Abstract
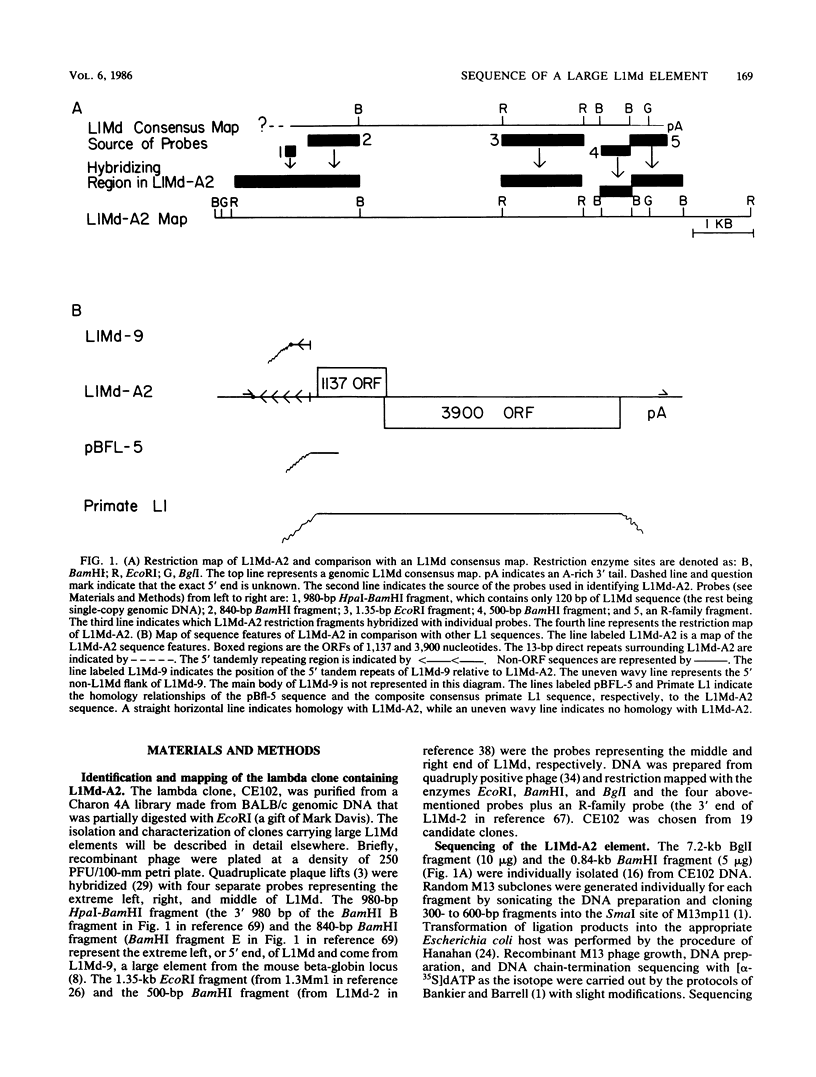
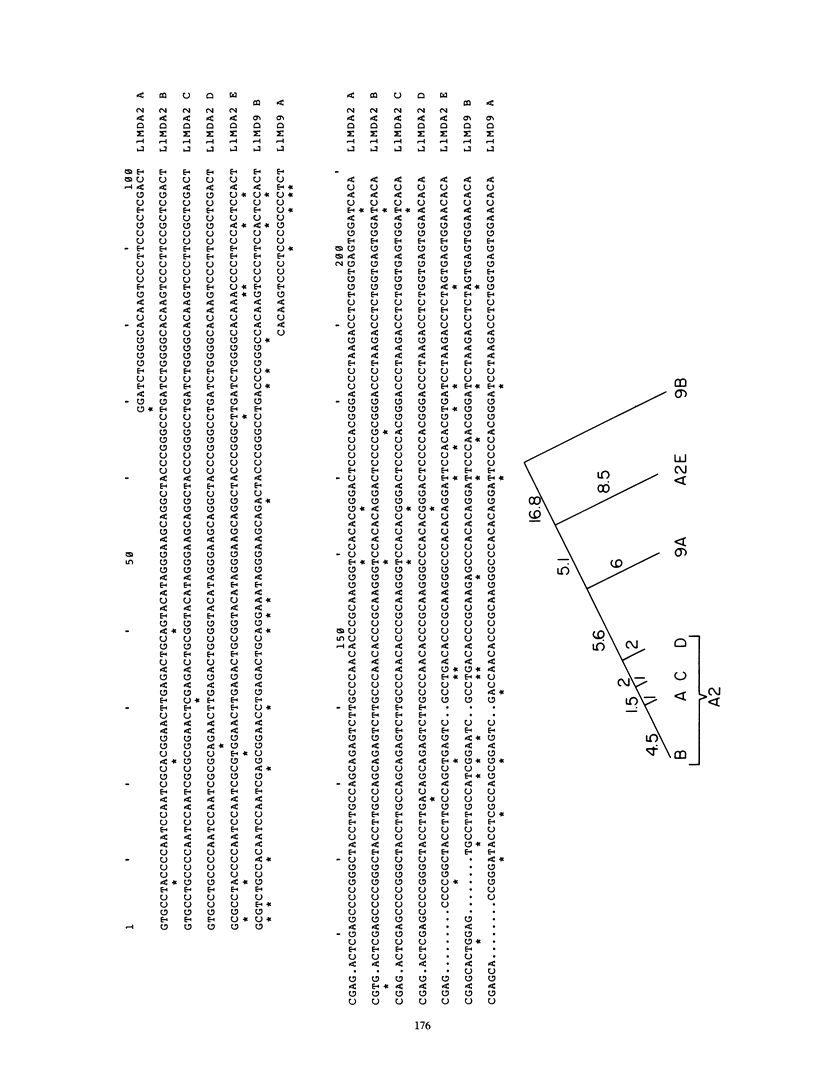
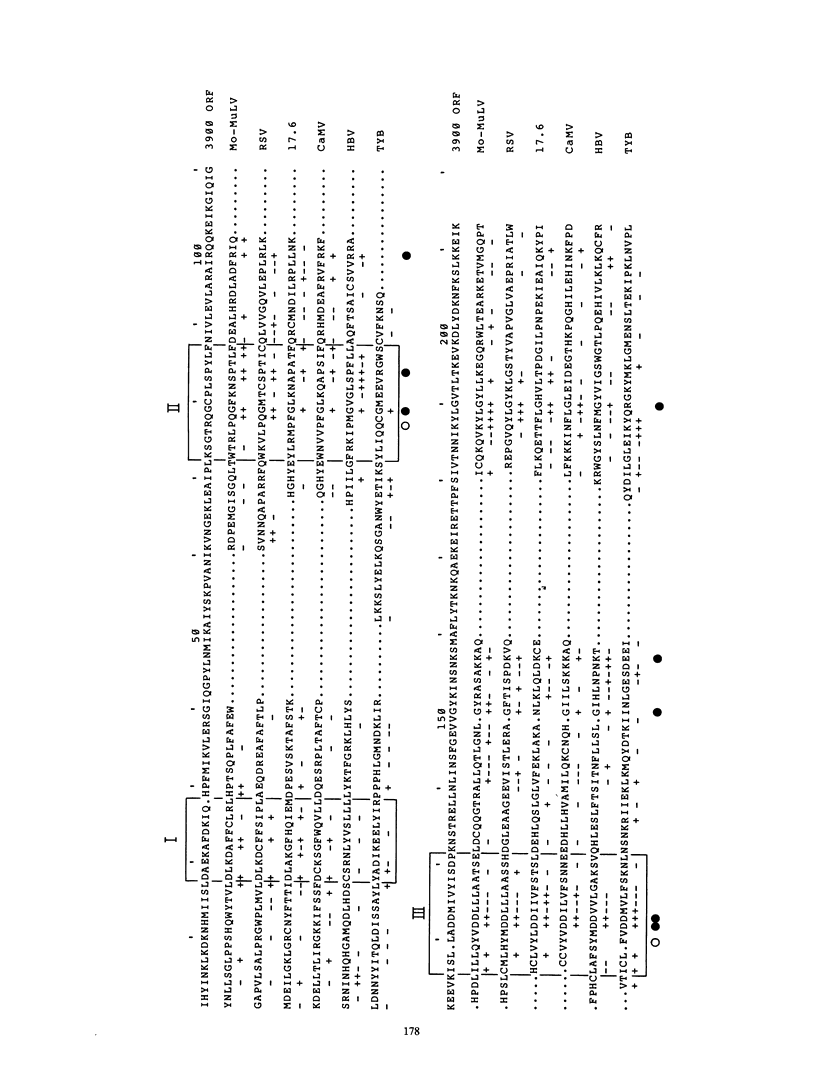
The complete nucleotide sequence of a 6,851-base pair (bp) member of the L1Md repetitive family from a selected random isolate of the BALB/c mouse genome is reported here. Five kilobases of the element contains two overlapping reading frames of 1,137 and 3,900 bp. The entire 3,900-bp frame and the 3' 600 bp of the 1,137-bp frame, when compared with a composite consensus primate L1 sequence, show a ratio of replacement to silent site differences characteristic of protein coding sequences. This more closely defines the protein coding capacity of this repetitive family, which was previously shown to possess a large open reading frame of undetermined extent. The relative organization of the 1,137- and 3,900-bp reading frames, which overlap by 14 bp, bears resemblance to protein-coding, mobile genetic elements. Homology can be found between the amino acid sequence of the 3,900-bp frame and selected domains of several reverse transcriptases. The 5' ends of the two L1Md elements described in this report have multiple copies, 4 2/3 copies and 1 2/3 copy, of a 208-bp direct tandem repeat. The sequence of this 208-bp element differs from the sequence of a previously defined 5' end for an L1Md element, indicating that there are at least two different 5' end motifs for L1Md.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battey J., Max E. E., McBride W. O., Swan D., Leder P. A processed human immunoglobulin epsilon gene has moved to chromosome 9. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5956–5960. doi: 10.1073/pnas.79.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Dover G. Organization and evolutionary progress of a dispersed repetitive family of sequences in widely separated rodent genomes. J Mol Biol. 1981 Aug 25;150(4):441–466. doi: 10.1016/0022-2836(81)90374-0. [DOI] [PubMed] [Google Scholar]
- Brown S. D., Piechaczyk M. Insertion sequences and tandem repetitions as sources of variation in a dispersed repeat family. J Mol Biol. 1983 Apr 5;165(2):249–256. doi: 10.1016/s0022-2836(83)80256-3. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Chao S. F., Hutchison C. A., 3rd, Edgell M. H. Transposition of a long member of the L1 major interspersed DNA family into the mouse beta globin gene locus. Nucleic Acids Res. 1985 Jul 25;13(14):5071–5084. doi: 10.1093/nar/13.14.