1 (SLC7A7, GeneID: 9056) | Liver, spleen, skeletal system, muscle, kidney, respiratory system, nervous system, immune system, pancreas (OMIM:222 700) | LPI is caused by defective cationic amino acid transport at the basolateral membrane of epithelial cells in kidney and intestine. LPI should be considered for the differential diagnosis of conditions associated with intestinal villous atrophy (blunting of villi with elongation of crypts) (112). |
| Cholesterol synthesis disorders | |||
| Conradi-Hunermann syndrome | 3-Beta-hydroxysteroid-Delta8, Delta7-isomerase (EBP, Gene ID: 10 682, E.C. 5.3.3.5) | Skeletal system (OMIM: 302 960) | Case report of a newborn male revealed significant delay in motility in small intestine and developmental delay (113). |
| Mevalonic aciduria | Mevalonate kinase (MVK, Gene ID: 4598, E.C. 2.7.1.36) | Nervous system, liver, spleen, muscles (OMIM: 251 170) | One case reported complete jejunal and ileal obstruction and failure to thrive in mevalonic aciduria (114). |
| SLOS | 7-Dehydrocholesterol reductase (DHCR7, Gene ID: 1717, E.C. 1.3.1.21) | Nervous system, muscles (OMIM: 270 400) | Infants with SLOS usually present GI problems that include dysmotility, hypomotility, GI reflux, constipation, formula intolerance and developmental delay (38). |
| Other lipid disorders | |||
| Medium chain acyl CoA dehydrogenase deficiency | Medium chain acyl CoA dehydrogenase (ACADM, Gene ID: 34, E.C. 1.3.99.3) | Liver, nervous system (OMIM: 201 450) | Clinical manifestation includes acute intestinal intussusceptions, i.e. enfolding of segment of intestine causing intestinal obstruction (115). |
| Trifunctional protein deficiency | Trifunctional enzyme subunit alpha, mitochondrial (HADHA, Gene ID: 3030, E.C. 4.2.1.17 and E.C. 1.1.1.211) | Muscle, liver, eye, nervous system (OMIM: 609 016) | Intestinal pseudo-obstruction reported in a mitochondrial tri-functional protein deficiency (116). |
| Long chain acyl-CoA dehydrogenase deficiency (LCHAD) | Trifunctional enzyme subunit beta, mitochondrial (HADHB, Gene ID: 3032, E.C. 2.3.1.16) | Heart, muscle, respiratory system (OMIM: 143 450) | Echogenic bowel syndrome reported in an LCHAD patient (117). |
| Ichthyosis prematurity syndrome (IPS) | Solute carrier family 27 (fatty acid transporter) (SLC27A4, GeneID: 10 999) | Skin, respiratory system | IPS is caused by a mutation in the gene encoding FATP4, a major fatty acid transporter in the enterocytes (OMIM: 608 649, OMIM: 604 194). |
| Miscellaneous disorders | |||
| Dihydrofolate reductase (DHFR) deficiency | Dihydrofolate reductase (DHFR, Gene ID: 1719, E.C. 1.5.1.3) | Nervous system (OMIM: 126 060) | DHFR deficiency results in megaloblastic anemia (OMIM: 613 839). Megaloblastic anemia leads to GI disturbances like malabsorption and diarrhea (118). |
| Glycerol kinase deficiency | Glycerol kinase (GK, GeneID: 2710, E.C. 2.7.1.30) | Digestive system, nervous system | GI symptoms (including gastroenteritis and decreased oral intake) are the initial features of this disease (OMIM: 307 030), (119). |
Figure 4.
Basolateral secretion profile obtained for different IEMs. Only highly affected secretion reactions are reported. Black bars where the secretion reaction could not carry any flux (flux block). Pink bars represent reduced flux through the secretion reaction. White bars represent no change. For full description of the IEMs and the corresponding references, please refer to Supplementary Material, Table S7.
We then categorized the IEMs according to the metabolic pathways they occurred in. The highest number of IEMs was associated with carbohydrate metabolism (23 IEMs), followed by amino acid metabolism (19 IEMs), lipid metabolism (18 IEMs), nucleotide metabolism (nine IEMs), heme biosynthesis (eight IEMs) and two further IEMs (see also Fig. 2). We then analyzed the computed effects of the IEMs on the defined metabolic tasks, which were either flux reduction (drop) or abolishment (block) (Fig. 3). A total of eight IEMs affected only one metabolic task. The hexose monophosphate shunt (HMP) metabolic task had the highest reduction in flux value in 27 IEMs (Fig. 3). Twelve IEMs caused reduction and five IEMs caused block in the cell maintenance/biomass function. Out of these 17 IEMs that affected the cell maintenance function, 8 were found to be associated with intestinal pathology or intestinal biochemical defect (Table 2). Interestingly, reduction in the flux through the trichloroacetic acid (TCA) cycle, synthesis of gamma-amino-butyric acid, and gluconeogenesis from glutamine were the other common effects observed in the simulation results for the IEMs affecting the cell maintenance task. Moreover, we predicted various IEMs to affect the transport function of the sIECs by causing reduction in alanine (10 IEMs), lactate (7 IEMs), proline (20 IEMs), arginine and ornithine secretion (6 IEMs each), or a block in arginine secretion (7 IEMs). The metabolic task for synthesis of phosphatidylcholine was affected by six IEMs, with all of them causing blocked flux (Fig. 3). Interestingly, three of these IEMs have been associated with small intestinal pathology (Table 2), but no changes in the phosphatidylcholine level have been reported.
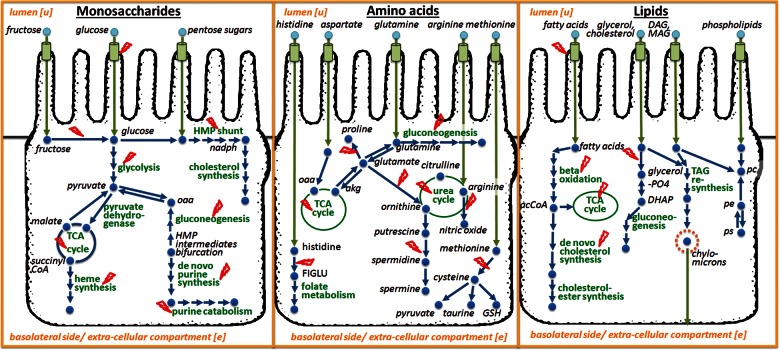
Figure 2.
Overview of enterocyte metabolism and its associated IEMs (in red). Only pathways that are associated with IEMs are shown. (A) Major pathways operating under carbohydrate metabolism. (B) Amino acid metabolism. (C) Lipid metabolism. The metabolic pathways appear in green and the metabolites in blue.
Our model also predicted disabled heme synthesis in eight different porphyrias (Fig. 3), which corresponds to the metabolic task to be eliminated by the highest number of IEMs. hs_sIEC611 also predicted a block in heme degradation in porphyrias, since; there is no dietary supply of heme in the form of heme proteins in the simulated average American and balanced diets. Additionally, we observed a block in the secretion of carbon monoxide and a reduction in urea synthesis and in urea secretion in the porphyria simulations (Fig. 4).
When simulating the average American diet, glucose–galactose malabsorption (OMIM: 606 824) affected in silico the highest number of metabolic tasks (i.e. 22 out of 50) (Fig. 3), including reduced fluxes through gluconeogenesis from glutamine and the synthesis of gamma-aminobutyric-acid, i.e. GABA from putrescine. The GABA synthesis, in particular, was affected in 23 IEMs (Fig. 3), with 17 of them exhibiting variable extents of neurological symptoms (Supplementary Material, Table S7). Two IEMs, gyrate atrophy of the choroid and retina (OMIM: 258 870) and ornithine translocase deficiency (OMIM: 238 970), showed the highest reduction in the flux value through a synthetic reaction for GABA (17% of the flux value of the healthy model).
