Abstract
Dipeptidyl peptidase-like proteins (DPLs) and Kv-channel interacting proteins (KChIPs) join Kv4 pore-forming subunits to form multi-protein complexes that underlie subthreshold A-type currents (ISA) in neuronal somatodendritic compartments. Here, we characterize the functional effects and brain distributions of N-terminal variants belonging to the DPL dipeptidyl peptidase 10 (DPP10). In the Kv4.2+KChIP3+DPP10 channel complex, all DPP10 variants accelerate channel gating kinetics; however, the splice variant DPP10a produces uniquely fast inactivation kinetics that accelerates with increasing depolarization. This DPP10a-specific inactivation dominates in co-expression studies with KChIP4a and other DPP10 isoforms. Real-time qRT-PCR and in situ hybridization analyses reveal differential expression of DPP10 variants in rat brain. DPP10a transcripts are prominently expressed in the cortex, whereas DPP10c and DPP10d mRNAs exhibit more diffuse distributions. Our results suggest that DPP10a underlies rapid inactivation of cortical ISA, and the regulation of isoform expression may contribute to the variable inactivation properties of ISA across different brain regions.
INTRODUCTION
Subthreshold transient K+ current (ISA) influences neuronal excitability and firing properties. The loss or reduction of ISA may contribute to increased excitability and susceptibility to induction and expression of epileptiform activity (Castro et al., 2001; Bernard et al., 2004). Localized to the somatodendritic compartments, ISA activity is also responsible for coordinating synaptic signal integration, controlling backpropagating action potentials, and regulating long-term potentiation (Hoffman et al., 1997; Schoppa and Westbrook, 1999; Johnston et al., 2000; Watanabe et al., 2002).
The physiological function of ISA depends on its unique kinetic properties, including rapid inactivation, rapid activation and inactivation in the subthreshold range of membrane potentials, and rapid recovery from inactivation at hyperpolarized potentials. These properties can vary significantly between ISA of different neuronal populations (reviewed by Jerng et al., 2004b). For example, at +50 mV ISA of hippocampal CA1 and CA3 pyramidal neurons inactivate with a time constant of 26-34 ms, whereas ISA of cortical pyramidal neurons inactivate with a time constant of 7-8 ms (Hoffman et al., 1997; Martina et al., 1998; Lien et al., 2002; Klee et al., 1995; Banks et al. 1996; Bekkers 2000; Korngreen and Sakmann, 2000). Moreover, in hippocampal CA1 and CA3 pyramidal neurons but not in cortical pyramidal neurons, ISA displays an unusual voltage dependence where the inactivation kinetics slow with increasing depolarization. Possible explanations for the variability of ISA properties include differences in 1) protein composition of the ISA channel, 2) enzymatic modifications of the proteins involved, and 3) intracellular environment.
The majority of neuronal I is mediated by the Kv4 subfamily of voltage-dependent K+ SA channels (Johns et al., 1997; Shibata et al., 2000; Malin and Nerbonne, 2000). Recent studies in heterologous expression systems have suggested that the combined effects of Kv4 modulatory β-subunits, K channel-interacting proteins (KChIPs) and dipeptidyl peptidase-like (DPL) proteins, are required to produce functional A-type currents from Kv4 ternary channel complexes that resemble native neuronal ISA (Nadal et al., 2003; Jerng et al., 2005). DPLs include the genetically-related dipeptidyl peptidase 6 (DPP6; also known as BSPL, DPL1, and DPPX) and dipeptidyl peptidase 10 (DPP10; also known as KIAA1492, DPRP3, DPL2, and DPPY) (Wada et al., 1992; de Lecea et al., 1994; Nagase et al., 2000; Chen et al., 2003; Qi et al., 2003; Nadal et al., 2003; Jerng et al., 2004a; Zagha et al., 2005; Ren et al., 2005). When expressed alone, Kv4 channels are transported poorly to the cell surface and express kinetic properties very different from those of native ISA channels (Shibata et al., 2003; reviewed by Jerng et al., 2004b). Co-expression with KChIPs allows Kv4 channels to reach the cell surface efficiently as well as reconstituting some of the kinetic properties of native ISA, such as rapid recovery from inactivation; however, the channels still behave differently from native ISA channels (An et al., 2000; Nadal et al., 2001). Only by the creation of ternary complexes between Kv4s, KChIPs and DPLs can other characteristic ISA functional properties such as rapid activation and rapid inactivation be reconstituted (Nadal et al., 2003; Jerng et al., 2005). The physiological significance of these observations are supported by gene knock-out, dominant negative, RNA interference, and co-immunoprecipitation studies that suggest that these three proteins are all part of the native channel complex responsible for producing ISA (Malin and Nerbonne, 2000; Nadal et al., 2003; Jerng et al., 2005; Hu et al., 2006; Lauver et al., 2006).
Although it is now clear that the Kv4, KChIP, and DPL subunits are all important components of native ISA channels, the mechanisms responsible for producing functional diversity in these channels between different brain regions is not known. To address this question, we examined the interplay between different KChIP and DPL in generating A-type currents with unique functional properties. We characterized the distribution of a number of alternative splice isoforms of DPP10 that are expressed in rat brain. Functional reconstitution studies in heterologous expression systems showed that the inactivation properties of reconstituted ISA channels depend upon the DPP10 isoform that is expressed. In summary, our studies suggest that differential inactivation properties produced by DPP10 alternative splice variants are an important determinant of the native ISA current functional variations between different neuronal populations. Some of the results presented here have appeared in abstract form (Jerng and Pfaffinger, 2006)..
RESULTS
Identification of DPP10 Exon 1 Variants
EST sequence databases, and an analysis of DPP10 transcripts in a study on asthma, suggest that DPP10 transcripts may express different 1st exons due to initiation from a variety of start sites within the DPP10 gene (Allen et al., 2003; Ren et al., 2005; Takimoto et al., 2006). To determine the variety of DPP10 transcripts made in rat brain, we performed rapid amplification of 5′ complementary DNA ends (5′ RACE) on mRNA isolated from rat brain and cloned the PCR products for DNA sequencing. Screening of colonies identified three alternative 1st exons named Exons 1a, 1c and 1d based on their order in the genomic map (Fig. 1A). (1a: 29/34; 1c: 1/34; 1d: 4/34). ESTs containing homologous sequences matching these alternative first exons have been described for DPP10 transcripts from other species, and we have identified the same three exons in a variety of mammalian genomic DPP10 sequences. In humans, ESTs identify another likely first exon variant positioned between Exons 1a and 1c, and thus called Exon 1b (hDPP10b: Fig. 1B). The 1b exon is conserved in the chimpanzee genome, but we did not detect this exon by 5′ RACE and were unable to identify a homologous sequence in the rat genome. To search for rarer Exon 1 variants, 5′ RACE was performed on cerebellar mRNAs, and RACE ligations were digested with Eco109i to selectively cut the most common 1a variant. However, no additional 5′ RACE products were isolated for DPP10. Of 12 clones examined, all were identified as the 1d variant. In contrast to our findings, 5′ race on human tissues by Allen et al. (2003) identified homologues to our 1a and 1d variants along with several 1st exon variants that did not contain an initiator AUG and therefore would produce a cytoplasmic DPP10 protein lacking a transmembrane domain. Our results suggest that such transcripts, if they exist at all in rat brain, are likely very rare.
Figure 1. Exon organization of the rat DPP10 gene.
(A) Exon structure within the DPP10 gene on rat chromosome 13. The 26 exons of DPP10 are shown schematically and to scale. Exons 1a, 1c, and 1d encode the variable cytoplasmic domain for DPP10a, DPP10c, and DPP10d. (B) Alignment of amino acid sequences for the variable domains of rat (r) and human (h) DPP10 variants. Exon 2 sequence is outlined in black. Sequences identical between the rat and human sequences are represented by dashes, and conserved residues are highlighted in grey. Grey-colored residues in exon 2 are part of the putative transmembrane domain. (C) Sequence alignment between rat and human DPP10 for the non-variable portion. Odd- and even-numbered exons show alternating black outlines, and grey highlights indicate residues conserved between rat and human DPP10. Asterisks represent the active site residues.
We next characterized the gene structure for the rat DPP10 gene using the genomic sequences made available by the National Center for Biotechnology Information (NCBI). The DPP10 gene spans approximately 1.77 million base pairs (1768716 bps) on rat chromosome 13 (Fig. 1A). The multiple Exon 1’s are scattered across approximately 0.93 million base pairs (932,696 bps). Exons 2 to 26, cover 34% of the gene’s total length, or approximately 0.7 million base pairs (674,401 bps). The overall DPP10 genomic structure and relative positions of Exons 1a, 1c and 1d are conserved among different species (see also Allen et al. (2003)).
Incorporation of Exons 1a, 1c or 1d into transcripts leads to the formation of distinct DPP10 peptides (DPP10a, DPP10c and DPP10d) with variable cytoplasmic N-termini. Exon 1a encodes a 20 amino acid peptide that is highly conserved from rodents to humans: its amino acid sequence is identical in all species examined except for a conservative substitution in the canine sequence (isoleucine 19 to valine). Exon 1c encodes a 24 amino acid peptide that is also highly conserved, but less so than Exon 1a, with 20/24 identical residues between human and rat (Fig. 1B). Exon 1d encodes a conserved 13 amino acid peptide with 12/13 residues identical between the rat and human sequences (Fig. 1B). The 776 amino acid residues encoded in DPP10 Exons 2-26 are also highly conserved from human to fish (Table 1). For example, in this region between rat and human, there are only 81 amino acid differences, resulting in ~90% sequence identity (Fig. 1C; Table 1). Two of the non-conserved substitutions between rat and human DPP10 may be traced to single nucleotide polymorphisms (rat to human: A88 to M88, and P340 to A340).
Table 1. Similarity and divergence between the non-variable portions of DPP10 amino acid sequences from various species.
The table is generated by the AlignX function of VectorNTI suite (Invitrogen Corporation, Carlsbad, CA). Similarity and divergence are represented as the percentage of corresponding residues that are identical and non-identical, respectively, between sequence pairs. The percentage of conserved residues therefore equals to 100 minus the similarity and divergence values.
| Similarity | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| DPP10 | human | cow | dog | rat | mouse | chicken | frog | fugu | |
| Divergence | human | 96 | 96 | 89 | 89 | 86 | 77 | 64 | |
| cow | 2 | 98 | 90 | 89 | 87 | 77 | 64 | ||
| dog | 2 | 2 | 90 | 89 | 87 | 77 | 65 | ||
| rat | 7 | 7 | 7 | 96 | 82 | 75 | 62 | ||
| mouse | 7 | 7 | 7 | 2 | 82 | 74 | 62 | ||
| chicken | 8 | 8 | 8 | 10 | 10 | 79 | 65 | ||
| frog | 14 | 14 | 14 | 14 | 15 | 12 | 64 | ||
| fugu | 17 | 18 | 17 | 18 | 18 | 16 | 16 | ||
In the Kv4.2+KChIP3+DPP10 Ternary Complex, DPP10a Variant Alone Confers Inactivation with Distinctly Rapid Kinetics and Voltage Dependence
In brain, K+ channels that underlie the majority of neuronal ISA are protein complexes containing Kv4 α-subunits and both DPL and KChIP auxiliary subunits (Jerng et al., 2005). Thus, it is physiologically compelling to investigate the functional effects of different DPP10 splicing variants in the ternary complex background. We micro-injected Kv4.2 and KChIP3 cRNAs with DPP10a, DPP10c, or DPP10d cRNAs in the same oocyte and examined the expressed currents using two-electrode voltage clamp. A molar ratio of 0.25:1:1 for Kv4.2, KChIP3, and DPP10 variant cRNAs was used to maximize the surface expression of ternary complex channels. Co-expression of Kv4.2 and KChIP3 alone yields macroscopic currents with kinetic and steady-state properties as previously described (Jerng et al., 2005). KChIP3 slows the inactivation kinetics of Kv4.2 channels at positive membrane potentials, shifts the voltage dependence of activation in hyperpolarizing direction, produces a slight rightward shift in the voltage dependence of steady-state inactivation, and dramatically accelerate recovery from inactivation (Table 2).
Table 2.
