Abstract
Given the anthropometric differences between men and women and previous evidence of sex-difference in genetic effects, we conducted a genome-wide search for sexually dimorphic associations with height, weight, body mass index, waist circumference, hip circumference, and waist-to-hip-ratio (133,723 individuals) and took forward 348 SNPs into follow-up (additional 137,052 individuals) in a total of 94 studies. Seven loci displayed significant sex-difference (FDR<5%), including four previously established (near GRB14/COBLL1, LYPLAL1/SLC30A10, VEGFA, ADAMTS9) and three novel anthropometric trait loci (near MAP3K1, HSD17B4, PPARG), all of which were genome-wide significant in women (P<5×10−8), but not in men. Sex-differences were apparent only for waist phenotypes, not for height, weight, BMI, or hip circumference. Moreover, we found no evidence for genetic effects with opposite directions in men versus women. The PPARG locus is of specific interest due to its role in diabetes genetics and therapy. Our results demonstrate the value of sex-specific GWAS to unravel the sexually dimorphic genetic underpinning of complex traits.
Author Summary
Men and women differ substantially regarding height, weight, and body fat. Interestingly, previous work detecting genetic effects for waist-to-hip ratio, to assess body fat distribution, has found that many of these showed sex-differences. However, systematic searches for sex-differences in genetic effects have not yet been conducted. Therefore, we undertook a genome-wide search for sexually dimorphic genetic effects for anthropometric traits including 133,723 individuals in a large meta-analysis and followed promising variants in further 137,052 individuals, including a total of 94 studies. We identified seven loci with significant sex-difference including four previously established (near GRB14/COBLL1, LYPLAL1/SLC30A10, VEGFA, ADAMTS9) and three novel anthropometric trait loci (near MAP3K1, HSD17B4, PPARG), all of which were significant in women, but not in men. Of interest is that sex-difference was only observed for waist phenotypes, but not for height or body-mass-index. We found no evidence for sex-differences with opposite effect direction for men and women. The PPARG locus is of specific interest due to its link to diabetes genetics and therapy. Our findings demonstrate the importance of investigating sex differences, which may lead to a better understanding of disease mechanisms with a potential relevance to treatment options.
Introduction
Height, fat mass, and fat distribution differ substantially between men and women, and these differences may, in part, explain the sex-specific susceptibilities to certain diseases [1], [2]. A subtle sexual dimorphism in body composition is already apparent during childhood, and emerges more prominently during adolescence as boys start exceeding girls with regard to height and muscle mass, while girls accumulate more fat mass [3]–[5]. These considerable differences in anthropometry may reflect sex-specific differences in steroid hormone regulation, adipogenesis, lipid storage, muscle metabolism, composition, and contractile speed, skeletal growth and maturation, or lipolysis, and suggest a genetic underpinning [1], [2], [6]–[10].
While direct measures of height or weight are easily obtained, measures of fat mass and fat distribution are more invasive and less frequently assessed in large-scale epidemiological studies. Instead, body mass index (BMI, computed as weight/height2) is used to assess overall adiposity, whereas waist-to-hip ratio (WHR) is a measure of fat distribution. Increased WHR is suggestive of more preferential accumulation of fat around the waist versus the hip. Obesity (defined as a BMI≥30 kg/m2) is a well-established risk factor for type 2 diabetes, cardiovascular disease, cancer and mortality [11]–[18]. Also the independent effect of WHR – derived by computing WHR adjusted for BMI - on morbidity and mortality has been demonstrated [19], [20]. Thus, anthropometric measures depict not only body size, but also fat distribution and consequently various facets of chronic disease risk.
Genome-wide association studies (GWAS) have successfully identified many genetic loci robustly associated with height [21]–[25], body mass index (BMI) [26]–[29], and waist-to-hip ratio (WHR) [30], . So far, all GWAS for anthropometric traits have been performed in men and women combined. However, in our most recent GWAS of WHR within the Genetic Investigation of ANthropometric Traits (GIANT) consortium, we found that seven of the 14 novel loci displayed more pronounced effects in women than in men, when we subsequently stratified analyses by sex [31]. In contrast, our GWAS for BMI or height genetic effects with GIANT, no sex-differences in the newly identified loci were noted [25], [29]. However, these GWAS did not specifically aim to identify genetic loci with sex-specific effects such that a systematic search for such sexually dimorphic loci was warranted.
Thus, given the obvious difference in physical appearance between men and women in body size and shape, together with the strong evidence of sex-specific effects of the recently identified WHR loci, we performed a systematic search for sex-specific loci for anthropometric traits. GWAS conducted separately in men and women not only improve power to identify sex-sensitive associations, but also allow testing for sex differences. Within the Genetic Investigation of ANthropometric Traits (GIANT) consortium, we performed sex-specific GWAS for six anthropometric traits involving a total of 270,775 subjects from 94 studies in order to investigate the extent and nature of sex-specific genetic effects on anthropometry.
Results
Discovery meta-analysis of sex-specific GWAS for anthropometric traits
In the discovery stage, sex-specific GWA analyses were conducted in 46 studies (Table S1), including up to 60,586 men and 73,137 women, testing ∼2.8 million autosomal SNPs for association with six anthropometric traits that are well established to represent body size and shape: i.e. height, weight, BMI, waist circumference (WC), hip circumference (HIP), and WHR. In order to capture body fat distribution independent of overall adiposity, the latter three traits were also analyzed with adjustment for BMI (WCadjBMI, HIPadjBMI, WHRadjBMI) yielding nine phenotypes n total (Methods). Study-specific information has been described previously [25], [29], [31] and details on study-specific analyses are given in Methods. All study participants were of European and European American descent. We performed an inverse-variance weighted fixed-effects meta-analysis for each of the 18 analyses (9 phenotypes, 2 sexes; Methods) yielding meta-analyzed sex-specific P-values for association (P-men, P-women) and corresponding sex-specific effect estimates. In order to account for multiple testing across SNPs genome-wide as well as across phenotypes, we applied a false-discovery-rate (FDR) approach [32].
Generally, in order to establish a sexually dimorphic association, we require both a significant SNP association with an anthropometric trait at least in one sex (P-men or P-women at 5% FDR across all SNPs and phenotypes tested) and a significant sex-difference of a SNP (P-value testing for difference in sex-specific effect estimates, P-diff, at 5% FDR). Sexually dimorphic SNPs could either show (i) concordant effect direction (CED), if the association is present in one sex (P-men or P-women at 5% FDR) and at least nominally significant and directionally concordant in the other (P-women or P-men<0.05, respectively), (ii) single sex effect (SSE), if the association is present in one sex and not significant in the other, or (iii) opposite effect direction (OED), if the association is present in one sex and at least nominally significant in the opposite direction in the other sex. We aimed to identify genetic loci with CED or SSE, which are biologically most plausible. Nevertheless, in this exploratory effort, we also searched for OED loci – which are biologically unlikely, but current lack of knowledge of such signals could be due to the fact that current GWAS of men and women combined cannot detect such signals.
We evaluated the power of two genome-wide approaches to screen for sex-sensitive genetic loci: (a) a scan for sex-specific association P-values in men and women separately (P-men, P-women, sex-specific scan) and (b) a scan for P-values testing for sex difference between effects of men and women (P-diff, sex-difference scan; details in Methods). Power calculations showed that the sex-specific scan had a higher probability to select SNPs with true underlying CED or SSE signal into follow-up, while the sex-difference scan had a higher probability to select SNPs with true underlying OED effect (details in Text S1). We thus conducted both scans.
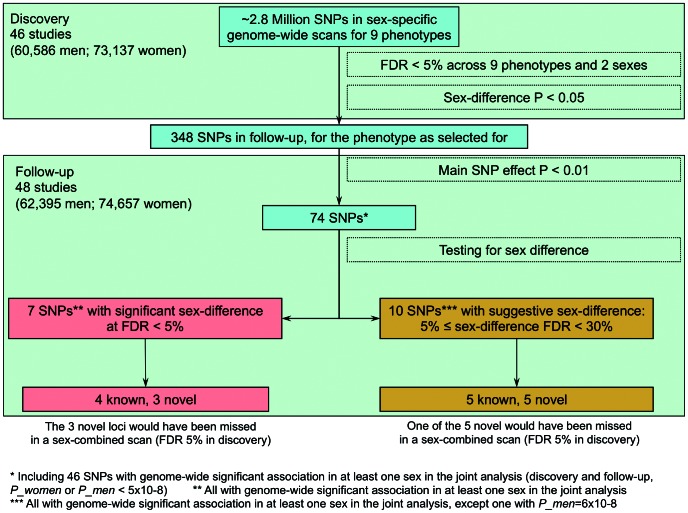
The sex-specific scan showed an excess of small P-values (Figure 1a, b). Controlling for 5% FDR (across all SNPs, nine phenotypes, two sexes; corresponding to a P-value of 2×10−5), this scan yielded 619 independent SNPs associated with at least one of the phenotypes in at least one of the sexes. Including a rough filter for sex-difference (nominal significant, P-diff<0.05), we took 348 out of these 619 SNPs forward for follow-up (73 SNPs for height, 28 for weight, 32 for BMI, 31 for WC, 46 for WCadjBMI, 33 for HIP, 38 for HIPadjBMI, 28 for WHR, and 39 for WHRadjBMI; Table S2). The sex-difference scan did not identify any SNPs at 5% FDR, despite the fact that the QQ-plot for all phenotypes combined as well as for phenotype-specific traits indicated some deviation of the observed P-diff distribution from the expected (under the null hypothesis of no sex-difference) for the waist phenotypes (WHRadjBMI, WHR, WCadjBMI) (Figure 2a,b). Indeed, even if we were to carry forward SNPs at 30% FDR, we would not have identified any significant OED effects. As such, no SNPs were taken forward from this second scan for follow-up.
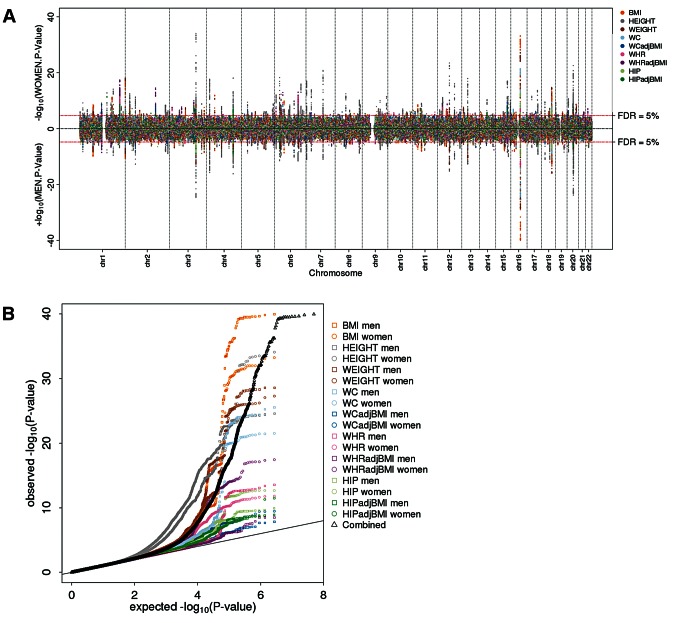
Figure 1. Genome-wide scan for sex-specific genome-wide association highlights numerous loci.
