Abstract
Autophagy is a cytoplasmic catabolic process that protects the cell against stressful conditions. Damaged cellular components are funneled by autophagy into the lysosomes, where they are degraded and can be re-used as alternative building blocks for protein synthesis and cellular repair. In contrast, aging is the gradual failure over time of cellular repair mechanisms that leads to the accumulation of molecular and cellular damage and loss of function. The cell’s capacity for autophagic degradation also declines with age, and this in itself may contribute to the aging process. Studies in model organisms ranging from yeast to mice have shown that single-gene mutations can extend lifespan in an evolutionarily conserved fashion, and provide evidence that the aging process can be modulated. Interestingly, autophagy is induced in a seemingly beneficial manner by many of the same perturbations that extend lifespan, including mutations in key signaling pathways such as the insulin/IGF-1 and TOR pathways. Here, we review recent progress, primarily derived from genetic studies with model organisms, in understanding the role of autophagy in aging and age-related diseases.
Keywords: Autophagy, Insulin/IGF signaling, TOR signaling, Dietary restriction, Germline removal, Mitochondrial respiration
Introduction to Aging
Aging is a complex process characterized by the progressive failure of maintenance and repair pathways important for cellular homeostasis, which results in a gradual accumulation of aberrant macromolecules and organelles [1]. The accumulation of such oxidized, misfolded, cross-linked, or aggregated molecules has deleterious effects on cellular homeostasis and on tissue and organ integrity. The defective molecules can disrupt homeostasis directly or by interfering with the activity of functional molecules and organelles, which leads to further dysfunction. This progressive decline in cellular integrity leads to aging, disease, and ultimately, to death. Although our understanding of the biology of aging has increased over the past century, the molecular events underlying this process have only recently begun to be explored. Interestingly, research in the last couple of decades focused on unraveling the molecular underpinnings of aging has shown that, in many model organisms, the rate of aging can be modulated by altering conserved signaling pathways and processes, suggesting that the aging process itself may ultimately be amenable to therapeutic manipulation. We provide a short overview of these conserved longevity paradigms below.
Conserved longevity paradigms
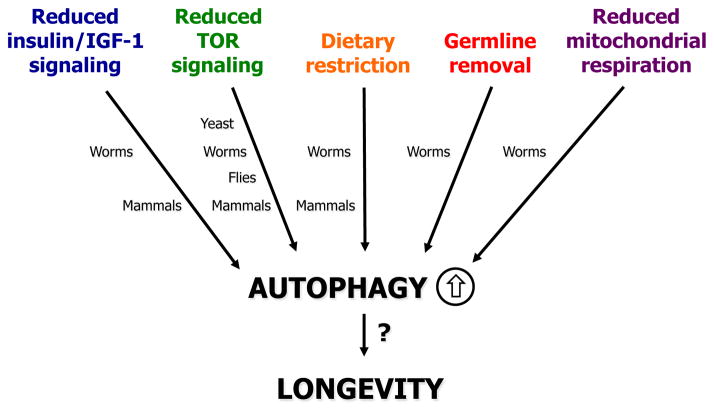
Over the last two decades, genes controlling metabolic functions have been shown to influence aging in multiple model organisms including the budding yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, and many mouse models. The initial discoveries were made in C. elegans, where single-gene mutations in the age-1 gene encoding phosphoinositide 3-kinase (PI3K) [2], and the daf-2 gene encoding the insulin/IGF-1–like receptor [3] were found to extend the lifespan. Since then, a focused effort has led to the discovery of numerous novel regulators of longevity in C. elegans and other organisms. For example, more than 150 C. elegans genes have been reported to increase lifespan when downregulated in genome-wide RNA interference (RNAi) approaches [4,5], and strikingly, many of them function in endocrine-related and metabolic signaling pathways and processes. Of these, the most prominent longevity-associated pathways are the insulin/IGF-1 and TOR signaling cascades, which play critical roles in nutrient sensing and metabolism. Other processes include dietary restriction, signals from the reproductive system, and mitochondrial respiration (Figure 1). In the following sections, we will briefly review these conserved longevity pathways and processes.
Figure 1.
Conserved longevity pathways and processes, which modulate aging via autophagy. Additional interventions not listed here, e.g., the polyamine spermidine may also engage autophagy as a possible effector mechanism to extend lifespan. For simplicity, the interactions between the pathways are not shown, whereas the degree of conservation is indicated. While the mechanisms by which autophagy affects aging through these longevity paradigms are not yet fully elucidated, several positive regulators of autophagy have been identified, including the forkhead transcription factor FOXO, the histone deacetylase SIRT1, the energy sensor AMPK, and the forkhead transcription factor PHA-4/FOXA.
Insulin/IGF-1 signaling
Insulin/IGF-1 signaling is tightly regulated by the nutritional status of the animal and is essential for growth. In response to the hormone insulin (or insulin-like growth factor; IGF-1), the insulin/IGF-1 receptor (InR) activates a cascade of kinases and phosphatases, including PI3K, PTEN, PDK, SGK and AKT, that results in inhibition of the forkhead transcription factor FOXO [6]. In worms, flies, and mice, perturbation of insulin/IGF-1 signaling has been shown to extend lifespan [7], and at least in worms and flies, these effects are mediated by FOXO [8–10]. FOXO regulates transcription of many genes involved in the stress response, bacterial resistance, and metabolism, which contribute to longevity in C. elegans [11]. Consistent with this, overexpression of FOXO is sufficient to extend lifespan in flies [10]. Interestingly, recent investigations have revealed that exceptionally long-lived humans, centenarians, display an overrepresentation of gene mutations in the insulin/IGF-1 pathway [7]. Thus, there is substantial genetic evidence from multiple model organisms and possibly in humans to suggest that the insulin/IGF-1 pathway modulates aging in a conserved fashion.
TOR signaling
The TOR pathway is another major signaling pathway that affects aging. TOR is a conserved member of the PI3K-related kinase family, and its nutrient-dependent activation results in a metabolic shift towards cell growth and division. The protein TOR was originally identified as the cellular target of rapamycin [12], a compound discovered in a soil bacterium on the Easter Island Rapa Nui [13]. TOR exists in two complexes, TORC1 and TORC2, which mediate distinct functions by signaling through different effector pathways. TORC1, which is inhibited by rapamycin, integrates nutrient-derived and mitogenic signals to regulate cell proliferation and cell size. In contrast, TORC2 is unaffected by rapamycin and controls cell shape. Inhibition of TORC1 by rapamycin leads to growth inhibition and immunosuppression [14], but has also been associated with delayed aging. TORC1 is activated by amino acids and insulin/IGF-1 through the kinase AKT, which represents a major point of convergence between the insulin/IGF-1 and the TOR pathways. Activated AKT phosphorylates and inhibits the tuberous sclerosis complex (TSC1/TSC2). The TSC1/TSC complex can also be regulated in a positive fashion by the energy sensor AMP-activated kinase (AMPK), which is activated by high AMP levels. The TSC complex is a negative regulator of the GTPase RHEB, which binds to and activates TORC1 in a GTP-dependent manner [14]. Once released, TORC1 activates multiple anabolic processes, including ribosome biogenesis, translation initiation, nutrient import, and inhibition of the catabolic process of autophagy.