Kinetic and voltage-dependent properties of currents mediated by Kv4.2 channel complexes.
| Conductance-voltage relationship | Steady-state inactivation | Half-inactivation time, t0.5 | Recovery from inactivation, τ at −100 mV (ms) |
||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| V0.5 (mV) | k (mV/e-fold) | V0.5 (mV) | k (mV/e-fold) | at −40 mV (ms) | at +50 mV (ms) | ||
| Kv4.2 + KChIP3 | −17.4 ± 0.9 (n = 10) | 20.4 ± 0.8 (n = 10) | −61.5 ± 0.5 (n = 22) | 3.6 ± 0.1 (n = 22) | 136 ± 7.6 (n = 8) | 107 ± 2.8 (n = 8) | 72 ± 2 (n = 14) |
| Kv4.2 ± KChIP3 + DPP10a | −25.7 ± 1.2 (n = 6)* | 19.8 ± 1.0 (n = 6) | −67.7 ± 1.1 (n = 6)* | 3.8 ± 0.8 (n = 6) | 27 ± 2.8 (n = 6)* | 10 ± 1.1 (n = 6)* | 28 ± 3.5 (n = 7)* |
| Kv4.2 + KChIP3 + DPP10c | −19.9 ± 1.1 (n = 4)# | 18.3 ± 1.0 (n = 4) | −62.1 ± 0.7 (n = 5)# | 4.8 ± 1.5 (n = 4)* | 30 ± 0.8 (n = 4)* | 40 ± 1.5 (n = 4)*# | 20 ± 6.0 (n = 5)* |
| Kv4.2 + KChIP3 + DPP10d | −23.9 ± 4.5 (n = 3)* | 18.4 ± 0.6 (n = 3) | −64.0 ± 1.4 (n = 3) | 4.3 ± 0.1 (n = 3)* | 35 ± 2.1 (n = 3)* | 42 ± 5.8 (n = 3)*# | 23.5 ± 4.9 (n = 4)* |
| ΔN2-40 | −14.4 ± 0.4 (n = 9) | 18.2 ± 0.3 (n = 9) | −61.7 ± 0.5 (n = 8) | 5.4 ± 0.5 (n = 8) | 221 ± 23 (n = 7) | 167 ± 3.8 (n = 7) | 167 ± 19 (n = 5) |
| ΔN2-40 + DPP10a | −24.9 ± 1.3 (n = 7)* | 20.4 ± 0.8 (n = 7)* | −70.2 ± 0.7 (n = 5)* | 4.1 ± 0.1 (n = 5) | 81 ± 20 (n = 7)* | 10 ± 0.9 (n = 7)* | 42 ± 0.5 (n = 4)* |
| ΔN2-40 + DPP10c | −34.3 ± 1.1 (n = 5)*# | 12.9 ± 0.6 (n = 5)*# | −73.0 ± 1.2 (n = 5)* | 4.0 ± 0.1 (n = 5) | 65 ± 2.8 (n = 4)* | 93 ± 1.7 (n = 4)*# | 50 ± 1.1 (n = 5)*# |
| ΔN2-40 + DPP10d | −30.4 ± 2.2 (n = 3)* | 17.1 ± 0.4 (n = 3)# | −71.4 ± 0.6 (n = 3)* | 4.4 ± 0.1 (n = 3) | 81+ 2.5 (n = 3)* | 111 ± 4.8 (n = 3)*# | 60 ± 6.8 (n = 4)*# |
| Kv4.2 | 0.5 ± 0.7 (n = 16) | 21.0 ± 0.5 (n = 16) | −64.1 ± 0.4 (n = 22) | 4.6 ± 0.2 (n = 22) | 293 ± 29 (n = 9) | 30.5 ± 1.0 (n =13) | 238 ± 7 (n = 21) |
| Kv4.2 + DPP10a | −26.9 ± 2.1 (n = 7)* | 19.7 ± 0.9 ( n = 7) | −74.1 ± 0.5 (n = 11)* | 4.7 ± 0.2 (n = 11) | 40 ± 4.2 (n = 8)* | 9 ± 0.9 (n = 8)* | 79 ± 4 (n = 9)* |
| Kv4.2 + DPP10c | −29.1 ± 1.0 (n = 5)* | 12.6 ± 3.3 (n = 5)*# | −71.0 ± 1.2 (n = 5)*# | 3.9 ± 0.0 (n = 5)*# | 48 ± 4.6 (n = 5)* | 16 ± 1.0 (n = 5)*# | 68 ± 3 (n = 6)* |
| Kv4.2 + DPP10d | −30.7 ± 1.7 (n = 3)* | 15.6 ± 0.5 (n = 3)*# | −72.9 ± 0.9 (n = 5)* | 4.4 ± 0.2 (n = 5) | 39 ± 2.3 (n = 3)* | 15 ± 0.2 (n = 3)*# | 76 ± 7 (n = 4)* |
| Kv4.2 + KChIP4a | 11.4n ± 0.7 (n = 5) | 22.9 ± 0.7 (n = 5) | −62.1 ± 0.8 (n = 3) | 3.9 ± 0.1 (n = 3) | 597 ± 32 (n = 6) | 466 ± 70 (n = 6) | 242 ± 14 (n = 5) |
| Kv4.2 + KChIP4a + DPP10a | −10.5 ± 1.1 (n = 5)* | 21.5 ± 0.3 (n = 5) | −72.7 ± 0.6 (n = 4)* | 4.1 ± 0.1 (n = 4) | 64 ± 8.8 (n = 4)* | 13 ± 1.4 (n = 4)* | 91 ± 3.9 (n = 4)* |
| Kv4.2 + KChIP4a + DPP6-S | −18.0 ± 0.9 (n = 3)*# | 24.2 ± 0.2 (n = 3)# | −68.2 (n = 2) | 4.5 (n = 2) | 176 ± 24 (n = 4)*# | 183 ± 32 (n = 3)*# | 119 ± 10 (n = 4)*# |
| ΔN2-40 + DPP10a/HA* | −26.9 ± 1.1 (n = 3) | 15.1 ± 0.5 (n = 3) | −71.2 ± 2.0 (n = 3) | 4.2 ± 0.1 (n = 3) | 38 ± 4.2 (n = 3) | 11 ± 0.4 (n = 3) | 41 ± 0.3 (n = 3) |
| ΔN2-40 + DPP10a/HA* + DPP10c/cMyc* (1:1) |
−29.5 ± 1.0 (n = 4) | 13.2 ± 0.3 (n = 4)# | −70.7 ± 0.3 (n = 3) | 3.8 ± 0.1 (n = 3)# | 45 ± 2.5 (n = 4) | 17 ± 1.2 (n = 4)# | 46 ± 4.2 (n = 4) |
| ΔN2-40 + DPP10a/HA* + DPP10c/cMyc* (1:3) |
−31.8 ± 1.4 (n = 4)# | 13.5 ± 0.4 (n = 4)# | −71.8 ± 0.9 (n = 3) | 3.7 ± 0.0 (n = 3)# | 47 ± 1.3 (n = 4) | 42 ± 1.2 (n = 4)# | 44 ± 2.8 (n = 4) |
| ΔN2-40 + DPP10c/cMyc* | −32.7 ± 3.0 (n = 3) | 14.1 ± 0.5 (n = 3) | −70.7 ± 0.9 (n = 3) | 3.7 ± 0.1 (n = 3)# | 55 ± 1.8 (n = 3) | 74 ± 3.7 (n = 3)# | 43 ± 3.9 (n = 3) |
Data were analyzed as described in Materials & Methods and presented as mean ± SEM.
(in data) = p < 0.05 when compared to respective background channel (i.e. Kv4.2, Kv4.2+KChIP3, ΔN2-40, Kv4.2+KChIP4a).
= p < 0.05 when compared to background channels with DPP10a or DPP10a/HA*. Two-tailed (independent) t-test was used for statistical comparison.
During channel inactivation, Kv4-mediated currents typically decay with a complex time course best described by sums of multi-exponential terms (Jerng et al., 1997; Böhring et al., 2001b; Beck et al., 2003). Complicating the matter further, the number of exponential terms varies depending on voltage and presence of auxiliary subunits. To simplify comparisons between channels with different subunit compositions, we index net changes in inactivation kinetics by measuring changes in the time at which half of the peak current is inactivated (half-inactivation time), which is independent of the number of exponentials required for a particular fitting. This model-independent method of describing inactivation kinetics has been previously used in Nadal et al. (2003) and Jerng et al. (2005) to compare these complex Kv4 current waveforms. The voltage dependence of the overall inactivation kinetics of Kv4.2+KChIP3 currents was studied by analysing the relationship between the half-inactivation time (t0.5) and membrane potential. As Fig. 2D shows, the t0.5 value notably decreases from −40 to −20 mV and then increases from 0 to 60 mV, illustrating a “U-shaped” voltage dependence. The slowing of inactivation kinetics with increasing depolarization is reminiscent of inactivation of ISA from hippocampal CA1 and CA3 pyramidal neurons.
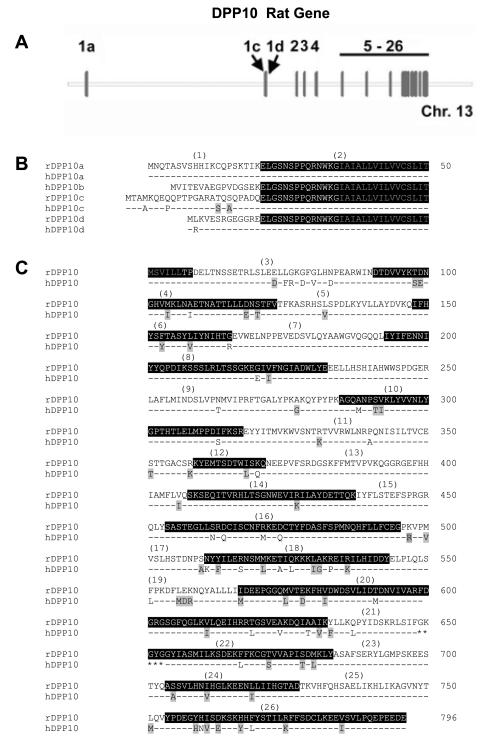
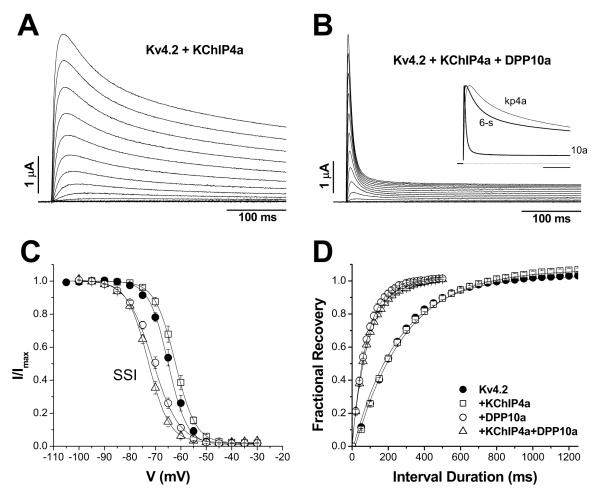
Figure 2. Incorporation of KChIP3 into Kv4.2 channel complex introduces differential voltage-dependence of inactivation, depending on the DPP10 variant present.
(A) DPP10a, but not DPP10c or DPP10d, introduces a distinct rapid inactivation to Kv4.2-mediated current at more depolarized potentials. Current traces are evoked by voltage-clamp protocol identical that used in Fig. 5A. Symbols are again assigned based on the DPP10 variant present. (B) Normalized and superimposed traces at −40 mV for 800 ms. (C) Normalized and superimposed traces at +60 mV for 800 ms. (D) Plot of half-inactivation time against voltage. With currents mediated by Kv4.2 plus KChIP3 channels, DPP10a confers inactivation that accelerates with depolarization, but not DPP10c or DPP10d. (E) Conductance-voltage relationship (g-V) and steady-state inactivation (SSI) of Kv4.2+KChIP3 with and without DPP10 variants. Fitting curves represent Boltzmann functions. Protocols are identical to those described in Fig. 5E.
As representative traces of Figure 2A show, the introduction of DPP10 variants to the Kv4.2+KChIP3 channel does not increase the peak current amplitude, but rather alters the gating kinetics of the channel. All DPP10 variants produce a dramatic acceleration of channel activation, but the primary difference between the splice variants is their effects on channel inactivation (Fig. 2B). While all DPP10 variants accelerate the apparent decay of Kv4.2+KChIP3 currents, at +60 mV DPP10a-containing channels inactivate dramatically faster than those with DPP10c or DPP10d (Fig. 2B). Curiously, at lower voltages, the overall inactivation associated with DPP10a co-expression is slower than those of DPP10c or DPP10d (Fig. 2C). We characterized the t0.5 over a wide voltage range and found that inclusion of DPP10a produces channels that accelerate inactivation with increasing depolarization, compared to channels containing DPP10c or DPP10d that slow inactivation with increasing depolarization, similar to Kv4.2+KChIP3 channels (Fig. 2D). A similar phenomenon was observed previously when comparing inactivation of ternary complex channels containing DPP10a with those containing DPP6-S and DPP6-L (Jerng et al., 2005).
To assess other biophysical effects of DPP10 variants on the Kv4.2+KChIP3+DPP10 ternary complex, we also characterized the voltage dependence of relative peak conductance, steady-state inactivation, and recovery from inactivation and found only minor or no significant differences between variants (Fig. 2E; Table 2). We therefore conclude that, although DPP10c and DPP10d accelerate channel activation and inactivation gating while preserving the U-shaped voltage dependence, DPP10a introduces a dominant inactivation pathway that overrides the unusual dependence of inactivation on voltage.
The N-terminal Domain of DPP10a, But Not Those of DPP10c or DPP10d, Mediates Rapid Inactivation at Depolarized Voltages in the Absence of Kv4.2 N-type Inactivation
The Kv4.2 N-terminus contains an inactivation particle that produces fast inactivation by blocking open channels (Gebauer et al., 2004). When co-expressed with KChIPs, the Kv4 N-terminus is sequestered by KChIP binding, eliminating the “ball-and-chain” (N-type) inactivation and revealing an underlying process with a U-shaped voltage dependence. The effect of DPP10a on inactivation of Kv4.2+KChIP3 channels is functionally similar to what would be seen if channels were reverting back to the an N-type inactivation. We were, therefore, interested in determining if the unique effects of DPP10a are produced by providing an alternative inactivation process that also override the U-shaped voltage dependence of inactivation or by de-repression produced by releasing the Kv4.2 N-terminus from its inactive KChIP-bound condition.