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. J., Shimada T., Moulton A. D., Harrison M., Nienhuis A. W. Intronless human dihydrofolate reductase genes are derived from processed RNA molecules. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7435–7439. doi: 10.1073/pnas.79.23.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. M., Schildkraut C. L. A family of moderately repetitive sequences in mouse DNA. Nucleic Acids Res. 1980 Sep 25;8(18):4075–4090. doi: 10.1093/nar/8.18.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Digan M. E., Dawid I. B. A family of oligo-adenylate-terminated transposable sequences in Drosophila melanogaster. J Mol Biol. 1983 Aug 25;168(4):715–727. doi: 10.1016/s0022-2836(83)80071-0. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Dush M. K., Sikela J. M., Khan S. A., Tischfield J. A., Stambrook P. J. Nucleotide sequence and organization of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that has a variable intron/exon arrangement. Proc Natl Acad Sci U S A. 1985 May;82(9):2731–2735. doi: 10.1073/pnas.82.9.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Characterization of a highly repetitive family of DNA sequences in the mouse. Nucleic Acids Res. 1982 Aug 25;10(16):5003–5013. doi: 10.1093/nar/10.16.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. C., Howarth A. J., Hahn P., Brown-Luedi M., Shepherd R. J., Messing J. The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 1981 Jun 25;9(12):2871–2888. doi: 10.1093/nar/9.12.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard W., Meitinger T., Höchtl J., Zachau H. G. A new family of interspersed repetitive DNA sequences in the mouse genome. J Mol Biol. 1982 May 25;157(3):453–471. doi: 10.1016/0022-2836(82)90471-5. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Skowronski J., Singer M. F. Defining the beginning and end of KpnI family segments. EMBO J. 1984 Aug;3(8):1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R., Arnheim N. Structure and organization of the highly repeated and interspersed 1.3 kb EcoRI-Bg1II sequence family in mice. Nucleic Acids Res. 1980 Nov 11;8(21):5031–5042. doi: 10.1093/nar/8.21.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis G. F., Hieter P. A., McBride O. W., Swan D., Leder P. Processed genes: a dispersed human immunoglobulin gene bearing evidence of RNA-type processing. Nature. 1982 Mar 25;296(5855):321–325. doi: 10.1038/296321a0. [DOI] [PubMed] [Google Scholar]