(a) Manhattan plot showing the men-specific (upward, up to 60,586 men) and women-specific (downward, up to 73,137 women) association P-values from the discovery with the 619 selected loci colored by the phenotype for which the locus was selected; (b) QQ-plot showing the sex-specific association P-values as observed against those expected under the null overall phenotypes (black) and for each phenotypes separately (colored).
Figure 2. Genome-wide sex-difference scan fails to pinpoint loci.
(a) Manhattan plot showing sex-difference P-values, (b) QQ plot for sex-difference P-values overall phenotypes (black) and for each phenotype separately (colored).
Follow-up of 348 SNPs reveals seven sexually dimorphic anthropometric trait loci
In the follow up, we examined the 348 SNPs for the phenotype that the SNP was selected for in 18 studies with in-silico genotype information (up to 20,340 men and 41,872 women) and in 30 studies with Metabochip data (up to 42,055 men and 32,785 women; which contained assays for 43% of selected SNPs prioritized for follow-up). Study-specific information is given in Tables S1, S3, S4A,S4B, S5 and Methods. Meta-analyses of the follow-up studies as well as jointly with discovery studies were conducted for each sex separately (P-women, P-men) and both combined (P, Methods).
As all 348 SNPs were derived from the sex-specific discovery scan, the follow-up was then used to establish unbiased estimates of sex-difference in an independent data set (Methods). We filtered SNPs with a main effect (P-value for association combined in men and women <0.01; Methods). This yielded 74 SNPs, which were subsequently tested for sex-difference. Seven of these 74 SNPs reached a significant sex-difference at 5% FDR (six for WHRadjBMI, one for WCadjBMI, Table 1). For these seven SNPs, the P-diff jointly for the discovery and follow-up ranged from 2.7×10−4 to 6.2×10−16 and the joint discovery and follow-up association P-value in the predominant sex – interestingly, all in women – was genome-wide significant (P-women<5×10−8). Effect sizes were similar when we restricted our follow-up analyses to population-based studies or control-only series in order to eliminate a potential bias by patient groups (Figure S1).
Table 1. Seven SNPs show sex difference.
| Discovery | Follow-up | Joint | ||||||||||||
| MEN | WOMEN | Sex-Diff | MEN | WOMEN | Sex-Diff | MEN | WOMEN | Sex-Diff | MEN | WOMEN | ||||
| SNP | Traita | Sexa | Geneb | P | P | P | Pc | Pc | Pc | P | P | P | N | N |
| rs6717858 | WHRadjBMI | W | GRB14/COBLL1 | 0.309 | 2.78E-15 | 6.49E-07 | 0.965 | 3.64E-16 | 1.08E-11 | 0.613 | 1.99E-29 | 6.18E-16 | 76,594 | 98,321 |
| rs2820443 | WHRadjBMI | W | LYPLAL1/SLC30A10 | 0.191 | 3.69E-18 | 1.25E-07 | 0.532 | 9.15E-21 | 2.60E-10 | 0.374 | 4.62E-37 | 6.95E-16 | 76,625 | 98,352 |
| rs1358980 | WHRadjBMI | W | VEGFA | 0.110 | 1.11E-13 | 3.02E-05 | 0.112 | 1.38E-19 | 4.53E-08 | 0.048 | 2.41E-31 | 2.46E-11 | 75,703 | 97,269 |
| rs11743303 | WCadjBMI | W | MAP3K1 | 0.974 | 2.27E-06 | 6.24E-04 | 0.172 | 7.15E-07 | 5.35E-05 | 0.570 | 2.69E-11 | 8.41E-07 | 85,136d | 107,403d |
| rs2371767 | WHRadjBMI | W | ADAMTS9 | 0.196 | 1.63E-08 | 1.85E-03 | 6.08E-03 | 8.55E-17 | 2.14E-04 | 8.34E-03 | 7.07E-23 | 1.91E-06 | 72,649 | 95,325 |
| rs10478424 | WHRadjBMI | W | HSD17B4 | 0.399 | 1.02E-05 | 9.84E-03 | 0.864 | 3.81E-05 | 1.67E-03 | 0.761 | 3.45E-09 | 2.66E-04 | 43,852e | 73,066e |
| rs4684854 | WHRadjBMI | W | PPARG | 0.955 | 2.36E-08 | 6.46E-05 | 0.132 | 1.48E-07 | 4.22E-03 | 0.411 | 4.17E-14 | 4.04E-06 | 74,652 | 96,472 |
Trait and sex for which the SNP was selected;
Gene labels state the nearest gene or the gene as published previously; details on all genes near the association signal can be found in the Figure S2;
One-sided P-Values.
larger sample size due to one additional study that did not have hip circumference, and therefore could not contribute to WHRadjBMI.
smaller sample size as this SNP was not on Metabochip.
Shown are the seven SNPs with significant (at 5% false discovery rate) sex difference in the follow-up data. These seven SNPs exhibit genome-wide significant association in women (joint discovery and follow-up P_women<5×10−8) and only two of these show nominally significant association in men (joint P_men<0.05). The three loci MAP3K1, HSD17B4, and PPARG are shown here for the first time for their anthropometric trait association as well as for sex-difference.
The seven confirmed sex-difference loci include three novel signals
We found that three of these seven identified loci describe novel associations with WCadjBMI (near MAP3K1) or WHRadjBMI (near HSD17B4 and PPARG) that were genome-wide significant in women (joint P-women: 3.4×10−9 to 4.2×10−14), but not in men (joint P-men: 0.41 to 0.76), whereas the remaining four had been established previously (ref). These three novel loci would have been missed by sex-combined scans at 5% FDR (equivalent to P>5.8×10−5).
Of particular interest is the PPARG region, which we identified for the first time as a locus for anthropometric traits (WHRadjBMI) in the context of a genome-wide study and with evidence for a women-specific association. PPARG is of considerable importance due to its function as a nuclear hormone receptor with specific known interaction with sex hormones, for example with estrogen receptors [33], and due to its role in type 2 diabetes development and therapy.
The remaining four loci were near (<1 cM) previously established sexually dimorphic loci for WHRadjBMI (GRB14/COBLL1, LYPLAL1/SLC30A10, VEGFA, and ADAMTS9; see Table 2) [31]. The further sexually dimorphic WHRadjBMI loci previously reported in that work were included among the ten additional SNPs at 30% FDR in our data (RSPO3, HOXC13, ITPR2-SSPN, see Table S6), which illustrates the pay-off between our power gain from this sex-specific approach and larger sample size with the increased multiple testing burden of interrogating nine phenotypes in comparison to one phenotype in our previous work. An overview of the SNP selection and findings is given in Figure 3 and the genes surrounding the seven signals are depicted in the region plots (Figure S2).
Table 2. Seven identified SNPs compared to previously published loci.
| SNP | CHR | POS_B36 | Effect allele | Other allele | EAFa | Discovery sex combined P | Published Geneb | Published SNP | Position of published SNP | Trait as published | Previous evidence for sex difference |
| rs6717858 | 2 | 165,247,907 | t | c | 0.581 | 5.85E-10 | GRB14 | rs10195252 | 165221337 | WHRADJ [31] | Yes |
| rs2820443 | 1 | 217,820,132 | t | c | 0.718 | 2.69E-12 | LYPLAL1; LYPLAL1 | rs11118346; rs4846567 | 217810342; 217817340 | HEIGHT [25]; WHRADJ [31] | Yes |
| rs1358980 | 6 | 43,872,529 | t | c | 0.474 | 1.42E-10 | VEGFA | rs6905288 | 43866851 | WHRADJ [31] | Yes |
| rs11743303 | 5 | 55,895,709 | g | a | 0.208 | 1.34E-03 | - | - | - | - | No |
| rs2371767 | 3 | 64,693,298 | g | c | 0.720 | 2.33E-06 | ADAMTS9 | rs6795735 | 64680405 | WHRADJ [31] | Yes |
| rs10478424 | 5 | 118,816,919 | a | t | 0.784 | 1.58E-04 | - | - | - | - | No |
| rs4684854 | 3 | 12,463,882 | c | g | 0.418 | 1.05E-04 | - | - | - | - | No |
The Effect allele refers to a positive effect direction in the discovery stage for the trait and gender, the SNP was selected for;
Gene near this SNP which was published previously from sex-combined analyses.
The seven SNPs with sex difference are considered to depict a known locus, if the index SNP is close to a published top SNP (<1 cM). These include four of the previously reported sexually dimorphic WHR loci (Heid et al., Nat Genet 2010).
Figure 3. Overview of design and findings.
Among the 7 identified loci, we defined those close to (<1 cM) published hits [25], [29], [31] as near published hits and novel otherwise. Novel loci with sex-combined discovery P-value<5.8×10−5, which is the P-value cut-off corresponding to 5% FDR, were declared as loci that could have been discovered also with sex-combined analysis, and otherwise that these would have been missed without the sex-stratified analyses. FDR = false discovery rate.
Although identifying sex-differences was our primary goal, we note that among the 348 SNPs chosen for follow-up, 46 SNPs exhibited genome-wide significant association in either men or women in the joint analysis of discovery and follow-up data (P-men or P-women≤5×10−8, 27 SNPs for height, 12 for WHRadjBMI, three for weight, three for BMI, one for WCadjBMI, zero for WC, HIP, HIPadjBMI, or WHR). Detailed information regarding P-values and effect estimates of these 46 SNPs are included in Table S2.
No opposite effect direction, but enrichment for genetic effects in women
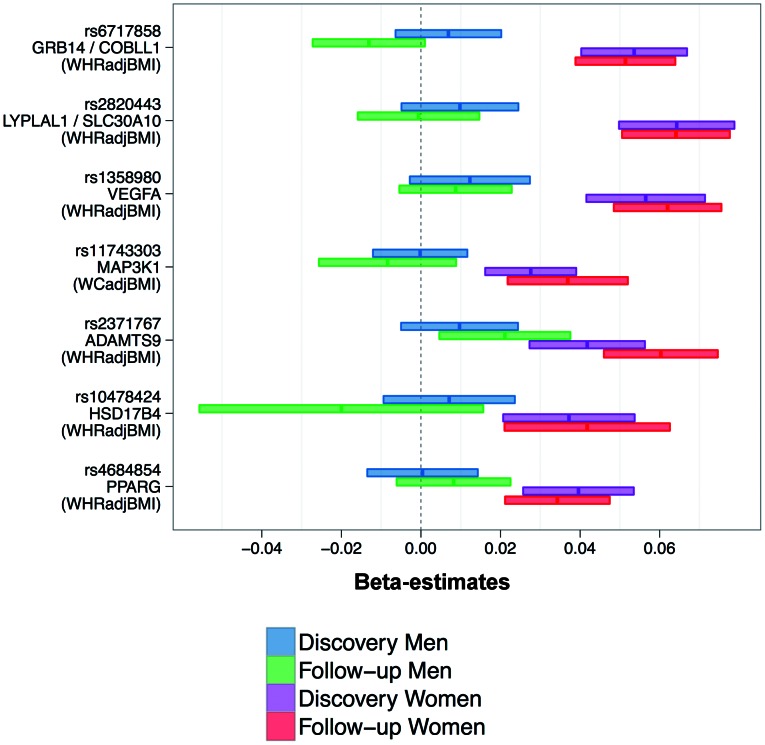
When examining the sex-specific effect estimates for the seven SNPs (Figure 4), we found that effect sizes were consistent in discovery and follow-up and that none of the seven loci showed OED. Furthermore, the associations for six of the seven SNPs were observed in women only (SEE), whereas for one SNP (ADAMTS9) we observed CED in both sexes, but the effect was more pronounced in women than in men. The absence of loci with OED together with the observation that the sex-difference scan did not detect any sex-difference, even at 30% FDR, our data does not support the existence of genetic loci that have opposite effect in men versus women.
