Abstract
Bacterial pili have long been recognized as mediators of initial host-pathogen interactions important for the progression of Gram-negative bacterial diseases. An appreciation for the role of pili on Gram-positive bacteria in virulence, as well as the unique properties of their biogenesis is a rapidly emerging area of research. In this review, we focus on recent advances in one of the longest-studied Gram-negative pilus systems, the chaperone/usher assembled pili, along with the newcomer to the field, the sortase-assembled pili of Gram-positive bacteria. In both systems, a wealth of new structural and molecular details has emerged recently. In light of this, we explore similarities between chaperone/usher and sortase-assembled pilus biogenesis and highlight paradigms unique to each, with an eye toward using knowledge of each system to raise new questions and inform future studies of the other.
Pili in bacteria
Bacterial pili are defined as non-flagellar, proteinaceous, multi-subunit surface appendages involved in adhesion to other bacteria, host cells, or environmental surfaces [1, 2]. Pili were first recognized by electron microscopy on Gram-negative bacteria more than 50 years ago [3] and on Gram-positive bacteria 40 years ago [4]. Since that time pili have been implicated in critical host-pathogen interactions, colonization, tropism determination, biofilm formation, invasion, and signaling events [5, 6]. Non-flagellar organelles of Gram-negative bacteria include the well-characterized chaperone/usher-assembled pili, type IV pili, and curli. Gram-positive bacteria express pili assembled by sortase enzymes as well as type IV pili similar to those of Gramnegative organisms. In each of these systems, bacteria have evolved mechanisms to efficiently assemble highly stable fibrillar structures on their surface while preventing the aggregation and/or premature assembly of highly interactive subunits within the cell interior. In this review we focus on one of the longest studied and best understood pilus systems, the chaperone/usher assembled pili of Gram-negative bacteria, along with the newest pilus system to be described, the sortase-assembled pili of Gram-positive bacteria. The diversity of chaperone/usher pathways across a number of bacteria provides an interesting parallel to the diversity of sortase-assembled pili now characterized. Both pilus pathways are pathogenic determinants for a variety of organisms and are proving to be promising targets for anti-virulence therapeutics [7, 8]. Thus, we highlight recent advances in understanding of the pilus structure and biogenesis of both pilus types and we stress paradigms that are similar between them, with the goal of highlighting how increased understanding of one system might shed light on the other (Table 1).
Table 1.
Pilus system attributes for CU and SA pili
| CU assembled Pili | SA pili | |
|---|---|---|
| Fiber initiation | Tip adhesins with highest binding affinity for the usher are incorporated first | Major subunit sufficient to initiate fiber polymerization by the pilin sortase |
| Fiber organization | Tip adhesin Tip fibrillum Major subunit backbone |
Minor subunit tip in some SA pili Minor subunits interspersed Major subunit backbone |
| Fiber organization | Non-covalent interactions via DSE | Covalent inter- and intra-subunit peptide bonds |
| Fiber termination | Incorporation of minor pilin subunit that cannot undergo DSE | Incorporation of minor pilin subunit that is attached to the cell wall by the housekeeping sortase |
| Receptor interactions | Ligand interactive sites mapped to tip adhesin | Minor subunits mediate adhesion, molecular basis undetermined |
| Assembly platform | Usher–mediated, location not examined | Secretion and sortase enzymes spatially restricted |
| Pilus fiber display | Peritrichous | Focal assembly; peritrichous display and/or absent from septum after cell wall attachment |
Chaperone/usher pili
In Gram-negative bacteria, the chaperone/usher (CU) pathway includes S and Dr pili, and is exemplified by the type 1 and Pap (P) pilus systems of uropathogenic Escherichia coli (UPEC) [9]. Although a single E. coli genome can encode up to 12 CU pili [10], the assembly machinery and fiber components are encoded by gene clusters distinct for each type of CU pilus, which contain the pilin subunits, the adhesin, and the dedicated chaperone and usher (Figure 1). We focus on P and type 1 pili as model CU pili in this review, based on the wealth of structural and molecular information available for each. The CU molecular machine functions to regulate the ordered secretion, folding and assembly of tens of thousands of pilin subunits into pilus fibers on the surface of the bacterium.
Figure 1. CU and SA pilus and fiber assembly components.