TORC1 (hereafter referred to as TOR) signaling has been shown to influence aging in many organisms. Reduced TOR activity extends lifespan in yeast, worms, flies, and mice [15]. Two processes regulated by TOR have been strongly linked to longevity. The first is protein synthesis regulated by the ribosomal protein S6 kinase (S6K), a downstream target of TOR. S6K inhibition reduces protein synthesis and has been shown to extend lifespan in yeast, worms, flies, and female mice [15,16]. The second TOR-regulated process involved in lifespan extension is the cellular recycling process of autophagy, which will be the focus of this review.
Dietary restriction
Dietary restriction, defined as the restriction of nutrients while avoiding malnutrition, is the most robust intervention currently known to delay aging. Dietary restriction was first observed to delay aging and disease in rats almost a century ago [17,18]. Since then, the effects of dietary restriction on aging has been studied extensively, and dietary restriction has been observed to extend the lifespan of yeast, invertebrates, fish, dogs, hamsters, mice, and apes [19]. Multiple molecular mechanisms have been proposed to mediate the effects of dietary restriction on longevity, including insulin/IGF-1 and TOR signaling [20]. However, it is currently unknown to what degree lifespan extension resulting from dietary restriction is mediated by these nutrient-sensing pathways.
Signals from the reproductive system
While there are numerous examples in nature of the inverse relationship between lifespan and fecundity, the trade-off between increased lifespan and decreased reproduction can, at least in certain cases, be uncoupled [21]. However, signals from the reproductive system can indeed influence aging, and may allow the animal to coordinate the timing of reproduction and aging. Specifically, the removal of the germline or germ precursor cells results in lifespan extension in both worms and flies [21]. In C. elegans, removing the entire reproductive system does not, however, extend lifespan, suggesting that the lifespan extension is not simply a consequence of sterility, but rather that specific signals from the germline and the somatic gonad may affect aging in opposing ways. Interestingly, newer studies have suggested that signals from the reproductive system may also affect longevity in mice, because lifespan can be extended by transplantation of ovaries from young into old mice [22,23].
Reduced mitochondrial respiration
The free-radical theory of aging proposes that aging results from the accumulation of oxidative damage over time, which leads to cellular dysfunction and organismal death [24]. The molecular damage is thought to be inflicted by reactive oxygen species (ROS), which are generated primarily during mitochondrial respiration. Although oxidative damage increases with age, it is unclear if this is causally related to aging; evidence exists both for and against this possibility. Importantly, reduced electron transport chain function decreases ROS levels and increases longevity in yeast [25], worms, flies, and mice [26]. In worms, this increased longevity is mediated, at least in part, by an upregulation of the mitochondrial unfolded protein response (UPR) [27].
In summary, it is clear that organismal aging is influenced in a conserved manner by a number of signaling cascades and processes. A key objective in aging research is to determine if these signaling pathways and processes that affect lifespan converge on common downstream mechanisms. Rapidly accumulating evidence suggests that the cellular recycling process of autophagy could be one such integrative mechanism, which maintains cellular homeostasis and plays a pro-survival role, and thus contributes to increased longevity. In the following sections, we introduce the autophagy process, and summarize the current literature on how this degradative process may not only be beneficial to aging, but also plays a critical role in age-related diseases such as neurodegeneration and cancer.
Introduction to Autophagy
Autophagy is an evolutionarily conserved intracellular recycling process that is induced in response to stresses such as nutrient deprivation, hyperthermia, and hypoxia [28–30]. Autophagy is responsible for the degradation of proteins, lipids, sugars, and nucleic acids as well as larger cellular components such as organelles. During the process, damaged cellular components are sequestered into a double-membrane structure called the autophagosome, which delivers its contents to the lysosome for subsequent degradation by acidic hydrolases.
Three types of autophagy have been identified: microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy [31]. Microautophagy is the least studied type and occurs when cytoplasmic components are degraded following direct invagination, protrusion, and/or septation of the lysosomal or endosomal membrane [32,33]. In CMA, a type of autophagy only present in higher organisms, the chaperone protein Hsc70 recognizes a specific pentapeptide motif (KFERQ) on unfolded proteins in the cytosol and escorts them directly to the lysosome-associated receptor protein type 2A (LAMP-2A). The cargo protein is then unfolded at the membrane and translocated into the lysosome for degradation [34]. Since the regulation of microautophagy and CMA in relation to aging is currently limited, we will primarily focus on macroautophagy in this review.
Macroautophagy – A multi-step process
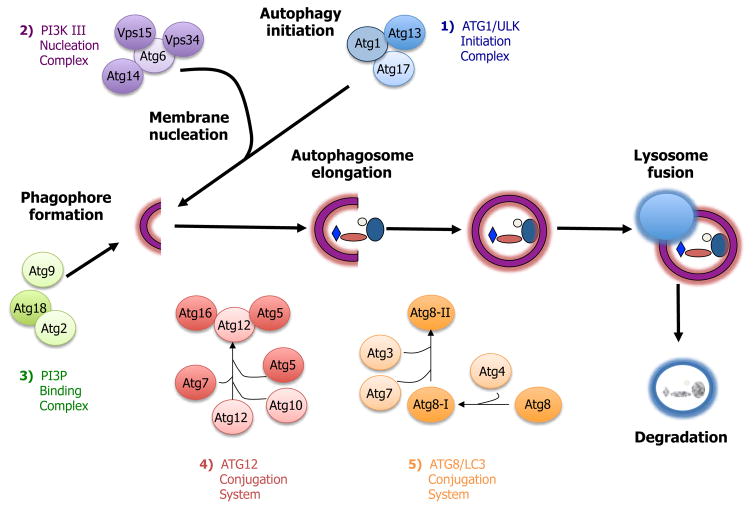
Macroautophagy (hereafter referred to as autophagy) is a multi-step process by which large macromolecules and organelles are degraded. This is currently the most extensively studied form of autophagy due to the interesting physiological and pathological consequences of a process that mediates the large-scale degradation of intracellular molecules. During autophagy, several autophagy-related (ATG) genes are engaged sequentially in a highly regulated manner. Genetic studies in yeast have identified more than 30 ATG genes that are required for autophagy [35], most of which are conserved from yeast to mammals. The essential ATG genes are organized into at least five functional groups that allow for the initiation, formation, elongation, and fusion of the autophagosome (Figure 2). These functional groups are 1) the Atg1/ULK initiation complex, 2) the Vps34/PI3-kinase nucleation complex, 3) the phosphatidylinositol 3-phosphate (PI3P)-binding Atg18/Atg2 complex, 4) the Atg5-Atg12 conjugation system, and 5) the Atg8/LC3-PE (Atg8/LC3-phosphatidylethanolamine) conjugation system.
Figure 2.