Figure 4. Consistently higher effect sizes for women for all seven loci.
Shown are beta-estimates and 95% confidence intervals for the seven identified SNPs (also stating the phenotype for which the SNP was selected for).
When comparing the effect sizes of the 46 SNPs with genome-wide significant sex-specific associations between women and men, we found again significant enrichment for larger effects in women for WHRadjBMI (Binomial test P = 1.1×10−4, Methods), but not for other phenotypes (P = 0.08, 0.08, 0.11, 0.16, for BMI, weight, height, or WCadjBMI, respectively). This underscores that our data does provide evidence for sexual dimorphism in the genetics, and thus biology, underlying WHRadjBMI, but not for height or BMI. This is consistent with the fact that the seven loci with confirmed sex-difference were for waist phenotypes only. Nevertheless, it should be noted that we identified suggestive sexually dimorphic genetic signals for height and BMI when applying a 30% FDR threshold (Table S6).
Age-stratified analyses and association with other traits for the seven SNPs
Hormonal changes during menopause affect a woman's body shape and composition, generally resulting in a more android body type. Therefore, we examined whether any of the seven confirmed sexually dimorphic loci showed evidence of age-specific effects (Methods). More specifically, we performed association analyses for the seven loci stratified by age with a cut-off at 50 years (i.e. average age of onset of menopause) and by sex. None of the loci showed evidence for age-specific effect among women (or men) (P for difference between age groups >0.135, Table S7).
When extending the investigation of the seven SNPs from the phenotype for which the SNP was selected (six for WHRadjBMI, one for WCadjBMI) to the other anthropometric phenotypes (Tables S8A–C), we found no nominally significant association with height (joint discovery and follow-up P-women and P-men from 0.065 to 0.86), except for one SNP (rs2820443, P-women = 2.8×10−6, P-men = 6.0×10−4). Four of the seven associations showed some evidence of BMI association (P-women or P-men 3.2×10−4 to 6.0×10−3). More specifically, we found – in women only – decreased HIPadjBMI (P-women from 2.7×10−27 to 0.015) and increased WCadjBMI (P-women from 7.6×10−22 to 3.82×10−4) for all WHRadjBMI increasing alleles, whereas no association with HIPadjBMI (P-women = 0.32) was observed for the SNP selected for WCadjBMI. This underscores that the seven sexually dimorphic SNPs are primarily waist- and WHR-related.
Using data from other GWAS consortia [34]–[36], we evaluated whether the seven SNPs showed associations with other metabolic traits consistent with the observed association with WHRadjBMI or WCadjBMI and whether the similar sex-specific pattern of association was also observed (Methods). We did indeed find directionally consistent enrichment (binomial P<0.05) for women-specific associations (P-women<0.05) with lipids, fasting insulin, type 2 diabetes, and HOMA-B (binomial P from 1.2×10−5 to 5.9×10−3; Tables S8D–G). Remarkable was the consistent women-specific association for the index SNP near the GRB14/COBLL1 with HDL-cholesterol, triglycerides, insulin, and type 2 diabetes (here though for a different SNP, but correlated with our index SNP, D′ = 1.0, r2 = 0.735) and for our SNP near MAP3K1 with triglycerides. Three of our novel SNP findings localize near well-known loci for type 2 diabetes (ADAMTS9, VEGFA, PPARG), although only our SNP near ADAMTS9 displayed a strong correlation with the published type 2 diabetes index SNP (rs4607103, r2 = 0.9, ∼0.001 cM), while the other two SNPs were uncorrelated with the reported type 2 diabetes SNPs (rs9472138, VEGFA, r2 = 0.008, ∼0.23 cM distant from our lead SNP; rs17036101, PPARG, r2 = 0.024, ∼0.15 cM). It should be noted that many of the studies that participated in GIANT also participated in the other consortia and given the correlation between the phenotypes, the sex-specific consistency is likely somewhat inflated. Taken together, our findings suggest common genetic influences on body fat distribution, lipids, and type 2 diabetes, particularly for women.
Pathway analyses
In order to summarize the biological pathways that are primarily depicted by our data on sex-difference, we examined whether the genes harbored by the seven confirmed loci showed enrichment for particular pathways or other units of the molecular networks (processes, functions) using MAGENTA (Methods). We found that PPARG Signaling, post-Golgi vesicle-mediated transport and kinase- and annexin-related molecular functions showed enrichment at 5% FDR (Table S9).
Potential functional or biological role of the seven loci
Regarding the biological role of the SNPs and genes in the proximity of the seven sex-specific SNPs, we searched literature and functional annotation data bases and catalogues (Methods). The genes inflicted in the seven regions of interest generally highlighted genes with a reported role in insulin sensitivity (PPARG, VEGFA, ADAMTS9, GRB14) and lipid-related traits (fatty liver: LYPLAL1; triglyceride concentrations: MAP3K1, HDL-C: GRB14). Among the index SNPs or their proxies (pairwise correlation, r2>0.8) in the immediate region (49 SNPs altogether), we found one SNP (rs10478424; r2 = 1 with lead SNP at HSD17B4) that was a predicted transcription factor binding site (TFBS). Interestingly, one of the transcription factors predicted to bind at this TFBS is PPARG, which itself is located near one of our other association signals. None of the other 48 SNPs tagged any known copy-number variant, was a non-synonymous coding variant, or was present in any of the other predicted functional classes. When extending this search to SNPs that were more moderately correlated (r2>0.5) with the lead index SNPs (146 SNPs altogether), these included several proxies of rs2820443 (near LYPLAL1/SLC30A10) that annotated as TFBSs as well as proxies of rs10478424 that disrupt predicted microRNA binding sites. These findings may indicate potential involvement in the regulation of gene transcription near those loci.
A specific description of potentially functionally elements in the association regions as indicated by UCSC and Ensembl genome browsers and more details from the literature and functional annotation data base searches can be found in the Text S1.
Effect of the seven sexually dimorphic loci on expression in relevant tissues
To localize the potentially causal gene at each locus, we examined the evidence for sex specific cis expression quantitative trait loci (eQTL) for genes near the seven identified SNPs in different types of human subcutaneous adipose tissue, lymphocytes, and whole blood (Methods). Although there was evidence for gene expression association of two (GRB14, ADAMTS9, Table S10) of the seven SNPs (SNP highly correlated with the peak SNP of the transcript, r2>0.8, the association of the peak SNP with the transcript expression vanished when adjusted for our SNP), the associations were not sex-specific (P-diff>0.05).
We also examined whether genes harbored by the seven sex-specific loci showed sex-specific expression in various tissues of mouse models using real-time PCR expression data (brown fat, inguinal and gonadal fat, and liver) and Illumina expression data (liver, inguinal and gonadal fat; Methods). We found significantly (at a significance level of 0.05/19 = 0.003) lower expression of GRB14 in brown fat of female mice (P-diff = 0.001); due to the role of brown fat in triglyceride catabolism [37], this is in-line with the previously described sexually dimorphic association of this SNP with HDL-C and triglycerides. For female compared to male mice, we found significantly lower expression of VEGFA (P-diff = 1×10−5) in inguinal fat, whereas in liver, we found higher expression for three genes (LYPLAL1, PPARG, MKRN1; P-diff from 0.002 to 0.003) with the latter two genes being located in the PPARG locus (Table S11).
Discussion
In our genome-wide search for sexually dimorphic associations including over 270,000 individuals from 94 studies from the GIANT consortium, we found evidence for seven loci with significant sex-difference including three novel anthropometric trait loci (near MAP3K1, HSD17B4, PPARG). Importantly, for all seven loci, the associations were observed for waist phenotypes with more prominent effects in women. These findings are consistent with our previous reports for sex-differences in the genetics of WHR [30], [31].
The waist phenotypes used in this study are well established proxy measures of body fat distribution. Women have more subcutaneous body fat, which is part of the skin, that is preferentially deposited at the hips and thighs whereas men have more visceral fat, which is fat in and around the inner organs and accumulates particularly around the waist [38]–[40]. It is well known that hormonal levels are associated with differences in fat distribution in men and women, distinctions that emerge early in childhood and subsequently amplify during puberty [41], [42]. Moreover, fat distribution in women changes as estrogen levels drop during menopause, leading to a more android shape, with greater visceral fat accumulation [43]. Subcutaneous and visceral fat has distinct morphological and functional properties that account in part for clinically relevant sex differences in a variety of metabolic phenotypes [40], [44].
Our findings of sex-specific genetic effects on waist-hip-ratio as a measure of fat distribution are consistent with a study of families in whom MC4R mutations segregate that demonstrated larger effects on obesity in female compared to male mutation carriers [45]. In animal models, microarray experiments have consistently demonstrated that adipose tissue mass, function, and distribution is regulated by networks of sexually dimorphic genes which are likely regulated by sex hormones [46]. In addition, sex-differences in mRNA and miRNA expression in abdominal and gluteal adipose tissue have been noted in humans [47], [48]. Lastly, animal studies demonstrate that exposure to sex hormones early in development is associated with long term, lifelong changes in adipose tissue distribution and function [49], [50]. Taken together, sex-differences in body shape appear to be determined by a complex interplay of genetic and hormonal factors.
Our data did not provide statistically significant evidence for sex-differences in the genetics of BMI and height. This is in-line with previous reports in a large twin heritability study [51], which shows differential heritability between sexes for waist-hip-ratio, but not for BMI or height. Interestingly, phenotypic differences in BMI between men and women are consistently smaller than those for waist-related traits and perhaps sex differences in BMI genetics are more subtle. This was not very surprising as our expectations for sex-differences were highest for waist-hip-ratio, given the previous report on sex-differences in waist-hip-ratio genetics, the strong link of the phenotype to body fat distribution, the change of body fat distribution by hormones, and the sex-specificity of fat distribution.
The lack of signals with opposite effect direction for men and women in our data is particularly intriguing. Given that no systematic search for sexually dimorphic associations with anthropometric traits has been done before in a large enough genome-wide effort, it was not really clear whether OED signals would exist. While our systematic search scanning the P-values testing for sex-difference between the sex-specific effect estimates was specifically designed to detect OED associations, we did not detect any. This is in-line with the prior believe that genetic variants do not affect anthropometry in one direction in men and in the opposite direction in women.
