Abstract
Background
One of the key genes that regulate human brain size, MCPH1 has evolved under strong Darwinian positive selection during the evolution of primates. During this evolution, the divergence of MCPH1 protein sequences among primates may have caused functional changes that contribute to brain enlargement.
Results
To test this hypothesis, we used co-immunoprecipitation and reporter gene assays to examine the activating and repressing effects of MCPH1 on a set of its down-stream genes and then compared the functional outcomes of a series of mutant MCPH1 proteins that carry mutations at the human- and great-ape-specific sites. The results demonstrate that the regulatory effects of human MCPH1 and rhesus macaque MCPH1 are different in three of eight down-stream genes tested (p73, cyclinE1 and p14ARF), suggesting a functional divergence of MCPH1 between human and non-human primates. Further analyses of the mutant MCPH1 proteins indicated that most of the human-specific mutations could change the regulatory effects on the down-stream genes. A similar result was also observed for one of the four great-ape-specific mutations.
Conclusions
Collectively, we propose that during primate evolution in general and human evolution in particular, the divergence of MCPH1 protein sequences under Darwinian positive selection led to functional modifications, providing a possible molecular mechanism of how MCPH1 contributed to brain enlargement during primate evolution and human origin.
Keywords: MCPH1, E2F1, Brain size, Primates, Evolution, Functional divergence
Background
A dramatic increase in brain size is one of the hallmarks of human evolution, but despite the significance of this trait, the causal molecular mechanism underlying this expansion is unclear [1]. Until recently, addressing this question with genetic tools has been difficult because the dramatically enlarged brain is a human-specific trait. Genetic studies of a rare brain developmental disorder, human autosomal primary microcephaly syndrome (MCPH, OMIM251200), have uncovered a set of genes that regulate brain development. To date, seven genes have been identified as being responsible for this syndrome: MCPH1, also known as BRIT1 (BRCT-repeat inhibitor of hTERT expression) [2,3], WDR62 (WD repeat domain 62; MCPH2) [4-6], CDK5RAP2 (cyclin-dependnet kinase 5 regulatory associated protein 2; MCPH3) [7], CEP152 (centrosomal protein 152 kDa; MCPH4) [8], ASPM (abnormal spindle like microcephaly associated protein; MCPH5) [9], and CENPJ (centromeric protein J; MCPH6) [7] and STIL (SCL/TAL1 interrupting locus; MCPH7) [10].
Previous evolutionary analyses of these microcephaly genes showed that four of them, ASPM, CDK5RAP2, CENPJ and MCPH1, evolved rapidly under Darwinian positive selection during the evolution of human and non-human primates [11-16]. ASPM also experienced positive selection across anthropoids [14-16], while CDK5RAP2 and CENPJ showed accelerated rates of non-synonymous substitutions over the course of primate evolution [11,16]. The signal of positive selection on MCPH1 was observed in the common ancestor of great apes and humans as well as in the human lineage [12], although another study on MCPH1 only detected positive selection in the anthropoids as a whole and no particular acceleration in the human lineage [16]. This rapid evolution suggests these genes may have had a key role in the evolutionary enlargement of the brain, although the link of CENPJ and MCPH1 to the evolution of gross brain size was not confirmed in the association analysis of absolute neonatal brain size among primates [16]. Among the four rapidly evolving microcephaly genes, only ASPM has been experimentally studied to detect the evolutionary consequence of protein sequence changes; mice carrying a truncated ASPM protein were shown to have reductions of both brain and testis size, while the transgenic mouse carrying human ASPM could rescue this phenotype, but did not cause any additional enlargement of the brain [17].
MCPH1 was the first gene identified as being responsible for autosomal recessive primary microcephaly, characterized by significantly reduced brain volume, mental retardation and premature chromosome condensation (PCC) syndrome [2,18]. The MCPH1 gene encodes a 2,508-bp-long coding sequence (CDS) with 14 exons, spanning about 240 kb at 8p23. The MCPH1 protein contains three BRCA1-Carboxyl Terminal (BRCT) domains, including one N terminal BRCT domain and a tandem pair at the C terminus. Numerous studies have implied that the BRCT domains of MCPH1 function as the key component for protein-protein interaction; this seems likely as the interaction of the MCPH1 tandem BRCT domains with proteins like E2F1 and r-H2AX is required for the activation of cell cycle checkpoint, DNA repair and apoptosis [19-23]. Several studies have likewise suggested that MCPH1 may also function as a tumor suppressor [3,23].
Evolutionary studies of MCPH1 have demonstrated a rapid change in protein sequence associated with the brain enlargement during primate evolution and human origin. Interestingly, during two key taxonomic transitions in primates, that is, between lesser apes and great apes, and between great apes and humans, absolute brain volume was greatly enlarged, and MCPH1 might be involved in this process [12]. Additionally, MCPH1 is also highly polymorphic in human populations and still carries the molecular signature of on-going positive selection [12]. Human population studies have reported a sex-specific association between a MCPH1 sequence variant and brain volume [24,25]. These results suggest that the protein sequence changes, especially the human-specific changes of MCPH1 may have caused the functional changes that explain the genetic basis for the evolution of brain size in primates.
Previously, the MCPH1 protein has been shown to play an essential role during cell cycle and cell apoptosis and it can physically interact with E2F1 to form a complex and bind the promoters of the target genes for regulating their transcriptional activities [19]. Beyond this, MCPH1 alone can also function as a transcriptional regulator, and we previously demonstrated MCPH1 could function as a transcriptional repressor [26]. Together, these regulatory mechanisms allow the experimental testing of the functional changes of MCPH1 during primate evolution.
To detect if the protein sequence divergence of MCPH1 among primates may confer any functional alterations, we selected eight known down-stream genes regulated by E2F1 and MCPH1: p73[19], p107[19], p18[27], p27[28], p14ARF[19], Caspase7[19], CyclinE1[19] and hTERT[26]. These genes are involved in cell proliferation and apoptosis, critical processes regulating brain development (see Additional file 1: Figure S1). We tested the activating effects (together with E2F1) and the repressing effects (MCPH1 alone) of MCPH1 on these genes’ promoter when introducing mutations at the sites containing human- and great-ape-specific amino acid changes. Our results demonstrated that most of the human-specific amino acid substitutions could influence the regulatory effects of MCPH1 on the down-stream genes, and a similar effect was also seen for one of the four great-ape-specific changes, suggesting that the species and lineage specific mutations of MCPH1 are indeed functional and potentially contributed to brain enlargement over the course of primate evolution.