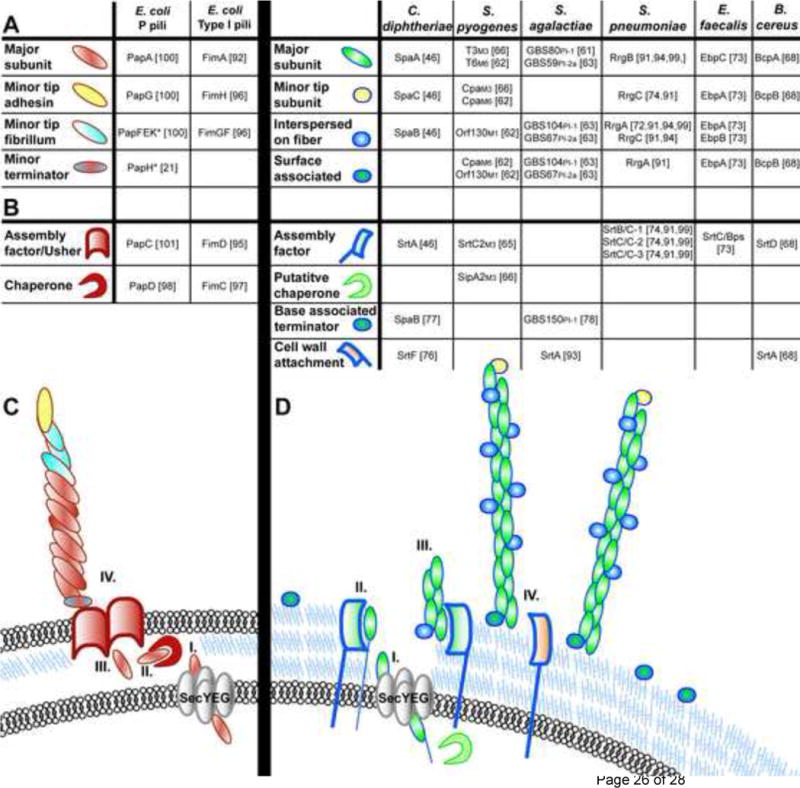
(a)–(b) E. coli prototype CU assembled pili. (a) Pilus subunit location within wild-type pilus fibers are depicted, as determined by immunoelectron microscopy. “*” indicates subunits whose location was deduced from biochemical and/or structural studies and not by immunoelectron microscopy. Empty boxes indicate no available data for that pilus biogenesis component. (b) Schematic of pilus biogenesis machinery for CU assembled pili. E. coli CU pilin subunits are translocated via the Sec machinery through the inner membrane (i), after which they associate with dedicated chaperone proteins in the periplasm (ii) which prevent subunit misfolding and facilitate delivery of the subunits to the outer membrane usher protein (iii) through which subunits are secreted and which serves as a platform for ordered pilus assembly (iv). (c)–(d) Gram-positive SA pili. (c) Table of pilus subunits for Gram-positive SA pili. Subscript for S. pyogenes indicates M1, M3 or M6 serotype and for S. agalactiae indicates the PI-1 or PI-2a pilus island. Slash marks (/) indicate differing names for the same protein in the literature. (d) Schematic of Gram-positive SA pili. Gram-positive pilin subunits are targeted for translocation across the cell membrane by the general secretion machinery via an N-terminal signal sequence (i). Presumably, subunits are then retained in the cell membrane by a transmembrane domain (ii) prior to LPXTG recognition and cleavage by sortase enzymes leading to ordered pilus fiber assembly (iii) and attachment to the cell wall (iv).
Chaperone-mediated donor strand complementation
Newly synthesized pilin subunits are translocated across the cytoplasmic membrane via the sec translocase where they form complexes with periplasmic chaperone proteins (PapD for P pili, FimC for type 1 pili; Figure 1). Pilus chaperones consist of two immunoglobulin-like (Ig-like) domains stabilized by a conserved salt bridge. This arrangement was first seen in the crystal structure of the prototype chaperone PapD [11] and later also shown for other chaperones including FimC [12, 13]. Each subunit of CU pili consists of an incomplete Ig-like fold and an N-terminal extension (Nte). Periplasmic chaperones for P and type 1 pili couple subunit folding with the capping of the interactive subunit assembly sites in a process termed donor strand complementation (DSC) [14–16]. In DSC, the G1 beta strand lining the interdomain cleft of the chaperone transiently serves as the C-terminal beta strand of the subunit, thus providing the steric information necessary for folding by completing the Ig-like fold of the subunit (Figure 2a).
Figure 2. CU pilin subunit structure.
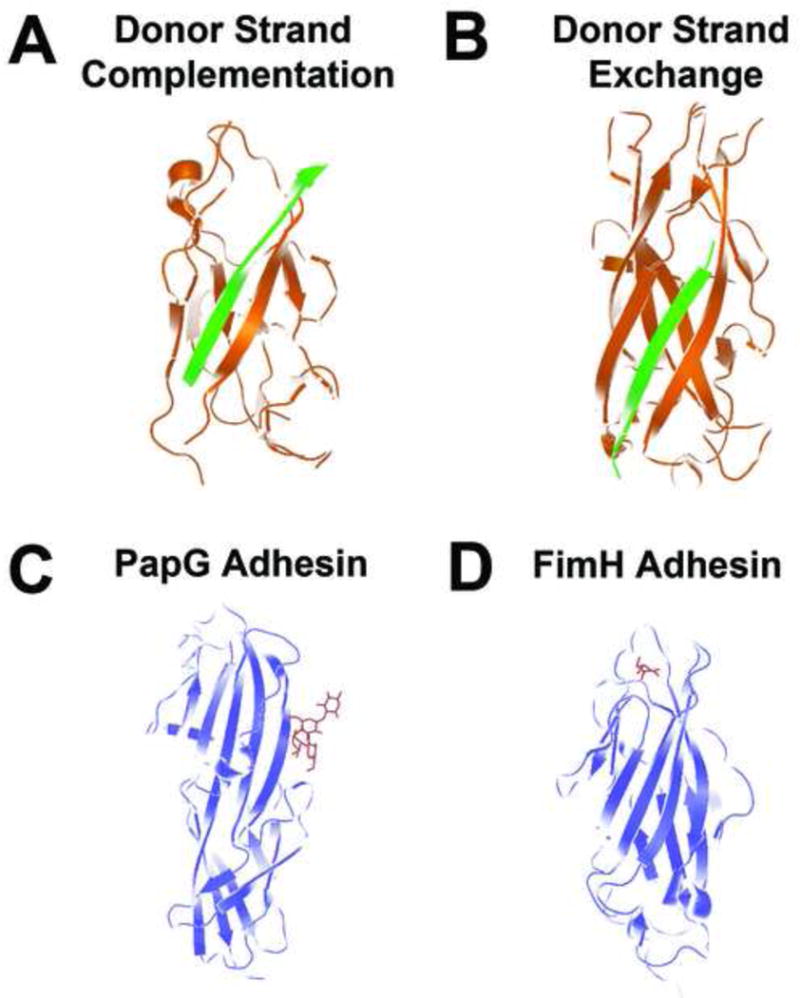
(a) Donor strand complementation. The interactive subunit assembly site created by the incomplete immunoglobulin fold of the PapE (orange) pilin subunit is capped by the G1 strand of the PapD chaperone (green), providing the steric information necessary to complement the Ig-like fold of the subunit and stabilize the subunit prior to pilus assembly (PDB code 1NOL). (b) Donor strand exchange. PapE (orange) pilin structure consists of an incomplete Ig fold, which is completed within the pilus structure by the N-terminal extension of the neighboring pilin subunit, PapK (green), to form a canonical Ig domain in the polymerized pilus fiber (PDB code 1N12). (c) PapG ligand-binding domain (blue) bound to GbO4 (red), which consists of the PapG ligand tetrasaccharide GalNAc beta 1–3Gal alpha 1–4Gal beta 1–4Glc linked to ceramide (PDB code 1J8R). (d) FimH ligand-binding domain interacting with its D-mannose ligand (red) (PDB code 1KLF).