The macroautophagy process. Macroautophagy (here referred to as autophagy) involves a series of steps: autophagy induction, membrane nucleation, phagophore formation, autophagosome elongation, lysosome fusion, and degradation. These steps are controlled by at least five different functional groups of proteins: 1) the Atg1/ULK initiation complex; 2) the PI3-kinase nucleation complex; 3) the PI3P-binding complex, which directs the distribution of Atg9, a transmembrane protein likely important for lipid delivery to the membrane; 4) the Atg5-Atg12 conjugation system; and 5) the Atg8/LC3 conjugation system. In the latter, Atg8 is cleaved by Atg4 to form Atg8-I and is then conjugated with phosphatidylethanolamine to become Atg8-II, which is incorporated into pre- and autophagosomal membranes.
Autophagy is induced when the Atg1/ULK initiation complex (functional group 1, consisting of the kinase Atg1/ULK, as well as Atg13 and Atg17) is activated. The next step involves membrane nucleation by the Class III Vps34/PI3-kinase I nucleation complex (functional group 2, consisting of Vps34, Atg6/Beclin1, and Vps15/p150) via production of PI3P, to start formation of a double-membrane structure that can engulf cargo for degradation. This structure is called a phagophore, or an isolation membrane. Multiple studies have investigated how and where in the cell the isolation membrane begins to form, and the results reveal another layer of complexity in the autophagy process. In mammalian cells, the autophagosomal membrane can be synthesized de novo [36] or can originate from multiple sources; the endoplasmic reticulum (ER) [37–40], golgi [41], mitochondria [42], or plasma membrane [43]. It is not yet known how varied the origin of this isolation membrane may be, nor if that variability has regulatory and/or mechanistic consequences. To start elongation, the phagophore recruits the PI3P-binding complex (functional group 3, consisting of Atg18/WIPI and Atg2), which regulates the distribution of Atg9, a transmembrane protein that has been proposed to deliver lipids to the phagophore and the growing autophagosome. During the next step, the phagophore expands into a double-membrane structure called the autophagosome. This occurs through the action of the Atg5-Atg12 conjugation system (functional group 4, in which Atg7 and Atg10 (E1- and E2-like enzymes, respectively) conjugate Atg12 to Atg5) and this complex then associates with Atg16 and the Atg8/LC3-phosphatidylethanolamine (PE) conjugation system (functional group 5, in which Atg8/LC3 is proteolyzed by the cysteine protease Atg4 and subsequently conjugated with PE in a process involving Atg7 and Atg3). During this process, PE-conjugated LC3 associates with the autophagosomal membrane and LC3 is therefore often used as an experimental marker of autophagosomes. The autophagosome eventually matures into a closed cargo-containing vesicle, which then fuses with the lysosome to become the autolysosome, and its contents are finally degraded for recycling [44].
Upstream regulators of autophagy
As a multi-step, multi-component, and highly complex process, autophagy must be tightly regulated to maintain its efficiency. Autophagy is regulated by the same conserved factors that regulate metabolism and aging. For example, autophagy is negatively regulated by amino acids, which activate inhibitory TOR-dependent signaling [45]. Inhibition of TOR in mammalian cells by starvation or rapamycin treatment results in dephosphorylation of Atg1 and Atg13, and leads to the formation of the Atg1/ULK initiation complex, consisting of Atg1, Atg13, and Atg17 [46]. Similarly, induction of autophagy is observed in yeast and worms following TOR inhibition [47,68]. TOR also regulates the localization of the helix-loop-helix transcription factor TFEB [48], which coordinately regulates the expression of many autophagosomal and lysosomal genes in mammals [49]. The formation of new autophagosomes requires the activity of the class III phosphatidylinositol 3-kinase (PI3K), Vps34. Vps34 is part of the autophagy-regulating complex (PI3K complex) consisting of Beclin1/Atg6, Atg14/Barkor, and p150/Vps-15 [50]. In addition to TOR, AMP-activated protein kinase (AMPK) was recently shown to directly regulate Atg1/ULK1/2 during starvation [51–53]. As mentioned above, AMPK restores cellular ATP levels by regulating metabolic enzymes and by inhibiting anabolic pathways, and thus acts in an opposing manner to TOR signaling [54]. Whether proteins other than AMPK are involved in regulating autophagy in a TOR-independent fashion remains to be shown; however, drugs known to induce autophagy in a TOR-independent manner have been identified [55,56].
The highly regulated degradative capacity of autophagy can have global effects on homeostasis as cell function declines with age. Damaged proteins and organelles accumulate during aging; this process is accelerated in many human disorders [57] and may result in cellular and organismal death. It is perhaps not surprising, therefore, that autophagy has been linked to multiple age-related diseases such as neurodegeneration, muscle disorders, and cancer [58,59]. Below, we discuss the current literature that proposes a role for autophagy in aging, and in age-related diseases in various model organisms.
Autophagy and Aging in S. cerevisiae
The lifespan of the budding yeast S. cerevisiae can be measured by two methods; replicative lifespan (RLS) and chronological lifespan (CLS) [60]. In RLS, the number of times a mother cell divides and gives rise to a daughter cell is measured by counting the number of scars on the cell surface after each division. In CLS, the survival time of a cell in stationary phase (without division, and therefore potentially mimicking aging of post-mitotic cells) is quantified. Recent investigations summarized below have suggested a link between autophagy and aging in yeast, most notably because autophagy genes are required for TOR inhibition to extend CLS [47,61], and because a pharmacological activator of autophagy, spermidine, increases lifespan via an autophagy-dependent mechanism [62].
Dietary restriction and TOR signaling
Both RLS and CLS can be modulated in S. cerevisiae by reducing nutrients in the growth media, which effectively induces dietary restriction [63]. One method to induce dietary restriction is by amino acid limitation, which not only extends CLS [64], but also induces autophagy [65]. Similarly, inhibition of the nutrient sensor TOR by rapamycin increases CLS, induces autophagy, and autophagy genes are required for rapamycin to extend lifespan [47]. Consistent with these observations, a large-scale screen for genes that regulate CLS in S. cerevisiae identified many autophagy gene mutants with reduced lifespan, and ATG16 mutants did not display lifespan extension in response to limitation of amino acids or tor mutation [61]. Taken together, these findings suggest that reduced TOR signaling modulates CLS in S. cerevisiae via autophagy upregulation. However, the role of autophagy in yeast aging is likely to be more complex, as a recent study showed that a majority of autophagy gene mutants have normal RLS in rich media, and deletion of only ATG15, but not other autophagy genes tested, blocks RLS extension induced by glucose limitation [66], another method of dietary restriction in yeast.
Pharmacological activation of autophagy
Autophagy can be manipulated pharmacologically in yeast with the natural polyamine compound, spermidine. Interestingly, spermidine not only increases CLS when added to yeast cultures, but also induces autophagy [62]. Moreover, spermidine-induced lifespan extension is not observed in autophagy-deficient mutants. Taken together, these data are consistent with autophagy mediating S. cerevisiae lifespan extension following spermidine treatment. Although not yet fully understood, the mechanisms underlying these observations may involve epigenetic changes in chromatin acetylation, which leads to upregulation of autophagy gene expression [62].