Results
Identification of lineage-specific MCPH1 amino acid substitutions
In order to identify lineage-specific amino acid substitutions, MCPH1 orthologs of representative primate and other mammalian species were obtained from the NCBI, EMBL and UniProt databases. We used a total of 11 species including 7 primates (human, great apes, lesser ape, Old World monkey and New World monkey) and 4 other representative mammalian species (mouse, rat, dog and cow) (Figure 1). Using MUSCLE and Clustal W, we aligned the MCPH1 protein sequences (see Additional file 2: Figure S2). During primate evolution, there are two key taxonomic transitions accompanied by brain enlargements (between lesser apes and great apes, and between great apes and humans). We identified nine sites containing substitutions considered as human-specific sites that occur in the human lineage but are relatively conserved in the other species (M96, S101, V310, H314, T377, Y425, L442, R485 and P835; Figure 1, Additional file 3: Table S1). We further selected four sites containing substitutions considered as great-ape-specific since they occurred in the ancestor of Hominidae (I161, E167, A510 and S841; Figure 1, Additional file 3: Table S1). The physicochemical properties of the 13 lineage-specific amino acid substitutions are shown in Additional file 3: Table S1 and the schematic map of the MCPH1 protein domains labeled with the lineage-specific substitutions are shown in Figure 1. All these lineage-specific sites were selected to generate mutant MCPH1 proteins for the reporter gene assays in order to test their functional effects.
Figure 1.
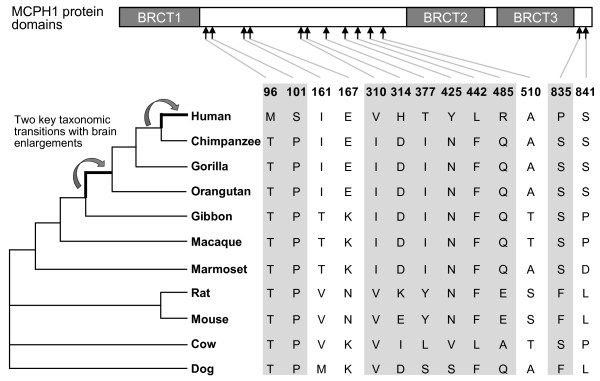
Schematic map of MCPH1 protein domains labeled with lineage-specific amino acid substitutions. Sites containing human specific substitutions are marked with shadows, and the two dramatic brain size enlargement events that coincided with the molecular signatures of Darwinian positive selection during primate evolution are indicated.
Test of protein-protein interaction between MCPH1 and E2F1
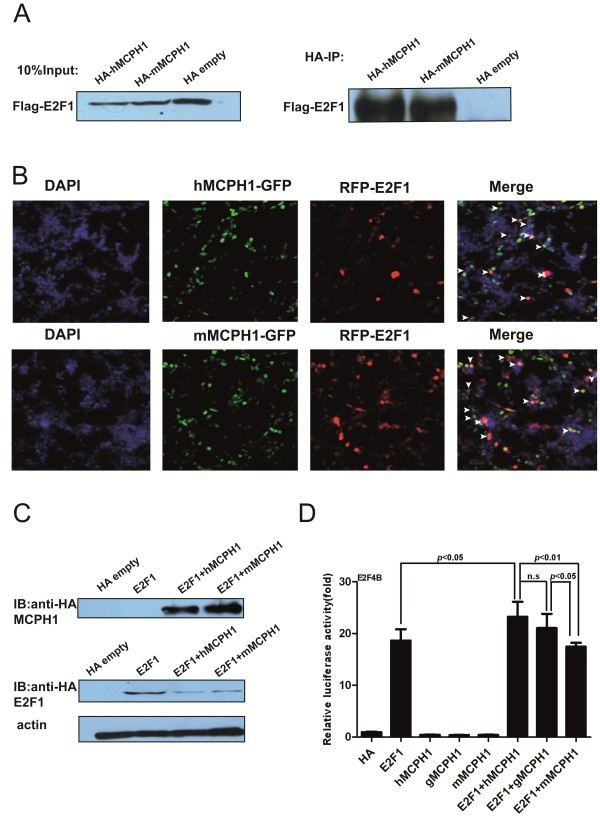
Previous studies suggested human MCPH1 (hMCPH1) could interact with E2F1in vitro and in vivo[19]. To test if the non-human primate MCPH1 can also interact with E2F1, we cloned the rhesus macaque MCPH1 (mMCPH1). The results of the co-immunoprecipitation assay showed that mMPCH1 can also directly interact with E2F1, and no difference was observed for the intensity of the interaction with E2F1 between hMCPH1 and mMCPH1 (Figure 2A). Given the established MCPH1-E2F1 interaction in human cell lines [19], this protein-protein interaction mechanism seems likely to have been conserved during primate evolution. We also conducted a cellular co-localization assay for both mMCPH1 and hMCPH1, and the results indicated that both were co-localized with E2F1 (Figure 2B), consistent with the results of the co-immunoprecipitation assay.
Figure 2.
Human and macaque MCPH1 can directly interact with E2F1. (A) Interaction of the transfected E2F1 and MCPH1 in HEK293T cells. (B) Co-localization of MCPH1-GFP and RFP-E2F1 in HEK293T cells. White arrows indicate co-staining areas. (C) Expression of the HA-tagged human and macaque MCPH1s were verified by Western blot using anti-HA monoclonal antibody (top panel), and the HA-tagged E2F1 was also verified by Western blot (middle panel). The bottom panel is the actin control. (D) Quantification of E2F1 and MCPH1 transcriptional activity using the E2F4B constructs. All histograms represent the mean ± SD of at least nine data points, including both biological and technical replicates (* P <0.05,** P <0.01, *** P <0.001, n.s, not significant).