Donor strand exchange
Once formed, each chaperone-subunit complex is targeted to the outer membrane usher (PapC for P pili, FimD for type 1 pili; Figure 1) where the chaperone-pilin complex then dissociates by an unknown mechanism and the subunit is added to the base of the growing pilus. The order of subunit assembly is based on affinity for the usher and for each other [17–20]. In the case of P pili, PapD-PapG (chaperone-adhesin) complexes bind first to empty PapC sites, followed by PapD-PapF (chaperone-minor pilin subunit) binding, which initiates tip assembly (Figure 1). The chaperone is then released from the PapD-PapG complex, as the Nte of PapF is zippered into the newly opened groove of PapG to complete the Ig-like fold of the PapG pilin domain (Figure 2a). This process is termed donor strand exchange (DSE) (Figure 2b), and results in a head to tail linear array of non-covalently assembled pilin subunits. The joining of PapG to PapF is then followed by multiple rounds of PapD-PapE (chaperone-minor pilin subunit) binding and DSE reactions to form the bulk of the pilus tip. The binding and incorporation of PapK terminates the addition of PapE subunits to the growing fibrillum and nucleates the incorporation of PapA major subunits into the fiber. Since PapH is unable to undergo DSE, pilus growth is terminated when PapH is incorporated at the base of the fiber because it lacks the open groove that would be necessary to join to any additional subunits to it [21, 22]. It has also been proposed that PapH mediates membrane attachment of the pilus fiber, although the mechanism for this is unknown [21, 22].
Usher-catalyzed assembly across the outer membrane
The PapC and FimD usher proteins serve as assembly sites and pores in the outer membrane allowing export of the growing fiber to the surface of the cell [23]. The N-terminal domain of the usher has been implicated in the initial binding of the chaperone-subunit complexes in the periplasm [24]. Once bound by initiating chaperone-subunit complexes, ushers undergo a conformational change, which is then maintained throughout fiber assembly [20]. Recently solved structures for the N-terminal domain of the usher FimD with a complex of the FimC chaperone and FimH adhesin pilin domain, as well as a FimD-FimC-FimF complex gave insight into usher surfaces responsible for binding to the chaperone-subunit complexes [25, 26]. A new structure of the central portion of PapC revealed that the usher is a dimer, in which each PapC monomer forms a 24 strand β barrel with a central channel able to export fully folded polymerized pilin subunits [27]. Cryo-electron microscopy (EM) studies showed that only one of the PapC dimer pores is used for extrusion of the assembled pilin subunits [27]. In the absence of subunits in the crystal structure, the pore in each monomer is blocked by a plug domain, which originates from within the PapC barrel. The plug is stabilized by interactions within the barrel and it is proposed that the conformational change that occurs upon binding of initiator chaperone-subunit complexes results in destabilization of those interactions and displacement of the plug, such that the polymerized subunits can transit the channel. It is also proposed that the dual usher structure facilitates ordered stepwise interaction and handoff of each subunit-chaperone complex at the periplasmic face of the usher during pilus biogenesis.
The process of polymerization and extrusion of the subunits through the outer membrane does not require energy input [28], rather it is likely driven by the energy stored in the ‘primed’ chaperone-subunit complexes [29–32]. Upon DSE, topological rearrangements of the subunit and condensation of the hydrophobic core form a polymer held together by non-covalent interactions, which are stronger than those in the stable chaperone-subunit complexes. In addition, once on the surface of the cell, the PapA pilus fiber packs into a tight right-handed helical cylinder, which is thought to provide additional force for outward growth of the organelle [23].
Chaperone/usher pilus adhesins
Bacterial attachment is a key event in the colonization of mucosal surfaces. CU pili contain adhesins that mediate attachment to a specific receptor. While the pilin subunits are single domain proteins, tip adhesins are composed of two domains. The C-terminal domain of tip adhesins is a typical Ig-like pilin domain that participates in DSE to join the adhesin to the tip of the pilus [12]. Crystal structures for the PapG, FimH and other CU adhesins show that each N-terminal ligand-binding domain also shares a similar β barrel jellyroll fold [12, 33–36]. However, each ligand-binding domain is variable with respect to its length and molecular organization. In addition, each adhesin uses a different surface to bind to its cognate ligand. The P pilus tip adhesin PapG binds to Galα1–4Gal-containing glycolipids [37] and is required for UPEC to cause pyelonephritis [38]. The PapG ligand site is on the long side of the ligand-binding domain (Figure 2c), but is made up of residues that are distal to the core Ig-like domain shared with the other receptor binding domains [33]. Type 1 pili contain the FimH adhesin at their tip which makes critical host-pathogen interactions necessary for UPEC pathogenesis [39–42]. FimH binds to mannosylated receptors on the surface of the bladder epithelium and the ligand site is located at the distal tip of the ligand-binding domain [34] (Figure 2d). Mannose is bound in a deep negatively charged pocket at the tip of the receptor-binding domain, which consists of an elongated eleven-stranded β barrel. Mutation of FimH residues essential for mannose-binding such as Q133 [34] results in attenuated virulence in a murine urinary tract infection model [43]. Scanning and high-resolution EM of UPEC in a mouse cystitis model revealed that adhesive type 1 piliated bacteria are able to interact with bladder superficial facet cells leading to binding, invasion, and formation of intracellular bacterial communities with biofilm properties [39, 44]. This process is dependent on the FimH adhesin and allows UPEC to gain a foothold in the acute stages of infection. Thus, these binding domains serve as the functional end of the pilus, providing specific affinity for host ligands and influencing bacterial tropism for host tissues.