Under an average American diet, the model for type I citrullinemia (OMIM: 215 700), which is defective in ASS1 (GeneID: 445, E.C.6.3.4.5), led to an increased flux through the citrulline basolateral secretion reaction, a single fumarate producing reaction (‘ADSL1’), and a decreased flux through the fumarase (‘FUM’). These predictions were consistent with the reported build-up of citrulline in patients (37). Moreover, the model predicted an increased flux through the luminal uptake reactions of two amino acid transporters, ASCT2 (SLC1A5, GeneID: 6510) and B(0)AT1 (SLC6A19, GeneID: 340 024), and through the alanine transaminase (E.C.2.6.1.2.). The latter converts alanine to pyruvate, which subsequently enters gluconeogenesis providing endogenous glucose. Furthermore, the type I citrullinemia model was able to utilize partially the urea cycle by synthesizing ornithine, which was then converted either to glutamate by the ornithine transaminase (E.C.2.6.1.13) or to citrulline by the ornithine carbamoyltransferase (E.C. 2.1.3.3).
Smith–Lemli–Opitz syndrome (SLOS, OMIM: 270 400) is a cholesterol synthesis disorder caused by a mutation in DHCR7 (GeneID: 1717, E.C.1.3.1.21). Under the average American diet, this enzymopathy led to an increased flux through ‘nadp’ and ‘nad’ utilizing reactions (i.e. ‘G6PDH2c’, ‘GNDc’, ‘AKGDm’, ‘SUCD1m’, and ‘ICDHxm’) and a decreased flux through the basolateral cholesterol secretion reaction (‘EX_chsterol(e)’) in the corresponding model. Additionally, we predicted increased fluxes through the reaction of electron transport chain, gluconeogenesis and purine salvage pathway (Supplementary Material, Table S11). These predicted changes were consistent with the reported low cholesterol level in patients (38).
Does the effect of IEMs on the metabolic capabilities of sIECs depend on diet? To address this question, we repeated the computations while simulating the balanced diet. We overall found the metabolic capabilities similar in enzyme-deficient sIECS under the balanced diet (Supplement Fig. S1). Fifity-four of the IEMs resulted in altered fluxes for various metabolic tasks (Supplement Table S10). For instance, while the biomass reaction was blocked in the cystinuria model under the average American diet, the flux value was identical to the healthy model under the balanced diet. In contrast, modeling the ribose 5-phosphate isomerase deficiency resulted in a block of the flux through the biomass reaction under balanced diet only. Moreover, IEMs of the amino acid metabolism [i.e. propionic acidemia, methylmalonic acidemia (MMA), cystathioninuria, and MMA type 3] and of the lipid metabolism (i.e. CACT deficiency and CPT-1 deficiency) showed the same flux value through the biomass reaction under balanced diet as the healthy model but had a lower flux value under the average American diet. The computed block or reduction in biomass reaction flux in these IEMs was due to limited uptake of methionine and proline in the case of cystinuria, and of alanine, asparagine, leucine, and phosphatidyl-ethanolamine for the IEMs of amino acid and lipid metabolism. The uptake limitation was a result of missing chloride ions in the in silico average American diet, as they were not listed in the corresponding diet chart (32). Consequently, one of the intestinal amino acid transporters (ATB0, SLC6A14, GeneID: 11 254), which has a broad substrate specificity for both neutral and cationic amino acids (39) and requires the presence of sodium and chloride for its activity, could not be used in this diet. In contrast, the limiting component in the simulated balanced diet was ribose. Additionally, blocked flux through HMP, reduced fluxes through reactions involved in pyrimidine synthesis, in the conversion of glutamine to carbon dioxide, and in the purine catabolism pathway contributed to a block in the biomass reaction for cystinuria.
DISCUSSION
In this study, we presented a comprehensive, predictive metabolic network for small intestinal enterocytes, which captures comprehensively its physiology and biochemistry. The key results include: (i) the average American diet resulted in higher secretion flux for metabolites involved in metabolic syndrome, (ii) enterocyte metabolism was perturbed by a large number of IEMs, (iii) porphyrias may not only perturb heme synthesis but also its degradation, (iv) a disturbed intestinal GABA synthesis may be linked to the neurological dysfunction observed in ornithine translocase deficient and gyrate atrophy patients, acting through the gut–brain axis and (v) identification of known and novel co-morbidities.
An objective of this work was to access the role of diet under physiological and pathological conditions. We chose to compare an average American diet with a balanced diet. The balanced diet modeled in this study represents an ideal diet composition (1), which may not be achieved by most of us on a daily basis. When we applied these dietary patterns, we observed a similar overall metabolic performance of the sIECs under both diets, but we also observed flux changes in the metabolic tasks for metabolites that have been shown to have implications in metabolic syndrome (40,41). For instance, the average American diet led to a significant increase of the fructose and glucose secretion flux into the extracellular compartment. The high consumption of fructose from added sugars has been linked to hypertension (42) and its increased levels in serum and urine have also been found in patients with diabetes type 2 (43). Additionally, it has been suggested that high glucose intake, mainly from sweetened beverages, could lead to weight gain (44). The absorption pathway of dietary lipid components is distinct from water-soluble nutrients as they are secreted into the lymph from the enterocytes. Once absorbed into the lymphatics, which drain into the left subclavian vein, dietary lipids reach finally the bloodstream (45). In the model, the computed higher secretion fluxes for cholesterol, cholesterol ester and triglycerides corresponded to the increased availability of dietary lipids in the average American diet compared with the balanced diet. It has been recommended to reduce the level of lipid in the average American diet (46) due to its close association with diseases, such as obesity and type-2 diabetes (47).
The epithelial cell surface of the small intestine consists of enterocytes and immune system cells, which are covered by a mucus layer, and represents a protective barrier between the gut microbiota and the host. Numerous diseases exhibit gastrointestinal (GI) signs, even though the small intestine may not be recognized as the primary site of disease. We investigated the potential consequences of IEMs on the sIEC metabolism and correlated them with known clinical features (Table 2). Surprisingly, one-third of the IEMs that are currently captured by the global human metabolic reconstruction (20) mapped onto hs_sIEC611. Even more astonishingly, almost two-thirds of the simulated IEMs perturbed significantly the enterocyte metabolism by deranging the function of at least one of the 50 metabolic tasks (Fig. 3). Almost half of the perturbing IEMs are known to result in intestinal pathology and thus, correspond to true positive predictions (Table 2), underlining again the value of computational modeling for elucidating genotype–phenotype relationships. The model's failure to correctly predict some known elevated reported biomarkers, e.g. citrulline and glycogen (Supplementary Material, Table S7), highlights that enterocyte metabolism is not the only cause for biomarkers but that other tissues, such as liver and muscle, also contribute significantly to plasma biomarkers. Our simulations represent isolated small intestinal enterocytes, but more accurate IEM modeling will require a comprehensive, multi-tissue model. Additionally, our study presents a qualitative but not a quantitative comparison between the changes in flux values for the basolateral secretion reactions and reported biomarker pattern. The presented work is a good starting point to investigate quantitative predictions for the flux ranges, once sufficient kinetic parameters for the reactions are known.