The seven loci shed new light on regions containing genes with a reported role for type 2 diabetes (PPARG, ADAMTS9, VEGFA), lipids (GRB14, MAP3K1) and hormone metabolism (HSD17B4). Particularly intriguing due to its relevance for type 2 diabetes and therapy is the PPARG, which showed association with WHR in women, but not in men. This is in-line with small candidate gene-by-environment interaction studies e.g. of saturated fat intake with PPARG variants for obesity-related traits [52], including differential effects by sex [53]. Although the index SNP is independent of the polymorphisms previously reported for type 2 diabetes [54]–[56], PPARG is a particularly interesting candidate as its encoded protein, PPARγ, is a nuclear hormone receptor that serves as a master regulator of adipocyte-specific genes contributing to adipocyte differentiation, susceptibility to obesity, and insulin sensitivity [33]. PPARg-agonists are used to treat type 2 diabetes by redistributing adipose tissue from abdominal visceral to subcutaneous compartment, which is thought to be preferable and improve insulin sensitivity. Interestingly, sex-differences in pioglitazone response have been described for nondiabetic overweight persons [57].
Furthermore, growth factor receptor-bound protein 14 (GRB14) binds to insulin receptors and inhibits their catalytic activity. GRB14 is a prerequisite for the development of insulin-sensitizing molecules to pathological states as obesity and type 2 diabetes [58]. Our current and previously reported findings [31], [34] suggest that the rs6717858 near GRB14 has a female-specific effect on insulin as well as on central obesity and lipids. Additionally, we highlight a region including the mitogen-activated protein kinase kinase kinase 1 (near MAP3K1), a locus known to be associated with triglyceride levels in sex-combined analyses [59], whereas we found this locus to be associated with WHR in women only. The MAP3K1 plays a pivotal role in a network of phosphorylating enzymes integrating cellular responses to a number of mitogenic and metabolic stimuli, including insulin and many growth factors [60]. Interestingly, mutations in MAP3K1 have recently been demonstrated to result in a 46XY disorder of sex development with varying manifestations of gonadal dysgenesis [61]. Thus this gene is implicated in normal sex development, which may be related to our observed sexually dimorphic genetic effect. Finally, the locus near HSD17B4 is of particular interest, because HSD17B4 is a multifunctional peroxisomal enzyme involved in steroid metabolism and fatty acid oxidation [62], [63]. It converts the more active hormone, estradiol, to the less active estrone. Sex differences in HSD17B4 expression with estradiol supplementation have been noted in zebra fish [64]. In addition, the position of our lead marker at a predicted transcription factor binding site located just upstream of a putative protein coding splice variant of HSD17B4 indicates a potential function for the association signal observed at this locus and may warrant additional follow-up work. Interestingly, among the genes that bind to this transcription factor binding site is PPARG.
A major strength of our study is that we were uniquely positioned to perform the analyses described, taking advantage of the highly efficient collaborative environment of the many study partners within the GIANT consortium, which allowed us to conduct the largest possible sex-difference GWAS for anthropometric phenotypes ever reported. As a consequence, we were well-powered (about 80% power) to detect sex-sensitive genetic effects of the same magnitude as those observed previously for WHR or to detect genetic effects as previously observed for height, but assuming these to appear only in one sex [25], [31]. Nevertheless, our statistical power to detect subtle sex differences in genetic effects was limited. Notably, we used a conservative approach to avoid false positive claims: (i) we used ranks instead of the absolute phenotypic values of anthropometric traits in order to avoid artefacts due to outliers, (ii) we applied double genomic control correction in order to avoid any artefact from possible population stratification, and (iii) we established sex-difference using our follow-up as opposed to the combined discovery+follow-up data sets to avoid overestimated sex-differences through winner's curse. It is a further strength that we were able to show associations of our sex-specific anthropometric trait signals also with other metabolic traits such as lipids, glucose and type 2 diabetes; however, we need to note the limitation that these associations were not adjusted for the anthropometric traits, so that some of the observed metabolic trait associations were expected due to the correlation of anthropometric traits with lipids, glucose and type 2 diabetes.
Overall, we found women-specific SNP effects for anthropometric traits, particularly for waist-related phenotypes. Our study findings lend support to distinct genetic effects on body shape by sex and argue for the importance of further integrative studies of sex differences of body shape. While the actual underlying genes and their mechanisms of action remain elusive, we hypothesize that such differences are hormonally regulated. Moreover, because body fat has a prominent endocrinological function and body fat distribution has a critical relevance for many metabolic pathways, understanding these differences could help improve our understanding of metabolic disease processes. Particularly the established sex-difference for the SNP near the therapy-relevant PPARG could impact treatment options. In the era of personalized medicine, which attempts to tailor treatment to fit the individual, a better differentiation between men and women in research and patient treatment could be an important start.
Summary and conclusion
While our data underscores a lack of genetic association in opposing direction in men versus women, we have highlighted female-specific effects in waist phenotypes. Our investigation underscores the importance of considering sex-differences when interrogating the genetic architecture of anthropometric traits. For those traits with strong a priori evidence for sex differences, the routine analysis of sex-specific genome-wide analyses may allow for numerous options for meta-analysis including a sex-combined scan optimally powered to detect the general association as well as sex-specific scans when searching for sexually dimorphic signals. Although our study focused on sex-differences for anthropometric traits, sex differences in genetic effects likely exist for other traits and diseases and should be taken into consideration in future genetic as well as translational studies.
Methods
Anthropometric phenotypes
The anthropometry of men and women differ in various aspects: Average height, waist circumference, and WHR, is higher for men than for women, whereas average BMI is similar. Variability for all phenotypes is similar for men and women, which can be seen on the example of the KORA study (Table S12) and specifically for WHR adjusted for BMI for all studies (Figure S3).
The anthropometric traits examined are height (cm), weight (kg), body mass index (BMI, kg/m2) computed as weight divided by meter of height squared, waist circumference (WC, cm), hip circumference (HIP, cm), and waist-hip-ratio (WHR). The last three traits were analyzed without and with adjustment for BMI, yielding nine phenotypes in total (height, weight, BMI, WC, HIP, WHR, WCadjBMI, HIPadjBMI, WHRadjBMI). For the further analyses, the traits were all transformed at the study-level by calculating age-adjusted residuals (including age and age2 into the regression model for trait creation) for men and women separately and adding BMI into the adjustment as indicated above; then – for all traits except height – the values were ranked and an inverse normal transformation was applied, whereas a z-score transformation was performed for height.
Study-specific analyses for discovery and follow-up
For discovery stage, we included 46 studies (up to 60,586 men, 73,137 women) on height, weight and BMI, 34 studies (up to 36,231 men, 45,192 women) on WC, 33 studies (up to 34,942 men, 43,316 women) on HIP and 32 studies (up to 34,629 men, 42,969 women) on WHR. Each study was genotyped using either Affymetrix or Illumina arrays. To enable meta-analyses across different SNP panels, each group performed genotype imputation using HapMap II CEU (build 21 or 22) via MACH [65], IMPUTE [66] or BimBam [67]. Details are given in Tables S1, S3, S4, S5 and Text S2.
For follow-up, we included (i) 30 studies (up to 42,055 men, 32,785 women) for height, weight and BMI and 27 studies (up to 36,671 men, 28,326 women) for WC, WCadjBMI, HIP, HIPadjBMI, WHR and WHRadjBMI that were genotyped using the custom iSELECT Metabochip array containing ∼195K SNPs designed to support large-scale follow-up of putative associations with metabolic and cardiovascular traits, and (ii) 18 studies (20,340 men, 41,872 women) for height, weight, and BMI and 14 studies (11,225 men, 32,610 women) for WC, WCadjBMI, HIP, HIPadjBMI, WHR and WHRadjBMI genotyped using genome-wide SNP chips with subsequent imputation for in silico follow up.
In each study, association was tested separately for men and women. The additive genetic effect for each SNP on each phenotype was estimated using a normal linear regression model using MACH2QTL [68], SNPTEST [66], ProbABEL [69], GenABEL [70], Merlin [71], or PLINK [72]. For studies with a case-control design, cases and controls were analyzed separately. Study-specific information was described previously [25], [29], [31] for discovery studies and in Tables S1, S3, S4, S5 for follow-up studies.
All involved studies were conducted according to the principles expressed in the Declaration of Helsinki. The studies were approved by the local Review Boards and all study participants provided written informed consent for the collection of samples and subsequent analysis.
Sex-specific discovery meta-analysis
All discovery study-specific files were processed in the meta-analysis centers through a standardized cleaning script that included checks of allele frequencies, compliance with Hapmap alleles, file completeness, number of markers, and ranges of test-statistics. We excluded monomorphic SNPs, SNPs with MAF*N≤3 (minor allele frequency multiplied by sample size) and SNPs with poor imputation quality, i.e. r2_hat <0.3 in MACH; observed/expected dosage variance <0.3 in BIMBAM; proper_info <0.4 in IMPUTE; information <0.8 in PLINK [65], [66], [72], [73].
Sex-specific standard errors and P-values from each participating study were genomic-control (GC) corrected [74] using the lambda factors as published [25], [29], [31], then beta-estimates were meta-analyzed using the inverse-variance weighted fixed effect model as implemented in METAL [75]. A sensitivity analysis using the sample-size weighted Z-score meta-analysis approach yielded the same results; only fixed effect model results are shown. The 2,971,914 SNPs in each of 18 analyses (nine phenotypes in two sexes) reduced to 2,846,694 SNPs with available chromosome and position annotation in dbSNP. The genetic position (cM) was extracted from HapMap release 22 (http://hapmap.ncbi.nlm.nih.gov/downloads/recombination/2008-03_rel22_B36/rates/) or - if unavailable - approximated by the inverse-distance weighted average of the genetic positions of the nearest HapMap SNPs (release 22) on each side.
SNP selection strategy
We conducted two types of genome-wide searches in the discovery stage: (a) In the sex-specific scan, we computed sex-specific association P-values for each SNP, concatenated these for all nine phenotypes totaling 50,586,560 P-values (i.e. 2 sexes×9 phenotypes×2.85 Million SNPs), and selected 20,215 SNPs at 5% FDR [32]. Pruning this list to independent SNPs (starting with the 20,215 SNPs sorted by increasing P-value and deleting SNPs within 0.2 cM of any of the SNPs above) yielded 619 independent SNPs. For each SNP and for the phenotype that the SNP was selected for, we also computed P-values (P-diff) testing for difference between the meta-analyzed men-specific and women-specific beta-estimates 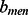 ,
, 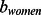 with corresponding standard errors
with corresponding standard errors 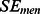 and
and 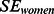 using the t statistic
using the t statistic
The correlation  between
between 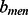 and
and 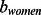 , computed as the Spearman rank correlation coefficient across all SNPs for each phenotype, ranged from 0.04 to 0.18 across phenotypes. From these 619 SNPs, we selected the 348 SNPs with nominally significant sex-difference (P-diff<0.05) to ensure some level of sex-difference in the discovery. Whether the sex-difference was significant was then evaluated in the follow-up stage (see below). (b) In the sex-difference scan, we computed P-diff for each of the ∼2.85 Million SNPs and each of the nine phenotypes and concatenated the totaling 25,293,280 P-values. We had planned to select SNPs for follow-up at an FDR of 5% across all SNPs and phenotypes, but there were none. Power considerations are provided in the Text S1 and Figure S4A.