To test whether DPP10a introduces its own inactivation process, rather than de-repression of Kv4.2 N-type inactivation, we removed Kv4.2 N-type inactivation by N-terminal truncation (ΔN2-40). This construct provides a simpler expression environment to test the differential effects of DPP10a as compared to DPP10c and DPP10d, independent of KChIP binding. In our study of ΔN2-40 expressed in Xenopus oocytes, we have determined that deleting the Kv4.2 N-terminal domain is qualitatively similar to co-expressing Kv4.2 with KChIP3: both conditions 1) increase Kv4.2 surface expression (ΔN2-40: ~ 14-fold; +KChIP3: ~ 12-fold), 2) slow Kv4.2 inactivation kinetics at positive potentials (Table 2), 3) confer inactivation kinetics that slow with depolarization (Fig. 2D and Fig. 3D), 4) significantly leftward shift the peak conductance-voltage (g-V) curve and moderately rightward shift the steady-state inactivation curve (Fig. 3E and 3F; Table 2), and 6) accelerate recovery from inactivation (Table 2). One significant difference that we have noted is that N-terminal deletion increases time to peak current, whereas KChIP3 co-expression does not (at +50 mV: Kv4.2 = 8.5 ± 0.5 ms (n = 12); Kv4.2+KChIP3 = 9.5 ± 0.4 ms (n = 5); ΔN2-40 = 19.9 ± 2.6 ms (n = 7)).
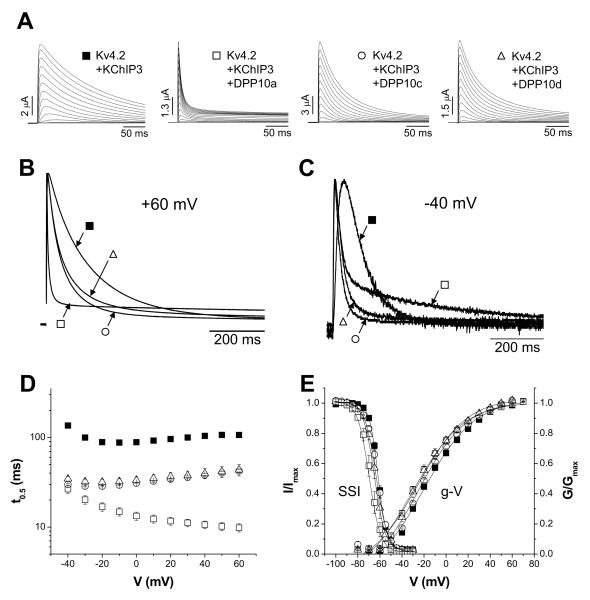
Figure 3. In the absence of Kv4.2 N-terminal domain, voltage-dependence of inactivation is also determined by the DPP10 variant present.
The N-terminal domain (residues 2-40) of Kv4.2 was genetically removed to eliminate the endogenous fast inactivation. (A) Outward currents from oocytes expressing ΔN2-40, ΔN2-40 plus DPP10a, ΔN2-40 plus DPP10c, and ΔN2-40 plus DPP10d were measured by depolarizing steps of 1 sec duration. Note that the scale bars for current amplitudes differ, indicating that all DPP10 variants increase the current expression of ΔN2-40 channels. (B) Overlapped 800-ms-long traces at +50 mV for ΔN2-40 with and without DPP10a, DPP10c, and DPP10d. (C) Normalize and overlapped traces at +50 mV for 500 ms. Note that, in both panels B and C, acceleration of inactivation by DPP10a does not require the Kv4.2 N-terminal domain. (D) Half-inactivation time as a function of voltage. The results are similar to those in Fig. 6D. (E) Measurement of channel activation by plotting the peak conductance-voltage relationship. Co-expression of DPP10 variants results in differential leftward shifts in the conductance-voltage relationship (g-V). (F) Voltage-dependence of steady-state inactivation for Kv4.2, ΔN2-40, and ΔN2-40 plus DPP10 variants. Both g-V and SSI were fitted with Boltzmann functions. Note that the relative order of leftward shifts observed in g-V curves (DPP10c > DPP10d > DPP10a) is preserved with the steady-state inactivation curves. (F) Recovery from inactivation associated with DPP10 variants. Fitting traces represent single exponential functions.
We next compared the effects of co-expressing DPP10 variants with ΔN2-40 to their effects when co-expressed into the ternary complex with Kv4.2+KChIP3. Our results show that the differential inactivation properties associated with DPP10 variants in ternary complexes are reproduced by co-expression of DPP10 variants with ΔN2-40. While all three DPP10 variants accelerate inactivation kinetics, the acceleration produced by DPP10a is dramatically faster than those of DPP10c or DPP10d. When DPP10a is co-expressed with ΔN2-40, the Kv4.2 current undergoes very rapid activation and inactivation similar to the current observed when wild-type Kv4.2 is co-expressed with KChIP3 plus DPP10a (compare Fig. 2A and 3A). In contrast, the currents expressed by ΔN2-40 + DPP10c or ΔN2-40 + DPP10d do not produce the same dramatic early faster inactivation and slower later inactivation (Fig. 3B). Furthermore, the voltage dependent effects on the inactivation kinetics are reproduced by expression with ΔN2-40. Similar to Kv4.2+KChIP3+DPP10a channels, inactivation of ΔN2-40 + DPP10a channels at −40 mV is overall slower than ΔN2-40 + DPP10c or ΔN2-40 + DPP10d (Fig. 3C). Interestingly, the effects of DPP10c and DPP10d on ΔN2-40 inactivation kinetics are not identical: the ΔN2-40 + DPP10c current inactivate significantly faster than ΔN2-40 + DPP10d current at both −40 mV and +50 mV (Figs. 3B and 3C). Although the effect is smaller, a similar trend is evident when these variants are co-expressed with Kv4.2 and KChIP3 (compare Fig. 2 and 3). The association of all DPP10 variants with ΔN2-40 accelerate recovery from inactivation to approximately the same degree (Fig. 3G; Table 2), but DPP10c causes 5 mV more leftward shift in the V0.5 of steady-state inactivation than DPP10a or DPP10d (Fig. 3F; Table 2). DPP10c also produces a ~5 mV leftward shift in the g-V curve over that of DPP10d and > −10 mV leftward shift over that of DPP10a (Fig. 3E; Table 2). These subtle differences suggest that the N-terminus of DPP10c may also have other functional interactions with the Kv4.2 channels that are different from those produced by DPP10a or DPP10d.
Because of the voltage dependence of inactivation depends upon the DPP10 variant expressed, DPP10c mediates the fastest inactivation at −40 mV while DPP10a mediates the fastest inactivation at +50 mV. To examine the voltage-dependence of inactivation more closely, we measured the half inactivation time (t0.5) at various voltages (Fig. 3D). As the figure shows, ΔN2-40 inactivation exhibits a U-shaped voltage dependence with t0.5 reaching minimum at ~ −15 mV where, according to the g-V relationship, approximately half of the channel population is activated. ΔN2-40 associated with DPP10c and DPP10d also show U-shaped voltage-dependence of inactivation, and throughout the voltage range, DPP10c accelerates inactivation more than DPP10d.
The voltage-dependence of inactivation in the presence of DPP10a is dramatically different, however. ΔN2-40 + DPP10a channels inactivate faster as the voltage increases from −50 mV to +50 mV (Fig. 3D). At −50 mV, ΔN2-40 + DPP10a channels have the same t0.5 value as ΔN2-40 + DPP10d channel, but by +50 mV DPP10a channels inactivate ~ 12 times faster (Table 2). Overall, our data indicates that the N-terminal domain of DPP10a is specifically involved in the acceleration of inactivation not by acting indirectly through the KChIP subunit or by de-repressing the Kv4.2 endogenous inactivation particle.
Kv4.2 N-type inactivation acts independently of DPP10 isoform expression
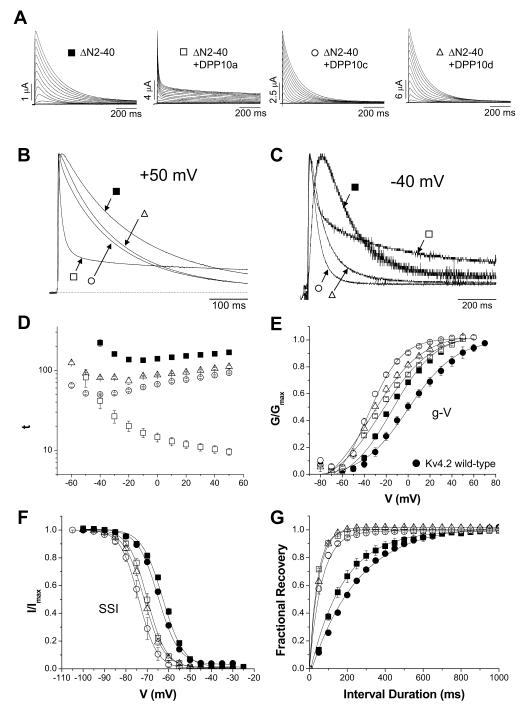
Since the DPP10a N-terminal domain functions similarly to the endogenous Kv4.2 N-terminal domain, we hypothesized that the dramatic differences in the kinetics and voltage dependence of inactivation observed with expression of different DPP10 isoforms would be reduced when co-expressed with wild-type Kv4.2 in the absence of KChIPs if the Kv4.2 N-terminal domain acts independently of the DPL proteins. Therefore, we examined the properties of Kv4.2 channels co-expressed with individual DPP10 variants without KChIP3. To determine the effects of the Kv4.2 N-terminus, oocytes were injected with Kv4.2 cRNA alone or with DPP10 variant cRNAs, giving rise to transient A-type currents that appeared in response to step depolarizations from a −100 mV holding potential. As the scale bars indicate, co-expression with DPP10a, DPP10c, or DPP10d without KChIP significantly increases the Kv4.2-mediated currents. The peak current at +50 mV increases by ~ 6-, ~ 13-, and ~ 19-fold respectively with DPP10a, DPP10c, and DPP10d. When Kv4.2 is expressed alone, the time to peak current is 8.5 ± 0.5 ms (n=12), and co-expression with either DPP10a, DPP10c, or DPP10d decreases time to peak current (DPP10a: 2.9 ± 0.1 ms (n=7); DPP10c: 4.2 ± 0.1 ms (n=5); DPP10d: 4.0 ± 0.3 ms (n=3)). The kinetics of current decay is accelerated by all DPP10 isoforms (Fig. 4A). While the effect at +60 mV is greater with DPP10a than with DPP10c or DPP10d, the difference is not as striking as when the isoforms were expressed in the Kv4.2+KChIP3 background (compare Fig. 4B with Fig. 2B).
Figure 4. In the absence of KChIP3, DPP10 variants similarly modulate Kv4.2-medated currents expressed in oocytes.
(A) Families of Kv4.2-mediated current traces in the absence or presence of DPP10a, DPP10c, and DPP10d. Currents were elicited by 1 sec-long depolarizing pulses from −100 to +60 mV in 10 mV increments from a holding potential of −100 mV. Only the first 250 ms is shown. (B) Normalized and overlapped traces at −40 mV of 800 ms duration. Type of trace is indicated by symbols assigned in A. (C) Same as in B, but at +60 mV for 250 ms. (D) Analysis of the voltage-dependence of half-inactivation time (t0.5). The y-axis is in log-scale to better illustrate the differences in t0.5 values at depolarized potentials. (E) Voltage-dependence of steady-state properties of activation and inactivation. Normalized conductance-voltage relationships (g-V) were calculated from peak conductances, using a K+ reversal potential of −90 mV. Normalized steady-state inactivation (SSI) was monitored using a 10 sec conditioning step that is followed by a 250-ms test pulse to +50 mV. The fitting traces represent Boltzmann functions.
Similar to KChIP3 co-expression and ΔN2-40 studies, at −40 mV Kv4.2+DPP10a current does not inactivate overall faster than Kv4.2+DPP10c or Kv4.2+DPP10d current. At this potential, the current produced by Kv4.2 alone is only beginning to emerge above baseline, activating and inactivating slowly (Fig. 4B). As the normalized overlapped traces show, all DPP10 variants dramatically accelerate the kinetics of activation and inactivation at −40 mV. Yet, the overall macroscopic inactivation of Kv4.2+DPP10a current is slower relative to the others (Fig. 4C). As for voltage dependence of inactivation, the value of t0.5 is decreased by all DPP10 variants over the voltage range studied. However, in contrast to earlier data using KChIP3 co-expression or ΔN2-40 channels, Kv4.2 expressed alone or with different DPP10 isoforms all inactivate more quickly with increasing depolarization (Fig. 4D).
Because inactivation kinetics are dependent on voltage, we examined the voltage dependence of activation and steady-state inactivation. Our results show that all DPP10 variants produce an ~ −28 mV hyperpolarizing shifts in the voltage dependence of activation and an ~ −9 mV hyperpolarizing shift in the half-inactivation voltage (V0.5) of steady-state inactivation (Fig. 4E; Table 2). The smaller amount of hyperpolarizing shift (~ 19 mV less) in the inactivation curve compared to the activation curve suggests that DPP10 variants accelerate the time course of recovery from inactivation. Indeed, the rate of recovery from inactivation of Kv4.2-mediated current at −100 mV is equally increased by co-expression of DPP10a, DPP10c, or DPP10d (Table 2). Therefore, the difference between inactivation time course of Kv4.2 associated with DPP10a compared to DPP10c and DPP10d are not due to differential effects on voltage dependence, but rather the combined effects of the Kv4.2 and DPP10a N-termini. Furthermore, the data clearly indicates that the Kv4.2 N-terminus, like the DPP10a N-terminus, suppresses the U-shaped voltage dependence of inactivation regardless of which DPP10 variant is expressed.