Divergent effects of hMCPH1 and mMCPH1 on transcriptional regulation
MCPH1 interacts with E2F1 to enhance its transactivation activity by forming a complex, which binds to the promoters of the E2F1 target genes, including p73, Caspase and RAD51, among others [19]. We, therefore, tested the enhancing effect of MCPH1 using the E2F4B reporter vector commonly used as the positive control in testing the transactivation activity of E2F1[29]. The expression of the HA-tagged MCPH1 and E2F1 were verified by Western blot using anti-HA monoclonal antibodies (Figure 2C). Our results showed that hMCPH1 could significantly enhance the transactivation activity of E2F1, but no significant effect was observed for mMCPH1 and gMCPH1 (the gibbon copy of MCPH1) (Figure 2D). In addition, the difference between E2F1 + hMCPH1 and E2F1 + mMCPH1 is significant (P <0.01, ANOVA test). The same trend was also seen when comparing E2F1 + hMCPH1 and E2F1 + gMCPH1, though not statistically significant (Figure 2D). Interestingly, the difference between E2F1 + gMCPH1 and E2F1 + mMCPH1 is significant (P <0.05, ANOVA test), suggesting continuum of functional divergence of MCPH1 during primate evolution with a major shift in function between Old World monkeys and apes and a probable further shift during human evolution (Figure 2D).
To check the conservation of the E2F1 binding sites (CGCGC), we aligned the promoter sequences of the target genes among representative primate species. We found that most of the E2F1 binding sites are highly conserved from humans to marmosets (see Additional file 4: Figure S3), ruling out the possibility of binding bias due to promoter sequence divergence. The E2F1 protein sequences are also highly conserved among primate species (Additional file 5: Figure S4), ruling out the possibility that the different effect of MCPH1 is caused by mutations in E2F1. Collectively, these results suggest a functional divergence of MCPH1 between humans and nonhuman primates.
We further tested a set of E2F1 target genes (p73, p107, p18, p27, p14ARF, Caspase7 and CyclinE1). The regulatory network of these genes is shown in Additional file 1: Figure S1. We found that the enhancing effect of hMCPH1 was significantly stronger than mMCPH1 for CyclinE1 and p73 (Figure 3A, B) and a similar trend was also observed for p14ARF, though not significantly (Figure 3C). No significant enhancing effects were detected for the other four genes, p18, p27, p107 and Caspase7 (see Additional file 6: Figure S5). Consistent with the result from the E2F4B assay, these data also suggest a functional divergence between hMCPH1 and mMCPH1 in enhancing the transactivation activity of E2F1.
Figure 3.
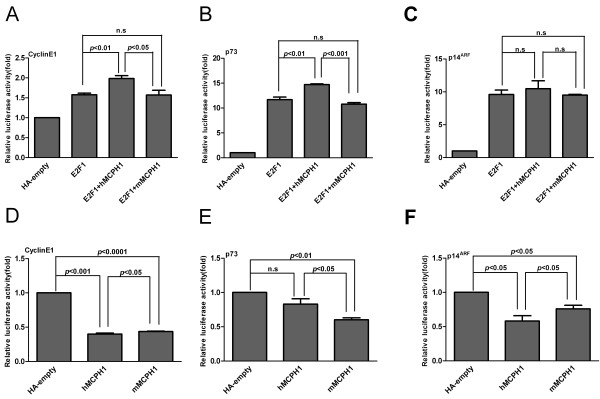
Comparison of MCPH1 enhancing and repressing effects between humans and macaques. (A) The enhancing effect difference between hMCPH1 and mMCPH1 on the CyclinE1 promoter. (B) The enhancing effect difference between hMCPH1 and mMCPH1 on the p73 promoter. (C) The enhancing effect difference between hMCPH1 and mMCPH1 on the p14ARF promoter. For enhancing effect assay, 0.2 ug target gene promoter construct was co-transfected with 0.2 ug HA tagged E2F1 per/well or 0.2 ug HA tagged E2F1 and 0.2 ug HA tagged hMCPH1 per/well or 0.2 ug HA tagged E2F1 and 0.2 ug HA tagged mMCPH1 per well in 24-well plates in HEK923T cells, Renilla was used as the internal control. The HA-tagged empty vector was used as the negative control. (D) The repressing effect difference between hMCPH1 and mMCPH1 on the CyclinE1 promoter. (E) The repressing effect difference between hMCPH1 and mMCPH1 on the p73 promoter. (F) The repressing effect difference between hMCPH1 and mMCPH1 on the p14ARF promoter. For repression assay, 0.2 ug target gene promoter construct was co-transfected with 0.2 ug HA tagged hMCPH1 per/well or 0.2 ug HA tagged mMCPH1 per/well in 24-well plates in HEK923T cells, Renilla was used as the internal control. The HA-tagged empty vector was used as the negative control. All histograms represent the mean ± SD of at least nine data points including both biological and technical replicates (* P <0.05,** P <0.01, *** P <0.001, n.s, not significant).
To further test the functional divergence between hMCPH1 and mMCPH1, we performed another assay to detect the regulatory effect of MCPH1 alone on eight down-stream genes (p73, p107, p18, p27, p14ARF, Caspase7, CyclinE1 and hTERT). Among the eight genes tested, seven showed a repressing effect for both hMCPH1 and mMCPH1, while only one, Caspase7, showed an activating effect. When comparing the effects between hMCPH1 and mMCPH1, three genes (CyclinE1, p73 and p14ARF) showed significant differences (P <0.05) of repressing effect (Figure 3D--F), the same three which showed enhancing differences between hMCPH1 and mMCPH1 in the E2F1-MCPH1 transactivation assay (Figure 3A-C). For CyclinE1 and p14ARF, hMCPH1 showed a stronger repressing effect than mMCPH1, while the opposite effect was observed for p73. We did not detect significant between-species differences for the other four genes with repressing effect (p18, p27, p107 and hTERT) though the trend was the same (see Additional file 7: Figure S6). Caspase 7 was the only gene with an activating effect, and we also observed a significant difference between hMCPH1 and mMCPH1 (P <0.05) (see Additional file 7: Figure S6D). Collectively, the data from the two reporter gene assays suggest a clear functional divergence of MCPH1 between humans and rhesus macaques.