Heme is essential for life and is often referred to as ‘pigment of life’ (48). It is synthesized from succinyl-CoA and glycine. Enzyme deficiencies in any of the steps along the biosynthetic pathway cause different forms of porphyrias (48). Consistently, the metabolic task ‘heme synthesis’ was blocked in the porphyria models (Fig. 3). Usually porphyrias show an accumulation of porphyrins, excreted in urine and feces, which is used for biochemical diagnosis (8). Possible GI signs include ileal infarction in acute intermittent porphyria and villous atrophy in variegate porphyria (49,50). These clinical signs may be attributed to the lack of heme in the diet, which can lead to non-optimal bilirubin concentrations within the enterocyte. Since neither of the analyzed diets contained heme supplementation, the porphyrias also resulted in a block in the metabolic task ‘heme degradation’, which produces bilirubin. This compound has potential antioxidant activity, comparable with that of beta-carotene and tocopherol, under low oxygen conditions (51) and, at very low concentrations, bilirubin tends to exert protective effects on the small intestine (52). The porphyria models were not able to secrete carbon monoxide (Fig. 4), which has various roles in the GI tract, including regulation of GI motility, activation of guanylate cyclase (suggested role in inflammation), function as a neurotransmitter, contribution to the membrane potential gradient and exerting protective role during GI injury (53,54). Particularly, the latter role suggests that this compound may contribute to the GI inflammation observed in chronic inflammatory bowel disease (IBD). Interestingly, IBD patients are prone to develop acute intermittent porphyria, i.e. AIP (55). The clinical manifestations of AIP patients also include neurological dysfunction (35). The predicted reduced urea secretion flux (Fig. 4) in the porphyria models was due to a reduced urea cycle, indicating ammonia toxicity. For instance, clinical features of hereditary coproporphyria and variegate porphyria include neurological dysfunction (35). The computed altered urea secretion flux for porphyrias could thus indicate that ammonia toxicity may also affect the brain of patients with types of other porphyrias. These computational results provide a possible mechanistic explanation for the extensive clinical manifestations observed in porphyria patients.
Gyrate atrophy (OMIM: 258 870) and ornithine translocase deficiency (OMIM: 238 970) are caused by mutations in ornithine aminotransferase (OAT, GeneID: 4942, E.C. 2.6.1.13) and mitochondrial ornithine transporter (SLC25A15, GeneID: 10 166), respectively. While gyrate atrophy patients usually have vision loss (OMIM: 258 870), ornithine translocase deficiency is associated with neurocognitive deficits, liver dysfunctions and lethargy (56). Interestingly, both disorders exhibit a variable degree of neurological manifestations. Gyrate atrophy patients have degenerative changes in the white matter and brain atrophy (57), whereas for ornithine translocase deficiency patients usually show developmental delay, ataxia, spasticity and learning disabilities (56). Although a disturbed creatine and phospho-creatine metabolism in gyrate atrophy, and intermittent hyperammonemia in ornithine translocase deficiency have been suggested to be partly responsible for the neurological symptoms, further clarification is needed (56,57). This is particularly true for the class of patients of ornithine translocase deficiency, who have deficiencies in executive function but never show symptoms of hyperammonemia (58). Interestingly, these two IEMs showed the highest reduction in flux through the synthesis of GABA, an inhibitory neurotransmitter (59). It has been proposed that the gut and the brain interact via the enteric nervous system through exchange of signaling molecules, which include cytokines and certain metabolites (60). This brain–gut axis is bidirectional and may have implications in various diseases (61). Recently, modulation of the GABA receptors by the probiotics was shown to interact via the brain–gut axis (62). To our knowledge, there have not been any studies analyzing these IEMs from the aspects of brain–gut axis interactions. Based on our computational results, one may speculate that neurological dysfunctions in gyrate atrophy and ornithine translocase-deficient patients may be caused by reduced GABA synthesis in the small intestine.
Metabolic networks have been used previously to study comorbidity, i.e. the co-occurrence of diseases (24). Our simulations revealed a flux block in the conversion of methionine to cysteine for MMA, propionic acidemia and cystathionuria (Fig. 3). This incapability agrees with reports of low cysteine levels observed in these IEMs (36,63,64), which differ from one another with respect to the deficient enzyme involved (Table 2). Interestingly, the co-existence of MMA and cystathionuria in patients has been reported in the literature (35). Additionally, MMA and propionic acidemia share common clinical features with respect to movement disorders, gastroenteritis and neurologic abnormalities (particularly vulnerable basal ganglia) (65). Our in silico analysis also predicted a failure to synthesize phosphatidylcholine from phosphatidylethanolamine in the latter two IEMs. This inability may also contribute to their similar clinical features, since phosphatidylcholine is an important membrane constituent and has been shown to exert anti-inflammatory roles, most highly beneficial in ulcerative colitis (66). Additionally, phosphatidylcholine acts in the mucus as a barrier against many inflammatory conditions (67). This example highlights again the promise of the computational systems biology approach to identify and investigate comorbidities.
Enzyme-deficient cells can bypass the missing functionality by employing alternate metabolic pathways (68). The sIEC model may be used to investigate potential effects of such alternate pathways and may explain the variability of clinical phenotypes within the same IEM. For instance, broad phenotypic variability is particularly evident for type I citrullinemia presenting the classic neonatal form, which causes severe neurological deficit or death without early treatment, the mild adult-onset form, which may not exhibit any symptoms, and a form usually seen in women during pregnancy or post partum (37). The predicted flux changes through citrulline utilizing, fumarate producing and amino acid-related reactions suggest that sIECs could switch to alternative pathways for production of energy precursors and to increased amino acid metabolism in order to maintain the biomass flux and energy state. Any mutation along these pathways could alter the observed phenotype and may contribute to the broad clinical features observed in type I citrullinemia patients. Another example is SLOS, which also has a broad clinical spectrum. While the severe end of the spectrum presents with life-threatening conditions (growth retardation, intellectual disability and multiple major and minor malformations), the milder forms have only minor physical abnormalities (38). The SLOS model utilized at a higher rate the NADH generating reactions of TCA cycle and reactions of the electron transport chain to meet the energy demands of the cell. Additionally, the reactions of the HMP were utilized to generate NADP, which was channeled into proline synthesis, and 5-phospho-alpha-d-ribose-1-diphosphate, which was used for the purine salvage pathway. Interestingly, key gluconeogenic reactions (i.e. ‘PCm’, ‘FBP’ and ‘G6PPer’) also had a higher flux in the SLOS model, indicating that when faced with enzyme deficiencies, sIECs could switch their dependency from amino acids to carbohydrate sources. These examples demonstrate how computational modeling of IEMs could provide biochemical explanations for adaptive mutations in alternative pathways and for phenotypic variability within the same IEM groups.
In conclusion, we want to emphasize that small intestinal enterocytes play a key role in whole body metabolism due to their involvement in enzymatic digestion and absorption of nutrients as well as inter-organ metabolism. Investigating whole body metabolism in silico will require a comprehensive organ- and tissue resolved metabolic model, which also includes small intestinal enterocyte and hepatocyte metabolism. Such a model could be employed to predict metabolic changes upon alteration of nutrition, genetic variations, and their impact onto human health. Since metabolic models connect genes with metabolic function, they are promising platforms for personalized systems biomedicine.
MATERIALS AND METHODS
Flux balance analysis (FBA) and flux variability analysis
The constraint-based modeling approach assumes a steady state for the biochemical transformations to occur (30). Under these conditions, the sum of input fluxes equals the sum of output fluxes since the change of metabolite concentration (dx) over time (dt) is zero. The steady-state equation can be written as S.v = 0, where S represents the stoichiometric matrix of size m × n (with ‘m’ being the number of metabolites and ‘n’ being the number of reactions in the network). v is a flux vector of size n× 1 containing a flux value vi for each reaction, i in the network. Flux balance analysis (FBA) aims to maximize a given objective function under a particular simulation condition as defined by the constraints, which are applied to the model. In this study, metabolic tasks were defined (see below) and employed individually as objective functions under different simulation conditions (e.g. dietary intake or IEM conditions). In FVA, the minimum and maximal flux values are computed for each reaction in the model, by choosing each reaction individually as objective function and performing FBA for that reaction (31,69).
Reconstructing a sIEC-specific metabolic network
A thorough literature search was performed to obtain an in-depth knowledge of all the major metabolic pathways known to occur in an epithelial cell of the small intestine. We then retrieved the corresponding reactions and genes from the human metabolic reconstruction (13), which is accessible through the BiGG database (70), to compile a draft reconstruction. Missing transport and metabolic reactions for peptides and for dietary fibers were added to the initial draft reconstruction upon detailed manual gap analysis and further review of the corresponding literature. Genome annotations from the EntrezGene database (71) as well as protein information from the UniProt (72) and BRENDA database (73) were used in addition to the information retrieved from the scientific literature to assign GPR associations to the reactions not present in Recon 1. For the reactions that were extracted from the Recon 1, GPR associations were kept as reported in Recon 1, since no comprehensive transcriptomic data are available for sIECs. The sIEC metabolic reconstruction was assembled and converted to a mathematical model using rBioNet as a reconstruction environment (74) and an established protocol (75).