, computed as the Spearman rank correlation coefficient across all SNPs for each phenotype, ranged from 0.04 to 0.18 across phenotypes. From these 619 SNPs, we selected the 348 SNPs with nominally significant sex-difference (P-diff<0.05) to ensure some level of sex-difference in the discovery. Whether the sex-difference was significant was then evaluated in the follow-up stage (see below). (b) In the sex-difference scan, we computed P-diff for each of the ∼2.85 Million SNPs and each of the nine phenotypes and concatenated the totaling 25,293,280 P-values. We had planned to select SNPs for follow-up at an FDR of 5% across all SNPs and phenotypes, but there were none. Power considerations are provided in the Text S1 and Figure S4A.
Follow-up and joint meta-analysis, establishing sex-difference
Study-specific follow-up data were quality-controlled in a similar manner as discovery data with increased attention towards strand-issues. We conducted sex-specific follow-up meta-analyses using the same statistical models as for the discovery. We combined (i) in silico studies and (ii) metabochip studies, and then combined results of (i) and (ii) implying a double genomic control correction (Text S1). Additionally, we conducted a joint meta-analysis combining the sex-specific association results of discovery and follow-up.
For SNPs selected for their small P-values of sex-specific association, the sex-difference estimates and corresponding P-values in the same data set would be inflated (see Figure S5) as the two tests are not independent. We therefore derived sex-difference estimates and corresponding P-values in the follow-up data alone.
As none of our selected SNPs stemmed from the scan targeted for OED signals (i.e. the sex-difference scan) or showed any evidence of OED in the discovery, we targeted our follow-up analysis for CED or SSE signals. We filtered for a main effect (P for both sexes combined <0.01) prior to testing for sex-difference (P-diff, as described above), since this increased the power to detect SSE and CED signals (Figure S4B, Text S1), while this filter did not introduce a bias such as a sex-stratified association filter would. SNPs with a P-diff in the follow-up at 5% FDR were considered as SNPs with significant sex difference.
Establishing genome-wide significance of anthropometric trait association and enrichment for female or male genetic effects
We considered a joint (discovery and follow-up combined) association P-men or P-women<5×10−8 as genome-wide significant. For each phenotype, we tested whether there were more male-specific or more female-specific associations among the associations with established genome-wide significance in at least one sex compared to the expected binomial distribution.
Age-stratified sex-specific meta-analysis and association with metabolic traits
For the identified signals with sex-difference, study partners of the discovery and the in silico follow-up re-analyzed their data stratified by sex and age group (≥50 years, <50 years) using the same models as described above. Age difference was tested within each sex using the same t statistic as applied for the sex-difference testing.
Sex-specific associations of the identified signals with metabolic traits were derived requesting a sex-stratified re-analysis from the Global Lipids Genetics Consortium (GLGC, Triglycerides, HDL-, LDL-, and Total cholesterol) [34], the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC, fasting insulin, fasting glucose, HOMA-B, HOMA-IR) [35], [76]; and, the DIAGRAM consortium (type 2 diabetes) [36]. We tested the overall number of SNPs with consistent nominally significant association for the sex that the SNP was selected for (P_women or P_men <0.05) compared to a binomial draw with an event rate of 0.05. It needs to be noted that this test does not account for the correlation between the traits nor for the fact that the consortia involve an overlap of studies.
Pathway analyses
In order to explore whether certain pathways are enriched among the genes depicted by loci with evidence for sex-difference, we applied MAGENTA [77]. Briefly, MAGENTA calculates gene-specific scores (for ∼18,000 genes) by combining the p-values (here: the sex-difference P-values from our discovery stage for a specific anthropometric phenotype) of SNPs in and around the genes (40 kb down-, 100 kb upstream). The genetic score is corrected for potential confounders, such as gene size, number of independent SNPs, LD pattern, length in genetic distance, and number of recombination hotspots. These scores are ranked and the genes within the top 5% of these scores are tested for enrichment in certain pathways (separately for each phenotype) as given by different databases (GO: http://www.geneontology.org/, KEGG: http://www.genome.jp/kegg/, Ingenuity: http://www.ingenuity.com/products/pathways_analysis.html, and PANTHER: http://www.pantherdb.org/). MAGENTA determines whether the genes among the 5% top scores link to certain pathways more often than expected by chance. FDR is controlled at 5% via 10,000 permutations (using a random set of genes with the same number of genes as those observed).
Search for biological and functional knowledge of the seven association regions
For the seven confirmed sex-difference loci (defined as the regions depicted by SNPs within 1.0 cM of the respective lead SNP showing a certain level of association, P≤100 * PleadSNP), we searched several catalogues and data bases to depict potential biologically relevant links or functional entities. We extended the regions of interest to +−500 kB around the lead SNP if the regions were very small and no gene was inflicted (as for the PPARG, VEGFA, MAP3K1 loci).
First, we performed an automated search for reported genes or variants in our regions in PubMed (http://www.pubmed.com) and OMIM (http://www.ncbi.nlm.nih.gov/omim) using Snipper (http://csg.sph.umich.edu/boehnke/snipper) or manually inspected UCSC (PMID: 22086951) and Ensembl (PMID: 21045057) genome browsers as well as the NHGRI GWAS catalog [78], [79]. Second, we explored whether SNPs known to provide reliable tags for Copy-Number-Variations (CNVs) in European-descent samples (combining four catalogues including 60167 CNV-tagging SNPs as described previously [31]) correlated with our lead SNPs. Third, we performed several online database searches to establish whether known variants within 500 kb of each lead SNP, that are correlated (r2>0.8 or 0.5) with our lead SNPs (using SNAP Proxy search [80]), might have putative or predicted function. (i) We searched the SIFT database [81] to determine whether any of these SNPs was predicted to affect protein function. (ii) We used SNPinfo [82] to investigate predicted and putative function in several functional classes, including splicing regulation, stop codons, polyphen predictions, SNPs3D predictions, transcription factor binding site (TFBS) prediction, and miRNA binding site prediction.
Expression QTL analyses in human and mouse tissue
We examined transcript expression of genes near each of the seven identified SNP. For human eQTL, we explored four different tissues (subcutaneous adipose tissue, whole blood, and lymphoblastoid cells; details on methodology and tissue samples in the Text S1). We computed sex-specific association including conditional analyses and r2 measures to identify cis eQTL signals that were likely to be coincident with the anthropometric trait signal. For mouse eQTL, we had four types of tissues (inguinal fat, gonadal fat, liver, brown fat) with expression derived by real-time RT-PCR as well as three types of tissue (liver, inguinal fat, gonadal fat) with Illumina assays (details in Text S1). The sex-specific association and a test for sex-differences were computed.
Supporting Information
Sensitivity Analysis excluding patient groups shows consistent results. Shown are sex-specific beta-estimates of the seven identified SNPs in the follow-up data without (original analysis) and with exclusion of patient groups. It can be seen that the results are robust and patient groups do not trigger the observed sex-differences.
(TIF)
Region plot of the 7 identified loci showing the (a) association P-value in women, (b) association P-value in men, and (c) the sex difference P-value.
(PDF)
Distribution of waist-to-hip ratio adjusted for BMI. Shown are the 5th, 25th, 50th (median), 75th, and 95th percentiles of the residuals of waist-hip-ratio (before inverse normal transform) adjusted for BMI (therefore zero mean) for each contributing study. It can be seen that the variability of the phenotype is symmetric and to a similar extent in men and women.
(TIFF)
Power comparison. (A) Discovery: Shown is the power for selecting a sex-sensitive SNP (42969 women, 34629 men; assuming a signal as PPARG in women, MAF = 0.42, R2 women = 0.00057, various effects for men) into follow-up at α = 2×10−5 for the sex-specific (orange) scan, the sex-difference scan (magenta), or the sex-combined scan (black). (B) Follow-up: Power (60936 women, 47896 men; assuming a signal such as PPARG as above in women, various effects for men) to establish sex-difference among the 348 SNPs in the follow-up by (i) testing all 348 SNPs for sex-difference (no prior filter for a main effect; blue) at 5% FDR (corresponding to a P-diff of 9.9×10−4; blue), or by (ii) testing first for a main effect (P-value combined for men and women <0.01) and then testing the remaining 74 SNPs for sex-difference at 5% FDR (here corresponding to a P-diff of 4.2×10−3; red).
(TIF)
Inflated P-values of the sex-difference test due to the selection on sex-specific association. We have simulated 1 Million SNPs under the null hypothesis of no sex-difference (and no association), selected 348 SNPs with the most extreme sex-specific association, and plotted the observed P-values of the sex-difference test compared to the expected. It can be seen that the observed sex-difference P-values are inflated (i.e. do not lie on the identity line), which indicates that the sex-difference test is not independent from the sex-specific association selection.
(TIFF)
List of studies including sample sizes.
(XLS)
Association results of the 348 loci put forward to follow-up, showing the results from 46 discovery studies including 60,556 men and 73,133 women and 48 follow-up studies including 62,397 men and 74,651 women. These include 46 SNPs with genome-wide significant association in men or women in the joint analysis of discovery and follow-up studies.
(XLS)
Study-specific designs and references. (follow-up stage only).
(XLS)
Study-specific methods. (follow-up stage only) (A) insilico studies, (B) metabochip studies.
(XLS)
Study-specific descriptives. (follow-up stage only).
(XLS)
Further ten SNP show sex-difference at 30% FDR. Shown are the SNPs with sex-difference in the follow-up according to 30% FDR additional to those shown in Table 1 . These include the further three loci previously reported for sexually dimorph waist-hip ratio associations, HOXC13, ITPR2/SSPN, and RPO3/C6orf173 [31].
(XLS)
No difference in any of the 7 associations between age groups. Age-stratified analyses results were conducted from 59 discovery and in silico follow-up studies for the seven identified SNPs.
(XLS)
Sex-specific association of the 7 SNPs with other traits.(A) BMI, height and weight: From the joint discovery and follow-up including 122,907 men and 147,746 women for height, 97,482 and 97,062 for weight, and 120,975 and 142,332 for BMI. It should be noted that results in Table 1 and Table S7 are only for the phenotype and sex for which the SNP was selected. (B) hip circumference (HIP), waist circumference (WC), waist-hip-ratio (WHR): Including 75,102 men and 96,383 women for hip, 86,196 and 108,303 for waist circumference, and 76,838 and 98,747 for waist-hip-ratio. (C) hip circumference adjusted for BMI (HIPadjBMI), waist circumference adjusted for BMI (WCadjBMI), waist-hip-ratio adjusted for BMI (WHRadjBMI). Including 74,949 men and 96,353 women for HIPadjBMI, 86,036 and 108,052 for WCadjBMI, and 76,625 and 98,352 for WHRadjBMI. (D) lipid traits: From the Global Lipids Consortium (Teslovich et al., 2010) including 39,104 men and 64,235 women. (E) glycaemic traits: From the MAGIC consortium (Prokopenko et al., 2009) including 54,046 men and 60,450 women. (F) type 2 diabetes: From the DIAGRAM consortium (Zeggini et al., 2008) including 18,786 men (4,451 cases and 14,335 controls) and 28,332 women (3,680 cases and 24,652 controls). (G) Summary of enrichment statistics: for sex-specific association of the 7 SNPs with the metabolic traits.