Sortase-assembled pili
Pili in Gram-positive bacteria are composed of a single major pilin subunit and usually 1–2 accessory subunits, the genes for which are all clustered in genetic loci. Pilin loci always contain a sortase gene (or genes), which encodes the enzyme that catalyzes the polymerization of these sortase-assembled (SA) pili. Several Gram-positive organisms such as Corynebacterium diphtheriae and Enterococcus faecium encode more than one pilin locus [45, 46], although genes of each locus encode components specific for the assembly of a unique pilus type [47].
Accessory factors for SA pili
Gram-positive organisms lack a membrane-bound periplasm but, nevertheless, secrete many virulence factors that require post-secretion modification [48]. It has been proposed that the space between the cell membrane and cell wall in Gram-positive bacteria provides a protected environment for folding and processing of secreted proteins [49–52], raising the possibility of a role for secreted chaperones in pilus biogenesis. In fact, in addition to pilin- and sortase-encoding genes, some pilus gene clusters of Streptococcus agalactiae, Streptococcus suis, Streptococcus pneumoniae, and Streptococcus pyogenes are also genetically linked to type I signal peptidase homologues. The pilin signal peptidase homologue in S. pyogenes lacks canonical signal peptidase active site residues and therefore does not act as a signal peptidase. Instead, it has been proposed to act as a chaperone to prevent premature subunit interactions during biogenesis, analogous to the chaperone activity required in CU pilus biogenesis, since it is required for pilus fiber formation and interaction with the major pilin subunit [53, 54]. However, the precise role for pilin locus-associated signal peptidase homologues in fiber formation remains to be determined.
Sortase-mediated pilus assembly
Each pilin locus contains at least one associated Class C sortase enzyme, termed the pilin sortase, which mediates pilus fiber formation. In addition, nearly every sequenced Gram-positive bacterial species encodes a Class A sortase, also called the housekeeping sortase. Sortases are transpeptidases that recognize and cleave the threonine-glycine bond within an LPXTG (or LPXTG-like) motif found within a C-terminal cell wall sorting signal (CWSS) of some secreted proteins, leading to the covalent linkage of proteins to the cell wall (by Class A housekeeping sortases) or to one another (by Class C pilin sortases) [55]. The paradigmatic Gram-positive pilus system for which we have the most information is in C. diphtheriae (see review by Ref. [56]). Studies in C. diphtheriae and S. pyogenes have demonstrated that sortase-mediated attachment of pilin subunits to one another during pilus biogenesis proceeds via isopeptide bond formation between the threonine and glycine of the cleaved LPXTG (or LPXTG-like) motif in one pilus subunit and a conserved lysine present within the neighboring pilin subunit [46, 57–59]. Sequential transpeptidation reactions result in a pilus fiber consisting of a linear array of pilin molecules, the bulk of which is composed of the major pilin subunit, as shown by immunogold-EM studies [46]. These pili range from 0.3–3 μm wide and can be as long as 10 μm [60]. The major pilin subunit is essential for pilus fiber formation [46, 61, 62]. In part, pilus length is determined by availability of the major pilus subunit, since overexpression leads to longer pili [46, 61, 63]. In comparison, minor subunits are generally dispensable for pilus formation and are typically incorporated into the pilus fiber in characteristic spatial locations along the pilus fiber: at the base, at the tip, or in patches along the length of the pilus as shown by immunogold-EM (Figure 1). The molecular details required for minor pilus subunit incorporation were first assessed in detail for C. diphtheriae. In this system, fiber incorporation of the minor tip-associated pilus subunit SpaC required the lysine of the major subunit’s YPKN pilin motif, whereas incorporation of the SpaB minor subunit instead utilizes a glutamic acid within the E-box conserved element in the major SpaA subunit [46, 58]. The presence of pilin motifs and E-boxes in many other Gram-positive pilin sequences suggests that there may be conserved elements for minor subunit incorporation; indeed the YPKN motif is also required for BcpA pilus polymerization in Bacillus cereus[64]. However, not all Gram-positive pilus systems studied thus far strictly adhere to these motif requirements. Pilus polymerization in S. pyogenes requires a non-canonical QVPTG motif within the CWSS of the major pilin subunit, as well as a lysine residue that is not associated with a canonical YPKN-type pilin motif. A similar non-canonical VPPTG motif within the S. pyogenes minor Cpa subunit is required, along with the conserved lysine of the major subunit for Cpa incorporation into the pilus [65, 66]. Further study of the sequence elements and mechanism(s) for minor pilin subunit incorporation, the basis for pilin sortase specificity, and the functional consequences of the localization pattern of subunit incorporation on individual pili will greatly increase our overall understanding of pilus biogenesis in Gram-positive bacteria.
Pilus structure of SA pili
Structural insights into the assembly of SA pili first emerged from crystallization of the minor pilin adhesive subunit from Streptococcus agalactiae and major pilin subunit from S. pyogenes, revealing a two-domain organization in each consisting of a modified Ig fold, similar to the Ig-like fold for CU pilin subunits [57, 67] (Figure 3a). An intermolecular isopeptide bond between the major pilin subunits of S. pyogenes formed between the invariant lysine of the pilin motif of one molecule with the threonine of the LPXTG in the next pilin molecule. Furthermore, two potentially self-generated intramolecular lysine-asparagine bonds were revealed within each subunit (Figure 3a). These covalent linkages, along with intermolecular bonding, provide a basis for the structural integrity necessary to maintain such long thin pili [57]. Based on sequence comparison, Kang et al. predicted the presence of intramolecular bonds in other Gram-positive pilin proteins [57]. Indeed, intramolecular amide bonds were recently demonstrated within the B. cereus BcpA major pilus subunit, one of which contributes to pilus fiber formation [68], as well as within the major SpaA pilin subunit of C. diphtheriae[59]. SpaA pilins also contain an additional intramolecular disulfide bond (Figure 3b), which is surprising since other pilin proteins examined lack cysteines and it was thought that Gram-positive bacteria lack disulfide bond-forming machinery [59, 69].