Rapid Inactivation Mediated by DPP10a, But Not DPP6-S, Overcomes the Inactivation Suppressing Effects of KChIP4a
KChIP4a, a splice variant of KChIP4, possesses a K+ channel inactivation suppressor (KIS) domain at its N-terminus and, as a result, dramatically slows down Kv4 channel inactivation (Holmqvist et al., 2001). Since DPP10a and KChIP4a both have potent modulatory domains located on the cytoplasmic surface of the channel, we examined the combinatorial effects of DPP10a and KChIPs on Kv4 inactivation by heterologously co-expressing Kv4.2, DPP10a, and KChIP4a and performing voltage-clamp studies. As Fig. 5A shows, KChIP4a indeed markedly slows Kv4.2 activation and inactivation kinetics, in agreement with the published report: the time to half-inactivation (t0.5) at +50 mV increases from ~30 ms for Kv4.2 channels to ~100 ms for Kv4.2+KChIP3 channels and to ~466 ms for Kv4.2+KChIP4a channels (Table 2). Association of KChIP4a with Kv4.2 channels additionally modifies the voltage dependence of channel activation, producing an ~ 11 mV depolarizing shift in the conductance-voltage relationship (Table 2). Measurements of steady-state inactivation show that KChIP4a does not significantly alter the half-inactivation voltage (V0.5) or voltage-sensitivity (slope) (Fig. 5C, Table 2). The kinetics of recovery from inactivation was also not significantly accelerated by KChIP4a (Fig. 5D, Table 2). The functional properties of Kv4.2+KChIP4a channels are consistent with those reported by Holmqvist et al. (2001).
Figure 5. Co-expression of DPP10a variant, but not DPP6-S, eliminates the dramatically slowed inactivation associated with KChIP4a.
(A) Families of Kv4.2-mediated current from Xenopus oocytes expressing Kv4.2+KChIP4a. Shown are current traces from −100 mV to +50 mV in 10 mV increments for the first 400 ms. (B) Currents from cells expressing Kv4.2+KChIP4a+DPP10a, elicited by a protocol same as one used in (A). For comparison, normalized current traces at +50 mV were overlapped for Kv4.2+KChIP4a, Kv4.2+KChIP4a+DPP6-S, and Kv4.2+KChIP4a+DPP10a (see inset). Note that DPP10a is able to markedly accelerate inactivation, even in the presence of KChIP4a. (C) Differential effects of DPP6-S (left panel) and DPP10a (right panel) on steady-state inactivation in the presence of KChIP4a. While DPP6-S and DPP10a both produce hyperpolarizing shifts in the steady-state inactivation, the magnitude of shift is greater for DPP10a. Fitting traces represent Boltzmann functions. (C) KChIP4a does not interfere with the ability of DPP10a to accelerate recovery from inactivation. The time course of recovery from inactivation, as fitted by single exponential functions, is identical between Kv4.2+DPP10a and Kv4.2+KChIP4a+DPP10a channels.
When DPP10a is co-expressed with Kv4.2+KChIP4a channels, the fast inactivation kinetics associated with DPP10a dominates, is similar to its co-expression with Kv4.2+KChIP3 and ΔN2-40 channels (Fig. 5B). The t0.5 value at +50 mV are similar between DPP10a co-expressed with Kv4.2, Kv4.2+KChIP3, ΔN2-40, and Kv4.2+KChIP4a (Table 2). The similar inactivation kinetics indicated by the t0.5 values were also confirmed by exponential fittings. Inactivation of DPP10a-containing channels occurs in three phases at +50 mV, and the early fast phase occupies more than half of total decay and decays with similar time constants (weight, time constant: Kv4.2+DPP10a = 69 ± 2 %, 6.3 ± 0.9 ms (n=9); Kv4.2+KChIP3+DPP10a = 69 ± 4 %, 7.3 ± 0.7 ms (n=7); ΔN2-40+DPP10a = 55 ± 1 %, 5.5 ± 0.3 ms (n=4); Kv4.2+KChIP4a+DPP10a = 73 ± 1 %, 6.4 ± 0.1 ms (n=5)). Like KChIP3, KChIP4a does not interfere with DPP10a’s ability to modulate voltage-dependent and kinetic properties of Kv4.2 channels. As compared to Kv4.2 + KChIP4a controls, Kv4.2 + KChIP4a + DPP10a channels show hyperpolarizing shifts in g-V relationships (~ −22 mV) and steady-state inactivation (~ −10 mV) (Fig. 5C; Table 2). DPP10a also dramatically accelerate the kinetics of recovery from inactivation by ~ 2.7 times (Fig. 5D). Our results also show that the ternary complex channels containing DPP10a exhibit different functional properties depending on whether KChIP3 or KChIP4a is present. Relative to KChIP3, incorporation of KChIP4a is associated with an activation mid-point depolarized by ~16 mV and a recovery from inactivation more than 3 times slower (Table 2).
In contrast to DPP10a, when DPP6-S (short variant) is co-expressed with Kv4.2 and KChIP4a, DPP6-S produces only a moderate acceleration of activation and inactivation (Fig. 5B, inset). This is detected as decreased values for time-to-peak (Fig. 5B, inset) and t0.5 (Table 2). When compared with Kv4.2+DPP6-S, this waveform represents a dramatic slow down and illustrates the dominance of KChIP4a’s effect over those of DPP6 on channel inactivation. Therefore, our main findings from studies using both KChIP3 and KChIP4a shows that channel complexes that contains the DPP10a variant, in spite of the type of KChIP variant present, will likely undergo rapid inactivation dictated by the DPP10a N-terminal domain. However, irrespective of the DPL present, KChIP4a determines a slower recovery from inactivation relative to that of KChIP3.
ΔN2-40 Channels Containing the DPP10a Subunit Feature DPP10a-mediated Fast Inactivation, Despite the Presence of Other DPP10 Variants
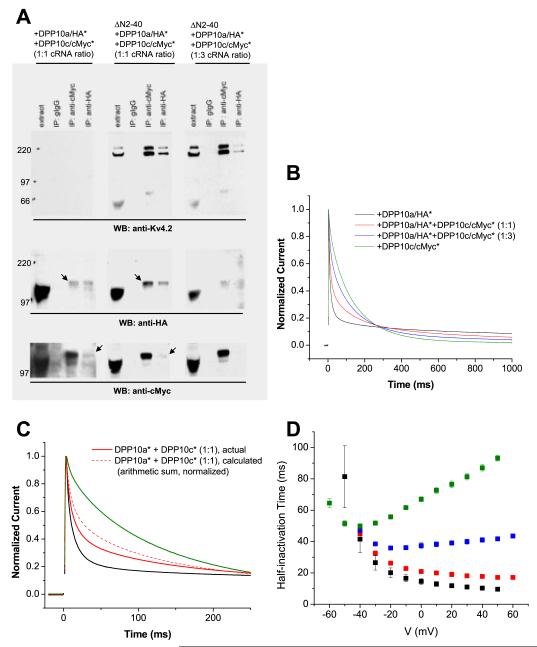
Previous studies have shown that DPL proteins can form homo- or hetero-oligomers, likely in the form of dimers (Ren et al., 2005; Strop et al., 2004). To examine how potential association of different DPP10 variants may affect the inactivation of Kv4.2-mediated ISA, we performed co-immunoprecipitation studies to confirm variant association and two-electrode voltage-clamp recording on oocytes co-expressing ΔN2-40, DPP10a, and DPP10c proteins to determine the functional effects of expressing mixed DPL populations (Fig. 6A). For co-immunoprecipitation, HA and cMyc tags were genetically inserted respectively into DPP10a and DPP10c immediately following the first residue of Exon 14 (DPP10a: serine 408; DPP10c: serine 412). This position in the extracellular beta-propeller domain is selected for its location in a loop away from the dimerization interfaces reported by Strop et al (2004).
Figure 6. DPP10a-mediated fast inactivation prevails in the presence of DPP10c.
HA and cMyc tags were inserted in an extracellular loop of DPP10a (DPP10a/HA*) and DPP10c (DPP10c/cMyc*), respectively, as to not interfere with the DPP10a N-terminal domain’s function. (A) Co-immunoprecipitation studies show that Kv4.2 does not interfere with DPP10 variants heteromerization. Oocytes extracts of were subjected to immunoprecipitation procedures as described in the experimental methods section. In the absence of Kv4.2, anti-HA antibody immunoprecipitate DPP10c/cMyc* along with DPP10a/HA*, and vice versa, indicating DPP10 variant heteromultimerization (left panels). When introduced, ΔN2-40 proteins binds to both variants and, most importantly, do not interfere with this co-immunoprecipitation of DPP10 variants (middle and right panels). (B) Normalized currents at +50 mV elicited from oocytes co-expressing ΔN2-40 with DPP10a/HA* alone, DPP10c/cMyc* alone, and DPP10a/HA* plus DPP10c/cMyc* at 1:1 and 1:3 cRNA molar ratios. (C) Comparison of ΔN2-40+DPP10a/HA*+DPP10c/cMyc* (1:1) current compared to arithmetic sum of ΔN2-40+DPP10a/HA-int and ΔN2-40/cMyc-int current. The two currents are significantly different, consistent with the formation of different DPP10 variants bound to same channel complexes. (D) Voltage-dependence of half-inactivation times for currents expressed by oocytes injected with ΔN2-40 plus DPP10a/HA* (black symbols), plus DPP10a/HA* and DPP10c/cMyc* at 1:1 cRNA ratio (red symbols), plus DPP10a/HA* and DPP10c/cMyc* at 1:3 cRNA ratio (blue symbols), and plus DPP10c/cMyc* (green symbols).
We first determined that the epitope tags function without preventing hetero-oligomerization (Fig. 6A, left panels). When the external HA-tagged DPP10a (DPP10a/HA*) and cMyc-tagged DPP10c (DPP10c/cMyc*) proteins are co-expressed without Kv4 proteins, either anti-HA or anti-cMyc antibodies can precipitate both tagged DPP10 variants. This shows that DPP10’s do indeed form homo-oligomers independent of Kv4, and that the epitope tags do not prevent interactions between monomers. To determine whether the hetero-oligomers can bind Kv4 channel proteins, a constant molar amount of ΔN2-40 cRNA was co-injected into oocytes with DPP10a/HA* and DPP10c/cMyc* in combination at a 1:1 or 1:3 molar ratio (Fig. 6A, middle and right panels). From extracts of oocytes containing DPP10a and DPP10c cRNA at 1:1 ratio, either anti-cMyc or anti-HA antibodies precipitates all three proteins, indicating that the hetero-oligomer can still bind Kv4.2 proteins (Fig. 6A, middle panels). By comparing the band intensities of the precipitated and co-precipitated DPP10 proteins, we see that the co-precipitation efficiency between DPP10a/HA* and DPP10c/cMyc* is not altered by the introduction of Kv4.2. At a 1:3 molar ratio of DPP10a/Ha* to DPP10c/cMyc* cRNA, as expected the amount of DPP10c/cMyc* protein recovered is increased by anti-cMyc antibody, but the amount of DPP10a/HA* and Kv4.2 protein precipitated is decreased (Fig. 6A, right panels).
Introduction of the external epitope tags also does not affect functional regulation of Kv4.2 by homomeric DPP10a or DPP10c. Both DPP10a/HA* and DPP10c/cMyc* modulate the gating kinetics and voltage-dependent properties of ΔN2-40 channels similarly to their untagged counterparts (Table 2). In particular, ΔN2-40 channels co-expressed with either DPP10a or DPP10a/HA* exhibit rapid inactivation that accelerates with depolarization. Both tagged DPP10 variants produce leftward shifts in conductance-voltage relationships, leftward shifts in steady-state inactivation, and accelerated recovery from inactivation. Similar to DPP10 facilitating the trafficking of Kv4 channels to the cell surface (Jerng et al., 2004a; Zagha et al., 2005), DPP10a/HA* and DPP10c/cMyc* both also dramatically enhance the surface expression of ΔN2-40 channels (DPP10a/HA*: ~17-fold; DPP10c/cMyc*: ~19-fold).
Since the modulatory effects of DPP10a and DPP10c on Kv4.2 gating differ primarily in the kinetics of inactivation, we focused on evaluating the current decays associated with the mixed 1:1 and 1:3 ratio injections. As Fig. 9B shows, inactivation kinetics vary depending on the ratio of DPP10a/HA* and DPP10c/cMyc* variants expressed. Consistent with DPP10 terminus’ restricted role in Kv4 inactivation, ΔN2-40 channels containing DPP10a/HA*, DPP10c/cMyc*, or a mixture of the two isoforms are similar in their conductance-voltage relationship, steady-state inactivation, and recovery from inactivation (Table 2). The observed inactivation waveform of ΔN2-40 channels co-expressed with DPP10a/HA* and DPP10c/cMyc* (1:1) was then compared with that of a 1-to-1 arithmetic sum of ΔN2-40+DPP10a/HA* and ΔN2-40+DPP10c/cMyc* currents (Fig. 6C). The absence of a match between these traces is consistent with DPP10a and DPP10c assembling into the same channel complex. The prominence of fast inactivation suggests that, in channel containing both DPP10a and DPP10c, the DPP10a variant has a significantly greater influence over inactivation kinetics. As Fig. 6D shows, the t0.5 values for ΔN2-40 channels co-expressed with both DPP10a/HA* and DPP10c/cMyc* at 1:1 ratio resemble those of channels co-expressed with only DPP10a/HA*. The relationship between t0.5 values and membrane potential indicates that, in addition to the fast inactivation typical of DPP10a-mediated currents, ΔN2-40 channels co-expressed with DPP10a/HA*+DPP10c/cMyc* at 1:1 cRNA molar ratio also inactivate more rapidly with increasing depolarization (Fig. 6D).