Autophagy and aging in C. elegans
The nematode C. elegans has been instrumental in gaining insights into the molecular mechanisms underlying aging [7], and the first direct link between autophagy and aging was made in C. elegans. Since then, autophagy has been shown to be induced in many C. elegans longevity models, such as in response to reduced insulin/IGF-1 signaling [67], dietary restriction [68–71], TOR inhibition [68,69], AMPK overexpression [51], germline removal [74], spermidine treatment [62], sirtuin overexpression [75], and reduced calcineurin signaling [76]. Consistent with a protective role for autophagy, mutation of autophagy genes in the worm is accompanied by lethality or a decrease in lifespan [69]1. Most of the autophagy-related genes, originally discovered in yeast, are conserved in C. elegans [78]. Our understanding of autophagy in C. elegans has increased greatly in recent years, and the nematode continues to be an important model organism in which to study the genetic basis of autophagy [79]. Below, we outline our current knowledge of the signaling pathways known to be involved in the autophagy process in C. elegans.
Dietary restriction and TOR signaling
Dietary restriction is the most well-established method of increased longevity that has been linked to autophagy. Dietary restriction can be accomplished by several methods in C. elegans. These include the genetic mutant eat-2, which expresses a defective acetylcholine receptor in the pharyngeal muscles that results in reduced food intake and increased longevity, and direct dilution of the bacterial food source which also extends lifespan [80]. Importantly, autophagy activity is increased in both the genetic and bacterial dilution methods of dietary restriction [68,69,71], and autophagy genes are required for the extended lifespan observed in the eat-2 mutant [68–70]. It is not yet known if autophagy genes are similarly required for lifespan extensions induced by other dietary restriction regimens, including bacterial dilution.
The longevity of dietary-restricted C. elegans has been shown to be dependent on the forkhead transcription factor pha-4/FOXA [81]. Interestingly, inhibition of pha-4/FOXA also decreases autophagy levels in eat-2 mutants, suggesting that pha-4/FOXA is required for increased autophagy in these animals [68]. These observations suggest that autophagy may be induced at the transcriptional level by pha-4/FOXA in dietary-restricted C. elegans.
In C. elegans, lifespan extension induced by dietary restriction may be at least partly mediated through TOR, because TOR inhibition in eat-2 mutants does not further extend lifespan [73]. Moreover, similar to dietary-restricted worms, inhibition of TOR extends lifespan in a pha-4/FOXA-dependent fashion [56], possibly via a transcriptional mechanism [74,82]. The GTPase RHEB-1 mediates longevity in C. elegans induced by intermittent fasting, a different dietary-restriction regimen that results in extended lifespan [82]. Importantly, animals carrying a mutation in daf-15/Raptor, a negative regulator of TOR, have an extended lifespan [83], display increased autophagy levels, and require autophagy genes to live long [68]. Collectively, these data suggest that lifespan extension induced by dietary restriction and reduced TOR signaling in C. elegans is at least partly mediated by autophagy, as is observed in yeast.
Insulin/IGF-1–like signaling
A direct link between autophagy and aging was first observed in the C. elegans daf-2 mutant, which has reduced InR activity. In 2003, it was observed that autophagy was induced in the daf-2/InR mutant and that the autophagy gene bec-1/Beclin1 is required for the long lifespan observed in these animals [67]. Since then, additional autophagy genes, such as atg-7, atg-12, and lgg-1/LC3 have been shown to be required for the longevity of daf-2/InR mutants [68,84]. Like autophagy genes, the transcription factor DAF-16/FOXO is also required for the long lifespan of daf-2/InR mutants [8,9]. Interestingly, autophagy induction appears to be maintained in daf-16; daf-2 double mutants [68], suggesting that daf-16/FOXO is not required for autophagy induction in C. elegans. However, another study has indicated that overexpression of DAF-16/FOXO induces autophagy in C. elegans [85], consistent with observations made in Drosophila [86,120] and in mammals [87]. While more research is clearly needed to clarify the role of DAF-16/FOXO in autophagy, these seemingly incompatible observations could be explained if other transcription factors compete with DAF-16/FOXO for targets and can compensate in the absence of daf-16/FOXO. The transcription factor PHA-4/FOXA has been shown to regulate autophagy genes in another C. elegans longevity mutant [74] (see section on germline signaling below) and may be a candidate for such a compensating factor.
Signals from the reproductive system
As outlined in the introduction, signals from the reproductive system have been implicated in modulating longevity. In C. elegans, removal of the germline can be genetically modeled, for example, by loss-of-function mutations in the Notch receptor GLP-1 [88]. Such glp-1 mutants lack a germline and have increased longevity, resembling worms in which the germline precursor cells have been ablated with a laser beam [89]. Interestingly, it was recently shown that glp-1 mutants have reduced TOR levels, increased autophagy levels, and require autophagy genes for their increased longevity [74]. These data suggest that germline-less glp-1 mutants display extended lifespan, in part, through upregulation of autophagy via reduced TOR signaling. Knockdown of the transcription factor PHA-4/FOXA in these mutants reduces longevity and autophagy gene expression [74], suggesting that autophagy is regulated at the transcriptional level in germline-less C. elegans. Germline-less mutants also have increased lipase activity [74], and the lipase LIPL-4 is required for their longevity [90]. Animals overexpressing LIPL-4 are long-lived [90], and these animals, similar to glp-1 mutants, have increased autophagy levels and also require autophagy genes to live long [74]. These observations are the first to suggest a possible molecular mechanism to explain how autophagy could modulate lifespan; namely, that lipid turnover by autophagy [91,92] could be relevant to aging.
Mitochondrial respiration
Reducing mitochondrial respiration by mutating multiple genes in the electron transport chain results in lifespan extension in C. elegans [26]. Autophagy levels are increased in isp-1 mutants, which lack a normally functioning iron sulfur protein of the mitochondrial complex III, and in animals subjected to RNAi knockdown of isp-1 or nuo-6, which encodes an NADH ubiquinone oxidoreductase of complex 1. Some indirect evidence exists that the increased longevity of mitochondrial mutants may also be dependent on autophagy. Specifically, RNAi knockdown of atp-3, encoding a mitochondrial ATP synthase of complex V, or clk-1, a coenzyme Q biosynthetic gene, extends the lifespan of wild-type worms but not of the autophagy mutants unc-51/Atg1, atg-18/WIPI, or bec-1/Beclin1 [69]. While a caveat to these experiments is that autophagy mutants are already short-lived, the results are indeed consistent with autophagy playing a critical role in the long lifespan of mitochondrial mutants.
Interestingly, the nematode ortholog of p53, cep-1, which is involved in maintenance of genomic integrity in response to stress, is required for the longevity of mitochondrial mutants atp-3 and isp-1 [93], suggesting that cep-1 and the mitochondrial mutants function by at least partially overlapping mechanisms. On the other hand, cep-1 depletion extends lifespan in C. elegans [94], and cep-1 mutants require the autophagy gene bec-1/Beclin for their long lifespan [95]. These findings suggest that cep-1 negatively regulates autophagy, and that the extended lifespan of these mutants is mediated by autophagy. Further studies are required to determine the exact role of cep-1–mediated autophagy, and autophagy in general, in lifespan extension in mitochondrial mutants.