Figure 3. SA pilin subunit structure.
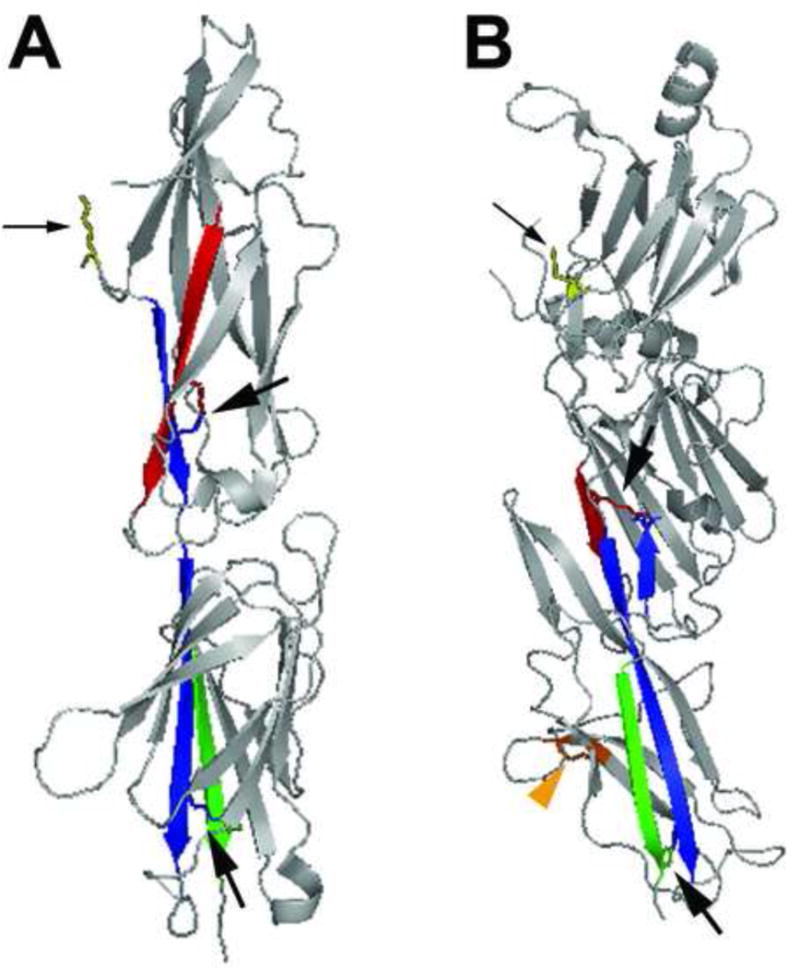
(a) The 32.5 kD two-domain S. pyogenes major pilin subunit structure in ribbon diagram (PDB code 3B2m). The N- and C-terminal β sheets of the molecule are colored red and green, respectively. The interacting β sheet for each amide bond is colored blue. Stick diagrams indicate Lys-Asn intramolecular bonds within each domain of the subunit (indicated by large black arrows) and the conserved lysine (yellow, small black arrow) that becomes covalently linked to the threonine of cleaved LPXTG-like motif in the neighboring subunit. (b) The 47 kD three-domain C. diphtheriae SpaA major pilin subunit structure in ribbon diagram (PDB code 3HR6). The N- and C-terminal β sheets that participate in intramolecular amide bonds are colored red and green (in the middle and C-terminal domain, respectively); the interacting β sheet for each amide bond is colored blue. Stick diagrams indicate Lys-Asn intramolecular bonds within each domain of the subunit (large black arrows), the conserved lysine of the YPKN pilin motif (yellow, small black arrow) that becomes covalently linked to the threonine of cleaved LPXTG motif in the neighboring subunit, and the disulfide bond in the C-terminal domain (orange, orange arrowhead).
The C-terminal pilin domain revealed in the X-ray crystal structure of the S. agalactiae minor pilin subunit mediates binding to host cells and the N-terminal domain is hypothesized to confer structural attributes, analogous to the two-domain makeup of the CU pilus tip adhesins [67]. Indeed, minor subunits in C. diphtheriae and S. pneumoniae are implicated in adhesion to host cells [70, 71]. In contrast to the well-characterized CU pilus adhesins, a molecular receptor has been identified for only one SA pilus: the S. pyogenes major pilin protein Spy0128 specifically interacts with the soluble scavenger receptor gp340 [72]. No receptors have been identified for SA pilus minor pilin subunits, although many contain MSCRAMM motifs predicted to mediate attachment to extracellular matrix molecules.
Sortase requirements for pilus growth
Pilus loci can encode 1–3 pilin sortases. In organisms bearing only one pilus sortase, the pilus-associated sortase is essential for fiber formation and incorporation of all minor subunits [46, 61, 62, 73]. Studies in organisms with pilus loci encoding more than one sortase have shown both functional specificity and redundancy among the enzymes. For instance, in a pilus locus in S. agalactiae (Group B Streptococcus, GBS) which contains two pilin sortases, each sortase was required for the incorporation of a single specific minor subunit [63]. S. pneumoniae encodes three pilus sortase enzymes (called SrtB-D or SrtC1–3), although there are conflicting reports as to the specific contribution of each pilin sortase to fiber formation. While deletion of any single pilin sortase does not abrogate pilus formation [74, 75], the absence of both SrtB/SrtC-1 and SrtC/SrtC-2 eliminates piliation and indicates that SrtD/SrtC02 is not sufficient for pilus polymerization [74]. Pneumococcal SrtB/SrtC-1 plays a specific role for incorporation of at least one minor pilin (RrgC) into the pilus fiber [74, 75]. SrtC/SrtC-2 is not sufficient for heterotrimeric pilus assembly, since SrtC/SrtC-2 alone cannot incorporate one of the minor subunits, RrgC, into the pilus [74].