We used the global human metabolic network, Recon 1 (13), as reaction database, but adjusted sub-cellular and extracellular location, reaction stoichiometry and directionality according to literature evidence. Only those reactions and pathways with literature evidence for their occurrence in human small intestinal enterocytes were incorporated into hs_sIEC611 from the global human metabolic reconstruction Recon 1, which captures metabolic capabilities known to occur in any human cell. Moreover, we added 262 transport and 50 metabolic reactions, which were not present in Recon 1, but for which supporting information for their presence in sIECs could be found in the scientific literature (Fig. 1E, Supplementary Material, Table S2). These reactions included many transport systems specific for enterocytes and metabolic pathways for sulfo-cysteine metabolism, dietary fiber metabolism, di- and tri-peptide degradation and cholesterol-ester synthesis (Fig. 1E, Supplementary Material, Table S2). In addition to these reactions, 73 reactions were added from our recently published acylcarnitine/fatty acid oxidation module for the human metabolic reconstruction (20). We added further 95 reactions, which were present in Recon 1, but for which the compartment was adjusted by placing them into the lumen compartment. The stoichiometry of the reactions catalyzed by the glucose 6-phosphate dehydrogenase (E.C. 1.1.1.49), the 6-phosphogluconolactonase (E.C. 3.1.1.31) and the phosphogluconate dehydrogenase (E.C. 1.1.1.44) was changed to three, since they were required to generate three molecules each of 6-phospho-d-glucono-1,5-lactone, 6-Phospho-d-gluconate and ribulose-5-phosphate simultaneously (76). The directionality of ATP requiring fatty acid activation reactions catalyzed by the fatty acyl-CoA ligase (E.C. 6.2.1.3) was changed in agreement with a recent report (76). Also, the cofactor requirement and sub-cellular localization of reactions included in the cholesterol synthesis pathway, which are catalyzed by the desmosterol reductase (E.C. 1.3.1.72) and HMG-CoA reductase (E.C. 2.3.3.10) reactions, were updated in accordance with the current literature evidence (76).
Cellular compartments
The sIEC reconstruction accounts for the following cellular compartments: cytoplasm, mitochondrion, endoplasmic reticulum, peroxisome and nucleus. Due to its specific anatomy of being a columnar cell, the sIEC possesses an apex and a base (see also Fig. 2). Both of these structural entities are specific with respect to their substrate uptake and product secretion and exhibit very distinct transporters. Therefore, an additional extra-cellular compartment (‘u’, lumen) was added to the sIEC reconstruction, which represents the gut lumen. Moreover, metabolites are released from the sIEC into either portal venous blood (non-fat nutrients) or lymph (fat nutrients). sIECs take up nutrients from the arterial supply (i.e. superior and inferior pancreatico-duodenal and superior mesenteric arteries) (77). These three biofluids are treated as one extra-cellular compartment (‘e’) in our metabolic reconstruction. Metabolic and transport reactions occurring at the lumen and at the basal side were added based on the available literature enabling the assignment of the transporter location, and thus, the luminal uptake and basal release of specific metabolites.
Formulation of biomass reaction
Metabolic reconstructions generally account for a biomass reaction, which captures all known precursors required for physiological and functional maintenance of the cell (75). We adopted the biomass reaction from Recon 1 (13), while making sIEC-specific modifications. For instance, phosphatidylinositol, phosphatidylglycerol, cardiolipin, phosphatidylserine and sphingomyelin were removed because the exact metabolic pathways involved in the synthesis and degradation of these molecules within the enterocytes are not known. However, the synthesis of phosphatidylethanolamine from phosphatidylserine in rat small intestine has been suggested (78). Deoxy-nucleotides have been also removed from the Recon 1 biomass reactions as sIECs cannot replicate (79). The sIEC biomass reaction thus comprises 20 amino acids, four nucleotides, cholesterol, phosphatidylcholine, phosphatidylethanolamine, folic acid and water (see Supplementary Material, Table S2 for a detailed list). In order to account for the energy required for the cell maintenance, ATP hydrolysis was also included in the biomass reaction.
GPR associations in the enterocyte reconstruction
The present metabolic reconstruction for human sIECs was assembled in a bottom-up manner based on the available literature data. A recent study has reported the proteomic composition of lipid droplets of large intestinal enterocytes, but it focused mainly on proteins participating in non-metabolic reactions (80). Since no comprehensive transcriptomic and proteomic dataset is currently available for human sIECs, GPR associations for all those reactions derived from the generic human metabolic network reactions in hs_sIEC611 were kept as reported in Recon 1. For the reactions that were derived from the IEM module, the GPR associations were retained as described in the module. For the newly added metabolic and transport reactions, the literature was thoroughly reviewed to derive the respective GPR associations (see Supplementary Material, Table S3 for details on GPR associations). Subsequently, the number of included metabolic genes (611) may be higher than the actual number of metabolic genes expressed in sIECs. However, only 201 reactions are associated with isozymes, while additional 48 reactions are associated with protein complexes as well as isozymes (Fig. 1C). Thus, 19% (249/1282) of the reactions of hs_sIEC611 may be affected by the current lack of comprehensive omics data for human sIECs, which corresponds to 396 unique genes. Further 19% (245/1282) of the reactions have no gene associated, which includes 61 diffusion and 16 spontaneous and non-enzymatic reactions. The remaining 168 reactions are orphan reactions.
Debugging of the sIEC model
Dead-end metabolites can only be produced or consumed within a metabolic reconstruction. A key step of the reconstruction process is to evaluate and eliminate these dead-end metabolites depending on available experimental support in the literature (75). Dead-end metabolites were identified using the COBRA toolbox (81). Blocked reactions, which cannot carry any flux value under the given simulation constraints, were identified using FVA (31). Dead-end metabolites and blocked reactions were evaluated manually and supporting evidence from the literature was assembled to connect them to the remainder of the network. Furthermore, we tested for the model's ability to produce each biomass component, as described elsewhere (75).
Secretion profile of the enterocyte model
Based on the data derived from the literature, we constrained 267 exchange reactions for luminal uptake secretion and basolateral uptake secretion (Supplementary Material, Table S5). Dietary fibers, triglycerides and other partially digested products cannot enter enterocytes and remain in the lumen, unless they are broken down into simpler units. For example, polysaccharides and disaccharides are hydrolyzed into monomeric forms prior to absorption. The bounds on the corresponding exchange reactions were kept unchanged.
Collection of dietary composition
Diet information was collected from the US Department of Agriculture, Agricultural research service (32). All nutrients were converted to a common unit of grams per day. An average American diet, for both males and females (≥2 years), was collected (32). A balanced diet representing the recommended dietary reference intakes (DRIs) of micronutrients as well as macronutrients (1,33) was also assembled (Supplementary Material, Table S8). The DRI reflects the minimum requirements advised to be taken daily to maintain a healthy lifestyle. The values were taken for males between 30 and 50 years of age. The total fat and cholesterol intake for balanced diet was also represented (34,82). We did not represent the energy value in food (i.e. caloric load) due to disparity between various sources and also due to lack of specific information. The uptake values of the various dietary nutrients in both diets were set as lower bounds on the corresponding luminal exchange reactions. The total carbohydrate content in the diet was converted to starch and dextrin uptake, since polysaccharides are most abundant carbohydrate component present in our diet (1). In the case of total sugars, disaccharides formed 60% and monosaccharides formed 40%, since free monosaccharides do not form significant part of the diet (1). Supplementary Material, Table S8 provides details of the dietary formulations, distribution and references used. Under the given simulation conditions, we assumed that all the available nutrients are completely taken up by the enterocytes.