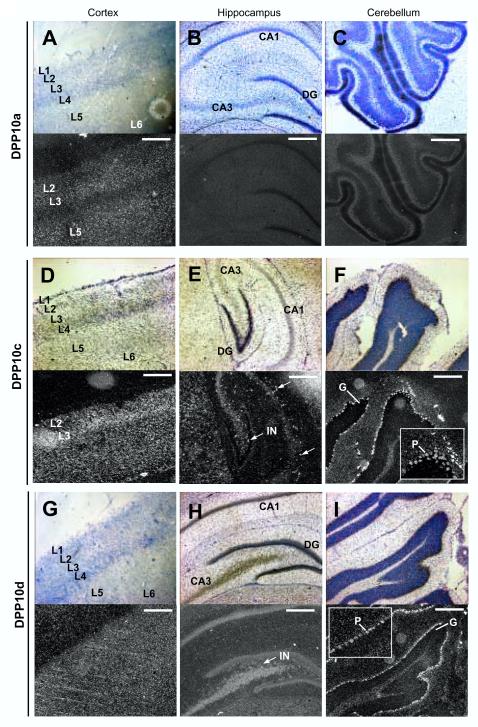
Figure 9. Cellular localization of DPP10 variant transcripts by in situ hybridization: Highlights.
Various sections were coated with photographic emulsion for autoradiography and Nissl stained, followed by bright- and darkfield microscopy. Shown are representative images for DPP10a (A, B, C), DPP10c (D, E, F), and DPP10d (G, H, I) signals in the cortex (left columns), the hippocampus (middle columns), and cerebellum (right columns). L1-L5 = cortical layers I-V; CA3 = Area CA3; CA1 = Area CA1; DG = dentate gyrus; G = granule cells; P = Purkinje cells; IN = interneurons. Scale bar = 500 μm.
When the ratio of injected DPP10a/HA*-to-DPP10c/cMyc* cRNAs is decreased to 1-to-3, the expressed current develops macroscopic inactivation with a U-shaped voltage dependence, as the t0.5 value initially decrease, reaches a low point at ~20 mV, and then subsequently increase with increasing depolarization (Fig. 6D). The slowing of overall inactivation and the appearance of U-shaped voltage dependence are likely due to the over-expression of the DPP10c/cMyc* protein, significantly increasing the number of channels with homomeric DPP10c/cMyc* and their contribution to the overall current decay.
Expression levels for DPP10 N-terminal Splice Variants in Rat Cerebral Cortex, Hippocampus, and Cerebellum
Since different DPP10 isoforms have different effects on the properties of Kv4.2 ternary complex and the corresponding ISA, we performed experiments to determine their mRNA distributions in the rat brain. To quantify putative differences in expression levels and to broadly examine the distribution patterns of different variants, we used quantitative real time fluorescent RT-PCR (qRT-PCR) following the SyberGreen protocol (Fig. 7). The efficiencies of different primers sets were determined by measuring the number of PCR cycles required to reach threshold detection for a larger range of starting template molecules (between 10 and 10,000,0000). The amplification efficiency was determined by linear regression fitting of a plot comparing the number of PCR cycles required to reach a critical threshold (Ct = 0.10) fluorescence verses the log of the number of starting template molecules (Morrison et al., 1998) to give cycles per 10 fold amplification which was converted to amplification efficiency per cycle.
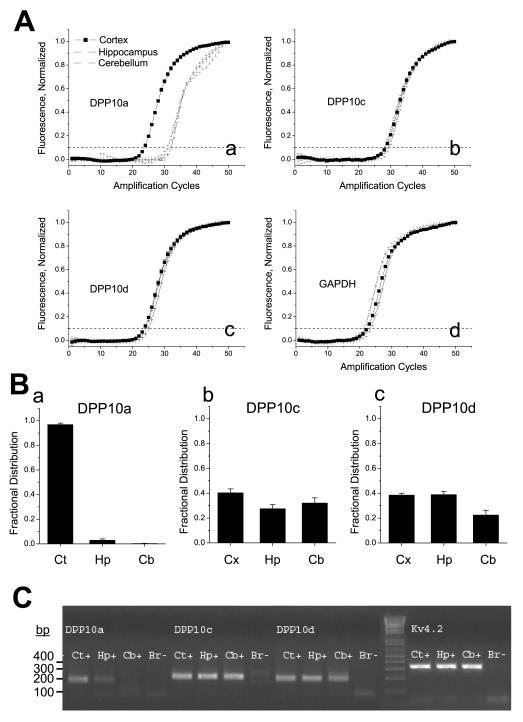
Figure 7. Regional distribution of DPP10 variants as analysed by quantitative RT-PCR.
(Aa-Ad) Quantitation of DPP10a, DPP10c, DPP10d, and GAPDH transcripts in rat cerebral cortex, hippocampus, and cerebellum by real-time fluorescent RT-PCR. The normalized fluorescence values are plotted as a function of amplification cycle number, and the cycle number at which the fluorescence value crosses the threshold value of 0.10 (dashed line) is defined as the Ct value. The number of molecules in the starting sample is determined from absolute standard curves generated specifically for each primer set on starting cDNA template dilutions from 10 to 10,000,000 molecules. (B) Fractional distribution of each splice variant in the cortex, hippocampus, and cerebellum following normalization by GAPDH. (C) Agarose gel analysis of products of DPP10a, DPP10c, DPP10d, and Kv4.2 amplifications after 50 cycles of qRT-PCR (See Experimental Methods for primer sequences). Note that the reactions amplified single products of the expected sizes, and no products are generated in the absence of starting material (Br-). Ct = cortex; Hp = hippocampus; Cb = cerebellum.
For quantification of DPP10 transcript levels in the brain, mRNA was isolated from rat cortex, hippocampus and cerebellum, reverse transcribed to cDNA using random priming, and amplified using primers specific for DPP10 variants, Kv4.2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The expression profile of GAPDH was characterized as an internal control. The number of amplification cycles required to reach Ct was determined for the different conditions. The amplified products were also separated out on agarose gels and stained with ethidium bromide to confirm the amplification of a single product (Fig. 7C). The data show that dramatically fewer amplification cycles for DPP10a are required for cortex than for hippocampus or cerebellum to reach Ct (Fig. 7Aa). In contrast, DPP10c, DPP10d, and the control GAPDH are amplified similarly in all three brain regions (Figs. 7Ab, 7Ac, 7Ad). Computing the fractional distribution of each splice variant following normalization by GAPDH, we see that DPP10a is only significantly expressed in the cortex (>95% of transcripts), compared to the flatter distributions for DPP10c and DPP10d (Figs. 7Ba, 7Bb, 7Bc).
Differential Distribution of DPP10 transcripts in rat brain
We next sought to determine if DPP10 variants are differential expressed in specific neuronal populations using in situ hybridization. Rat brain slices were labelled by hybridization with radiolabelled variant-specific probes against Exon 1a, 1c, and 1d. Overall, the results confirm and extend our qRT-PCR results. The mRNA transcripts containing Exon 1a (DPP10a) appear highly expressed in the cerebral cortex, with the radioactive signals absent from a cortical mid-layer (Figs. 8A and 8D). Darkfield microscopy of the cortical region shows that cells expressing DPP10a are broadly distributed in Layers 2-3 and 5, but not Layer 4 (Fig. 9A). DPP10a is also highly expressed in the anterior portion of the thalamus that contains the reticular thalamic nucleus (Fig. 8A). Low signals are seen in the hippocampus and cerebellum, respectively (Figs. 8A, 8D, and 8G; Figs. 9B and 9C).
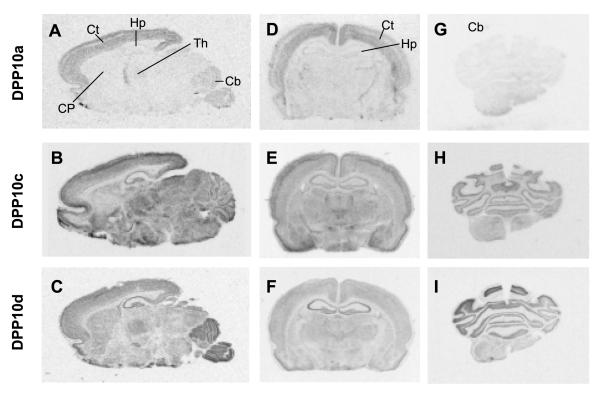
Figure 8. Physical distribution of DPP10a, DPP10c, and DPP10d transcripts in rat brain.
In situ hybridization using radiolabelled antisense RNA probes was used to detect DPP10a, DPP10c, and DPP10d mRNA transcripts. Autoradiographs from sagittal sections (A, B, and C) and coronal sections from the hippocampal (D, E, and F) and cerebellar (G, H, and I) regions are shown. Sections were hybridized with probes against DPP10a (A, D, and G), DPP10c (B, E, and H), and DPP10d (C, F, and I). The results show that DPP10a is preferentially expressed in the cortex, while DPP10c and DPP10d show more generalized distribution, with notable levels in the cerebellum and hippocampus. Ct = cortex; Hp = hippocampus; Th = thalamus; Cb = cerebellum; CP = caudate putamen.
Transcripts containing Exon 1c (DPP10c) are broadly distributed in rat brain (Fig. 8B). In particular, DPP10c is expressed at relatively high levels in the cortex, hippocampus, cerebellum, and thalamus (Fig. 8B, 8E, and 8H). DPP10c expression is especially notable in Layer 3 of the cortex and is weakest in the caudate putamen region (Fig. 8B, Fig. 9D). In the hippocampus, DPP10c is poorly expressed in pyramidal cells but prominently in interneurons, many of which are located between CA3 and dentate gyrus (Fig. 9E). In the cerebellum, DPP10c is highly expressed in the Purkinje cells and weakly expressed in the granule cells (Fig. 9F).
Exon 1d shows a distinct expression pattern compared to Exon 1a or 1c (Figs. 8C, 8F, and 8I). Like DPP10c, DPP10d transcripts are widely expressed with apparently high levels in the hippocampus and cerebellum, and with modest levels in the cortex and thalamus. An even distribution of DPP10d transcripts is detected in the cortical layers (Fig. 9G). And like DPP10c, DPP10d transcripts in the hippocampus are detected mostly in the interneurons (Fig. 9H). In the cerebellum DPP10d also shows prominent expression in the Purkinje cells but weak expression in the granule cells (Fig. 9I). However, unlike DPP10c, DPP10d shows higher expression in the caudate putamen relatively to other brain regions (Fig. 8C). In summary, by both qRT-PCR and in situ hybridization, DPP10 variants show differential expression in brain regions; in particular, DPP10a is strongly expressed in the cerebral cortex but weakly expressed in the hippocampus and cerebellum.
DISCUSSION
DPP10 N-terminal variants are differentially expressed in the brain and produce different functional effects
5′RACE analysis performed on mRNAs extracted from whole rat brain as well as rat cerebellum clearly establishes the existence of three DPP10 splicing variants: DPP10a, DPP10c, and DPP10d. These variants are generated by alternative splicing of the first DPP10 exon (Exon 1) and differ only by their cytoplasmic N-terminal domain. The N-terminal variants of DPP10 are expressed with distinct patterns in rat brain: DPP10a expression is largely restricted to Layer 2/3 and Layer 5 of the cerebral cortex, whereas DPP10c and DPP10d exhibits a more diffuse distribution throughout the brain, with significant levels in hippocampal interneurons and cerebellar Purkinje cells.
A critical finding in our study is that ternary complex channels formed from Kv4.2, KChIP3, and different DPP10 variants display dramatically distinct kinetics and voltage dependence of inactivation at depolarized potentials. This indicates that the cytoplasmic portion of DPP10 proteins determines inactivation properties at positive voltages, consistent with our previous findings (Jerng et al., 2004a). In the presence of KChIPs, Kv4 channel inactivation shows a U-shaped inactivation profile, with slower inactivation kinetics the further the membrane potential is from the midpoint for activation. While the precise molecular mechanism responsible for this U-shaped inactivation is not known, it is clear from our previous studies with DPP6-L, DPP6-S, and our studies here with DPP10c and DPP10d, that this inactivation mechanism remains intact in the presence of DPLs. Therefore, it is remarkable that DPP10a expression specifically overrides the U-shaped inactivation mechanism by introducing its own inactivation mechanism that is fast and relatively voltage-independent.
Interestingly, regardless of the DPP10 variant present, inactivation kinetics do not slow down with depolarization when Kv4.2 is expressed without KChIPs, due to an intrinsic pore-blockade (N-type) inactivation mechanism of Kv4 channels (Gebauer et al., 2004). In fact, previous investigators examining DPL variants in co-expression with Kv4 subunits did not find the dramatic differential effects of the DPP10a variant precisely because they did not study the a-variant or failed to include KChIPs in their expression systems. These results have been interpreted to suggest that N-terminal variants of DPLs do not differentially regulate Kv4 channel functional properties, in contradiction to our previous conclusions (Nadal et al., 2006; Takimoto et al., 2006; Li et al., 2006). However, here we have shown that once intrinsic N-type inactivation is removed from Kv4 channels, either by KChIP co-expression or mutation, the differential effects of the DPP10a N-terminus are clearly seen.