(XLS)
Pathway analysis reveals enrichment of PPAR signaling among genes with evidence for sex-difference. Shown are P-values for phenotype-specific enrichment using MAGENTA to determine whether certain pathways/biological processes/molecular functions are enriched among genes harboring SNPs with evidence for sex-difference. Only categories with false discovery rate (FDR)<10% are reported in the table and those with FDR<5% are considered statistically significant (bold). In short, for a certain phenotype in the discovery data, MAGENTA assigns a gene-specific score based on the sex-difference P-values of SNPs in or nearby (40 kb down-, 100 kb upstream) the respective gene. The score is calculated for each o the ∼18 000 annotated genes and corrected for gene-size, number of independent SNPs, LD pattern, length in genetic distance, and number of recombination hotspots. Then MAGENTA determines whether the genes with a score in the top 5% (∼900 genes for each phenotype) overlap with certain pathways more than expected by chance (i.e. as compared to a random gene set including the same number of genes).
(XLS)
Associations with cis gene expression (eQTL) in human subcutaneous adipose tissue (SAT) and lymphocytes. Transcripts of genes near each of the identified seven SNPs were examined in four differenet data sets (DeCode, MoIOBB, childhood asthma study, HapMap, details in Text S1). Sex-specific associations with the transcript are shown for the identified SNP with and without conditioning on the most significant SNP for that transcript (peak SNP) and also for the peak SNP with and without conditioning on the identified SNP, if FDR<5% (DeCode) or FDR<1% (MolOBB, childhood asthma study, HapMap) in one sex. For GRB14 and ADAMTS9, our identified SNP was highly correlated (r2>0.8) with the peak transcript SNP in men. No SNP showed nominally significant sex difference (Pdiff>0.05).
(XLS)
Gene expression in mice. Shown are expression levels in female and male mice, if expression was nominally significant in at least on sex, and the P-value testing for sex difference (P-diff<0.05) from the mouse experiments in three different center: (i) 21 male and 21 female mice with Illumina array for inguinal or gonadal fat (Houston, H), (ii) 139 male and 133 female mice with Illumina array for liver (Oxford, O), (iii) 7 male and 7 female mice with PCR analysis for brown fat, inguinal or gonadal fat, and liver (Regensburg, R). Examined genes are listed in Table S13.
(XLS)
Sex-specific phenotype description. Shown are mean and standard deviation (std.) of the investigated traits in a German general population study (KORA-S3 and KORA-S4). Age range is 25–75 years of age with mean of 53.6 for men and 52.9 for women. All phenotypes are adjusted for age and age2 as in the genetic association analyses.
(XLS)
Genes included in mouse eQTL investigations. (A) Regensburg mouse eQTL. Also shown are primers used for analysis of gene expression. (B) Oxford and Houston mouse eQTL. We explored all genes containing SNPs with at least some level of association in the initial discovery (P_men or P_women smaller than hundred times the index SNP P-value), but at least 3 genes and at maximum all genes within ±1 cM of the index SNP (annotation using genome browser Ensembl build 54).
(XLS)
Supplementary note.
(PDF)
Extended acknowledgments.
(PDF)
Funding Statement
Funding for this study was provided by the Aarno Koskelo Foundation; the Academy of Finland (40758, 41071, 77299, 102318, 104781, 110413, 117787, 117844, 118065, 118590, 120315, 120386, 121584, 123885, 124243, 124282, 125876, 126925, 129269, 129293, 129322, 129378, 129494, 129680, 139635, 211497, 213506, 250207, 1114194); the Academy of Finland Centre of Excellence in Complex Disease Genetics; the ALF/LUA research grant in Gothenburg; the Agency for Science, Technology, and Research of Singapore (A*STAR); Althingi (the Icelandic Parliament); the American Heart Association (0730094N); the Australian Research Council (ARC; grants DP0770096 and DP1093900); Biocentrum Helsinki; the Boston Medical Center; Boston University School of Medicine; the British Diabetes Association; BDA Research; the British Heart Foundation (BHF; PG/02/128, PG/07/133/24260, RG2008/08, RG2008/014); Cancer Research UK (C490/A8339, C490/A10124); the Central Norway Health Authority; Centre Hospitalier Universitaire Vaudois; the Chief Scientist Office of Scotland; the City of Kuopio; Deutsche Forschungsgemeinschaft; Diabetes Hilfs- und Forschungsfonds Deutschland (DHFD); Diabetes UK; the Donald W. Reynolds Foundation; Dresden University of Technology Funding Grant; the Dutch Brain Foundation; the Dutch Diabetes Research Foundation; the Dutch Kidney Foundation (E033); Dutch Inter University Cardiology Institute Netherlands (ICIN); the EMGO Institute for Health and Care Research (EMGO+); the Emil Aaltonen Foundation; the Erasmus Medical Center; Erasmus University; the Estonian Government (SF0180142s08); the Estonian Biocentre Centre of Excellence in Genomics; European Commission (FP5-QLG2-CT-2002-00896, FP5-QLG2-CT-2002-01254, FP5-QLRT-2001-01254, FP6-LSHM-CT-2004-005272, FP6-LSHM-CT-2004-503485, FP6-LSHG-CT-2004-518153, FP6-LSHG-CT-2006-018947, FP6-LSHM-CT-2006-037593, FP6-LSHM-CT-2006-037697, FP6-LSHM-CT-2007-037273, FP7-HEALTH-201413, FP7-HEALTH-201536, FP7-HEALTH-201550, FP7-HEALTH-201668, FP7-HEALTH-201865, FP7-HEALTH-230374, FP7-HEALTH-245536, FP7-HEALTH-261433), the Directorate C-Public Health (grant 2004310), the Marie Curie Intra-European Fellowship, EURO-BLCS, Framework 5 award (QLG1-CT-2000-01643); the European Regional Development Fund; the European Science Foundation (ESF); Federal State of Mecklenburg-West Pomerania; the Finnish Cultural Foundation; the Finnish Diabetes Research Foundation; the Finnish Foundation for Cardiovascular Research; the Finnish Funding Agency for Technology and Innovation (TEKES; 40058/07 and 70103/06); the Finnish Heart Association; Finnish Medical Society; the Finnish Ministry of Education and Culture (OKM; 627:2004–2011); the Finnish Ministry of Health and Social Affairs (5254); the Finnish National Institute for Health and Welfare; the Finnish Slot Machine Association (RAY); the Folkhälsan Research Foundation; the Fondation LeDucq; the Foundation for Life and Health in Finland; the Genetic Association Information Network (GAIN); the German Diabetes Association; the German Federal Ministry of Education and Research (BMBF; 01KU0908A, 01AK803A-H, 01IG07015G, 01GS0823, 01GS0834, 01ZZ0103, 01ZZ0403, 01ZZ9603, 03ZIK012, 01ER1206); the German Research Center for Environmental Health; GlaxoSmithKline; Göteborg Medical Society; The Great Wine Estates of the Margaret River region of Western Australia; Greek General Secretary of Research and Technology (PENED 2003); Healthway, Western Australia; the Heinz Nixdorf Foundation; Helmholtz Zentrum München; Helsinki University Central Hospital; Hjartavernd (the Icelandic Heart Association); Hospital district of Pirkanmaa, South Ostrobothnia, and Central Finland; the International Stichting Alzheimer Onderzoek (ISAO); Italian Ministry of Health; John D. and Catherine T. MacArthur Foundation; Juho Vainio Foundation; Juvenile Diabetes Research Foundation International (JDRF); Karolinska Institutet; Stockholm County Council (560183, 562183); Knut and Alice Wallenberg Foundation; Lundberg Foundation; March of Dimes (6-FY-09-507); Medical Research Foundation of Umeå University; Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano; Ministry of Science, Education and Sports of the Republic of Croatia (108-1080315-0302); Montreal Heart Institute Foundation; The Municipal Health Care Center and Hospital in Jakobstad; Municipality of Nijmegen; Municipality of Rotterdam; Närpes Health Care Foundation; National Alliance for Research on Schizophrenia and Depression (NARSAD); National Health and Medical Research Council of Australia (NHMRC; 241944, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 496688, 552485, 613672, 613601 and 1011506); Netherlands Brain Foundation; Netherlands Centre for Medical Systems Biology (CMSB); Netherlands Ministry of Health, Welfare and Sport; Netherlands Organization for Scientific Research (NWO; 014-93-015, 050-060-810, 40-0056-98-9032, 175-010-2005-011, 175-010-2007-006, 184-021-007, 311-60-008, 400-05-717, 480-04-004, 480-05-003, 904-61-090, 904-61-193, 911-03-012, 916-761-70, 985-10-002, SPI 56-464-14192); Neuroscience Campus Amsterdam (NCA); NHS Support for Science Funding Programme; Nord-Trøndelag County Council; Nordic Center of Cardiovascular Research (NCCR); Nordic Centre of Excellence in Disease Genetics, Nordic Centre of Excellence in Systems Biology (070014); Northern Netherlands Collaboration of Provinces (SNN); Norwegian Institute of Public Health; Norwegian Research Council (project number 185764); Norwegian University of Science and Technology (NTNU); Novo Nordisk Foundation; Ollqvist Foundation; Orion-Farmos Research Foundation; Paavo Nurmi Foundation; Päivikki and Sakari Sohlberg Foundation; Paul Michael Donovan Charitable Foundation; Perklén Foundation; Petrus and Augusta Hedlunds Foundation; Province of Groningen; Radboud University Nijmegen Medical Centre; Royal Swedish Academy of Science; Sardstedt AG & Co.; Siemens Healthcare; Signe and Ane Gyllenberg Foundation; Sigrid Juselius Foundation; Social Insurance Institution of Finland (4/26/2010); South Tyrolean Sparkasse Foundation; Susan G. Komen Breast Cancer Foundation; Swedish Cancer Society, Swedish Cultural Foundation in Finland; Swedish Diabetes Association; Swedish Foundation for Strategic Research (SSF); Swedish Heart-Lung Foundation; Swedish Research Council (0593, 8691, 2006-3832, K2007-66X-20270-01-3, K2010-52X-20229-05-3, and K2010-54X-09894-19-3), Swedish Society for Medical Research; Swedish Society of Medicine; Swiss National Science Foundation (310000-112552, 33CSCO-122661); Tampere Tuberculosis Foundation; Torsten and Ragnar Söderberg Foundation; UK Economic and Social Research Council (ESRC); UK Medical Research Council (MRC; G0000649, G0601261, G9521010D, G0000934, G0500539, G0600705), Centre for Obesity and Related Metabolic Disease, Centre of Epidemiology for Child Health, PrevMetSyn/SALVE, MRC-GSK pilot programme (grant ID 85374); UK National Institute for Health Research (NIHR); Cambridge Biomedical Research Centre, CamStrad, Comprehensive Biomedical Research Centre, London Cardiovascular Biomedical Research Unit, Oxford Biomedical Research Centre, (RP-PG-0310-1002); University Hospital Essen; University Hospital Kuopio; University Hospital Oulu; University Hospital Tampere; University Hospital Turku; University of Groningen; University of Lausanne; University of Leipzig; University of Tromsø; University Medical Center Groningen; University of Tartu; US Department of Agriculture (USDA) National Research Initiative Competitive Grant (2007-35205-17883); US National Cancer Institute (NCI); US National Heart, Lung, and Blood Institute (NHLBI; 268200625226C, 268201100005C, 268201100006C, 268201100007C, 268201100008C, 268201100009C, 268201100010C, 268201100011C, 268201100012C, 5R01HL087679-02, and 1RL1MH083268-01); US National Human Genome Research Institute (NHGRI), US National Institute of Allergy and Infectious Diseases (NIAID), US National Institute of Child Health and Human Development (NICHD), US National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), US National Institutes of Health (NIH; 5R01MH63706:02, K23DK080145, M01RR16500, N01AG12100, N01AG12109, N01HC25195, N01HC35129, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85084, N01HC85085, N01HC85086, N01HC85239, N01HG65403, N02HL64278, P01CA87969, P30DK063491, P30DK072488, R01AA007535, R01AA014041, R01AA013320, R01AA013321, R01AA013326, R01AG004563, R01AG008724, R01AG008861, R01AG010175, R01AG013196, R01AG015928, R01AG020098, R01AG023629, R01AG027058, R01AG028555, R01CA047988, R01CA049449, R01CA050385, R01CA065725, R01CA067262, R01DA12854, R01DK062370, R01DK072193, R01DK073490, R01DK075681, R01DK075787, R01DK089256, R01HD042157, R01HG002651, R01HL036310, R01HL043851, R01HL059367, R01HL071981, R01HL075366, R01HL086694, R01HL087641, R01HL087647, R01HL087652, R01HL087676, R01HL087679, R01HL087700, R01HL088215, R01HL105756, R01HS006516, R01LM010098, R01MH059565, R01MH059566, R01MH059571, R01MH059586, R01MH059587, R01MH059588, R01MH060870, R01MH060879, R01MH061675, R01MH067257, R01MH079469, R01MH079470, R01MH081800, RL1MH083268, U01CA098233, U01DK062418, U01HG004402, U01HL069757, U01HL072515, U01HL080295, U01HL084729, U01HL084756, U24MH068457, U54RR020278, UL1RR025005, UL1RR033176, Z01HG000024); US National Institute of Neurological Disorders and Stroke (NINDS); Vasa, Närpes and Korsholm Health Care Centers; Västra Götaland Foundation; VU University Amsterdam; Walter E. Nichols, M.D. and Eleanor Nichols endowments at Stanford University; Wellcome Trust (068545/Z/02, 069224, 072960, 075491, 076113/B/04/Z, 076113/K/04/Z, 079557, 079895, 081682, 083270, 084183/Z/07/Z, 085301, 086596/Z/08/Z, 089061/Z/09/Z, 089062/Z/09/Z, 090532/Z/09/Z, 098051), Go-DARTS, WT Research Career Development Fellowship, WT Career Development Award, WT Centre for Human Genetics, WT Clinical Research Facility, WT Sanger Institute, Scottish Health Informatics Programme; and the Yrjö Jahnsson Foundation (56358). More details of acknowledgements can be found in Text S2. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Legato M (2004) Principles of Gender-Specific Medicine, Vol. 1 & Vol. 2: New York: Elsevier Academic Press.