Conversion of the dietary components into exchange reaction constraints
All dietary components were converted to moles/human/day (Supplementary Material, Table S8). To obtain the number of enterocytes per small intestine in a human, we used data reported by Chougule et al. (83). The authors reported that a 10 cm segment of the small intestine contained 7.5 × 106 cells, of which 92% were enterocytes. The wet weight of the small intestine was derived from their median regional wet weights (79 g for duodenum, 411 g for jejunum and 319 g for ileum) (84). We assumed that the average length of the human small intestine was 20 foot (609.6 cm) (77) and that the enterocytes consist of 70% water (7). The dry weight of the small intestine enterocytes per human was thus 809 × 0.3 × 0.552 = 133.9704 gDW_enterocytes. The conversion factor for all the dietary inputs from moles/human/day to mmol/gDW_enterocytes/hr was thus R = 24 × 133.9704 × 10−3 = 3.2152896 (see also Supplementary Material, Table S8).
Mapping of IEMs onto the enterocyte metabolic network
The IEM information, i.e. affected genes, reactions, organs and identified biomarkers, was taken from our previous work (20). For the simulations of each IEM condition on an average American diet, we only considered those IEMs with one gene-one reaction and one gene-multiple reaction associations. We constrained the corresponding reactions to be inactive (i.e. lower and upper reaction bounds were set to be zero) and performed FBA for each of the metabolic tasks (see Supplementary Material, Table S7 for the agreement of the in silico results with the literature for the diagnostically relevant biomarkers as well as biochemical defects reported for each simulated IEM.)
Capturing adaptive mechanisms under IEM conditions
The affected reaction(s) for an IEM was set as objective function in the healthy model and maximized using FBA. Then, 25% of this maximized value was used to assign the lower bound to this reaction in the healthy model and FVA was performed to obtain the minimal and maximal flux through each model reaction i (FVAmini and FVAmaxi). The IEM condition was simulated by constraining the lower and the upper bounds of the affected reaction(s) to zero and FVA was performed. The outputs obtained for the healthy model were compared with the output of the corresponding IEM model. The ratio of flux span (FSr) was calculated, using the following formula FSr = abs (FVAmaxi,Healthy-FVAmini,Healthy)/abs(FVAmaxi,IEM-FVAmini,IEM), where FVAmax is the maximal flux value and FVAmin is the minimal flux value under healthy and IEM conditions. The candidate reactions were identified using a 5% cutoff on the FSr (i.e. reactions with at least 5% difference in flux span between healthy and IEM were chosen).
All fluxes were computed and given in mmol/gDW_enterocytes/h. All simulations were carried out with MatLab (MathWork Inc.) as programming environment, using Tomlab (TomOpt Inc.) as a linear programming solver, and the COBRA toolbox (81).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Icelandic Research Fund (No. 100406022). Funding to pay the Open Access publication charges for this article was provided by Icelandic Research Fund (No. 100406022).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof J.J. Jonson, Prof L. Franzson, Prof R.M.T. Fleming, Mrs A. Heinken and Mrs H. Haraldsdottir for valuable discussions as well as Mrs M. Galhardo and Mr Y. Sun for assistance in figure preparation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gropper S.A.S., Smith J.L., Groff J.L. Advanced nutrition and human metabolism. Australia; United States: Wadsworth/Cengage Learning; 2009. [Google Scholar]
- 2.Goodman B.E. Insights into digestion and absorption of major nutrients in humans. Adv. Physiol. Educ. 2010;34:44–53. doi: 10.1152/advan.00094.2009. [DOI] [PubMed] [Google Scholar]
- 3.Mithieux G. New data and concepts on glutamine and glucose metabolism in the gut. Curr. Opin. Clin. Nutr. Metab. Care. 2001;4:267–271. doi: 10.1097/00075197-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Dietschy J.M., Siperstein M.D. Cholesterol synthesis by the gastrointestinal tract: localization and mechanisms of control. J. Clin. Invest. 1965;44:1311–1327. doi: 10.1172/JCI105237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J.H., Chiba M., Baillie T.A. Is the role of the small intestine in first-pass metabolism overemphasized. Pharmacol. Rev. 1999;51:135–158. [PubMed] [Google Scholar]
- 6.Pang K.S. Modeling of intestinal drug absorption: roles of transporters and metabolic enzymes (for the Gillette Review Series) Drug Metab. Dispos. 2003;31:1507–1519. doi: 10.1124/dmd.31.12.1507. [DOI] [PubMed] [Google Scholar]
- 7.Guyton A.C., Hall J.E. Text book of medical physiology. W.B. Saunders company; 2000. [Google Scholar]
- 8.Fernandes J. Inborn metabolic diseases: diagnosis and treatment. Heidelberg: Springer; 2006. [Google Scholar]
- 9.Mootha V.K., Hirschhorn J.N. Inborn variation in metabolism. Nat. Genet. 2010;42:97–98. doi: 10.1038/ng0210-97. [DOI] [PubMed] [Google Scholar]
- 10.Roy C.N., Andrews N.C. Recent advances in disorders of iron metabolism: mutations, mechanisms and modifiers. Hum. Mol. Genet. 2001;10:2181–2186. doi: 10.1093/hmg/10.20.2181. [DOI] [PubMed] [Google Scholar]
- 11.Martins A.M. Inborn errors of metabolism: a clinical overview. Sao Paulo Med. J. 1999;117:251–265. doi: 10.1590/s1516-31801999000600006. [DOI] [PubMed] [Google Scholar]
- 12.Palsson B. Systems biology : properties of reconstructed networks. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 13.Duarte N.C., Becker S.A., Jamshidi N., Thiele I., Mo M.L., Vo T.D., Srivas R., Palsson B.O. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigurdsson M.I., Jamshidi N., Steingrimsson E., Thiele I., Palsson B.O. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst. Biol. 2010;4:140. doi: 10.1186/1752-0509-4-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gille C., Bolling C., Hoppe A., Bulik S., Hoffmann S., Hubner K., Karlstadt A., Ganeshan R., Konig M., Rother K., et al. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol. Syst. Biol. 2010;6:411. doi: 10.1038/msb.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang R.L., Xie L., Bourne P.E., Palsson B.O. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol. 2010;6:e1000938. doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordbar A., Lewis N.E., Schellenberger J., Palsson B.O., Jamshidi N. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol. Syst. Biol. 2010;6:422. doi: 10.1038/msb.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordbar A., Jamshidi N., Palsson B.O. iAB-RBC-283: a proteomically derived knowledge-base of erythrocyte metabolism that can be used to simulate its physiological and patho-physiological states. BMC Syst. Biol. 2011;5:110. doi: 10.1186/1752-0509-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y., Huang J. Reconstruction and analysis of human heart-specific metabolic network based on transcriptome and proteome data. Biochem. Biophys. Res. Commun. 2011;415:450–454. doi: 10.1016/j.bbrc.2011.10.090. [DOI] [PubMed] [Google Scholar]
- 20.Sahoo S., Franzson L., Jonsson J.J., Thiele I. A compendium of inborn errors of metabolism mapped onto the human metabolic network. Mol. Biosyst. 2012;8:2545–2558. doi: 10.1039/c2mb25075f. [DOI] [PubMed] [Google Scholar]
- 21.Heinken A., Sahoo S., Fleming R.M., Thiele I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes. 2012;4:28–40. doi: 10.4161/gmic.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlomi T., Cabili M.N., Ruppin E. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol. Syst. Biol. 2009;5:263. doi: 10.1038/msb.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamshidi N., Vo T.D., Palsson B.O. In silico analysis of SNPs and other high-throughput data. Methods Mol. Biol. 2007;366:267–285. doi: 10.1007/978-1-59745-030-0_15. [DOI] [PubMed] [Google Scholar]
- 24.Lee D.S., Park J., Kay K.A., Christakis N.A., Oltvai Z.N., Barabasi A.L. The implications of human metabolic network topology for disease comorbidity. Proc. Natl Acad. Sci. USA. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folger O., Jerby L., Frezza C., Gottlieb E., Ruppin E., Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011;7:517. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frezza C., Zheng L., Folger O., Rajagopalan K.N., MacKenzie E.D., Jerby L., Micaroni M., Chaneton B., Adam J., Hedley A., et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 27.Thiele I., Price N.D., Vo T.D., Palsson B.O. Candidate metabolic network states in human mitochondria: Impact of diabetes, ischemia, and diet. J. Biol. Chem. 2005;280:11683–11695. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 28.Sigurdsson M.I., Jamshidi N., Jonsson J.J., Palsson B.O. Genome-scale network analysis of imprinted human metabolic genes. Epigenetics. 2009;4:43–46. doi: 10.4161/epi.4.1.7603. [DOI] [PubMed] [Google Scholar]
- 29.Le Gall M., Tobin V., Stolarczyk E., Dalet V., Leturque A., Brot-Laroche E. Sugar sensing by enterocytes combines polarity, membrane bound detectors and sugar metabolism. J. Cell. Physiol. 2007;213:834–843. doi: 10.1002/jcp.21245. [DOI] [PubMed] [Google Scholar]
- 30.Orth J.D., Thiele I., Palsson B.O. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahadevan R., Schilling C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 32.U.S. Department of Agriculture, A.R.S. (NHANES 2007–2008)
- 33.U.S. Department of Agriculture, A.R.S.U.N.N.D.f.S.R., Release 24. Nutrient Data Laboratory Home Page http://www.ars.usda.gov/nutrientdata .