Detection of regulatory changes of MCPH1 for the human-specific sites
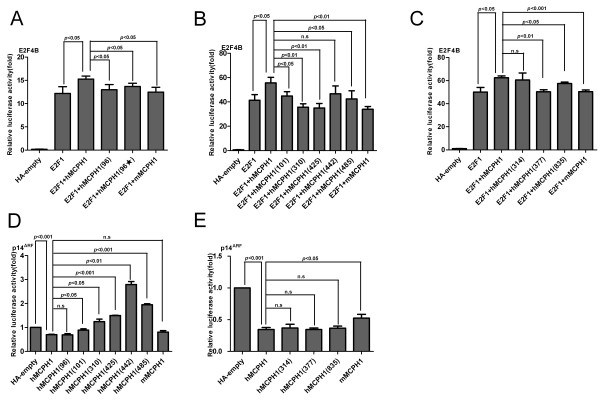
In order to test the functional effect of the human specific amino acids changes, we first constructed two mutant hMCPH1s by introducing a backward mutation of changing the human-specific amino acid to an ancestral amino acid (Met96Thr) and introducing a random mutation (Met96Leu) at Site-96 (Figure 1). With the use of the E2F4B transactivation reporter assay, we measured the enhancing effect of the two mutant hMCPH1s. As shown in Figure 4A, both mutant hMCPH1s showed a significantly decreased enhancing effect as compared with the wild-type hMCPH1, similar to the difference between hMCPH1 and mMCPH1, suggesting that the human-specific amino acid change at Site-96 can influence the enhancing effect of MCPH1.
Figure 4.
Results of assays testing transcriptional regulations of mutant MCPH1s containing mutations at human-specific sites. (A-C) The results of the E2F4B assay testing the changes of enhancing effect. (D-E) The results of the p14ARF assay testing the repressing effect. All histograms represent the mean ± SD of at least nine data points including both biological and technical replicates (* P <0.05,** P <0.01, *** P <0.001, n.s, not significant).
We next generated a series of mutant hMCPH1s carrying backward mutations at the other eight human-specific sites (101, 310, 314, 377, 425, 442, 485 and 835), and tested their enhancing effects. Five of the eight mutant hMCPH1s (101, 310, 377, 425 and 835) showed significantly decreased enhancing effects compared with the wild-type hMCPH1 (P <0.05) (Figure 4B, C) and the three non-significant sites (314, 442 and 485) showed the same trend. Among the nine human specific sites tested, the majority (6/9) showed a significantly decreased enhancing effect when mutated to the ancestral amino acids, suggesting most of the human-specific MCPH1 amino acid changes are in fact functional.
We also tested the repressing effect on p14ARF for the human-specific sites. As shown in Figure 4D, E, we detected a significant decrease of the p14ARF repressing effect for five sites (101, 310, 425, 442 and 485), and the same trend was observed for the other four, though it was not significant. Among the five significant sites, three (101, 310 and 425) overlapped with the six sites showing decreased enhancing effect. Taken together, we detected regulatory changes for most (eight out of nine) human-specific sites, suggesting hMCPH1 has acquired functional modifications during the origin of humans.
Detection of regulatory changes of MCPH1 for the great-ape-specific sites
To detect the functional consequences of MCPH1’s four great-ape-specific mutations (Figure 1), we conducted analyses similar to that used for the human-specific sites. The results indicated that for the enhancing assay, although all four great-ape-specific sites showed a decreased level as compared with the wild-type of hMCPH1, none were statistically significant (P >0.05; Figure 5A). In contrast, in the repressing assay, one great-ape-specific site (Site-167) showed a significant decrease of the repressing effect (Figure 5B), suggesting that this great-ape-specific mutation may have also caused functional modifications of MCPH1 during the evolution of the ancestor of Hominidae.
Figure 5.
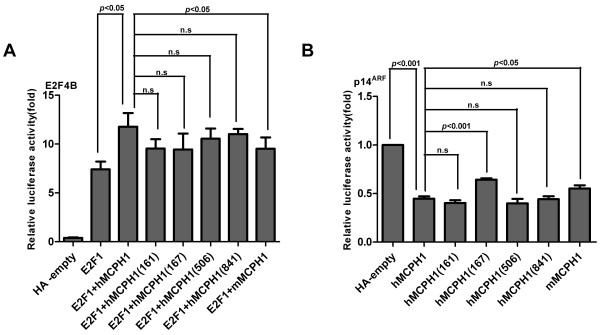
Results of assays testing transcriptional regulations of mutant MCPH1s containing mutations at human-specific sites. (A) Results of the E2F4B assay testing the changes of enhancing effect. (B) Results of the p14ARF assay testing the repressing effect. All histograms represent the mean ± SD of at least nine data points, including both biological and technical replicates (* P <0.05,** P <0.01, *** P <0.001, n.s, not significant).
Discussion
MCPH1 has experienced strong Darwinian positive selection during primate evolution, but there are no data showing functional divergence. Here we demonstrated evidence of functional divergence of MCPH1 between humans and nonhuman primates. Most of the human-specific amino acid changes could alter the regulatory effects of MCPH1 on the transcription of the down-stream genes, and a similar effect was observed for one of the four great-ape-specific amino acid changes. Accordingly, our data support the hypothesis that selection on MCPH1 has resulted in functional divergence at the protein level, which potentially contributes to changes in the development and evolution of brain size.
Absolute brain size has increased in parallel across primate evolution. Along the two branches we focused on in this analysis, absolute brain volume increased from 70 to 152 ml to 230 to 565 ml during the transition between lesser apes and great apes [30,31], and from 230 to 565 ml to 1,129 to 1,685 ml during the transition between great apes and humans [30,31]. Previous studies indicated that there were accelerated amino acid substitutions during both the origin of Hominidae’s ancestor and of our own species, paralleling the two brain enlargements [12], suggesting that the amino acid substitutions of MCPH1 were probably adaptive and may have contributed to the brain expansion during primate evolution. In addition, the gradient change of MCPH1’s transcription regulation from macaque to gibbon, and to humans (Figure 2D) seems to imply a continuum of functional divergence rather than a number of discrete shifts, which calls for further functional tests in extensive primate lineages.