Sirtuins
Sirtuins are a group of histone deacetylases that modulate transcriptional regulation. Sirtuin activity is regulated by NAD (+) levels and their function is therefore closely linked to cellular energy consumption [96,97]. Recently, the role of sirtuins in longevity has been under debate. In the original report, overexpression of the SIRT1 homolog sir-2 in C. elegans extended lifespan in a daf-16/FOXO–dependent manner [98]. Of particular relevance to this review, the same overexpressing strain was reported to have increased autophagy levels and to require the autophagy gene bec-1/Beclin1 for its lifespan extension [99]. However, data from a recent publication challenges the 2001 report by showing that the strain used in that study had an additional mutation, and when outcrossed, the longevity of the sir-2 overexpressing strain was greatly decreased [100]. While further studies with the newly outcrossed strain are needed to clarify whether autophagy is indeed induced by sir-2 overexpression, the mammalian homolog SIRT1 has indeed been shown to regulate autophagy [101].
AMP-activated protein kinase
The alpha-subunit of the energy sensor AMP-activated protein kinase (AMPK), aak-2 was originally shown to be required for the long lifespan of C. elegans daf-2/InR mutants [102]. Importantly, overexpression of aak-2/AMPK is sufficient to induce lifespan extension [102,103], at least in part, by inhibition of a downstream transcription factor CRTC-1 (CREB-regulated transcriptional coactivator) [103]. Recently, it has been shown that overexpression of aak-2/AMPK results in increased autophagy that is dependent on the autophagy gene unc-51/Atg1/ULK1, a direct phosphorylation target of AMPK [51]. However, it is not yet known if autophagy genes are required for the lifespan extension observed in these AAK-2/AMPK-overexpressing animals.
Pharmacological activation of autophagy
If the beneficial effects of autophagy on health and longevity extend to higher organisms, it may be possible to manipulate the pathway pharmacologically for therapeutic purposes. Along these lines, the polyphenol compound resveratrol, present in red wine, has been shown to increase autophagy levels in C. elegans [104]. The resveratrol-dependent autophagy induction requires the sirtuin gene sir-2 [75], which may induce autophagy at least in mammalian cells by deacetylation of Atg5, Atg7, and Atg8 autophagy proteins [101]. Resveratrol has also been shown to extend the lifespan of obese mice, but had no such effect in mice fed a normal diet [105]. Despite these observations, it remains unclear if the resveratrol-induced effects on lifespan and autophagy are interlinked.
The polyamine spermidine has also been linked to aging and autophagy in C. elegans, as well as in other species. As was described above with yeast, addition of spermidine to C. elegans cultures increases autophagy levels and extends lifespan [62], and intracellular spermidine levels decrease with age in humans [62]. These exciting findings add to the prospects of developing drug interventions designed to induce autophagy artificially.
In summary, while the mechanisms remain largely unclear, the evidence that autophagy is beneficial for health and opposes aging continues to mount, especially from research performed in the nematode C. elegans. These studies suggest that pharmacological activation of autophagy may be a realistic and promising treatment for aging and for age-related diseases. Below, we discuss research on the role of autophagy in age-related disorders, as modeled in C. elegans.
Age-related disease models
The risk of many diseases, including neurodegenerative disorders, increases with age. In many neurodegenerative diseases, damaged proteins accumulate in the cell and form highly toxic aggregates. The autophagy process may play an important role in the removal of these aggregated proteins, and C. elegans has proven to be a valuable tool in modeling such protein aggregation diseases. One example is the C. elegans model for Huntington’s disease, in which polyglutamine (polyQ) fragments containing 40 or more glutamine residues are expressed in various tissues [106]. Aggregation of the polyQ fragment in the cytoplasm of muscle cells in the body wall leads to defects in locomotion and paralysis [106]. Notably, RNAi-mediated knockdown of autophagy genes in these animals accelerates disease onset [107,108]. In a second Huntington’s disease model, human huntingtin fragments containing 150 glutamine residues are expressed in the ASH neurons (a bilaterial pair of environmental-sensing neurons located in the head) of C. elegans [107,109]. Here, too, knockdown of autophagy genes accelerates toxicity [107,109]. These data suggest that autophagy may be important to prevent accumulation of aggregated polyQ-expanded proteins, and that a reduction in autophagy may accelerate the progression of Huntington’s disease.
A model for Alzheimer’s disease also exists in C. elegans in which human Aβ-amyloid peptide is expressed in the muscle. As in humans with Alzheimer’s diseases, Aβ causes aggregation that, in the C. elegans model, results in paralysis with age. Interestingly, this mutant also accumulates autophagosomes due to defects in their maturation into degradative autolysosomes [110,111]. Moreover, autophagy genes are required for the degradation of Aβ in this C. elegans model [112]. Thus, autophagy seems to play a role in both Huntington’s and Alzheimer’s neurodegenerative disease models in C. elegans. These observations lend further support to a role for autophagy in aging and suggest that targeting this cellular process may be a novel approach to developing therapeutics against age-related diseases.
Autophagy and Aging in Drosophila
As in S. cerevisiae and C. elegans, autophagy has been implicated in the aging process in Drosophila. Autophagosome formation and autophagy gene expression decrease with age in the fly [113,120], and mutations in the autophagy genes Atg7 or Atg8 decrease their lifespan while increasing their sensitivity to stress [86]. Conversely, increases in lifespan are observed when the autophagy gene Atg8 is overexpressed specifically in neurons in adult flies [113]. Notably, lifespan extension was not seen when Atg8 was overexpressed using an early neuronal driver line [113]. Taken together, these observations suggest that neurons are important cells for autophagy-dependent longevity, and raise the interesting possibility that induction of autophagy in a specific tissue or cell type may benefit the entire organism. Moreover, the timing of Atg8 expression in the central nervous system may be critical for the longevity-associated effects. In contrast to the Atg8 studies, ubiquitous overexpression of the autophagy gene Atg1 induces autophagy but also increases cell death [114]. The increased cell death observed here may be a result of excessive autophagic activity. Taken together, these observations bring attention to the complexity of autophagy induction at the organismal level, and highlight the need for further investigation into the mechanism by which induction of autophagy may be sufficient to increase longevity.
TOR/Insulin signaling
Flies with reduced TOR levels, or expressing dominant negative forms of S6K, TSC1, or TSC2 (suppressors of the TOR pathway) are long-lived [115,116]. Consistent with these results, rapamycin treatment results in a modest lifespan extension, and this effect requires the autophagy gene Atg5 [117], suggesting that TOR extends lifespan in Drosophila at least partially through autophagy. In addition, mutation of the gene dSesn (Sestrin), a negative regulator of TOR, results in muscle degeneration, which is an age-dependent phenotype also observed in Atg1-silenced flies [118]. These data suggest that knockout of Sestrin phenocopies silencing of autophagy, which occurs with increased TOR activity. While rapamycin does not extend the lifespan of dFOXO mutants (unpublished data in Bjedov and Partridge, 2011), dFOXO is essential and sufficient for autophagy induction [86,119,120]. dFOXO expression specifically in the adult head fat body results in increased longevity [10]; however, it is not yet known if this increased lifespan is dependent on autophagy.