- 34.Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids (Macronutrients) Washington, DC: The National Academies Press; 2005. [DOI] [PubMed] [Google Scholar]
- 35.Stanbury J.B., Wyngaarden J.B., Fredrickson D.S., Goldstein J.L., Brown M.S. The Metabolic Basis of Inherited Disease. McGraw-Hill Book Company, New York, NY; 1983. [Google Scholar]
- 36.Treacy E., Arbour L., Chessex P., Graham G., Kasprzak L., Casey K., Bell L., Mamer O., Scriver C.R. Glutathione deficiency as a complication of methylmalonic acidemia: response to high doses of ascorbate. J. Pediatr. 1996;129:445–448. doi: 10.1016/s0022-3476(96)70080-x. [DOI] [PubMed] [Google Scholar]
- 37.Thoene J.G. 1993. Citrullinemia Type I. In Pagon, R.A., Bird, T.D., Dolan, C.R., Stephens, K., Adam, M.P. (eds), GeneReviews™ [Internet], University of Washington, Seattle; 1993–2004 Jul 07 [updated 2011 Aug 11]
- 38.Irons M. 1993. Smith-Lemli-Opitz syndrome. In Pagon, R.A., Bird, T.D., Dolan, C.R., Stephens, K., Adam, M.P. (eds), GeneReviews™ [Internet], University of Washington, Seattle; 1993–1998 Nov 13 [updated 2007 Oct 24]
- 39.Broer S. Apical transporters for neutral amino acids: physiology and pathophysiology. Physiology (Bethesda) 2008;23:95–103. doi: 10.1152/physiol.00045.2007. [DOI] [PubMed] [Google Scholar]
- 40.Grundy S.M., Brewer H.B., Jr., Cleeman J.I., Smith S.C., Jr., Lenfant C. Definition of metabolic syndrome: report of the National Heart, Lung and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109:433–438. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 41.Zivkovic A.M., German J.B., Sanyal A.J. Comparative review of diets for the metabolic syndrome: implications for nonalcoholic fatty liver disease. Am. J. Clin. Nutr. 2007;86:285–300. doi: 10.1093/ajcn/86.2.285. [DOI] [PubMed] [Google Scholar]
- 42.Jalal D.I., Smits G., Johnson R.J., Chonchol M. Increased fructose associates with elevated blood pressure. J. Am. Soc. Nephrol. 2010;21:1543–1549. doi: 10.1681/ASN.2009111111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawasaki T., Akanuma H., Yamanouchi T. Increased fructose concentrations in blood and urine in patients with diabetes. Diabetes Care. 2002;25:353–357. doi: 10.2337/diacare.25.2.353. [DOI] [PubMed] [Google Scholar]
- 44.Ainscough R., Bardill S., Barlow K., Basham V., Baynes C., Beard L., Beasley A., Berks M., Bonfield J., Brown J., et al. Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium . Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 45.Lehninger A.L., Nelson D.L., Cox M.M. Principles of biochemistry. New York: Worth Publishers; 2000. [Google Scholar]
- 46.Flock M.R., Kris-Etherton P.M. Dietary Guidelines for Americans 2010: implications for cardiovascular disease. Curr. Atheroscler. Rep. 2011;13:499–507. doi: 10.1007/s11883-011-0205-0. [DOI] [PubMed] [Google Scholar]
- 47.Grundy S.M., Cleeman J.I., Daniels S.R., Donato K.A., Eckel R.H., Franklin B.A., Gordon D.J., Krauss R.M., Savage P.J., Smith S.C., Jr., et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung and Blood Institute Scientific Statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 48.Thunell S. Porphyrins, porphyrin metabolism and porphyrias. I. update. Scand. J. Clin. Lab. Invest. 2000;60:509–540. doi: 10.1080/003655100448310. [DOI] [PubMed] [Google Scholar]
- 49.Budhoo M.R., Mitchell S., Eddlestone J., MacLennan I. Sickle cell trait and acute intermittent porphyria leading to small bowel infarction. J. R. Coll. Surg. Edinb. 1999;44:130–131. [PubMed] [Google Scholar]
- 50.Twaddle S., Wassif W.S., Deacon A.C., Peters T.J. Case report: celiac disease in patients with Variegate porphyria. Dig. Dis. Sci. 2001;46:1506–1508. doi: 10.1023/a:1010600206867. [DOI] [PubMed] [Google Scholar]
- 51.Stocker R., Yamamoto Y., McDonagh A.F., Glazer A.N., Ames B.N. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 52.Oates P.S., West A.R. Heme in intestinal epithelial cell turnover, differentiation, detoxification, inflammation, carcinogenesis, absorption and motility. World J. Gastroenterol. 2006;12:4281–4295. doi: 10.3748/wjg.v12.i27.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibbons S.J., Farrugia G. The role of carbon monoxide in the gastrointestinal tract. J. Physiol. 2004;556:325–336. doi: 10.1113/jphysiol.2003.056556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuckerbraun B.S., Otterbein L.E., Boyle P., Jaffe R., Upperman J., Zamora R., Ford H.R. Carbon monoxide protects against the development of experimental necrotizing enterocolitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G607–G613. doi: 10.1152/ajpgi.00055.2005. [DOI] [PubMed] [Google Scholar]
- 55.Sieg I., Beckh K., Kersten U., Doss M.O. Manifestation of acute intermittent porphyria in patients with chronic inflammatory bowel disease. Z. Gastroenterol. 1991;29:602–605. [PubMed] [Google Scholar]
- 56.Camacho J., Rioseco-Camacho N. Hyperornithinemia–hyperammonemia–homocitrullinuria Syndrome. 1993.