Fully polymerized fibers are attached to the cell wall by the housekeeping sortase in C. diphtheriae, with deletion of this gene resulting in the secretion of pilus fibers into the culture supernatant [76]. However, deletion of the housekeeping SrtA in S. pneumoniae did not result in an increase in shed pilus fibers, suggesting that a role for the housekeeping sortase in pilus attachment to the cell wall may not be universal among Gram-positive bacteria [75]. An accessory pilin protein in C. diphtheriae, SpaB, is also important for efficient anchoring of the pilus fiber to the cell wall, leading to the proposal that SpaB can serve as the pilus terminator when it is attached to the cell wall by the housekeeping sortase [77]. Similarly, the housekeeping sortase of S. agalactiae appears to act in concert with a specific minor subunit, GBS150, for cell wall anchoring of the pilus [78]. Pilus length termination by minor pilin subunits in Gram-positive bacteria may be analogous to PapH termination of pilus growth of Pap CU pili, since deletion of either spaB or papH results in longer pili presumably due to a termination defect [21, 77]. Consistent with a role in pilus termination, C. diphtheriae SpaB was observed at the base of the pilus fiber, but was also observed along the length of the pilus fiber (Figure 1). Thus, it will be important to specifically determine the mechanistic basis underlying differential locations and roles for SpaB in pilus biogenesis, the range of mechanisms by which pilus termination and cell wall attachment can occur in other Gram-positive pilus systems, and the different roles for sortases and minor pilins in pilin polymerization and function.
Localized pilus assembly
Like their Gram-negative counterparts, pilus subunits in Gram-positive bacteria are targeted to the membrane for Sec-mediated translocation by the presence of a Sec signal sequence. Recent studies in a variety of Gram-positive bacteria show that protein translocation can be coordinated at distinct sites on the cell surface. In Bacillus subtilis, SecY and SecA proteins of the general secretory pathway are colocalized at multiple discrete sites, forming a punctate helical pattern along the cell membrane of this rod-shaped bacterium [79]. The ATPase of the Sec machinery, SecA, in the Gram-positive cocci S. pyogenes, Streptococcus mutans, and Enterococcus faecalis is reported to localize to distinct domains leading to the proposal that protein secretion may be spatially restricted [80–82]. This localized secretion domain has been termed the ExPortal in S. pyogenes[51, 81]. However, a separate study reported homogenous SecA localization throughout the S. pyogenes cell membrane, leading to an alternative model for localization in which domains within the N-terminus of secreted proteins themselves, and not the location of the Sec machinery, are proposed to direct the localization of secreted proteins in Gram-positive cocci [83]. A YSIRK domain within the N-terminal secretion signal differentially influenced the sites of several sortase substrates on the cell surface in both S. pyogenes and Staphylococcus aureus, although it is unclear is if this is a consequence of localized secretion, sortase function, or due to another factor [83, 84].
After translocation across the cell membrane, pilus subunits must efficiently interact with their cognate sortase(s) for fiber assembly and cell wall attachment. This process may be facilitated by spatially coordinating secretion and sortase processing, whereby SecA and sortases are co-localize at a single site, as has been observed in S. mutans and E. faecalis[80, 82]. Consistent with this notion, the housekeeping sortase of S. pyogenes localizes to distinct membrane foci that are most often found near the division septum, sites of nascent cell wall synthesis and thus the predicted sites for sortase-mediated attachment of pilus fibers to lipid II precursors [85]. The pilus-associated sortase of E. faecalis also localizes to distinct foci found predominantly at the division plane, and the pilus subunits accumulate in foci in the absence of the pilus sortase suggesting they are secreted and processed at focal membrane domains [82]. Similarly, pilin subunits in C. diphtheriae can be detected in clusters on the cell surface [70, 86]. In S. pneumoniae, pili viewed by immunofluorescence are topologically localized to symmetric foci at the equatorial plane, supporting the hypothesis that pili are assembled and attached to the nascent cell wall in a spatially restricted manner [74]. Interestingly, the focal deposition of pneumococcal pili required the presence of either the SrtB/SrtC-1 or the SrtD/SrtC-3 pilin sortase, but not the SrtC/SrtC-1 pilin sortase, leading to the speculation that SrtB/SrtC-1 and/or SrtD/SrtC-3 may also be found focally localized at sites of pilus biogenesis. Together these reports suggest a localized pilus assembly complex whereby pilin subunits are secreted at focal membrane domains where both pilin and housekeeping sortases are also spatially restricted. Clustering of related pathways in this way may enable newly secreted pilin proteins to quickly come into contact with their cognate sortase for efficient pilus polymerization prior to cell wall attachment and subsequent distribution around the cell surface. A positively charged cytoplasmic amino acid motif has recently been shown to be important for focal localization of the pilin sortase of E. faecalis[82]. Mislocalization of the pilin sortase by mutation of the positively charged motif resulted in a decrease in piliation, supporting the idea that localized sortase action enhances efficient pilus production. It remains to be seen how localized pilus secretion and assembly platforms extends to other Gram-positive organisms, especially the rod-shaped bacilli; how pilus systems utilizing multiple pilin sortases organize their assembly machinery; whether a hierarchy of sortase interaction occurs; and what molecular and cellular factors are involved on mediating localization.
There are no reports examining the localization of CU pilus secretion or assembly sites, although conflicting reports exist regarding whether Sec components display any subcellular organization in Gram-negative organisms [87–89]. It will be interesting to know whether other Gram-negative pilus assembly apparatuses, such as that of the CU pili, localize to discrete sites, and if so, how localized biogenesis occurs in these cells, and how this translates into pilus display around the cell surface. The approaches used and mechanisms unveiled in the study of SA pilus localization may be useful in guiding future investigation of localization in Gram-negative pilus systems.