We propose that the specific effects of DPP10a on inactivation are produced because the DPP10a N-terminus functionally acts like a ball peptide for N-type inactivation. Thus the apparent contradiction is resolved. In the presence of Kv4 N-type inactivation, DPP10a appears to act similar to other DPL variants, but in the absence of Kv4 N-type inactivation, as produced by KChIP co-expression, DPP10a re-introduces an N-type mechanism to the channel. Although the functional effects of DPP10a N-terminus are similar to that of the Kv4.2 N-terminus, they share no evident sequence similarity. Our future studies will determine whether the inactivation produced by the DPP10a N-terminus uses a pore-blocking mechanism similar to other characterized N-terminal inactivation peptides or whether it uses a distinct mechanism to promote channel inactivation.
Functional Roles of DPP10 Transmembrane and Extracellular Domains
Even though different N-terminal variants mediate different inactivation kinetics, all DPP10 variants produce comparable changes in other Kv4 properties: enhanced surface expression, accelerated activation kinetics, accelerated recovery from inactivation, leftward shift in g-V relationships, and leftward shift in steady-state inactivation. These results suggest that the single transmembrane segment and the extracellular domains are important for the changes common to all DPP10s. This is consistent with findings by Zagha et al. (2005), Ren et al. (2005), and Li et al. (2005) that showed the transmembrane domain and the surrounding region is sufficient for the DPL interaction with and the modulation of Kv4 channels. The extracellular domain of DPLs, based on C-terminal truncation studies, has been suggested to contribute in stabilizing the Kv4-DPL interaction (Zagha et al., 2005). Our lab also has observed that, upon cMyc-tagging of the DPP10 distal C-terminus, the co-immunoprecipitation between DPP10 and Kv4.2 is diminished, consistent with role of the extracellular domain in complex stabilization (H Jerng, unpublished observations).
The mechanism underlying the DPL-mediated changes in voltage-dependent properties of Kv4 channels was examined recently by Dougherty and Covarrubias (2006), and their results offer an explanation as to why, compared to controls, all DPP10 variants accelerate the kinetics of inactivation at all membrane potentials and under all co-expression conditioned tested. Co-expression of DPP6-S with Kv4.2 causes faster outward and inward movements of the channel voltage sensor during depolarization and repolarlization, respectively, resulting in a −26 mV shift in the gating charge-voltage relationship. The same effect likely occurs with DPP10, as co-expression of DPP10 produces a dramatic hyperpolarizing shift (~28 mV) in the voltage-dependent activation (Fig. 4E). This shift results in the acceleration of the time course of activation at a given voltage, and since activation and inactivation are interdependent or “coupled” (Hille, 2001), acceleration of inactivation rate occurs as well. Nevertheless, our results show that, in addition to this activation-coupled acceleration of inactivation, the cytoplasmic N-terminus of DPP10a confers an additional rapid inactivation pathway that is preferred with increasing voltage.
Exclusively Expressed in the Brain, the DPP10a Variant Distributes Mainly in the Cerebral Cortex and Promotes Uniquely Fast Inactivation
The overall finding of this study indicates that DPL splice variants likely contribute to the diverse kinetics and voltage-dependence of inactivation expressed by Kv4-mediated ISA in neurons. Previous studies have shown that neuronal ISA channel functional properties are matched by a combination of Kv4, KChIP, and DPL protein co-assembled into a functional channel (Nadal et al., 2003; Jerng et al., 2005). Given the differential, but overlapping expression of DPL variants in the brain, we also need to consider the functional impact of DPL protein heteromultimeric assembly into the channel. Our co-expression studies mixing DPP10a and DPP10c variants suggest that, in neurons where variant expressions overlap, Kv4 channel complexes likely contain hetero-oligomers of DPP10 subunits. Kv4.2 channel complexes with hetero-oligomers of DPP10a and DPP10c inactivate with kinetics that are distinct from DPP10a or DPP10c alone. The precise inactivation kinetics depend upon the expression ratio of DPP10a and DPP10c. Reducing the ratio of DPP10a to DPP10c changes the overall inactivation kinetics from fast to slow, and transforms the voltage dependence from monotonic decaying, to “voltage-independent”, to U-shaped. Similar results would likely be produced by co-expression with other DPL with DPP10a. Thus, we predict that the precise kinetic and voltage dependence of inactivation seen for any specific neuron will depend upon the precise expression levels for the different DPL variants.
The challenge then is to relate these observations to the functional composition of ISA currents in particular neurons. Because our in situ hybridizations and qRT-PCR show that DPP10a is preferentially expressed in the cortex, we hypothesize that cortical neurons are a likely place to observe neurons with accelerated ISA current inactivation kinetics that show little to no U-shaped voltage dependence. In Table 3, we summarize what is known about the inactivation kinetics of ISA in different cortical neuron populations. The published data have been obtained at similar temperature, and the reported junction potential errors have been corrected whenever possible. Importantly, the table shows that in certain cortical neuronal populations, such as Layer 2/3 and Layer 5 cortical pyramidal neurons, the ISA shows rapid inactivation kinetics with little or no U-shaped voltage dependence (Zhou and Hablitz, 1996; Bekkers, 2000; Korngreen and Sakmann, 2000). In contrast, in other cortical cortical neurons, such as bitufted interneurons of Layer 2/3, ISA shows pronounced slowing of inactivation with increasing depolarization (Korngreen et al., 2005). Based on our guiding hypothesis, we would suggest that the relative fraction of DPP10a subunits compared to other DPLs is likely lower in bitufted interneurons compared to cortical pyramidal neurons. Given the oomplex expression environment of native neurons there are clearly other factors that could explain these kinetic differences. However, given what we do know about functional effects of known Kv4 channel subunits in heterologous expression we can likely rule out differences in Kv4 subunits or KChIPs as being responsible. Although cortical pyramidal cells and interneurons express both Kv4.2 and Kv4.3, the impact of Kv4.2/Kv4.3 heterotetramer formation on inactivation kinetics is likely to be minimal (Burkhalter et al., 2006; Guo et al., 2002). Furthermore, we have shown that although different KChIPs, such as KChIP3 and KChIP4a, have distinct effects on channel functional properties the robust DPP10a effect on inactivation properties overrides these differences. Our future studies are aimed at directly testing the potential role of DPP10a in shaping cortical ISA currents, by combining voltage clamp, single-cell RT-PCR, and RNA interference to establish a clear functional connection between DPP10a expression and ISA functional properties.
Table 3.
Biophysical properties of cortical ISA and Kv4.2+KChIP3+DPP10a current expressed in oocytes
| Recording conditions and Voltage-dependence Parameters | ||||||
|---|---|---|---|---|---|---|
| Cell type | Temperature | E-junction | Activation | Inactivation | ||
|
|
||||||
| (°C) | (mV) | V0.5 (mV) |
slope factor (mV) |
V0.5 (mV) |
slope factor (mV) |
|
| Cortical pyramidal cell, Layer 2/3 | 21-23 | 4 | −4 | 12 | −51 | 7 |
| Cortical interneuron, Layer 2/3 | 20-22 | 2 | −15 | 18 | −77 | 11 |
| Cortical pyramidal cell, Layer 5 | 22-25 | 7 | −19 | 17 | −82 | 7 |
| Cortical pyramidal cell, Layer 5 | 20-22 | 2 | −5 | 20 | −68 | 10 |
| XO, Kv4.2 + KChIP3 + DPP10a | 21-23 | NA | −26 | 20 | −68 | 4 |
| XO, Kv4.2 + KChIP3 + DPP10c | 21-23 | NA | −20 | 18 | −62 | 5 |
| XO, Kv4.2 + KChIP3 + DPP10d | 21-23 | NA | −24 | 18 | −64 | 4 |
| Time to Peak and Kinetic Parameters | ||||
|---|---|---|---|---|
| Cell type | Inactivation at +30 mV (t0.5, ms) |
Inactivation Volt-dependence |
Recovery at −100 mV (τ, ms) |
Reference |
| Cortical pyramidal cell, Layer 2/3 |

|
NA | Zhou & Hablitz, 1996 | |
| Cortical interneuron, Layer 2/3 | 60 | Korngreen et al., 2005 | ||
| Cortical pyramidal cell, Layer 5 | 41 | Bekkers, 2000a | ||
| Cortical pyramidal cell, Layer 5 | 28 | Korngreen & Sakmann, 2000 | ||
| XO, Kv4.2 + KChIP3 + DPP10a | 28 | this study | ||
| XO, Kv4.2 + KChIP3 + DPP10c | 36 | U-shaped | 20 | this study |
| XO, Kv4.2 + KChIP3 + DPP10d | 38 | U-shaped | 24 | this study |
For comparison with published ISA data, only the mean values for experimental data are shown. Experimental data from this study were analyzed as described in Experimental Methods. All experiments were both conducted at approximately room temperature. When possible, V0.5 of activation and inactivation have been corrected for the indicated junction potentials (E-junction). The time constants (τ) of inactivation kinetics from published data have been converted to t0.5 by using the relationship: t0.5 = ln(2) * t. The voltage-dependence of inactivation is described as “monotonic” when the literature clearly states that inactivation time course at depolarized potential is “voltage independent” or “weakly voltage dependent”. Box outlines the critical similarity between Kv4.2+KChIP3+DPP10a current in oocytes when compared to cortical ISA. NA = not available. XO = Xenopus oocytes.
It is unlikely that differential DPP10a expression alone explains all differences in ISA functional properties throughout the brain. Indeed, neuronal populations with fast, voltage-independent ISA kinetic properties that express little to no DPP10a have clearly been described, such as cerebellar granule neurons and cortical neurons of the piriform cortex (Bardoni and Belluzzi, 1993; Banks et al., 1996). Recently, we have identified another DPP6 N-terminal variant that is expressed in cerebellar granule neurons and piriform cortical neurons, and also seems to accelerate inactivation similarly to DPP10a (H Jerng, unpublished data). This variant shows distinct sequence similarity in the N-terminus with DPP10a, and preliminary studies suggest that it is differentially expressed in distinct populations of neurons that do not express DPP10a. Other studies have shown that KChIP variations, such as KChIP4a, also differentially regulate Kv4 functional properties (Holmqvist et al., 2002). Thus, an important goal for the future is to understand the contribution of specific auxiliary proteins in shaping the unique functional properties of ISA in different neurons.
Relevance of Auxiliary Subunit Isoforms to Neuronal Physiology
We are finally left with the question of why differential ISA kinetic properties are important for the function of different neuronal populations. The dominance of DPP10a on inactivation kinetics, along with its specific expression in the brain (Takimoto et al., 2006) and its particularly elevated expression in the cortex, suggests that the DPP10a variant is likely important to the normal function of cortical neurons. At this time, however, it is unknown precisely why cortical neurons require ISA with rapid inactivation that accelerates with voltage (Banks et al., 1996; Bekkers, 2000; Korngreen and Sakmann, 2000), rather than ISA with inactivation that slows with depolarization such as is observed in hippocampal neurons (Hoffman et al., 1997; Martina et al., 1998; Lien et al., 2002; Klee et al., 1995). Published reports have shown that ISA is important in regulating neuronal firing frequency and patterns (Kim et al., 2005; Yuan et al., 2005), and a correlation between ISA inactivation kinetics and AP firing frequency has also been implicated by a study of superior colliculus neurons (Saito and Isa, 2005). Thus, the DPP10a variant being expressed may be important for the fine-tuning of excitability and rapid AP firing frequency in cortical neurons. Other studies have shown that ISA current are important for the regulation of synaptic integration and suppression of back-propagating action potentials, and a study of DPL contribution to dendritic ISA would require an exmaination of the subcellular trafficking and compartmentalization of the DPP10 and DPP6 variants. Nonetheless, it seems likely that the differential voltage dependence of inactivation produced by different DPL variants would show differential regulation in response to subthreshold signals versus spikes. The difficulty in making specific functional connections to different DPL variants is that it is likely that a number of channel genes are differentially expressed in different neuronal populations. To overcome this problem, our future studies will alter native DPL expression using RNA interference and viral vector expression to directly examine the functional connection between DPL variant expression and neurophysiological function.