- 57.Valtonen M., Nanto-Salonen K., Jaaskelainen S., Heinanen K., Alanen A., Heinonen O.J., Lundbom N., Erkintalo M., Simell O. Central nervous system involvement in gyrate atrophy of the choroid and retina with hyperornithinaemia. J. Inherit. Metab. Dis. 1999;22:855–866. doi: 10.1023/a:1005602405349. [DOI] [PubMed] [Google Scholar]
- 58.Lanpher B.C., Gropman A., Chapman K.A., Lichter-Konecki U., Urea Cycle Disorders C., Summar M.L. In: GeneReviews™ [Internet] Pagon R.A., Bird T.D., Dolan C.R., Stephens K., Adam M.P., editors. Seattle: University of Washington; 1993. Urea cycle disorders overview 1993–2003 Apr 29 [updated 2011 Sep 01] [Google Scholar]
- 59.Wang F.Y., Zhu R.M., Maemura K., Hirata I., Katsu K., Watanabe M. Expression of gamma-aminobutyric acid and glutamic acid decarboxylases in rat descending colon and their relation to epithelial differentiation. Chin. J. Dig. Dis. 2006;7:103–108. doi: 10.1111/j.1443-9573.2006.00247.x. [DOI] [PubMed] [Google Scholar]
- 60.Collins S.M., Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology. 2009;136:2003–2014. doi: 10.1053/j.gastro.2009.01.075. [DOI] [PubMed] [Google Scholar]
- 61.Prins A. The brain–gut interaction: the conversation and the implications. S. Afr. J. Clin. Nutr. 2011;24:s8–s14. [Google Scholar]
- 62.Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G., Bienenstock J., Cryan J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl Acad. Sci. USA. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mani S., Yang G., Wang R. A critical life-supporting role for cystathionine γ-lyase in the absence of dietary cysteine supply. Free Radic. Biol. Med. 2011;50:1280–1287. doi: 10.1016/j.freeradbiomed.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 64.Heil S.G., Hogeveen M., Kluijtmans L.A., van Dijken P.J., van de Berg G.B., Blom H.J., Morava E. Marfanoid features in a child with combined methylmalonic aciduria and homocystinuria (CblC type) J. Inherit. Metab. Dis. 2007;30:811. doi: 10.1007/s10545-007-0546-6. [DOI] [PubMed] [Google Scholar]
- 65.Leonard J.V. The management and outcome of propionic and methylmalonic acidaemia. J. Inherit. Metab. Dis. 1995;18:430–434. doi: 10.1007/BF00710054. [DOI] [PubMed] [Google Scholar]
- 66.Stremmel W., Hanemann A., Ehehalt R., Karner M., Braun A. Phosphatidylcholine (lecithin) and the mucus layer: evidence of therapeutic efficacy in ulcerative colitis? Dig. Dis. 2010;28:490–496. doi: 10.1159/000320407. [DOI] [PubMed] [Google Scholar]
- 67.Schneider H., Braun A., Fullekrug J., Stremmel W., Ehehalt R. Lipid based therapy for ulcerative colitis-modulation of intestinal mucus membrane phospholipids as a tool to influence inflammation. Int. J. Mol. Sci. 2010;11:4149–4164. doi: 10.3390/ijms11104149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fenton W.A., Gravel R.A., Rosenblatt D.S. The metabolic and molecular bases of inherited disease. Vol. 2. New York: McGraw-Hill Companies; 2001. pp. 2165–2193. [Google Scholar]
- 69.Gudmundsson S., Thiele I. Computationally efficient flux variability analysis. BMC Bioinformatics. 2010;11:489. doi: 10.1186/1471-2105-11-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schellenberger J., Park J.O., Conrad T.M., Palsson B.O. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics. 2010;11:213. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Consortium T.U. Reorganizing the protein space at the universal protein resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheer M., Grote A., Chang A., Schomburg I., Munaretto C., Rother M., Sohngen C., Stelzer M., Thiele J., Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thorleifsson S.G., Thiele I. rBioNet: a COBRA toolbox extension for reconstructing high-quality biochemical networks. Bioinformatics. 2011;27:2009–2010. doi: 10.1093/bioinformatics/btr308. [DOI] [PubMed] [Google Scholar]
- 75.Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nature Protocols. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murray R.K., Bender D.A., Botham K.M., Kennelly P.J., Rodwell V.W., Weil P.A. Harper's Illustrated Biochemistry. McGraw Hill Medical, New York, NY; 2009. [Google Scholar]
- 77.Lockhart R.D., Hamilton G.F., Fyfe F.W. Anatomy of the human body. London: R. MacLehose & company limited; 1974. [Google Scholar]
- 78.Wise E.M., Jr., Elwyn D. Rates of reactions involved in phosphatide synthesis in liver and small intestine of intact rats. J. Biol. Chem. 1965;240:1537–1548. [PubMed] [Google Scholar]
- 79.McDevitt J., Feighery C., O'Farrelly C., Martin G., Weir D.G., Kelleher D. Effects of tumour necrosis factor-alpha on BrdU incorporation in cultured human enterocytes. Mediators Inflamm. 1995;4:31–37. doi: 10.1155/S0962935195000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bouchoux J., Beilstein F., Pauquai T., Guerrera I.C., Chateau D., Ly N., Alqub M., Klein C., Chambaz J., Rousset M., et al. The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol. Cell. 2011;103:499–517. doi: 10.1042/BC20110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schellenberger J., Que R., Fleming R.M., Thiele I., Orth J.D., Feist A.M., Zielinski D.C., Bordbar A., Lewis N.E., Rahmanian S., et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernandez M.L., Calle M. Revisiting dietary cholesterol recommendations: does the evidence support a limit of 300 mg/d. Curr. Atheroscler. Rep. 2010;12:377–383. doi: 10.1007/s11883-010-0130-7. [DOI] [PubMed] [Google Scholar]
- 83.Chougule P., Herlenius G., Hernandez N.M., Patil P.B., Xu B., Sumitran-Holgersson S. Isolation and characterization of human primary enterocytes from small intestine using a novel method. Scand. J. Gastroenterol. 2012;47:1334–1343. doi: 10.3109/00365521.2012.708940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paine M.F., Khalighi M., Fisher J.M., Shen D.D., Kunze K.L., Marsh C.L., Perkins J.D., Thummel K.E. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J. Pharmacol. Exp. Ther. 1997;283:1552–1562. [PubMed] [Google Scholar]
- 85.Crenn P., Messing B., Cynober L. Citrulline as a biomarker of intestinal failure due to enterocyte mass reduction. Clinical Nutr. 2008;27:328–339. doi: 10.1016/j.clnu.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 86.Kohler E.S., Sankaranarayanan S., van Ginneken C.J., van Dijk P., Vermeulen J.L., Ruijter J.M., Lamers W.H., Bruder E. The human neonatal small intestine has the potential for arginine synthesis; developmental changes in the expression of arginine-synthesizing and -catabolizing enzymes. BMC Dev. Biol. 2008;8:107. doi: 10.1186/1471-213X-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamada E., Wakabayashi Y., Saito A., Yoda K., Tanaka Y., Miyazaki M. Hyperammonaemia caused by essential aminoacid supplements in patient with short bowel. Lancet. 1993;341:1542–1543. doi: 10.1016/0140-6736(93)90687-c. [DOI] [PubMed] [Google Scholar]
- 88.Lecha M., Puy H., Deybach J.C. Erythropoietic protoporphyria. Orphanet J. Rare Dis. 2009;4:19. doi: 10.1186/1750-1172-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barnes H.D., Hurworth E., Millar J.H. Erythropoietic porphyrin hepatitis. J. Clin. Pathol. 1968;21:157–159. doi: 10.1136/jcp.21.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meerman L., Koopen N.R., Bloks V., van Goor H., Havinga R., Wolthers B.G., Kramer W., Stengelin S., Müller M., Kuipers F., et al. Biliary fibrosis associated with altered bile composition in a mouse model of erythropoietic protoporphyria. Gastroenterology. 1999;117:696–705. doi: 10.1016/s0016-5085(99)70464-6. [DOI] [PubMed] [Google Scholar]