Age-related disease models
Drosophila research has resulted in numerous discoveries on age-related diseases such as protein aggregation disorders and neurodegeneration, and in some of these instances, autophagy has been shown to play a regulatory role. For example, in the Drosophila model of Huntington’s disease, inhibition of autophagy enhances polyQ protein accumulation, aggregation, and toxicity [121]. Moreover, in a model of the neurodegenerative disease spinobulbar muscular atrophy (SBMA), autophagy gene knockdown promotes cellular degeneration in the fly eye due to increased polyQ expansion of the androgen receptor [122]. Conversely, the severity of these neurodegeneration phenotypes is reduced by rapamycin-mediated induction of autophagy [123]. These data strongly support a cytoprotective role for autophagy, as was observed in the C. elegans models of neurodegeneration. Consistent with this notion, overexpression of Atg8 in adult fly neurons, which is sufficient to extend lifespan, is similarly sufficient to protect against age-associated accumulation of insoluble ubiquitinated protein aggregates [113].
Autophagy and Aging in Mammals
Understanding the longevity role of autophagy in model organisms such as yeast, worms, flies, and mice should ultimately allow the information to be harnessed to improve human health. In this regard, evidence is mounting that aging and autophagy are also linked in mammals, in support of observations made in invertebrate model organisms. First, autophagy function has been shown to decrease with age in rodent livers [124] and autophagic activity decreases with age in hypothalamic neurons in mice [125]. Second, chaperone-mediated autophagy (CMA) activity decreases in senescent cells in culture [126], and in many organs of old rodents [127]. Third, impairment of CMA in mammalian cells increases their sensitivity to stress, an effect that is associated with decreased longevity in invertebrate model organisms [128]. Lastly, overexpression of lysosomal-associated membrane protein 2A (LAMP-2A), a gene required for CMA, in transgenic mice allows liver function to be maintained into old age at a level comparable to that seen in young mice [129]. Although direct evidence is currently lacking, these observations are consistent with the notion that autophagy could be beneficial to longevity in mammals, as is observed in invertebrate model organisms.
Age-related disease models
Many models of age-related diseases have been developed in mammals, and they have generated useful tools and information regarding the relationship between autophagy and disease. The loss of autophagy genes with age correlates with the accumulation of damaged proteins [124]. In addition, lower levels of the autophagy gene Beclin1 have been observed in aging human brains [130]. These data reflect the importance of basal levels of autophagy for normal homeostasis, while the need for autophagy may be an amplified under disease conditions. In the sections below, we will focus on links between autophagy and two major age-related diseases, namely neurodegenerative diseases and cancer.
Neurodegenerative disease
The rate of neurological degeneration, and therefore dysfunction, increases with age. One of the hallmarks of neurodegenerative diseases is the accumulation of aggregated proteins. For example, tauopathies and amyloid beta (Aβ) plaques are caused by an accumulation of tau and Aβ proteins, respectively, in Alzheimer’s disease (AD), while α-synuclein protein forms Lewy bodies in Parkinson’s disease (PD), and polyQ expansion in specific proteins causes protein aggregation in Huntington’s disease (HD). Notably, autophagy is necessary for the removal of each of these aggregation-prone proteins in mammals [121,123,131,132]. Moreover, induction of autophagy by TOR-dependent and -independent mechanisms in mammalian cell culture and mouse models increases the clearance and reduces the toxicity of proteins prone to aggregation [55,121,123,132–134]. More specifically, Beclin1 mRNA and protein levels are reduced in a mouse AD model in which neuronal populations are specifically affected by the AD pathology. In addition, overexpression of Beclin1 in this model reduces Aβ accumulation [135]. Mice deficient in Atg5, Atg7, or FIP200 (FAK family-interacting protein of 200 kDa) autophagy genes in the central nervous system show a progressive accumulation of ubiquitinylated proteins and inclusion bodies in the neurons [136–138]. Taken together, these data support the observations made in invertebrate models that reduced autophagy levels lead to an acceleration of disease states, while overexpression of an autophagy gene may result in decreased toxicity, presumably by enhancing the entire autophagy process. Importantly, these observations also suggest that neuron-specific overexpression of autophagy genes may have potential as a therapy to slow the progression of neurodegenerative diseases.
Cancer
The incidence of many cancers increases with age, making it an age-related disease. The role of autophagy in cancer is very complex, and has been associated with both tumor suppression and tumor survival, dependent on the types of tumors [29,58]. In tumor suppression, autophagy likely acts through turnover of damaged proteins and organelles such as mitochondria. If mitochondria are not degraded efficiently, ROS levels can rise and cause further damage and genomic instability [139,140]. In tumor survival, autophagy preserves tumor cells under conditions of limited nutrition and hypoxia, and can increase their resistance to chemotherapy and radiation therapy [141–143].
There is ample evidence to support autophagy as a mechanism for tumor suppression. For example, the autophagy genes Beclin1, Atg5, Atg4, and BIF-1/endophilin B1 (Bax-interacting factor) act as tumor-suppressor genes in mice, as shown by the formation of tumors in mice deficient in these genes [144–148]. Beclin1 is mutated in some human cancers, as are the autophagy-related genes UVRAG and Bif1 [148–153]. In addition, a number of oncogenes inhibit autophagy [154,155], while autophagic activity is reduced upon oncogenesis in murine pancreatic cancer models [156,157]. Further studies are needed to determine how autophagy plays a protective role in tumor progression as well as to address if autophagy induction is sufficient to prevent tumor formation.
As mentioned above, autophagy can also be used as a survival mechanism by tumor cells, as illustrated by the observation that complete removal of the autophagy genes Beclin1 or Atg5 reduces proliferation and induces apoptotic death of cancer cells [158–161]. This finding suggests that autophagy inhibition may be a good therapeutic target to combat at least some cancers, which is supported by a study in colorectal tumors in which pharmacological inhibition of autophagy caused death of the tumor in response to nutrient deprivation [143].
Overall, these data support a role for autophagy as a pro-survival mechanism for both cancerous and normal cells. Many questions remain to be addressed, including whether autophagy plays a cell non-autonomous role in normal or cancerous cells. This information will be relevant to designing therapies that modulate autophagy in targeted tissues while preserving its normal levels in others.
Conclusion and Future Perspectives
Aging results from the gradual decline in cellular repair and housekeeping mechanisms, which leads to an accumulation of damaged cellular constituents and ultimately to the degeneration of tissues and organs. Decades of research on the subject have revealed that the aging process is influenced by genetics and that many metabolic signaling genes can affect aging by mechanisms still to be fully elucidated. Importantly, many of these genes, including the nutrient sensor TOR and AMP-dependent kinase (AMPK), are emerging as important regulators of the process of autophagy. The upregulation of autophagy in long-lived mutants would likely enable the cell to endure stressful conditions by increasing the rate of turnover of damaged macromolecules. It is possible that autophagy promotes cell maintenance by removing accumulated toxic material and by using recycled components as an alternative nutrient resource. This suggests that autophagy favors longevity because an organism can recover more quickly from stress-induced cellular damage. Evidence that autophagy influences the aging process has been observed in multiple model organisms, from yeast to multicellular organisms such as worms and flies, and a more recent and exciting finding is that autophagy is implicated in neurodegenerative diseases that affect humans.