Concluding remarks
In the past several years, highly resolved molecular and structural details of the well-studied Gram-negative CU pilus system have been reported, important not only for their insights into our understanding of pilus biogenesis, but as a detailed model of protein secretion and translocation across biological membranes. From a practical standpoint, understanding the fine structural details of the CU pilus biogenesis pathway has allowed for the creation of rationally designed compounds which inhibit pilus formation by interfering with chaperone-subunit-usher interactions [90], inhibit biofilm formation in vitro, and attenuate virulence in vivo[7], paving the way for future virulence factor-specific therapeutics. In contrast, SA pili of Gram-positive bacteria have only recently been appreciated, during which time we have seen rapid advancement in our understanding of these ranging from genetics to biochemistry to pathogenesis. The search for housekeeping sortase-inhibitors as potential anti-infective therapies is an active area of research and several candidate inhibitors have been identified [8]. A deeper understanding of pilin sortase structure and function relationships will be important for extending inhibitor searches to pilin sortases. While bacterial pili are thought to function in adherence, the biogenesis pathways of these extracellular fibers are specialized to circumvent inherent physiological differences between Gram-negative and Grampositive bacteria, such as the presence of an oxidizing periplasmic space to a thick cell wall, respectively. It will be of interest to determine which aspects of pilus biogenesis are conserved across pilus types and organisms (Box 1). Consideration of increasingly refined details of well-established CUr pilus systems along with new paradigms emerging from the world of Gram-positive pili will facilitate our understanding of conserved and dedicated aspects of bacterial pilus biogenesis.
Box 1. Outstanding questions CU assembled pili.
CU assembled pili
How does the pilin/chaperone complex dissociate at the usher?
How does the usher facilitate productive DSE reactions and extrusion of polymerized pili to the surface of the bacterium?
What are the molecular details governing the specificity between the different pilin subunits during DSE?
How is the pilus fiber anchored to the cell?
Do Gram-negative bacteria employ spatially localized secretion and assembly strategies for efficient pilus polymerization?
SA pili
How does the putative chaperone protein act to facilitate pilus polymerization?
Do Gram-positive bacteria utilize a dual platform system at the membrane to facilitate secretion, substrate recognition, and assembly?
What mediates the incorporation and location of minor subunits in the pilus fiber?
How do Gram-positive bacteria spatially and temporally coordinate secretion with sorting?
What are the molecular receptors for the pilus adhesins?
General questions
How are multiple pilus operons regulated to efficiently display one particular pilus type at the appropriate time?
What determines the specificity for assembly in organisms with multiple, distinct pili?
Acknowledgments
We thank Ashley Nenninger, Stefan Fälker, and Fabrice Neiers for critical reading of this manuscript and helpful discussion. This work was supported by National Institutes of Health and Office of Research on Women’s Health Specialized Center of Research DK64540, DK51406, AI48689, AI29549, and AI49950 (to S.J.H.); NIH grant AI38273 (M.G.C.); and an AHA Postdoctoral Fellowship 0625736Z (K.A.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fronzes R, et al. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. Embo J. 2008;27:2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 3.Duguid JP, et al. Non-flagellar filamentous appendages (fimbriae) and haemagglutinating activity in Bacterium coli. J Pathol Bacteriol. 1955;70:335–348. doi: 10.1002/path.1700700210. [DOI] [PubMed] [Google Scholar]
- 4.Yanagawa R, et al. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn J Vet Res. 1968;16:31–37. [PubMed] [Google Scholar]
- 5.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria – structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kline KA, et al. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Cegelski L, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 9.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren A, Branden CI. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature. 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 12.Choudhury D, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 13.Pellecchia M, et al. NMR solution structure of the periplasmic chaperone FimC. Nat Struct Biol. 1998;5:885–890. doi: 10.1038/2325. [DOI] [PubMed] [Google Scholar]
- 14.Bann JG, et al. Catalysis of protein folding by chaperones in pathogenic bacteria. Proc Natl Acad Sci USA. 2004;101:17389–17393. doi: 10.1073/pnas.0408072101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung DL, et al. Structural basis of chaperone self-capping in P pilus biogenesis. Proc Natl Acad Sci USA. 1999;96:8178–8183. doi: 10.1073/pnas.96.14.8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer FG, et al. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 17.Dodson KW, et al. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YM, et al. P pilus assembly motif necessary for activation of the CpxRA pathway by PapE in Escherichia coli. J Bacteriol. 2004;186:4326–4337. doi: 10.1128/JB.186.13.4326-4337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YM, et al. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J Bacteriol. 2007;189:5276–5283. doi: 10.1128/JB.01648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saulino ET, et al. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. Embo J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baga M, et al. Biogenesis of E. coli Pap pili: papH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- 22.Verger D, et al. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 2006;7:1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thanassi DG, et al. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng TW, et al. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J Bacteriol. 2004;186:5321–5331. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eidam O, et al. Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD. FEBS Lett. 2008;582:651–655. doi: 10.1016/j.febslet.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama M, et al. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. Embo J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remaut H, et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob-Dubuisson F, et al. Chaperone-assisted self-assembly of pili independent of cellular energy. J Biol Chem. 1994;269:12447–12455. [PubMed] [Google Scholar]
- 29.Puorger C, et al. Infinite kinetic stability against dissociation of supramolecular protein complexes through donor strand complementation. Structure. 2008;16:631–642. doi: 10.1016/j.str.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Sauer FG, et al. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 31.Vitagliano L, et al. A molecular dynamics study of pilus subunits: insights into pilus biogenesis. J Mol Biol. 2007;367:935–941. doi: 10.1016/j.jmb.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 32.Zavialov AV, et al. Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem J. 2005;389:685–694. doi: 10.1042/BJ20050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodson KW, et al. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 34.Hung CS, et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. 2002;44:903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- 35.Buts L, et al. The fimbrial adhesin F17-G of enterotoxigenic Escherichia coli has an immunoglobulin-like lectin domain that binds N-acetylglucosamine. Mol Microbiol. 2003;49:705–715. doi: 10.1046/j.1365-2958.2003.03600.x. [DOI] [PubMed] [Google Scholar]
- 36.Merckel MC, et al. The structural basis of receptor-binding by Escherichia coli associated with diarrhea and septicemia. J Mol Biol. 2003;331:897–905. doi: 10.1016/s0022-2836(03)00841-6. [DOI] [PubMed] [Google Scholar]