- 91.Donovan A., Roy C.N., Andrews N.C. The ins and outs of iron homeostasis. Physiology. 2006;21:115–123. doi: 10.1152/physiol.00052.2005. [DOI] [PubMed] [Google Scholar]
- 92.Vassallo M.J., Camilleri M., Sullivan S.N., Thomforde G.M. Effects of erythromycin on gut transit in pseudo-obstruction due to hereditary coproporphyria. J. Clin. Gastroenterol. 1992;14:255–259. doi: 10.1097/00004836-199204000-00013. [DOI] [PubMed] [Google Scholar]
- 93.Migchielsen A.A., Breuer M.L., van Roon M.A., te Riele H., Zurcher C., Ossendorp F., Toutain S., Hershfield M.S., Berns A., Valerio D. Adenosine-deaminase-deficient mice die perinatally and exhibit liver-cell degeneration, atelectasis and small intestinal cell death. Nat. Genet. 1995;10:279–287. doi: 10.1038/ng0795-279. [DOI] [PubMed] [Google Scholar]
- 94.Pita A.M., Fernandez-Bustos A., Rodes M., Arranz J.A., Fisac C., Virgili N., Soler J., Wakabayashi Y. Orotic aciduria and plasma urea cycle-related amino acid alterations in short bowel syndrome, evoked by an arginine-free diet. JPEN J. Parenter. Enteral. Nutr. 2004;28:315–323. doi: 10.1177/0148607104028005315. [DOI] [PubMed] [Google Scholar]
- 95.Hendriksz C.J., Gissen P. Glycogen storage disease. Paediatr. Child Health. 2011;21:84–89. [Google Scholar]
- 96.Guery M.J., Douillard C., Marcelli-Tourvieille S., Dobbelaere D., Wemeau J.L., Vantyghem M.C. Doctor, my son is so tired … about a case of hereditary fructose intolerance. Ann. Endocrinol. 2007;68:456–459. doi: 10.1016/j.ando.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 97.Buchman A.L. Clinical nutrition in gastrointestinal disease. USA: SLACK incorporated; 2006. [Google Scholar]
- 98.Chung-Faye G., Hayee B., Maestranzi S., Donaldson N., Forgacs I., Sherwood R. Fecal M2-pyruvate kinase (M2-PK): a novel marker of intestinal inflammation. Inflamm. Bowel Dis. 2007;13:1374–1378. doi: 10.1002/ibd.20214. [DOI] [PubMed] [Google Scholar]
- 99.Savilahti E., Launiala K., Kuitunen P. Congenital lactase deficiency. A clinical study on 16 patients. Arch. Dis. Child. 1983;58:246–252. doi: 10.1136/adc.58.4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walker W.A. Pediatric gastrointestinal disease: pathophysiology, diagnosis, management. 2004. [Google Scholar]
- 101.Visser D., Heijnen J.J. The mathematics of metabolic control analysis revisited. Metab. Eng. 2002;4:114–123. doi: 10.1006/mben.2001.0216. [DOI] [PubMed] [Google Scholar]
- 102.Milla P.J., Atherton D.A., Leonard J.V., Wolff O.H., Lake B.D. Disordered intestinal function in glycogen storage disease. J. Inherit. Metab. Dis. 1978;1:155–157. doi: 10.1007/BF01805585. [DOI] [PubMed] [Google Scholar]
- 103.Bali D.S., Chen Y.T., Goldstein J.L. 1993. Glycogen storage disease Type I. In Pagon, R.A., Bird, T.D., Dolan, C.R., Stephens, K., Adam, M.P. (eds), GeneReviews™ [Internet], University of Washington, Seattle; 1993–2006 Apr 19 [updated 2010 Dec 23].
- 104.Pascual J.M., Wang D., Lecumberri B., Yang H., Mao X., Yang R., De Vivo D.C. GLUT1 deficiency and other glucose transporter diseases. Eur. J. Endocrinol. 2004;150:627–633. doi: 10.1530/eje.0.1500627. [DOI] [PubMed] [Google Scholar]
- 105.Golachowska M.R., van Dael C.M., Keuning H., Karrenbeld A., Hoekstra D., Gijsbers C.F., Benninga M.A., Rings E.H., van Ijzendoorn S.C. MYO5B mutations in patients with microvillus inclusion disease presenting with transient renal Fanconi syndrome. J. Pediatr. Gastroenterol. Nutr. 2012;54:491–498. doi: 10.1097/MPG.0b013e3182353773. [DOI] [PubMed] [Google Scholar]
- 106.Jacob R., Zimmer K.P., Schmitz J., Naim H.Y. Congenital sucrose–isomaltase deficiency arising from cleavage and secretion of a mutant form of the enzyme. J. Clin. Invest. 2000;106:281–287. doi: 10.1172/JCI9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iyer R.K., Yoo P.K., Kern R.M., Rozengurt N., Tsoa R., O'Brien W.E., Yu H., Grody W.W., Cederbaum S.D. Mouse model for human arginase deficiency. Mol. Cell. Biol. 2002;22:4491–4498. doi: 10.1128/MCB.22.13.4491-4498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Claes D.J., Jackson E. Cystinuria: mechanisms and management. Pediatr. Nephrol. 2012 doi: 10.1007/s00467-011-2092-6. [DOI] [PubMed] [Google Scholar]
- 109.Wadman S.K., Sprang F.J.V., Stekelenburg G.J.V., de Bree P.K. Three new cases of histidinemia. Acta Paediatr. 1967;56:485–492. [Google Scholar]
- 110.Cathelineau L., Briand P., Rabier D., Navarro J. Ornithine transcarbamylase and disaccharidase activities in damaged intestinal mucosa of children—diagnosis of hereditary ornithine transcarbamylase deficiency in mucosa. J. Pediatr. Gastroenterol. Nutr. 1985;4:960–964. doi: 10.1097/00005176-198512000-00018. [DOI] [PubMed] [Google Scholar]
- 111.Janne J., Alhonen L., Keinanen T.A., Pietila M., Uimari A., Pirinen E., Hyvonen M.T., Jarvinen A. Animal disease models generated by genetic engineering of polyamine metabolism. J. Cell Mol. Med. 2005;9:865–882. doi: 10.1111/j.1582-4934.2005.tb00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reinoso M.A., Whitley C., Jessurun J., Schwarzenberg S.J. Lysinuric protein intolerance masquerading as celiac disease: a case report. J. Pediatr. 1998;132:153–155. doi: 10.1016/s0022-3476(98)70502-5. [DOI] [PubMed] [Google Scholar]
- 113.Fairbanks T., Emil S. Colonic perforation in the first few hours of life associated with rhizomelic chondrodysplasia punctata. Pediatr. Surg. Int. 2005;21:662–664. doi: 10.1007/s00383-005-1426-5. [DOI] [PubMed] [Google Scholar]
- 114.Nimubona L., Laloum D., Rolland M.O., Read M.H., Guillois B., Duhamel J.F. An intestinal obstruction in an eight-month-old child suffering from mevalonic aciduria. Acta Paediatr. 2002;91:714–716. doi: 10.1080/080352502760069179. [DOI] [PubMed] [Google Scholar]
- 115.Hoflack M., Caruba C., Pitelet G., Haas H., Mas J.C., Paquis V., Berard E. Infant coma in the emergency department: 2 cases of MCAD deficiency. Arch. Pediatr. 2010;17:1074–1077. doi: 10.1016/j.arcped.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 116.Gilbert J., Ibdah J.A. Intestinal pseudo-obstruction as a manifestation of impaired mitochondrial fatty acid oxidation. Med. Hypotheses. 2005;64:586–589. doi: 10.1016/j.mehy.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 117.Lee M., Cook C.R., Wilkins I. A new association of second-trimester echogenic bowel and metabolic disease of the neonate. J. Ultrasound Med. 2007;26:1119–1122. doi: 10.7863/jum.2007.26.8.1119. [DOI] [PubMed] [Google Scholar]
- 118.Shinton N.K. Vitamin B 12 and folate metabolism. Br. Med. J. 1972;1:556–559. doi: 10.1136/bmj.1.5799.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Klein R.D., Thorland E.C., Gonzales P.R., Beck P.A., Dykas D.J., McGrath J.M., Bale A.E. A multiplex assay for the detection and mapping of complex glycerol kinase deficiency. Clin. Chem. 2006;52:1864–1870. doi: 10.1373/clinchem.2006.072397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.