Studies in C. elegans in particular have contributed greatly to the current evidence linking autophagy to aging. Using mostly steady-state measurements, autophagic events are increased in many longevity mutants. In many cases, the long lifespan is dependent on autophagy genes that are involved in multiple steps of the process, suggesting that flux through the process is critical. Further experiments are needed to measure if the autophagosome flux is indeed increased in longevity mutants. Recent work in C. elegans has also provided clues about potential novel regulators of autophagy such as the transcription factor PHA-4/FOXA and the energy sensor AMPK; both can also be linked to aging in multiple longevity mutants. It will be interesting to identify additional molecules that mediate autophagy induction and may have anti-aging effects. These effects may be tied to the type of cargo that is turned over in long-lived mutants. While the identity and regulation of the cargo remain to be determined, studies in germline-less C. elegans suggest that lipids could be aging-relevant cargo [74]. As such, turnover of lipids via lipophagy could play a critical role in the aging process, perhaps because the accumulation of lipids that occurs with age disrupts cellular homeostasis by interfering with the efficiency of autophagy [162]. Future biochemical research is needed to investigate this interesting hypothesis and its relevance in other longevity models.
Specific types of cargo have previously been identified in yeast and mammalian cells, such as mitochondria (mitophagy), ribosomes (ribophagy), peroxisomes (pexophagy), and endoplasmic reticulum (reticulophagy) [163,164]. The turnover of damaged mitochondria may be beneficial for longevity because removal of these leaky organelles can prevent further damage to the cells from ROS. Pharmacologic and genetic tools that specifically block mitophagy, for example yeast mutants deficient specifically in mitophagy [165,166], should prove useful in determining if mitochondrial turnover is essential for lifespan extension.
If the autophagic cargo relevant to aging is indeed selective and is not just bulk degradation of cytosolic components, the identification of the receptor(s) required for cargo recognition will be a great challenge. A prominent example of an autophagy receptor is the ubiquitin recognition protein p62 [167]. This protein has been shown in vitro to bind not only ubiquitin but also LC3, a protein localized to the pre-autophagosomal and autophagosomal membranes. These interactions are thought to be the mechanism by which p62 brings selective, ubiquitinated cargo to the autophagosome for degradation. Systematic studies are needed to determine if other p62-like proteins such as NBR1, NDP52 and NIX play a role in selective autophagy, and if such receptor proteins recognize and destine aging-relevant cargo for degradation via autophagy.
Studies in the model organism Drosophila have been informative in addressing tissue specificity of autophagy induction, which could be important in targeting autophagy for therapeutic purposes, as well as to suggest that induction of autophagy can be sufficient to extend lifespan. In the fly, autophagy has been successfully induced by single gene overexpression of either Atg1 or Atg8. Importantly, overexpression of Atg8 specifically in neurons during adulthood, but not during development, is sufficient for profound lifespan extension [113]. These studies highlight the need to understand which tissues are most important for autophagy-dependent longevity. If autophagy induction is beneficial in some tissues but not others, it will be necessary to design autophagy-targeting therapies that act in a tissue-specific manner.
In summary, we have reviewed here some of the key regulators and mediators of autophagy, and described the accumulating evidence that this complex process plays an important role in aging. It is clear that we do not yet completely understand all the single steps in the process, including how a membrane source for autophagosome formation is selected, and it is likely that many regulators in the process of autophagy have yet to be discovered. We can speculate that autophagy is selective for certain cargos, but how this might relate to aging remains unclear. These gaps in knowledge illustrate the need to identify the key players involved in autophagy regulation before we can begin to fully understand this complex process, and how it might be harnessed to promote healthy human aging, and to develop therapies for age-related diseases in humans.
Acknowledgments
We would like to thank Drs. Andrew Hellman, Caroline Kumsta, Louis Lapierre, and Anne O’Rourke for feedback on the manuscript. MH is funded by NIH/NIA (Grant numbers: R01 AG038664 and R01 AG039756). MH is also an Ellison Medical Foundation New Scholar in Aging (Grant number: AG-NS-0481-08).
Footnotes
Paradoxically, one report has suggested that RNAi suppression of several autophagy genes results in lifespan extension [77]. However, we note that this study was not well controlled.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Rajawat YS, Bossis I. Autophagy in aging and in neurodegenerative disorders. Hormones (Athens) 2008;7:46–61. doi: 10.14310/horm.2002.1111037. [DOI] [PubMed] [Google Scholar]
- 2.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 4.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 7.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 8.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 9.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 10.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 11.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 12.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 13.Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 14.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Hansen M, Kapahi P. TOR Signaling and Aging. In: Hall MN, Tamanoi F, editors. The Enzymes. Burlington: Academic Press; 2010. pp. 279–299. [Google Scholar]
- 16.Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rous P. The influence of diet on transplanted and spontaneous mouse tumor. J Exp Med. 1914;20:433–451. doi: 10.1084/jem.20.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCay CM, Maynard LA, Sperling G, Barnes LL. The Journal of Nutrition. Volume 18 July--December, 1939. Pages 1--13. Retarded growth, life span, ultimate body size and age changes in the albino rat after feeding diets restricted in calories. Nutr Rev. 1975;33:241–243. doi: 10.1111/j.1753-4887.1975.tb05227.x. [DOI] [PubMed] [Google Scholar]
- 19.Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 21.Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- 22.Cargill SL, Carey JR, Müller HG, Anderson G. Age of ovary determines remaining life expectancy in old ovariectomized mice. Aging Cell. 2003;2:185–190. doi: 10.1046/j.1474-9728.2003.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason JB, Cargill SL, Anderson GB, Carey JR. Transplantation of young ovaries to old mice increased life span in transplant recipients. J Gerontol A Biol Sci Med Sci. 2009;64:1207–1211. doi: 10.1093/gerona/glp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Raamsdonk JM, Hekimi S. Reactive Oxygen Species and Aging in Caenorhabditis elegans: Causal or Casual Relationship? Antioxid Redox Signal. 2010;13:1911–1953. doi: 10.1089/ars.2010.3215. [DOI] [PubMed] [Google Scholar]
- 25.Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hur JH, Cho J, Walker DW. Aging: Dial M for Mitochondria. Aging (Albany NY) 2010;2:69–73. doi: 10.18632/aging.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu CT. Eaten alive: autophagy and neuronal cell death after hypoxia-ischemia. Am J Pathol. 2008;172:284–287. doi: 10.2353/ajpath.2008.071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strømhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 36.Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 38.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 39.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonsen A, Stenmark H. Self-eating from an ER-associated cup. J Cell Biol. 2008;182:621–622. doi: 10.1083/jcb.200807061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longatti A, Orsi A, Tooze SA. Autophagosome formation: not necessarily an inside job. Cell Res. 2010;20:1181–1184. doi: 10.1038/cr.2010.132. [DOI] [PubMed] [Google Scholar]
- 44.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 45.Meijer AJ. Amino acid regulation of autophagosome formation. Methods Mol Biol. 2008;445:89–109. doi: 10.1007/978-1-59745-157-4_5. [DOI] [PubMed] [Google Scholar]