Interestingly, all the human- and great-ape-specific mutations are located in the non-BRCT domains (Figure 1). Since the three BRCT domains of MCPH1 are critical for protein-protein interaction, the amino acid changes during primate evolution seems not to have caused drastic functional alteration, but rather a modification of the existing function.
Inferring the exact functional alterations of the human-specific and great-ape-specific mutations is difficult. Previous studies have shown that the middle domain (residues 367 to 485) of MCPH1, where the four human-specific mutations are located, is the binding domain by Condensin II for homologous recombination repair [32,33], an important mechanism for cell cycle checkpoints and genome integrity. Concurrently, all four human-specific sites located in this middle domain showed altered regulatory effects when mutated into ancestral amino acids, suggesting that the human-specific mutations may have changed the binding property with Condensin II. Additionally, all four human-specific mutations caused changes in physicochemical properties of amino acids (see Additional file 3: Table S1).
We also found that for the regulatory changes of the down-stream genes, almost half (three out of eight) of the tested genes (p73, CyclinE1 and p14ARF) had significant differences between humans and rhesus macaques, either in the enhancing assay with MCPH1-E2F1 or in the repressing assay with MCPH1 alone (Figure 3), indicating a functional divergence between humans and non-human primates. The protein p73 is involved in both cell cycle regulation and induction of apoptosis [34]. E2F1 is an important regulator of p73, especially during brain development [35]. CyclinE1, meanwhile, is involved in cell cycle and is a key target gene of E2F1[36] and has been shown to take part in the determination of the number of neurons during mouse corticogenesis by regulating the G1 mode of cell division [37]. p14ARF is an alternate reading frame product of CDKN2A involved in cell cycle regulation that is also involved in self-renewal of neural stem cells and neural development [38-41]. Additionally, human population studies have reported that the MCPH1 sequence variants were associated with brain volume in a sex-specific manner [24,25]. Recently, it was also reported that MCPH1 might have contributed to the evolution of sexual dimorphism in brain mass across anthropoid primates [42]. In fact, two of the down-stream genes regulated by MCPH1, p73 and cyclinE1, were reported to be associated with sex dimorphism during germ line development [43,44], suggesting that the regulation of MCPH1 on brain development may differ between males and females. Taken together, the strengthened transactivation effect of human MCPH1 on these down-stream genes may contribute to the greatly enlarged neuro-progenitor pool in the human brain during neurogenesis, which is in line with recent studies that suggest MCPH1’s functional role in neuro-progenitor cells through the Chk1-Cdc25-Cdk1 pathway [45,46].
Conversely, when MCPH1 acts alone as a transcription repressor, there were also differences between humans and rhesus macaques on the repressing effect of the down-stream genes (p73, CyclinE1 and p14ARF), implying that a homeostasis of gene expression regulation by MCPH1 is required during neurogenesis.
Although we observed functional divergence of MCPH1 due to its protein sequence changes during primate evolution and human origin, it should be stressed that we did not establish a direct link between the adaptive changes of MCPH1 and the ever-increasing brain size in primates. As shown in the MCPH1 knock-out mice analysis, the truncated MCPH1 not only caused a reduction in brain size, but also resulted in a reduction of testis size [45], suggesting that MCPH1 may also play a role during testis development. Accordingly, we cannot rule out the possibility that the adaptive evolution of MCPH1 in primates may be caused by selection on other phenotypes, though the current evidences mostly favor enlargement of the brain.
Initially proposed by King and Wilson [47], the importance of cis-regulatory changes in human evolution has recently been tested and confirmed [48]. However, our functional data of MCPH1 suggests that protein sequence changes may also have significant phenotypic effects. Hence, the evolution of an important trait like brain function may require genetic alterations at multiple regulatory levels.
Conclusions
We demonstrated the existence of functional alterations caused by the lineage-specific mutations of MCPH1 during the evolution of primates, especially during the origin of humans. The functional changes of MCPH1 are likely executed by regulating several key down-stream genes.
Methods
Ethical statement
The research protocol of this study was approved by the internal review board of Kunming Institute of Zoology, Chinese Academy of Sciences (Approval ID: SYDW-2012011).
Cell culture
The HEK293T cell line was obtained from ATCC. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Rockville, MD, USA) with 10% fetal bovine serum (Hyclone, Logan, UT, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Cloning of the macaque and gibbon MCPH1 gene
To clone the cDNA of the macaque and gibbon MCPH1 gene, we extracted the total RNA from macaque and gibbon brain tissue using TRIzol (Invitrogen, Carlsbad, CA, USA). RACE was carried out using a SMARTTM RACE cDNA amplification kit (Clontech, Palo Alto, CA, USA). The nested PCR was used and the primer sequences are:
macaMCPH1-3RACE: 5GAGAAAGAGGAGCATCAGGAGATCTATCA3;
macaMCPH1-5RACE: 5GGATTCCTCAGAAGTCACGCAACTGA3;
macaMCPH1-nest_3RACE: 5GAAAGAGGAGCATCAGGAGATCTATCAT3;
macaMCPH1-nest_5RACE: 5CAGAAGTCACGCAACTGAAAGTTGCA3
gibbonMCPH1-3RACE: 5TTAGCTGTGGGGAGTCTTCATATGATGAC3;
gibbonMCPH1-5RACE: 5GCGGGGTCCTCAATGGTGTAAGA3;
gibbonMCPH1-nest_3RACE: 5AGCTGTGGGGAGTCTTCATATGATGAC3;
gibbonMCPH1-nest_5RACE: 5GGTCCTCAATGGTGTAAGAAAAGCCA3;
The macaque MCPH1 PCR products were cloned into the pMD-19 simple-T-vector (Takara, Tokyo, Japan), then digested with BamH I and EcoR I, and cloned into a pCGN-HAM vector. The gibbon MCPH1 PCR products were cloned into the pMD-19 simple-T-vector (Takara, Tokyo, Japan), then digested with HindIII and AgeI, and cloned into a pCGN-HAM vector. The final constructs were confirmed by sequencing (ABI-3130 automatic sequencer).