- 2.Wizemann TM, Pardue ML (2001) Exploring the biological contributions to human health: does sex matter? Washington, DC: National Academies Press. [PubMed]
- 3.Malina RM (2005) Variation in body composition associated with sex and ethnicity. In: Heymsfield S, editor. Human body composition. 2nd ed. Champaign, IL: Human Kinetics. pp. 271–298.
- 4. Taylor RW, Grant AM, Williams SM, Goulding A (2010) Sex differences in regional body fat distribution from pre- to postpuberty. Obesity 18: 1410–1416. [DOI] [PubMed] [Google Scholar]
- 5. Wells JC (2007) Sexual dimorphism of body composition. Best practice & research Clinical endocrinology & metabolism 21: 415–430. [DOI] [PubMed] [Google Scholar]
- 6. Green HJ, Fraser IG, Ranney DA (1984) Male and female differences in enzyme activities of energy metabolism in vastus lateralis muscle. Journal of the neurological sciences 65: 323–331. [DOI] [PubMed] [Google Scholar]
- 7. Komi PV, Karlsson J (1978) Skeletal muscle fibre types, enzyme activities and physical performance in young males and females. Acta physiologica Scandinavica 103: 210–218. [DOI] [PubMed] [Google Scholar]
- 8. Simoneau JA, Lortie G, Boulay MR, Thibault MC, Theriault G, et al. (1985) Skeletal muscle histochemical and biochemical characteristics in sedentary male and female subjects. Canadian journal of physiology and pharmacology 63: 30–35. [DOI] [PubMed] [Google Scholar]
- 9. Trotter M, Peterson RR (1970) Weight of the skeleton during postnatal development. American journal of physical anthropology 33: 313–323. [DOI] [PubMed] [Google Scholar]
- 10. Garn SM, Nagy JM, Sandusky ST (1972) Differential sexual dimorphism in bone diameters of subjects of European and African ancestry. American journal of physical anthropology 37: 127–129. [DOI] [PubMed] [Google Scholar]
- 11. Vazquez G, Duval S, Jacobs DR Jr, Silventoinen K (2007) Comparison of body mass index, waist circumference, and waist/hip ratio in predicting incident diabetes: a meta-analysis. Epidemiol Rev 29: 115–128. [DOI] [PubMed] [Google Scholar]
- 12. Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, et al. (2003) Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA : the journal of the American Medical Association 289: 76–79. [DOI] [PubMed] [Google Scholar]
- 13. Must A, Spadano J, Coakley EH, Field AE, Colditz G, et al. (1999) The disease burden associated with overweight and obesity. Jama-Journal of the American Medical Association 282: 1523–1529. [DOI] [PubMed] [Google Scholar]
- 14. Yusuf S, Hawken S, Ounpuu S, Bautista L, Franzosi MG, et al. (2005) Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet 366: 1640–1649. [DOI] [PubMed] [Google Scholar]
- 15. Canoy D, Boekholdt SM, Wareham N, Luben R, Welch A, et al. (2007) Body fat distribution and risk of coronary heart disease in men and women in the European Prospective Investigation Into Cancer and Nutrition in Norfolk cohort: a population-based prospective study. Circulation 116: 2933–2943. [DOI] [PubMed] [Google Scholar]
- 16. Welborn TA, Dhaliwal SS, Bennett SA (2003) Waist-hip ratio is the dominant risk factor predicting cardiovascular death in Australia. Medical Journal of Australia 179: 580–585. [DOI] [PubMed] [Google Scholar]
- 17. Borugian MJ, Sheps SB, Kim-Sing C, Olivotto IA, Van Patten C, et al. (2003) Waist-to-hip ratio and breast cancer mortality. American journal of epidemiology 158: 963–968. [DOI] [PubMed] [Google Scholar]
- 18. Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M (2008) Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet 371: 569–578. [DOI] [PubMed] [Google Scholar]
- 19. Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, et al. (2008) General and abdominal adiposity and risk of death in Europe. The New England journal of medicine 359: 2105–2120. [DOI] [PubMed] [Google Scholar]
- 20. Langenberg C, Sharp SJ, Schulze MB, Rolandsson O, Overvad K, et al. (2012) Long-term risk of incident type 2 diabetes and measures of overall and regional obesity: the EPIC-InterAct case-cohort study. PLoS medicine 9: e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, et al. (2008) Genome-wide association analysis identifies 20 loci that influence adult height. Nature genetics 40: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Estrada K, Krawczak M, Schreiber S, van Duijn K, Stolk L, et al. (2009) A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Human molecular genetics 18: 3516–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tonjes A, Koriath M, Schleinitz D, Dietrich K, Bottcher Y, et al. (2009) Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Human molecular genetics 18: 4662–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, et al. (2010) A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Human molecular genetics 19: 2303–2312. [DOI] [PubMed] [Google Scholar]
- 25. Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, et al. (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. (2007) A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. (2008) Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nature genetics 40: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics 41: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature genetics 42: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, et al. (2009) Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS genetics 5: e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, et al. (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nature genetics 42: 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini YaH, Y (1995) Controlling The False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Statist Soc Ser B 57: 289–300. [Google Scholar]
- 33. Lehrke M, Lazar MA (2005) The many faces of PPARgamma. Cell 123: 993–999. [DOI] [PubMed] [Google Scholar]
- 34. Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, et al. (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature genetics 40: 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, et al. (2011) Brown adipose tissue activity controls triglyceride clearance. Nature Medicine 17: 200–205. [DOI] [PubMed] [Google Scholar]
- 38. Jackson AS, Stanforth PR, Gagnon J, Rankinen T, Leon AS, et al. (2002) The effect of sex, age and race on estimating percentage body fat from body mass index: The Heritage Family Study. Int J Obes Relat Metab Disord 26: 789–796. [DOI] [PubMed] [Google Scholar]
- 39. McQuaid SE, Manolopoulos KN, Dennis AL, Cheeseman J, Karpe F, et al. (2010) Development of an arterio-venous difference method to study the metabolic physiology of the femoral adipose tissue depot. Obesity 18: 1055–1058. [DOI] [PubMed] [Google Scholar]
- 40. Pi-Sunyer FX (2004) The epidemiology of central fat distribution in relation to disease. Nutr Rev 62: S120–126. [DOI] [PubMed] [Google Scholar]
- 41. Maynard LM, Wisemandle W, Roche AF, Chumlea WC, Guo SS, et al. (2001) Childhood body composition in relation to body mass index. Pediatrics 107: 344–350. [DOI] [PubMed] [Google Scholar]
- 42. Hattori K, Tahara Y, Moji K, Aoyagi K, Furusawa T (2004) Chart analysis of body composition change among pre- and postadolescent Japanese subjects assessed by underwater weighing method. Int J Obes Relat Metab Disord 28: 520–524. [DOI] [PubMed] [Google Scholar]
- 43. Lovejoy JC, Champagne CM, de Jonge L, Xie H, Smith SR (2008) Increased visceral fat and decreased energy expenditure during the menopausal transition. International journal of obesity 32: 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cinti S (2001) The adipose organ: morphological perspectives of adipose tissues. Proc Nutr Soc 60: 319–328. [DOI] [PubMed] [Google Scholar]
- 45. Dempfle A, Hinney A, Heinzel-Gutenbrunner M, Raab M, Geller F, et al. (2004) Large quantitative effect of melanocortin-4 receptor gene mutations on body mass index. J Med Genet 41: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, et al. (2009) Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 150: 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rantalainen M, Herrera BM, Nicholson G, Bowden R, Wills QF, et al. (2011) MicroRNA expression in abdominal and gluteal adipose tissue is associated with mRNA expression levels and partly genetically driven. PloS one 6: e27338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Min JL, Nicholson G, Halgrimsdottir I, Almstrup K, Petri A, et al. (2012) Coexpression network analysis in abdominal and gluteal adipose tissue reveals regulatory genetic loci for metabolic syndrome and related phenotypes. PLoS genetics 8: e1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alexanderson C, Eriksson E, Stener-Victorin E, Lystig T, Gabrielsson B, et al. (2007) Postnatal testosterone exposure results in insulin resistance, enlarged mesenteric adipocytes, and an atherogenic lipid profile in adult female rats: comparisons with estradiol and dihydrotestosterone. Endocrinology 148: 5369–5376. [DOI] [PubMed] [Google Scholar]
- 50. Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, et al. (2011) Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 6: 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zillikens MC, Yazdanpanah M, Pardo LM, Rivadeneira F, Aulchenko YS, et al. (2008) Sex-specific genetic effects influence variation in body composition. Diabetologia 51: 2233–2241. [DOI] [PubMed] [Google Scholar]
- 52. Robitaille J, Despres JP, Perusse L, Vohl MC (2003) The PPAR-gamma P12A polymorphism modulates the relationship between dietary fat intake and components of the metabolic syndrome: results from the Quebec Family Study. Clinical genetics 63: 109–116. [DOI] [PubMed] [Google Scholar]
- 53. Morini E, Tassi V, Capponi D, Ludovico O, Dallapiccola B, et al. (2008) Interaction between PPARgamma2 variants and gender on the modulation of body weight. Obesity 16: 1467–1470. [DOI] [PubMed] [Google Scholar]
- 54. Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, et al. (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- 57. Shea MK, Nicklas BJ, Marsh AP, Houston DK, Miller GD, et al. (2011) The effect of pioglitazone and resistance training on body composition in older men and women undergoing hypocaloric weight loss. Obesity 19: 1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goenaga D, Hampe C, Carre N, Cailliau K, Browaeys-Poly E, et al. (2009) Molecular determinants of Grb14-mediated inhibition of insulin signaling. Molecular endocrinology 23: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, et al. (2010) Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arteriosclerosis, thrombosis, and vascular biology 30: 2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research 35: D61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pearlman A, Loke J, Le Caignec C, White S, Chin L, et al. (2010) Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. American journal of human genetics 87: 898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leenders F, Tesdorpf JG, Markus M, Engel T, Seedorf U, et al. (1996) Porcine 80-kDa protein reveals intrinsic 17 beta-hydroxysteroid dehydrogenase, fatty acyl-CoA-hydratase/dehydrogenase, and sterol transfer activities. J Biol Chem 271: 5438–5442. [DOI] [PubMed] [Google Scholar]