- 46.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, et al. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 51.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang L, Chen S, Du F, Li S, Zhao L, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 55.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–64. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morimoto RI. Stress, aging, and neurodegenerative disease. N Engl J Med. 2006;355:2254–2255. doi: 10.1056/NEJMcibr065573. [DOI] [PubMed] [Google Scholar]
- 58.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mijaljica D, Prescott M, Devenish RJ. Autophagy in disease. Methods Mol Biol. 2010;648:79–92. doi: 10.1007/978-1-60761-756-3_5. [DOI] [PubMed] [Google Scholar]
- 60.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 63.Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 64.Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, et al. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang F, Watkins JW, Bermudez M, Gray R, Gaban A, et al. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy. 2008;4:874–886. doi: 10.4161/auto.6556. [DOI] [PubMed] [Google Scholar]
- 67.Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 68.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tóth ML, Sigmond T, Borsos E, Barna J, Erdélyi P, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 70.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 71.Mörck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 73.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 74.Lapierre LR, Gelino S, Meléndez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dwivedi M, Song HO, Ahnn J. Autophagy genes mediate the effect of calcineurin on life span in C. elegans. Autophagy. 2009;5:604–607. doi: 10.4161/auto.5.5.8157. [DOI] [PubMed] [Google Scholar]
- 77.Hashimoto Y, Ookuma S, Nishida E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes Cells. 2009;14:717–726. doi: 10.1111/j.1365-2443.2009.01306.x. [DOI] [PubMed] [Google Scholar]
- 78.Meléndez A, Levine B. Autophagy in C. elegans. WormBook; 2009. pp. 1–26. [DOI] [PubMed] [Google Scholar]
- 79.Kovacs AL, Zhang H. Role of autophagy in Caenorhabditis elegans. FEBS Lett. 2010;584:1335–1341. doi: 10.1016/j.febslet.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 80.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 82.Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- 83.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 84.Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 85.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 88.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 89.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 90.Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rodriguez-Navarro JA, Cuervo AM. Autophagy and lipids: tightening the knot. Semin Immunopathol. 2010;32:343–353. doi: 10.1007/s00281-010-0219-7. [DOI] [PubMed] [Google Scholar]
- 93.Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, et al. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arum O, Johnson TE. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J Gerontol A Biol Sci Med Sci. 2007;62:951–959. doi: 10.1093/gerona/62.9.951. [DOI] [PubMed] [Google Scholar]
- 95.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- 96.Batta K, Das C, Gadad S, Shandilya J, Kundu TK. Reversible acetylation of non histone proteins: role in cellular function and disease. Subcell Biochem. 2007;41:193–212. [PubMed] [Google Scholar]
- 97.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 98.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 99.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 100.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–970. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia K, Hart AC, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3:21–25. doi: 10.4161/auto.3528. [DOI] [PubMed] [Google Scholar]
- 108.Khan LA, Yamanaka T, Nukina N. Genetic impairment of autophagy intensifies expanded polyglutamine toxicity in Caenorhabditis elegans. Biochem Biophys Res Commun. 2008;368:729–735. doi: 10.1016/j.bbrc.2008.01.150. [DOI] [PubMed] [Google Scholar]
- 109.Voisine C, Varma H, Walker N, Bates EA, Stockwell BR, et al. Identification of potential therapeutic drugs for huntington’s disease using Caenorhabditis elegans. PLoS One. 2007;2:e504. doi: 10.1371/journal.pone.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Askanas V, Engel WK. Inclusion-body myositis: a myodegenerative conformational disorder associated with Abeta, protein misfolding, and proteasome inhibition. Neurology. 2006;66:S39–S48. doi: 10.1212/01.wnl.0000192128.13875.1e. [DOI] [PubMed] [Google Scholar]
- 111.Murphy MP, Golde TE. Inclusion-body myositis and Alzheimer disease: two sides of the same coin, or different currencies altogether? Neurology. 2006;66:S65–S68. doi: 10.1212/01.wnl.0000192108.02654.ac. [DOI] [PubMed] [Google Scholar]
- 112.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 113.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 114.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 116.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–8. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu H, Wang MC, Bohmann D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev. 2009;126:624–637. doi: 10.1016/j.mod.2009.06.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Demontis F, Perrimon N. FOXO/4E-BP Signaling in Drosophila Muscles Regulates Organism-wide Proteostasis during Aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 122.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 123.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 124.Vittorini S, Paradiso C, Donati A, Cavallini G, Masini M, et al. The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J Gerontol A Biol Sci Med Sci. 1999;54:B318–B323. doi: 10.1093/gerona/54.8.b318. [DOI] [PubMed] [Google Scholar]
- 125.Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, et al. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 2012;13:258–265. doi: 10.1038/embor.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dice JF. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- 127.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 128.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 131.Montie HL, Cho MS, Holder L, Liu Y, Tsvetkov AS, et al. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18:1937–1950. doi: 10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 133.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 135.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 137.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 138.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2:80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- 140.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ogier-Denis E, Houri JJ, Bauvy C, Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem. 1996;271:28593–28600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 142.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, et al. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 143.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 144.Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 145.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 148.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 149.Coppola D, Khalil F, Eschrich SA, Boulware D, Yeatman T, et al. Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer. 2008;113:2665–2670. doi: 10.1002/cncr.23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 151.Kim SY, Oh YL, Kim KM, Jeong EG, Kim MS, et al. Decreased expression of Bax-interacting factor-1 (Bif-1) in invasive urinary bladder and gallbladder cancers. Pathology. 2008;40:553–557. doi: 10.1080/00313020802320440. [DOI] [PubMed] [Google Scholar]
- 152.Liang C, Feng P, Ku B, Dotan I, Canaani D, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 153.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 155.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 156.Réz G, Tóth S, Pálfia Z. Cellular autophagic capacity is highly increased in azaserine-induced premalignant atypical acinar nodule cells. Carcinogenesis. 1999;20:1893–1898. doi: 10.1093/carcin/20.10.1893. [DOI] [PubMed] [Google Scholar]
- 157.Tóth S, Nagy K, Pálfia Z, Réz G. Cellular autophagic capacity changes during azaserine-induced tumour progression in the rat pancreas. Up-regulation in all premalignant stages and down-regulation with loss of cycloheximide sensitivity of segregation along with malignant transformation. Cell Tissue Res. 2002;309:409–416. doi: 10.1007/s00441-001-0506-7. [DOI] [PubMed] [Google Scholar]
- 158.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Høyer-Hansen M, Bastholm L, Mathiasen IS, Elling F, Jäättelä M. Vitamin D analog EB1089 triggers dramatic lysosomal changes and Beclin 1-mediated autophagic cell death. Cell Death Differ. 2005;12:1297–1309. doi: 10.1038/sj.cdd.4401651. [DOI] [PubMed] [Google Scholar]
- 161.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 167.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]