- Jackson M., Heller D., Leinwand L. Transcriptional measurements of mouse repeated DNA sequences. Nucleic Acids Res. 1985 May 10;13(9):3389–3403. doi: 10.1093/nar/13.9.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn C. L., Hutchison C. A., 3rd, Phillips S. J., Weaver S., Haigwood N. L., Voliva C. F., Edgell M. H. DNA sequence organization of the beta-globin complex in the BALB/c mouse. Cell. 1980 Aug;21(1):159–168. doi: 10.1016/0092-8674(80)90123-3. [DOI] [PubMed] [Google Scholar]
- Katzir N., Rechavi G., Cohen J. B., Unger T., Simoni F., Segal S., Cohen D., Givol D. "Retroposon" insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1054–1058. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B. K., Dawid I. B. The 1723 element: a long, homogeneous, highly repeated DNA unit interspersed in the genome of Xenopus laevis. J Mol Biol. 1983 Nov 5;170(3):583–596. doi: 10.1016/s0022-2836(83)80122-3. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Biro P. A. Genomic representation of the Hind II 1.9 kb repeated DNA. Nucleic Acids Res. 1982 May 25;10(10):3221–3239. doi: 10.1093/nar/10.10.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Nucleotide sequence definition of a major human repeated DNA, the Hind III 1.9 kb family. Nucleic Acids Res. 1982 May 25;10(10):3211–3219. doi: 10.1093/nar/10.10.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Burton F. H., Edgell M. H., Hutchison C. A., 3rd A large interspersed repeat found in mouse DNA contains a long open reading frame that evolves as if it encodes a protein. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2308–2312. doi: 10.1073/pnas.81.8.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Hardies S. C., Edgell M. H., Hutchison C. A., 3rd Tempo and mode of concerted evolution in the L1 repeat family of mice. Mol Biol Evol. 1985 Mar;2(2):127–140. doi: 10.1093/oxfordjournals.molbev.a040340. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Melton D. W., Konecki D. S., Brennand J., Caskey C. T. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2147–2151. doi: 10.1073/pnas.81.