EXPERIMENTAL METHODS
Molecular biology
Plasmids containing rat Kv4.2, human KChIP3, human DPP10a, and human DPP6-S cDNAs were obtained as described previously (Jerng et al., 2004a; Jerng et al., 2005). Kv4.2/ΔN2-40 plasmid was constructed by subcloning a PCR fragment encoding Kv4.2 starting methionine, residues 41-630, and stop codon into pBluescript. KChIP4a cDNA was generated by purchasing a human hippocampal KChIP4b clone (IMAGE #4817099) from American Type Culture Collection (ATCC, Manassas, VA, USA) and substituting in the 5′ sequences that codes for the K-channel inactivation suppressor (KIS) domain of KChIP4a. A 5′ partial cDNA containing Exon I of human DPP10d (IMAGE #5299985) was purchased from ATCC. The complete DPP10d cDNA in pBSR was generated by incorporating the missing 3′ sequences from the DPP10a construct. Exon I of human DPP10c, flanked 5′ by the Kozak consensus sequence, was generated by overlap extension PCR using multiple oligonucleotide primers. Primers were chosen based on published sequences. Replacement of DPP10a 5′ sequence with that of DPP10c led to the full-length DPP10c cDNA. Tagging the extracellular domains of DPP10a, DPP10c, and DPP10d with HA and c-Myc epitopes were performed by the PCR-based overlap extension method. Terminal HA- or cMyc-tagging strategies were abandoned because they either disrupted DPP10 mediation of inactivation acceleration (N-terminal tag) or severely affected DPP10-Kv4.2 association (C-terminal tag) (Jerng et al., 2004a; H Jerng, unpublished observations). For DPP10a and DPP10d, the probes for in situ hybridization were synthesized by overlap extension PCR and subcloned into pBluescript II KS(+) (Stratagene, La Jolla, CA). The DPP10c probe was made from DPP10 5′RACE products by PCR and subcloned as described for other variants. Variant-specific probes contain sequences spanning from 5′ UTR to the end of Exon I. Sequences for all DNA constructs were confirmed and verified by automated sequencing (DNA sequencing facility, Baylor College of Medicine, Houston, TX).
RNA transcript for oocyte expression were synthesized from linearized DNAs using mMessage mMachine high-yield capped RNA transcription kit (Ambion, Austin, TX). Radiolabelled riboprobes for in situ hybridization were produced by in vitro transcription (Riboprobe in vitro transcription systems: Promega Corporation, Madison, WI) of linearized DNAs in the presence of [35S]-UTP (PerkinElmer Life and Analytical Sciences, Boston, MA).
Brain mRNA harvest and cDNA synthesis
Sprague-Dawley rats were anaesthetized by isofluorane inhalation and rapidly killed by cervical dislocation, following regulations set by the Animal Welfare Act, Public Health Services Animal Welfare Policy, National Institute of Health Guide for Care and Use of Laboratory Animals, and Baylor College of Medicine. The same procedure was performed to prepare tissues for in situ hybridization. Tissue samples of hippocampus, cortex, and cerebellum (10 mg each) were collected from P21 rats, and total RNA was isolated using the RNAqueous-Micro kit (Ambion, Austin, TX). Reverse transcription (RT) reactions were conducted with the Superscript III kit (Invitrogen) using random hexamers and following manufacturer’s instructions.
5′-RACE and quantitative real-time PCR
5′RACE reactions were conducted using the First Choice RLM RACE Kit (Ambion, Austin, TX) following manufacturer’s instructions. Briefly, RNA was extracted from one whole P28 rat brain. Un-capped RNA species are dephosphorylated at their 5′ ends by calf intestinal phosphatase. Subsequently, mRNA molecules are treated with tobacco acid pyrophosphatase to remove 5′ cap structure, expose 5′ phosphate, and allow ligation of an adapter RNA oligonucleotide. RT-PCR was conducted on the resulting modified mRNA molecule using adaptor-directed forward primers and DPP10-specific reverse primers, which are described below as Pan-DPP10-r1 and Pan-DPP10-r2. All resulting PCR products were cloned using the pGEM-T Easy TA cloning kit (Promega, Madison, WI). RNAs were also separately extracted from hippocampal, cortical, and cerebellar regions for 5′ RACE studies of DPP10 variants present in specific brain regions.
Procedures for quantitative real-time PCR were adapted from Liss et al. (2001). Synthesized by RT, the cDNAs for three of the known rat DPP10 variants were amplified using the following sets of forward (f) and reverse (r) primers: DPP10a-f1 = 5′-TGGTTTGTCTTGGAACTCTG-3′; DPP10c-f1 = 5′-GAGGAAGTGTGAGCTCCGA-3′; DPP10d-f1 = 5′-ACCCAGCAGGAACTTAGAG-3′ and Pan-DPP10-r1 = 5′-CTCTAGTGACAGTCTTGTTTC-3′ (Exon III). GAPDH primer pairs were also designed to serve as input controls for the different cDNA preparations: GAPDH-f1 = 5′-GTCTTCACCACCATGGAGA-3′ and GAPDH-r1 = 5′-ATGACCTTGCCCACAGCCT-3′. Twenty cycles of PCR amplification using Pfu Ultra polymerase (Stratagene) were performed using the external primer pairs. Subsequent nested quantitative PCR amplifications were conducted using pairs of internal primers with the following sequences: DPP10a-f2 = 5′-CCATCACATCAAGTGTCAGC-3′; DPP10c-f2 = 5′-GATGACAGCCATGAAGCAG-3′; DPP10d-f2 = 5′-GAACTTAGAGCATCCCAGTG-3′; Pan-DPP10-r2 = 5′-GGATGACAGACATTGTGATG-3′ (Exon II); GAPDH-f2 = 5′-CCAAAAGGGTCATCATCTC-3′ and GAPDH-r2 = 5′-ATCCACAGTCTTCTGAGTG-3′. Quantitative real-time PCR reactions were carried out using IQ SYBR Green Supermix (Bio-Rad) on a PTC-200 thermal cycler with a chromo-4 detection system (Bio-Rad). The amplification protocol consists of an one-time, 3 min incubation at 95°C and the following PCR program: 40 cycles of 95°C for 15 sec, 53°C for 1 min, 80°C for 10 sec, and a plate read. After amplification, a melting curve analysis was conducted where SYBR Green Fluorescence was measured at 1°C intervals between 40-90°C. It is based on this analysis that 80°C was determined to be the optimal temperature for the fluorescent quantitation of PCR products.
Tissue preparation and in situ hybridization
Following decapitation, whole brains of P12 Sprague-Dawley rats were quickly removed and frozen in isopentane (−30°C, 30 s). Frozen tissues were cut using a Jung CM3000 cryostat (Leica, Heidelberger, Germany), and tissue sections (20 μm) were mounted onto slides with additional coating of poly-L-lysine and postfixed with 4% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS, pH 7.4) for 1 hr at room temperature. Afterwards, the sections underwent 5-min washes (three times in PBS, then once in water), air dried, and stored at −70°C until use.
In situ hybridizations with [35S]-labelled riboprobes were performed as described previously (Broides et al., 1996). Briefly, slide-mounted sections were treated with 1 mg/ml proteinase K for 10 min, acetylated with acetic anhydride for 10 min, dehydrated quickly in ascending concentrations (50%-100%) of ethanol, and air dried. The sections were then incubated with the [35S]-labelled riboprobes (1 × 107 cpm/ml) for 16-20 hrs at 60°C in a hybridization solution containing 50% formamide, 10% dextran sulfate, 0.02% Ficoll, 0.02% polyvinyl pyrolidone, 0.02% bovine serum albumin, 500 μg/ml tRNA, 10 mM dithiothreitol, 0.3 M NaCl, 10 mM Tris (pH 8.0), and 1 mM EDTA (pH 8.0). Afterwards, the sections were treated with 20 μg/ml RNase A for 35 min to remove non-specifically bound probes, followed by four high-stringency washes of decreased salinity (2X-0.1X SSC buffer) at room temperature and one wash with 0.1X SSC at high temperature (65°C) for 35 min. After dehydration through graded ethanol, the sections were dried and apposed to BioMax MR film (Eastman Kodak Company, Rochester, NY) for 3 days at 4°C. The slides were also dipped in photographic emulsion (NTB2, Kodak) and left to expose for an appropriate period of time before being developed and counter-stained with cresyl-violet. The bright- and darkfield images were viewed using a Olympus BX60 microscope (Olympus America Inc., Central Valley, PA) and captured using C-View V2.1 imaging software (Digital Video Camera Company Inc., Austin, TX).
Heterologous expression in Xenopus oocytes
In accordance with a protocol approved by the Institutional Animal Care and Use Committees (IACUC), Xenopus laevis frogs were anaesthetized with 0.1% Tricane solution absorbed through the skin, and stage V-VI oocyte were surgically harvested through a small incision and defolliculated by collagenase I treatment. The incision was sutured following surgery. Using a Nanoinjector (Drummond, Broomall, PA), oocytes were injected with Kv4.2 cRNA (typically 2-4 ng/oocyte) with or without auxiliary subunit cDNAs. For electrophysiological recordings, injected oocytes were incubated at 18°C for 1-3 days in standard ND96 solution (in mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.4 adjusted with NaOH) supplemented with 5 mM Na-pyruvate and 5 μg/ml gentamycin. The oocytes were incubated for 4-5 days for immunoprecipitation procedures.
Immunoprecipitation and Western blotting
Our procedures for immunoprecipitation and western blotting have been described in detail elsewhere (Jerng et al., 2004a). The oocytes of interest were incubated in homogenization buffer with non-denaturing detergent (1% Triton X-100) and subsequently ruptured by repeated passage through small-gauge needles. The protein extracts were treated overnight with various antibodies: goat IgG (ICN Biomedical, Irvine, CA), goat anti-Kv4.2 antibody (C20, Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-HA antibody (Y-11-G, Santa Cruz Biotechnology), and goat anti-c-Myc antibody (A-11-G, Santa Cruz, Biotechnology). Precipitated proteins were recovered by antibody binding to immobilized protein A/G beads (Pierce, Rockford, IL), eluted under denaturing and reducing conditions (10% SDS and 100 mM DTT), and separated on gel matrix by SDS-PAGE. After their transfer onto Immobilon membranes (Millipore, Bedford, MA), the resulting blots were probed with anti-Kv4.2, anti-HA, and anti-c-Myc antibodies, followed by secondary HRP-conjugated antibodies. Bound antibody complexes were detected by chemiluminescence using an ECL kit (Pierce).
Solutions and electrophysiological recordings
Whole-cell currents from injected oocytes were elicited using the two-electrode voltage clamp technique. The microelectrodes, with <1 MΩ tip resistance, were filled with 3 M KCl solutions. The voltage-clamp amplifier (Oocyte Clamp, Warner Instruments, Hamden, CT) operated under the control of the pClamp6 software (Axon instruments, Foster City, CA). The data were digitized and low-pass filtered (Frequency Devices, Haverhill, MA) at various frequencies in accordance with the sampling rate. The capacitative transients were subtracted either on-line using P/4 subtraction or off-line by scaling up transients at voltages without ionic currents (at −90 mV) and subtracting them from total currents. Only recordings with offsets <2 mV were analysed, and the average leak current was <0.2 μA and subtracted off-line, assuming Ohmic leak.
Data acquisition and analysis
Digitized voltage-clamp data were acquired using pClamp6 (Axon Instruments) and analysed with Clampfit (Axon Instruments) and Origin software (OriginLab Corp., Northampton, MA). Peak conductance (Gp) was calculated as Gp = Ip/ (Vc-Vrev), where Ip is the peak current, Vc is the command voltage, and Vrev is the reversal potential (−90 mV in ND96). Peak conductance-voltage (g-V) curves were described using the first-order Boltzmann function: Gp/Gpmax = 1/ (1 + exp(Vm – Va)/ k)), where Gp/Gmax is the fraction of maximal conductance, Vm is the membrane potential, Va is the potential for half-maximal activation, and k is the slope factor. Steady-state inactivation is also described using a single Boltzmann function. The time courses of recovery from inactivation were measured using a two-pulse protocol. Briefly, two pulses to +50 mV (inactivating pulse and test pulse) were separated by a variable recovery interval at −100 mV to determine the time required for channel recovery. The results for recovery from inactivation were fitted with single exponential functions. Time course of inactivation was described with the sum of two or three exponential terms using Clampfit. The left cursor was positioned at the point of inflection as determined by differentiating the current. Data are presented as mean ± standard error of the mean (SEM).