- 63. Peltoketo H, Isomaa V, Poutanen M, Vihko R (1996) Expression and regulation of 17 beta-hydroxysteroid dehydrogenase type 1. J Endocrinol 150 Suppl: S21–30. [PubMed] [Google Scholar]
- 64. Thompson JB, Dzubur E, Wade J, Tomaszycki M (2011) The effects of estradiol on 17beta-hydroxysteroid dehydrogenase type IV and androgen receptor expression in the developing zebra finch song system. Brain research 1401: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li Y, Willer C, Sanna S, Abecasis G (2009) Genotype imputation. Annual review of genomics and human genetics 10: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics 39: 906–913. [DOI] [PubMed] [Google Scholar]
- 67. Guan Y, Stephens M (2008) Practical issues in imputation-based association mapping. PLoS genetics 4: e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aulchenko YS, Struchalin MV, van Duijn CM (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC bioinformatics 11: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- 71. Abecasis GR, Wigginton JE (2005) Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. American journal of human genetics 77: 754–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Servin B, Stephens M (2007) Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS genetics 3: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 75. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. (2009) Variants in MTNR1B influence fasting glucose levels. Nature genetics 41: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D (2010) Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS genetics 6: e1001058 doi:10.1371/journal.pgen.1001058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hindorff LA, MacArthur JEBI, Wise A, Junkins HA, Hall PN, et al.. (2010) A Catalog of Published Genome-Wide Association Studies. [DOI] [PMC free article] [PubMed]
- 80. Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 82. Xu Z, Taylor JA (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic acids research 37: W600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity Analysis excluding patient groups shows consistent results. Shown are sex-specific beta-estimates of the seven identified SNPs in the follow-up data without (original analysis) and with exclusion of patient groups. It can be seen that the results are robust and patient groups do not trigger the observed sex-differences.
(TIF)
Region plot of the 7 identified loci showing the (a) association P-value in women, (b) association P-value in men, and (c) the sex difference P-value.
(PDF)
Distribution of waist-to-hip ratio adjusted for BMI. Shown are the 5th, 25th, 50th (median), 75th, and 95th percentiles of the residuals of waist-hip-ratio (before inverse normal transform) adjusted for BMI (therefore zero mean) for each contributing study. It can be seen that the variability of the phenotype is symmetric and to a similar extent in men and women.
(TIFF)
Power comparison. (A) Discovery: Shown is the power for selecting a sex-sensitive SNP (42969 women, 34629 men; assuming a signal as PPARG in women, MAF = 0.42, R2 women = 0.00057, various effects for men) into follow-up at α = 2×10−5 for the sex-specific (orange) scan, the sex-difference scan (magenta), or the sex-combined scan (black). (B) Follow-up: Power (60936 women, 47896 men; assuming a signal such as PPARG as above in women, various effects for men) to establish sex-difference among the 348 SNPs in the follow-up by (i) testing all 348 SNPs for sex-difference (no prior filter for a main effect; blue) at 5% FDR (corresponding to a P-diff of 9.9×10−4; blue), or by (ii) testing first for a main effect (P-value combined for men and women <0.01) and then testing the remaining 74 SNPs for sex-difference at 5% FDR (here corresponding to a P-diff of 4.2×10−3; red).
(TIF)
Inflated P-values of the sex-difference test due to the selection on sex-specific association. We have simulated 1 Million SNPs under the null hypothesis of no sex-difference (and no association), selected 348 SNPs with the most extreme sex-specific association, and plotted the observed P-values of the sex-difference test compared to the expected. It can be seen that the observed sex-difference P-values are inflated (i.e. do not lie on the identity line), which indicates that the sex-difference test is not independent from the sex-specific association selection.
(TIFF)
List of studies including sample sizes.
(XLS)
Association results of the 348 loci put forward to follow-up, showing the results from 46 discovery studies including 60,556 men and 73,133 women and 48 follow-up studies including 62,397 men and 74,651 women. These include 46 SNPs with genome-wide significant association in men or women in the joint analysis of discovery and follow-up studies.
(XLS)
Study-specific designs and references. (follow-up stage only).
(XLS)
Study-specific methods. (follow-up stage only) (A) insilico studies, (B) metabochip studies.
(XLS)
Study-specific descriptives. (follow-up stage only).
(XLS)
Further ten SNP show sex-difference at 30% FDR. Shown are the SNPs with sex-difference in the follow-up according to 30% FDR additional to those shown in Table 1 . These include the further three loci previously reported for sexually dimorph waist-hip ratio associations, HOXC13, ITPR2/SSPN, and RPO3/C6orf173 [31].
(XLS)
No difference in any of the 7 associations between age groups. Age-stratified analyses results were conducted from 59 discovery and in silico follow-up studies for the seven identified SNPs.
(XLS)
Sex-specific association of the 7 SNPs with other traits.(A) BMI, height and weight: From the joint discovery and follow-up including 122,907 men and 147,746 women for height, 97,482 and 97,062 for weight, and 120,975 and 142,332 for BMI. It should be noted that results in Table 1 and Table S7 are only for the phenotype and sex for which the SNP was selected. (B) hip circumference (HIP), waist circumference (WC), waist-hip-ratio (WHR): Including 75,102 men and 96,383 women for hip, 86,196 and 108,303 for waist circumference, and 76,838 and 98,747 for waist-hip-ratio. (C) hip circumference adjusted for BMI (HIPadjBMI), waist circumference adjusted for BMI (WCadjBMI), waist-hip-ratio adjusted for BMI (WHRadjBMI). Including 74,949 men and 96,353 women for HIPadjBMI, 86,036 and 108,052 for WCadjBMI, and 76,625 and 98,352 for WHRadjBMI. (D) lipid traits: From the Global Lipids Consortium (Teslovich et al., 2010) including 39,104 men and 64,235 women. (E) glycaemic traits: From the MAGIC consortium (Prokopenko et al., 2009) including 54,046 men and 60,450 women. (F) type 2 diabetes: From the DIAGRAM consortium (Zeggini et al., 2008) including 18,786 men (4,451 cases and 14,335 controls) and 28,332 women (3,680 cases and 24,652 controls). (G) Summary of enrichment statistics: for sex-specific association of the 7 SNPs with the metabolic traits.
(XLS)
Pathway analysis reveals enrichment of PPAR signaling among genes with evidence for sex-difference. Shown are P-values for phenotype-specific enrichment using MAGENTA to determine whether certain pathways/biological processes/molecular functions are enriched among genes harboring SNPs with evidence for sex-difference. Only categories with false discovery rate (FDR)<10% are reported in the table and those with FDR<5% are considered statistically significant (bold). In short, for a certain phenotype in the discovery data, MAGENTA assigns a gene-specific score based on the sex-difference P-values of SNPs in or nearby (40 kb down-, 100 kb upstream) the respective gene. The score is calculated for each o the ∼18 000 annotated genes and corrected for gene-size, number of independent SNPs, LD pattern, length in genetic distance, and number of recombination hotspots. Then MAGENTA determines whether the genes with a score in the top 5% (∼900 genes for each phenotype) overlap with certain pathways more than expected by chance (i.e. as compared to a random gene set including the same number of genes).
(XLS)
Associations with cis gene expression (eQTL) in human subcutaneous adipose tissue (SAT) and lymphocytes. Transcripts of genes near each of the identified seven SNPs were examined in four differenet data sets (DeCode, MoIOBB, childhood asthma study, HapMap, details in Text S1). Sex-specific associations with the transcript are shown for the identified SNP with and without conditioning on the most significant SNP for that transcript (peak SNP) and also for the peak SNP with and without conditioning on the identified SNP, if FDR<5% (DeCode) or FDR<1% (MolOBB, childhood asthma study, HapMap) in one sex. For GRB14 and ADAMTS9, our identified SNP was highly correlated (r2>0.8) with the peak transcript SNP in men. No SNP showed nominally significant sex difference (Pdiff>0.05).
(XLS)
Gene expression in mice. Shown are expression levels in female and male mice, if expression was nominally significant in at least on sex, and the P-value testing for sex difference (P-diff<0.05) from the mouse experiments in three different center: (i) 21 male and 21 female mice with Illumina array for inguinal or gonadal fat (Houston, H), (ii) 139 male and 133 female mice with Illumina array for liver (Oxford, O), (iii) 7 male and 7 female mice with PCR analysis for brown fat, inguinal or gonadal fat, and liver (Regensburg, R). Examined genes are listed in Table S13.
(XLS)
Sex-specific phenotype description. Shown are mean and standard deviation (std.) of the investigated traits in a German general population study (KORA-S3 and KORA-S4). Age range is 25–75 years of age with mean of 53.6 for men and 52.9 for women. All phenotypes are adjusted for age and age2 as in the genetic association analyses.
(XLS)
Genes included in mouse eQTL investigations. (A) Regensburg mouse eQTL. Also shown are primers used for analysis of gene expression. (B) Oxford and Houston mouse eQTL. We explored all genes containing SNPs with at least some level of association in the initial discovery (P_men or P_women smaller than hundred times the index SNP P-value), but at least 3 genes and at maximum all genes within ±1 cM of the index SNP (annotation using genome browser Ensembl build 54).
(XLS)
Supplementary note.
(PDF)
Extended acknowledgments.
(PDF)