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier-Rotival M., Soriano P., Cuny G., Strauss F., Bernardi G. Sequence organization and genomic distribution of the major family of interspersed repeats of mouse DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):355–359. doi: 10.1073/pnas.79.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T. Rapidly evolving mouse alpha-globin-related pseudo gene and its evolutionary history. Proc Natl Acad Sci U S A. 1981 Jan;78(1):450–453. doi: 10.1073/pnas.78.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Onda H., Sasada R., Igarashi K., Sugino Y., Nishioka K. The complete nucleotide sequences of the cloned hepatitis B virus DNA; subtype adr and adw. Nucleic Acids Res. 1983 Mar 25;11(6):1747–1757. doi: 10.1093/nar/11.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarca R., Haseltine W. A. Sequence similarity among retroviruses--erratum. Nature. 1984 Jun 21;309(5970):728–728. doi: 10.1038/309728b0. [DOI] [PubMed] [Google Scholar]
- Potter S. S. Rearranged sequences of a human Kpn I element. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1012–1016. doi: 10.1073/pnas.81.4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Rinehart F. P., Ritch T. G., Deininger P. L., Schmid C. W. Renaturation rate studies of a single family of interspersed repeated sequences in human deoxyribonucleic acid. Biochemistry. 1981 May 26;20(11):3003–3010. doi: 10.1021/bi00514a003. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Saigo K., Kugimiya W., Matsuo Y., Inouye S., Yoshioka K., Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984 Dec 13;312(5995):659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Brown F. L., Zavodny P. J., Maio J. J. Transcription of the KpnI families of long interspersed DNAs in human cells. Nature. 1983 Jul 21;304(5923):277–280. doi: 10.1038/304277a0. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Singer M. F., Thayer R. E., Grimaldi G., Lerman M. I., Fanning T. G. Homology between the KpnI primate and BamH1 (M1F-1) rodent families of long interspersed repeated sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5739–5745. doi: 10.1093/nar/11.16.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J., Singer M. F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P., Meunier-Rotival M., Bernardi G. The distribution of interspersed repeats is nonuniform and conserved in the mouse and human genomes. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984 Apr;1(3):269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Valerio D., Duyvesteyn M. G., Dekker B. M., Weeda G., Berkvens T. M., van der Voorn L., van Ormondt H., van der Eb A. J. Adenosine deaminase: characterization and expression of a gene with a remarkable promoter. EMBO J. 1985 Feb;4(2):437–443. doi: 10.1002/j.1460-2075.1985.tb03648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva C. F., Jahn C. L., Comer M. B., Hutchison C. A., 3rd, Edgell M. H. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res. 1983 Dec 20;11(24):8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva C. F., Martin S. L., Hutchison C. A., 3rd, Edgell M. H. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984 Oct 5;178(4):795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. Potentiator effect of the SV40 72-bp repeat on initiation of transcription from heterologous promoter elements. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):921–934. doi: 10.1101/sqb.1983.047.01.106. [DOI] [PubMed] [Google Scholar]
- Weaver S., Comer M. B., Jahn C. L., Hutchison C. A., 3rd, Edgell M. H. The adult beta-globin genes of the "single" type mouse C57BL. Cell. 1981 May;24(2):403–411. doi: 10.1016/0092-8674(81)90330-5. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wilde C. D., Crowther C. E., Cripe T. P., Gwo-Shu Lee M., Cowan N. J. Evidence that a human beta-tubulin pseudogene is derived from its corresponding mRNA. Nature. 1982 May 6;297(5861):83–84. doi: 10.1038/297083a0. [DOI] [PubMed] [Google Scholar]
- Wilson R., Storb U. Association of two different repetitive DNA elements near immunoglobulin light chain genes. Nucleic Acids Res. 1983 Mar 25;11(6):1803–1817. doi: 10.1093/nar/11.6.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witney F. R., Furano A. V. Highly repeated DNA families in the rat. J Biol Chem. 1984 Aug 25;259(16):10481–10492. [PubMed] [Google Scholar]
- Yang J. K., Masters J. N., Attardi G. Human dihydrofolate reductase gene organization. Extensive conservation of the G + C-rich 5' non-coding sequence and strong intron size divergence from homologous mammalian genes. J Mol Biol. 1984 Jun 25;176(2):169–187. doi: 10.1016/0022-2836(84)90419-4. [DOI] [PubMed] [Google Scholar]