Cloning of fluorescent MCPH1 plasmids
The full length cDNAs of human MCPH1 and macaque MCPH1 were PCR amplified, and the PCR products were digested with Age I and EcoR I and cloned into frame for N-terminal fusions into the cFUGW plasmids. The final constructs were confirmed by sequencing (ABI-3130 automatic sequencer).
Transient transfection and luciferase reporter assays
All transfections were carried out in triplicates in the 24-well plates (Corning, Corning, NY, USA). About 2 × 105 cells were seeded for 24 h prior to transfection. Briefly, equal numbers of cells were plated in the 24-well and 6-well plates and grown to 80% confluence. The indicated amounts of vectors were mixed in OPTI-MEM medium (Gibco) with Lipofectamine 2000 (Invitrogen). The solution was incubated for about 30 minutes at room temperature and then placed on the cultured cells. After four to six hours, the medium was changed into DMEM (Gibco) with 10% fetal bovine serum (Hyclone). For luciferase assay, cells were grown in the 24-well plates and transfected with the indicated amounts of vectors, including pTK-Renilla as an internal control, and Lipofectamine 2000 (Invitrogen) was used. Luciferase activity was assayed 28 to 32 h after transfection. The luciferase activity in cell extracts was determined by the Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The relative light units were measured using a luminometer.
The wide range of relative luciferase activity seen in different panels was likely due to the different amount of cells used at each biological replicate, which did not influence the measurement of relative activity. At least three technical replicates were conducted for each experiment. To avoid transfection efficiency bias, we also performed at least three biological replicates for the luciferase assay. The promoter constructs of p73, p107, p18, p27, p14ARF, Caspase7 and CyclinE1 were kindly provided by Dr. Wuhan Xiao from Institute of Hydrobiology, Chinese Academy of Sciences, and these constructs were published before [26,49,50].
To test the co-localization of MCPH1 and E2F1, the GFP tagged MCPH1 expression vector was co-transfected with the RFP tagged E2F1 expression vector into the HEK293T cells. After 24 to 48 h, the cells were checked under fluorescent microscopy.
Generating MCPH1 mutants
The human MCPH1 gene copy was used to generate mutants carrying mutations at the sites with human-specific and great-ape-specific mutations. A total of 13 sites were tested, including the 9 sites containing human-specific mutations and the 4 sites containing great-ape-specific mutations. The mutant MCPH1s were prepared using the QuickChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and specific oligodeoxynucleotide primer sets. The intended mutations were confirmed by sequencing. The oligodeoxynucleotide primers used for generating the mutant MCPH1s are shown in Additional file 8: Table S2 and Additional file 9: Table S3.
Co-immunoprecipitation
Flag-E2F1 and HA-MCPH1 were co-transfected into the HEK293T cells in the six-well plates by Lipofectamine 2000 with the total amount of 16 ug DNA. After 36 to 38 h of transfection, cells were lysed with 400 ul lysis buffer (50 ml Tris–HCl, pH = 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton-100; 1 Mm Na3VO4) containing a cocktail of protease inhibitors (Sigma Chemical St. Louis, MO, USA). Cell lyses were incubated with HA agarose beads (Sigma Chemical) overnight at 4°C with lysis buffer, and boiled in 2 × protein loading buffer. Blots were blocked in 0.05% Tris-buffered saline (TBS), 20% Tween and 5% non-fat milk followed by incubation with the indicated primary (anti-Flag (M2) from Sigma or anti-HA from Covance, Princeton, NJ, USA) and secondary antibody (anti-mouse from KPL, Inc. Maryland, Washington, D.C, USA) in this buffer. Immunoreactivity was detected with an enhanced chemiluminescence system (Pierce Protein Biology, Rockford, IL) with colored markers (Fermentas, Pittsburgh PA, USA) as the molecular size standard.
Western blotting
Proteins from the HEK293T cells were homogenized in RIPA lysis buffer (50 mM Tris–HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton-100; 1 mM Na3VO4) containing a cocktail of protease inhibitor (Sigma Chemical). Extracted proteins (15 to 20 μg) were separated by SDS-polyacrylamide gel electrophoresis and electrophoreticly transferred to a membrane incubated with anti-HA monoclonal antibody (Covance). Immunoreactivity was detected with an enhanced chemiluminescence system (Pierce Protein Biology) with colored markers (Fermentas) as the molecular size standard.
MCPH1 protein sequence comparison among representative mammalian species
The MCPH1 protein sequences of human, non-human primates (chimpanzee, gorilla, orangutan, gibbon, macaque and marmoset) and representative vertebrate species (rat, mouse, cow and dog) were obtained from the NCBI [51], EMBL [52] and UniProt [53] databases. Orthologous sequences were aligned using Muscle in MEGA5 [54] and Clustal W 7.0.5.2 (BioEdit software. http://www.mbio.ncsu.edu/bioedit/bioedit.html. Tom Hall Ibis Bioscience Carlsbad, CA, USA) (see Additional file 2: Figure S2).
Statistical analysis
Statistical analysis was performed using Prism 5 (GraphPad Software, Inc. 2236 Avenida de la Playa La Jolla, CA 92037 USA), the data were analyzed using the two-tailed Student’s t test. ANOVA test analysis using R program [55]. A P-value of <0.05 was considered statistically significant.
Database
Nucleotide sequences have been deposited to the NCBI GeneBankTM database [56] with accession numbers JX194162 and JX861895 for the MCPH1 coding region sequences of rhesus macaque and gibbon.
Abbreviations
ASPM: Abnormal spindle like microcephaly associated protein; BRCT: BRCA1-carboxyl terminal; CDK5RAP2: Cyclin-dependnet kinase 5 regulatory associated protein 2; CDS: Coding sequence; CENPJ: Centromeric protein J; CEP152: Centrosomal protein 152 kDa; DMEM: Dulbecco modified eagle medium; IP: Immunoprecipitation; MCPH: Primary microcephaly; PCC: Premature chromosome condensation; STIL: SCL/TAL1 interrupting locus; TBS: Tris-buffered saline; TSS: Transcriptional start site; WDR62: WD repeat domain 62.