ACKNOWLEDGEMENTS
We thank Dr. Lily Jan for providing the Kv4.2 cDNA and Drs. Ramiro Salas and Mariella DeBiasi for their invaluable assistance and advice on the in situ hybridization experiments. We also thank Dr. Covarrubias for critical reading of the manuscript and Alison Prince for assisting in harvesting quality oocytes for the functional studies. This work was supported by a grant from the National Institute of Health (P01 NS37444).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen M, Heinemann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, Tinsley J, Zhang Y, Holt R, Jones EY, Lench N, Carey A, Jones H, Dickens NJ, Dimon C, Nicholls R, Baker C, Xue L, Townsend E, Kabesch M, Weiland SK, Carr D, von Mutius E, Adcock IM, Barnes PJ, Lathrop GM, Edwards M, Moffatt MF, Cookson WO. Positional cloning of a novel gene influencing asthma from Chromosome 2q14. Nat. Genet. 2003;35:258–263. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Böhring R, Dannenberg J, Peters HC, Leicher T, Pongs O, Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. J. Biol. Chem. 2001a;276:23888–23894. doi: 10.1074/jbc.M101320200. [DOI] [PubMed] [Google Scholar]
- Böhring R, Boland LM, Varghese A, Gebauer M, Pongs O, Isbrandt D. Kinetic analysis of open- and closed-state inactivation transition in human Kv4.2 A-type potassium channels. J. Physiol. 2001b;535:65–81. doi: 10.1111/j.1469-7793.2001.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks MI, Haberly LB, Jackson MB. Layer-specific properties of the transient K current (IA) in piriform cortex. J. Neurosci. 1996;16:3862–3876. doi: 10.1523/JNEUROSCI.16-12-03862.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranauskas G. Cell-type-specific splicing of KChIP4 mRNA correlates with slower kinetics of A-type current. Eur. J. Neurosci. 2004;20:385–391. doi: 10.1111/j.1460-9568.2004.03494.x. [DOI] [PubMed] [Google Scholar]
- Beck EJ, Bowlby M, An WF, Rhodes KJ, Covarrubias M. Remodeling inactivation gating of Kv4 channels by KChIP1, a small-molecular-weight calcium-binding protein. J. Physiol. 2002;538:691–706. doi: 10.1113/jphysiol.2001.013127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkers JM. Distribution and activation of voltage-gated potassium channels in cell-attached and outside-out patches from large layer 5 cortical pyramidal neurons of the rat. J. Physiol. 2000;525:611–620. doi: 10.1111/j.1469-7793.2000.t01-2-00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, Anderson A, Becker A, Poolos NP, Beck H, Johnston D. Acquired dendritic channelopathy in temporal lobe epilepsy. Science. 2004;305:532–535. doi: 10.1126/science.1097065. [DOI] [PubMed] [Google Scholar]
- Broide RS, Robertson RT, Leslie FM. Regulation of alpha7 nicotinic acetylcholine receptor in the developing rat somatosensory cortex by thalamocortical afferents. J. Neurosci. 1996;16:2956–2971. doi: 10.1523/JNEUROSCI.16-09-02956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhalter A, Gonchar Y, Mellor RL, Nerbonne JM. Differential expression of IA channel subunits Kv4.2 and Kv4.3 in mouse visual cortex neurons and synapses. J. Neurosci. 2006;26:12274–12282. doi: 10.1523/JNEUROSCI.2599-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PA, Cooper EC, Lowenstein DH, Baraban SC. Hippocampal heterotopia lack functional Kv4.2 potassium channels in the methylazoxymethanol model of cortical malformation and epilepsy. J. Neurosci. 2001;21:6626–6634. doi: 10.1523/JNEUROSCI.21-17-06626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ajami K, McCaughan GW, Gorrell MD, Abbott CA. Dipeptidyl peptidase IV gene family. Adv. Exp. Med. Biol. 2003;524:79–86. doi: 10.1007/0-306-47920-6_10. [DOI] [PubMed] [Google Scholar]
- de Lecea L, Soriano E, Criado JR, Steffensen SC, Henriksen SJ, Sutcliffe JG. Transcripts encoding a neural membrane CD26 peptidase-like protein are stimulated by synaptic activity. Mol. Brain. Res. 1994;25:286–296. doi: 10.1016/0169-328x(94)90164-3. [DOI] [PubMed] [Google Scholar]
- Dougherty K, Covarrubias M. A dipeptidyl aminopeptidase-like protein remodels gating charge dynamics in Kv4.2 channels. J. Gen. Physiol. 2006;128:745–753. doi: 10.1085/jgp.200609668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer M, Isbrandt D, Sauter K, Callsen B, Nolting A, Pongs O, Böhring R. N-type inactivation features of Kv4.2 channel gating. Biophys. J. 2004;86:210–223. doi: 10.1016/S0006-3495(04)74097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Li H, Aimon F, Johns DC, Rhodes K, Trimmer JS, Nerbonne JM. Role of heteromultimers in the generation of myocardial transient outward K+ currents. Circ. Res. 2002;90:586–593. doi: 10.1161/01.res.0000012664.05949.e0. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 3rd Ed Sinauer Associates; Sunderland, MA: 2001. pp. 622–624. [Google Scholar]
- Holmqvist MH, Cao J, Hernandez-Pineda R, Jacobson MD, Carroll KI, Sung MA, Betty M, Ge P, Gilbride KJ, Brown ME, Jurman ME, Lawson D, Silos-Santiago I, Xie Y, Covarrubias M, Rhodes KJ, Distefano PS, An WF. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. USA. 2002;99:1035–1040. doi: 10.1073/pnas.022509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- Hough RB, Lengeling A, Bedian V, Lo C, Bucan M. Rump white inversion in the mouse disrupts dipeptidyl aminopeptidase-like protein 6 and causes dysregulation of Kit expression. Proc. Natl. Acad. Sci. USA. 1998;95:13800–13805. doi: 10.1073/pnas.95.23.13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HJ, Carrasquillo Y, Karim F, Jung WF, Nerbonne JM, Schwarz TL, Gereau RW. The Kv4.2 potassium channel subunit is required for pain plasticity. Neuron. 2006;50:89–100. doi: 10.1016/j.neuron.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Jerng HH, Covarrubias M. K+ channel inactivation mediated by the concerted action of the cytoplasmic N- and C-terminal domains. Biophys. J. 1997;72:163–174. doi: 10.1016/S0006-3495(97)78655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Qian Y, Pfaffinger PJ. Modulation of Kv4.2 channel expression and gating by dipeptidyl peptidase 10 (DPP10) Biophys. J. 2004a;87:2380–2396. doi: 10.1529/biophysj.104.042358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Pfaffinger PJ, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol. Cell. Neurosci. 2004b;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Jerng HH, Kunjilwar K, Pfaffinger PJ. Multiprotein assembly of Kv4.2, KChIP3, and DPP10 produces ternary channel complex with ISA-like properties. J. Physiol. 2005;568:767–788. doi: 10.1113/jphysiol.2005.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Pfaffinger PJ. Variants of DPP10 are differentially localized in brain and distinctly modulate Kv4.2 channel properties. Biophys. J. 2006;90:B248. (Abstr) [Google Scholar]
- Johns DC, Nuss HB, Marban E. Suppression of neuronal and cardiac transient outward currents by viral gene transfer of dominant-negative Kv4.2 constructs. J. Biol. Chem. 1997;272:31598–31603. doi: 10.1074/jbc.272.50.31598. [DOI] [PubMed] [Google Scholar]
- Johnston D, Hoffman DA, Magee JC, Poolos NP, Watanabe S, Colbert CM, Migliore M. Dendritic potassium channels in hippocampal pyramidal neurons. J. Physiol. 2000;525:75–81. doi: 10.1111/j.1469-7793.2000.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wei D-S, Hoffman DA. Kv4 potassium channel subunits control action potential repolarization and frequency-dependent broadening in rat hippocampal CA1 pyramidal neurones. J. Physiol. 2005;569:41–57. doi: 10.1113/jphysiol.2005.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee R, Ficker E, Heinemann U. Comparison of voltage-dependent potassium currents in rat pyramidal neurons acutely isolated from hippocampal regions CA1 and CA3. J. Neurophysiol. 1995;74:1982–1995. doi: 10.1152/jn.1995.74.5.1982. [DOI] [PubMed] [Google Scholar]
- Korngreen A, Sakmann B. Voltage-gated K+ channels in layer 5 neocortical pyramidal neurones from young rats: subtypes and gradients. J. Physiol. 2000;525:621–639. doi: 10.1111/j.1469-7793.2000.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korngreen A, Kaiser KMM, Zilberter Y. Subthreshold inactivation of voltage-gated K+ channels modulates action potentials in neocortical bitufted interneurones from rats. J. Physiol. 2005;562:421–437. doi: 10.1113/jphysiol.2004.077032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauver A, Yuan L-L, Jeromin A, Nadin BM, Rodriguez JJ, Davies HA, Stewart MG, Wu G-Y, Pfaffinger PJ. Manipulating Kv4.2 expression identifies a specific component of hippocampal pyramidal neuron A-current that depends upon Kv4.2 expression. J. Neurochem. 2006;99:1207–1223. doi: 10.1111/j.1471-4159.2006.04185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-L, Qu Y-J, Lu YC, Bondarenko VE, Wang S, Skerrett IM, Morales MJ. DPP10 is an inactivation modulatory protein of Kv4.3 and Kv1.4. Am. J. Physiol. Cell. Physiol. 2005;291:C966–C976. doi: 10.1152/ajpcell.00571.2005. [DOI] [PubMed] [Google Scholar]
- Lien C-C, Martina M, Schultz JH, Ehmke H, Jonas P. Gating, modulation and subunit composition of voltage-gated K+ channels in dendritic inhibitory interneurones of rat hippocampus. J. Physiol. 2002;538:405–419. doi: 10.1113/jphysiol.2001.013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss B, Franz O, Sewing S, Bruns R, Neuhoff H, Roeper J. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChIP3.1 transcription. EMBO J. 2001;20:5715–5724. doi: 10.1093/emboj/20.20.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SA, Nerbonne JM. Elimination of the fast transient in superior cervical ganglion neurons with expression of Kv4.2W362F: Molecular dissection of IA. J. Neurosci. 2000;20:5191–5199. doi: 10.1523/JNEUROSCI.20-14-05191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina M, Schultz JH, Ehmke H, Monyer H, Jonas P. Functional and molecular differences between voltage-gated K+ channels of fast-spiking interneurons and pyramidal neurons of rat hippocampus. J. Neurosci. 1998;18:8111–8125. doi: 10.1523/JNEUROSCI.18-20-08111.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:964–958. [PubMed] [Google Scholar]
- Nadal MS, Amarillo Y, Vega-Saenz de Miera E, Rudy B. Evidence for the presence of a novel Kv4-mediated A-type K+ channel-modifying factor. J. Physiol. 2001;537:801–9. doi: 10.1111/j.1469-7793.2001.00801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal MS, Ozaita A, Amarillo Y, Vega-Saenz de Miera E, Ma Y, Mo W, Goldberg EM, Misumi Y, Ikehara Y, Neubert TA, Rudy B. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channel. Neuron. 2003;37:449–461. doi: 10.1016/s0896-6273(02)01185-6. [DOI] [PubMed] [Google Scholar]
- Nadal MS, Amarillo Y, Vega-Saenz de Miera E, Rudy B. Differential characterization of three alternative spliced isoforms of DPPX. Brain Res. 2006 doi: 10.1016/j.brainres.2006.03.106. doi:10.1016/j.brainres.2006.03.106. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Ishikawa K, Hirosawa M, Ohara O. Prediction of the coding sequences of unidentified human genes. XVII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:143–150. doi: 10.1093/dnares/7.2.143. [DOI] [PubMed] [Google Scholar]
- Qi SY, Riviere PJ, Trojnar J, Junien J-L, Akinsanya KO. Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine proteases. Biochem. J. 2003;373:179–189. doi: 10.1042/BJ20021914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hayashi Y, Yoshimura N, Takimoto K. Transmembrane interaction mediates complex formation between peptidase homologues and Kv4 channels. Mol. Cell. Neurosci. 2005;29:320–332. doi: 10.1016/j.mcn.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, Burke SL, Strassle BW, Buchwalder L, Menegola M, Cao J, An WF, Trimmer JS. KChIPs and Kv4 α subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Isa T. Voltage-gated transient outward currents in neurons with different firing patterns in rat superior colliculus. J. Physiol. 2000;528:91–105. doi: 10.1111/j.1469-7793.2000.00091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppa NE, Westbrook GL. Regulation of synaptic timing in the olfactory bulb by an A-type potassium current. Nat. Neurosci. 1999;2:1106–1113. doi: 10.1038/16033. [DOI] [PubMed] [Google Scholar]
- Shibata R, Nakahira K, Shibasaki K, Wakazono Y, Imoto K, Ikenaka K. A-type K+ current mediated by the Kv4 channel regulates the generation of action potential in developing cerebellar granule cells. J. Neurosci. 2000;20:4145–4155. doi: 10.1523/JNEUROSCI.20-11-04145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Misonou H, Campomanes CR, Anderson AE, Schrader LA, Doliveira LC, Carroll KI, Sweatt JD, Rhodes KJ, Trimmer JS. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J. Biol. Chem. 2003;278:36445–54. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- Strop P, Bankovich AJ, Hansen KC, Garcia KC, Brunger AT. Structure of a human A-type potassium channel interacting protein DPPX, a member of the dipeptidyl aminopeptidase family. J. Mol. Biol. 2004;343:1055–1065. doi: 10.1016/j.jmb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Takimoto K, Hayashi Y, Ren X, Yoshimura N. Species and tissue differences in the expression of DPPY splicing variants. Biochem. Biophys. Res. Comm. 2006;348:1984–1100. doi: 10.1016/j.bbrc.2006.07.157. [DOI] [PubMed] [Google Scholar]
- Tkatch T, Baranausakas G, Surmeier DJ. Kv4.2 mRNA abundance and A-type K+ current amplitude are linearly related in basal ganglia and basal forebrain neurons. J. Neurosci. 2000;20:579–588. doi: 10.1523/JNEUROSCI.20-02-00579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Hoffman DA, Migliore M, Johnston D. Dendritic K+ channels contribute to spike-timing dependent long-term potentiation in hippocampal pyramidal neurons. Proc. Natl. Acad. Sci. USA. 2002;99:8366–8371. doi: 10.1073/pnas.122210599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Burkhalter A, Nerbonne JM. Functional role of the fast transient outward K+ current IA in pyramidal neurons in (rat) primary visual cortex. J. Neurosci. 2005;25:9185–9194. doi: 10.1523/JNEUROSCI.2858-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagha E, Ozaita A, Chang SY, Nadal MS, Lin U, Saganich MJ, McCormack T, Akinsanya KO, Qi SY, Rudy B. Dipepidyl peptidase 10 modulates Kv4-mediated A-type potassium channels. J. Biol. Chem. 2005;280:18853–18861. doi: 10.1074/jbc.M410613200. [DOI] [PubMed] [Google Scholar]
- Zhou F-M, Hablitz JJ. Layer I neurons of the rat neocortex. II. Voltage-dependent outward currents. J. Neurophysiol. 1996;76:668–682. doi: 10.1152/jn.1996.76.2.668. [DOI] [PubMed] [Google Scholar]