- 37.Stromberg N, et al. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Gal alpha 1–4Gal-containing isoreceptors. Embo J. 1990;9:2001–2010. doi: 10.1002/j.1460-2075.1990.tb08328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts JA, et al. The Gal(alpha 1–4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 40.Bahrani-Mougeot FK, et al. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol Microbiol. 2002;45:1079–1093. doi: 10.1046/j.1365-2958.2002.03078.x. [DOI] [PubMed] [Google Scholar]
- 41.Martinez JJ, et al. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. Embo J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu XR, et al. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc Natl Acad Sci USA. 1996;93:9630–9635. doi: 10.1073/pnas.93.18.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen SL, et al. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0902179106. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson GG, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 45.Hendrickx AP, et al. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology. 2008;154:3212–3223. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- 46.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 47.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyon WR, et al. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. Embo J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matias VR, Beveridge TJ. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol. 2005;56:240–251. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 50.Matias VR, Beveridge TJ. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J Bacteriol. 2006;188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- 52.Zuber B, et al. Granular layer in the periplasmic space of Gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J Bacteriol. 2006;188:6652–6660. doi: 10.1128/JB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahner D, Scott JR. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. J Bacteriol. 2008;190:527–535. doi: 10.1128/JB.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakata M, et al. Mode of expression and functional characterization of FCT-3 pilus region encoded proteins in the Streptococcus pyogenes serotype M49. Infect Immun. 2009;77:32–44. doi: 10.1128/IAI.00772-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marraffini LA, et al. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandlik A, et al. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang HJ, et al. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 58.Ton-That H, et al. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 59.Kang HJ, et al. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Telford JL, et al. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 61.Lauer P, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 62.Mora M, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosini R, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 64.Budzik JM, et al. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci USA. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barnett TC, Scott JR. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J Bacteriol. 2002;184:2181–2191. doi: 10.1128/JB.184.8.2181-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quigley BR, et al. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol Microbiol. 2009;72:1379–1394. doi: 10.1111/j.1365-2958.2009.06727.x. [DOI] [PubMed] [Google Scholar]
- 67.Krishnan V, et al. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Budzik JM, et al. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- 69.Dutton RJ, et al. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mandlik A, et al. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson AL, et al. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol Microbiol. 2007;66:329–340. doi: 10.1111/j.1365-2958.2007.05908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edwards AM, et al. Scavenger receptor gp340 aggregates group A streptococci by binding pili. Mol Microbiol. 2008;68:1378–1394. doi: 10.1111/j.1365-2958.2008.06220.x. [DOI] [PubMed] [Google Scholar]
- 73.Nallapareddy SR, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falker S, et al. Sortase-mediated assembly and surface topology of adhesive pneumococcal pili. Mol Microbiol. 2008;70:595–607. doi: 10.1111/j.1365-2958.2008.06396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.LeMieux J, et al. Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J Bacteriol. 2008;190:6002–6013. doi: 10.1128/JB.00379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swaminathan A, et al. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mandlik A, et al. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci USA. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nobbs AH, et al. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun. 2008;76:3550–3560. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campo N, et al. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- 80.Hu P, et al. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol. 2007;53:150–154. doi: 10.1016/j.archoralbio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 81.Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 82.Kline KA, et al. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carlsson F, et al. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- 84.DeDent A, et al. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. Embo J. 2008;27:2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci USA. 2008;105:18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guttilla IK, et al. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in Gram-positive bacteria. J Bacteriol. 2009;191:5603–5612. doi: 10.1128/JB.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brandon LD, et al. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol Microbiol. 2003;50:45–60. doi: 10.1046/j.1365-2958.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- 88.Espeli O, et al. SetB: an integral membrane protein that affects chromosome segregation in Escherichia coli. Mol Microbiol. 2003;50:495–509. doi: 10.1046/j.1365-2958.2003.03736.x. [DOI] [PubMed] [Google Scholar]
- 89.Shiomi D, et al. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol Microbiol. 2006;60:894–906. doi: 10.1111/j.1365-2958.2006.05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pinkner JS, et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc Natl Acad Sci USA. 2006;103:17897–17902. doi: 10.1073/pnas.0606795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barocchi MA, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brinton CC., Jr The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans N Y Acad Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 93.Dramsi S, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 94.Hilleringmann M, et al. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of RrgA. PLoS Pathog. 2008;4:e1000026. doi: 10.1371/journal.ppat.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jones CH, et al. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jones CH, et al. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klemm P, Christiansen G. The fimD gene required for cell surface localization of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1990;220:334–338. doi: 10.1007/BF00260505. [DOI] [PubMed] [Google Scholar]
- 98.Kuehn MJ, et al. Immunoglobulin-like PapD chaperone caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc Natl Acad Sci USA. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.LeMieux J, et al. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun. 2006;74:2453–2456. doi: 10.1128/IAI.74.4.2453-2456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lindberg F, et al. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature. 1987;328:84–87. doi: 10.1038/328084a0. [DOI] [PubMed] [Google Scholar]
- 101.Norgren M, et al. Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol Microbiol. 1987;1:169–178. doi: 10.1111/j.1365-2958.1987.tb00509.x. [DOI] [PubMed] [Google Scholar]