Competing interests
The authors declare that no competing interests exist.
Authors’ contributions
LS and BS designed the study. LS performed experiments. ML, QL and XBQ contributed analytic tools. LS and BS analyzed data. LS and BS wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Summary of the E2F1 regulatory pathway. E2F1 could up-regulate cell apoptosis associated genes p73, p14ARF, Caspase7 and cell proliferation associated genes CyclinE1, p107, p18 and p27 promoters’ activity. E2F1 also represses the hTERT promoter activity.
Alignment of the full length MCPH1 protein sequences among different species including human, chimpanzee, gorilla, orangutan, gibbon, macaque, marmoset, rat, mouse, cow and dog. The framed sites are the human- and great-ape-specific sites.
The physicochemical properties of MCPH1 lineage specific amino acids.
Alignment of the promoter sequences of p73, p107, p18, p27, p14ARF, Caspase7 and hTERT (genes’ transcriptional start site (TSS) downstream 500bp and TSS upstream 2000bp) among primate species including human, chimpanzee, gorilla, orangutan, macaque and marmoset. The aligned sequences are the E2F1-specific binding sites located in the promoter region. The promoter sequences of CyclinE1 were not available for most of the nonhuman primate species. For Caspase 7, the promoter sequences of chimpanzee and marmoset were not available. The numbers for positions are the distances from the transcriptional start site.
Alignment of the full length E2F1 protein sequences among different primate species including human, chimpanzee, gorilla and macaque.
The results of the enhancing assay for the E2F1 target genes p18, p27, p107 and Caspase7.
The results of the repressing assay for the target genes including p18, p27, p107, Caspase7 and TERT.
Primers used for the generation of human-specific mutants.
Primers used for the generation of great-ape-specific mutants.
Contributor Information
Lei Shi, Email: yuansishi@gmail.com.
Ming Li, Email: liming@mail.kiz.ac.cn.
Qiang Lin, Email: linqiang@mail.kiz.ac.cn.
Xuebin Qi, Email: qixuebin@mail.kiz.ac.cn.
Bing Su, Email: sub@mail.kiz.ac.cn.
Acknowledgements
We would like to thank Dr. Wuhan Xiao from Institute of Hydrobiology, Chinese Academy of Sciences, for kindly providing wild type E2F1 expression plasmid and E2F4B, p27, p73, p107, p18, p14ARF, Caspase7, CyclinE1 and hTERT promoters and we also wish to thank Hui Zhang for her technical assistance in this study.
This work was supported by grants from the National 973 project of China (2011CBA00401), the National Natural Science Foundation of China (31130051), and the Natural Science Foundation of Yunnan Province, China (2009CD107) and the West light Doctoral program.
References
- Gilbert SL, Dobyns WB, Lahn BT. Genetic links between brain development and brain evolution. Nat Rev Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, Karbani G, Jafri H, Rashid Y, Mueller RF, Markham AF, Woods CG. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-Y, Elledge SJ. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113:881–889. doi: 10.1016/S0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- Bilguvar K, Oztürk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tüysüz B, Caglayan AO, Gökben S, Kaymakçalan H, Barak T, Bakircioğlu M, Yasuno K, Ho W, Sanders S, Zhu Y, Yilmaz S, Dinçer A, Johnson MH, Bronen RA, Koçer N, Per H, Mane S, Pamir MN, Yalçinkaya C, Kumandaş S, Topçu M, Ozmen M, Sestan N. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Désir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, Passemard S, Misson JP, Lindsay S, Gergely F, Dobyns WB, Roberts E, Abramowicz M, Woods CG. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topcu M, McDonald MT, Barry BJ, Felie JM, Sunu C, Dobyns WB, Folkerth RD, Barkovich AJ, Walsh CA. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga S, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, Costa SM, Baralle D, Raponi M, Karbani G, Rashid Y, Jafri H, Bennett C, Corry P, Walsh CA, Woods CG. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. Erratum in: Nat Genet 2005, 37:555. Lizarraga, Sophia [corrected to Lizarraga, Sofia B] [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, Ferguson M, Matsuoka M, Macgillivray C, Nightingale M, Patry L, Rideout AL, Thomas A, Orr A, Hoffmann I, Michaud JL, Awadalla P, Meek DC, Ludman M, Samuels ME. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, Walsh CA, Woods CG. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD, Vallender EJ, Lahn BT. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–79. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Wang YQ, Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum Mol Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon Y-H, Collura R, Ruvolo M, Barrett JC, Woods CG, Walsh CA, Jurka J, Larionov V. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:e126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F, Meier R. Positive selection in ASPM is correlated with cerebral cortex evolution across primates but not with whole brain size. Mol Biol Evol. 2008;25:2247–2250. doi: 10.1093/molbev/msn184. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Venditti C, Barton BA, Mundy NI. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 2011;28:625–638. doi: 10.1093/molbev/msq237. [DOI] [PubMed] [Google Scholar]
- Pulvers JN, Bryk J, Fish JL, Wilsch-Bräuninger M, Arai Y, Schreier D, Naumann R, Helppi J, Habermann B, Vogt J, Nitsch R, Tóth A, Enard W, Pääbo S, Huttner WB. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A. 2010;107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, Neitzel H, Jackson AP. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–266. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-Z, Lin F-T, Lin W-C. MCPH1/BRIT1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep. 2008;9:907–915. doi: 10.1038/embor.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282:35416–35423. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers LJ, Coull BJ, Stack SJ, Morrison CG. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 2008;27:139–144. doi: 10.1038/sj.onc.1210595. [DOI] [PubMed] [Google Scholar]
- Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gao H, Lin SY, Peng G, Huang X, Zhang P, Goss JA, Brunicardi FC, Multani AS, Chang S, Li K. BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet. 2010;6:e1000826. doi: 10.1371/journal.pgen.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JK, Li Y, Su B. A common SNP of MCPH1 is associated with cranial volume variation in Chinese population. Hum Mol Genet. 2008;17:1329–1335. doi: 10.1093/hmg/ddn021. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Agartz I, Djurovic S, Brown AA, Roddey JC, Kähler AK, Mattingsdal M, Athanasiu L, Joyner AH, Schork NJ, Halgren E, Sundet K, Melle I, Dale AM, Andreassen OA. Alzheimer's Disease Neuroimaging Initiative. Sex-dependent association of common variants of microcephaly genes with brain structure. Proc Natl Acad Sci U S A. 2010;107:384–388. doi: 10.1073/pnas.0908454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Li M, Su B. MCPH1/BRIT1 represses transcription of the human telomerase reverse transcriptase gene. Gene. 2012;495:1–9. doi: 10.1016/j.gene.2011.12.053. [DOI] [PubMed] [Google Scholar]
- Blais A, Monte D, Pouliot F, Labrie C. Regulation of the human cyclin-dependent kinase inhibitor p18INK4c by the transcription factors E2F1 and Sp1. J Biol Chem. 2002;277:31679–31693. doi: 10.1074/jbc.M204554200. [DOI] [PubMed] [Google Scholar]
- Iwanaga R, Komori H, Ishida S, Okamura N, Nakayama K, Nakayama KI, Ohtani K. Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene. 2006;25:1786–1798. doi: 10.1038/sj.onc.1209210. [DOI] [PubMed] [Google Scholar]
- Helin K, Wu CL, Fattaey AR, Lees JA, Dynlacht BD, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Barton RA, Mundy NI. Reconstructing the ups and downs of primate brain evolution: implications for adaptive hypotheses and Homo floresiensis. BMC Biol. 2010;8:9. doi: 10.1186/1741-7007-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Mekel-Bobrov N, Lahn BT. Genetic basis of human brain evolution. Trends Neurosci. 2008;31:637–644. doi: 10.1016/j.tins.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JL, Liang Y, Li K, Chen J. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283:29586–29592. doi: 10.1074/jbc.M804080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita D, Shintomi K, Ono T, Gavvovidis I, Schindler D, Neitzel H, Trimborn M, Hirano T. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol. 2011;194:841–854. doi: 10.1083/jcb.201106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocati N, Di Ilio C, De Laurenzi V. p63/p73 in the control of cell cycle and cell death. Exp Cell Res. 2012;318:1285–1290. doi: 10.1016/j.yexcr.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Meyer G, Cabrera Socorro A, Perez Garcia CG, Martinez Millan L, Walker N, Caput D. Developmental roles of p73 in Cajal-Retzius cells and cortical patterning. J Neurosci. 2004;24:9878–9887. doi: 10.1523/JNEUROSCI.3060-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, Kennedy H, Dehay C. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci U S A. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, Khochbin S, Gazzeri S. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006;26:4339–4350. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23:3386–3396. doi: 10.1038/sj.emboj.7600328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI. Microcephaly genes and the evolution of sexual dimorphism in primate brain size. J Evol Biol. 2013;26:906–911. doi: 10.1111/jeb.12091. [DOI] [PubMed] [Google Scholar]
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2:a001198. doi: 10.1101/cshperspect.a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endo Rev. 2006;27:398–426. doi: 10.1210/er.2005-0017. [DOI] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- Passemard S, El Ghouzzi V, Nasser H, Verney C, Vodjdani G, Lacaud A, Lebon S, Laburthe M, Robberecht P, Nardelli J, Mani S, Verloes A, Gressens P, Lelièvre V. VIP blockade leads to microcephaly in mice via disruption of Mcph1-Chk1 signaling. J Clin Invest. 2011;121:3071–3087. doi: 10.1172/JCI43824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Somel M, Liu X, Khaitovich P. Human brain evolution: transcripts, metabolites and their regulators. Nat Rev Neurosci. 2013;14:112–127. doi: 10.1038/nrn3372. [DOI] [PubMed] [Google Scholar]
- Ji W, Zhang W, Xiao W. E2F-1 directly regulates thrombospondin 1 expression. PLoS One. 2010;5:e13442. doi: 10.1371/journal.pone.0013442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Su B. Identification and functional characterization of a primate-specific E2F1 binding motif regulating MCPH1 expression. FEBS J. 2012;279:491–503. doi: 10.1111/j.1742-4658.2011.08441.x. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov.
- e! Ensembleast. http://www.ensembl.org.
- UniProt. http://www.uniprot.org/
- MEGA: Molecular Evolutionary Genetics Analysis. http://www.megasoftware.net.
- The R Project for Statistical Computing. http://www.r-project.org/
- GenBank. http://www.ncbi.nlm.nih.gov/genbank/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of the E2F1 regulatory pathway. E2F1 could up-regulate cell apoptosis associated genes p73, p14ARF, Caspase7 and cell proliferation associated genes CyclinE1, p107, p18 and p27 promoters’ activity. E2F1 also represses the hTERT promoter activity.
Alignment of the full length MCPH1 protein sequences among different species including human, chimpanzee, gorilla, orangutan, gibbon, macaque, marmoset, rat, mouse, cow and dog. The framed sites are the human- and great-ape-specific sites.
The physicochemical properties of MCPH1 lineage specific amino acids.
Alignment of the promoter sequences of p73, p107, p18, p27, p14ARF, Caspase7 and hTERT (genes’ transcriptional start site (TSS) downstream 500bp and TSS upstream 2000bp) among primate species including human, chimpanzee, gorilla, orangutan, macaque and marmoset. The aligned sequences are the E2F1-specific binding sites located in the promoter region. The promoter sequences of CyclinE1 were not available for most of the nonhuman primate species. For Caspase 7, the promoter sequences of chimpanzee and marmoset were not available. The numbers for positions are the distances from the transcriptional start site.
Alignment of the full length E2F1 protein sequences among different primate species including human, chimpanzee, gorilla and macaque.
The results of the enhancing assay for the E2F1 target genes p18, p27, p107 and Caspase7.
The results of the repressing assay for the target genes including p18, p27, p107, Caspase7 and TERT.
Primers used for the generation of human-specific mutants.
Primers used for the generation of great-ape-specific mutants.