Abstract
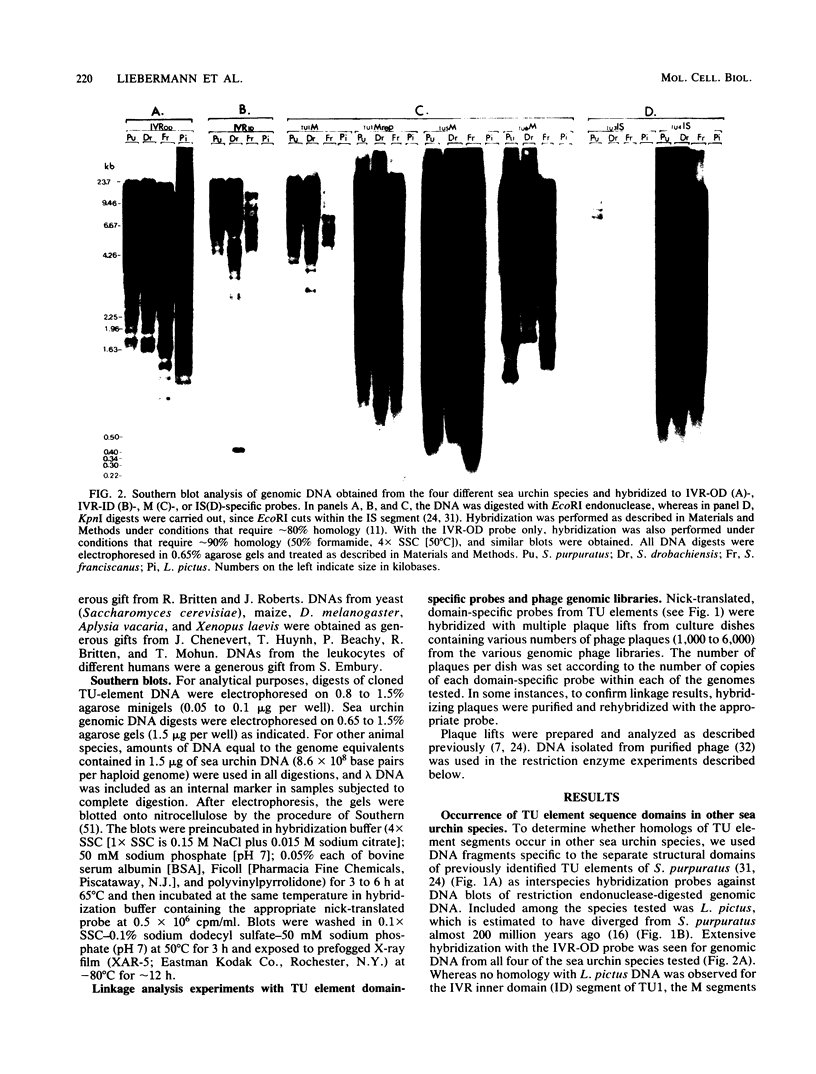
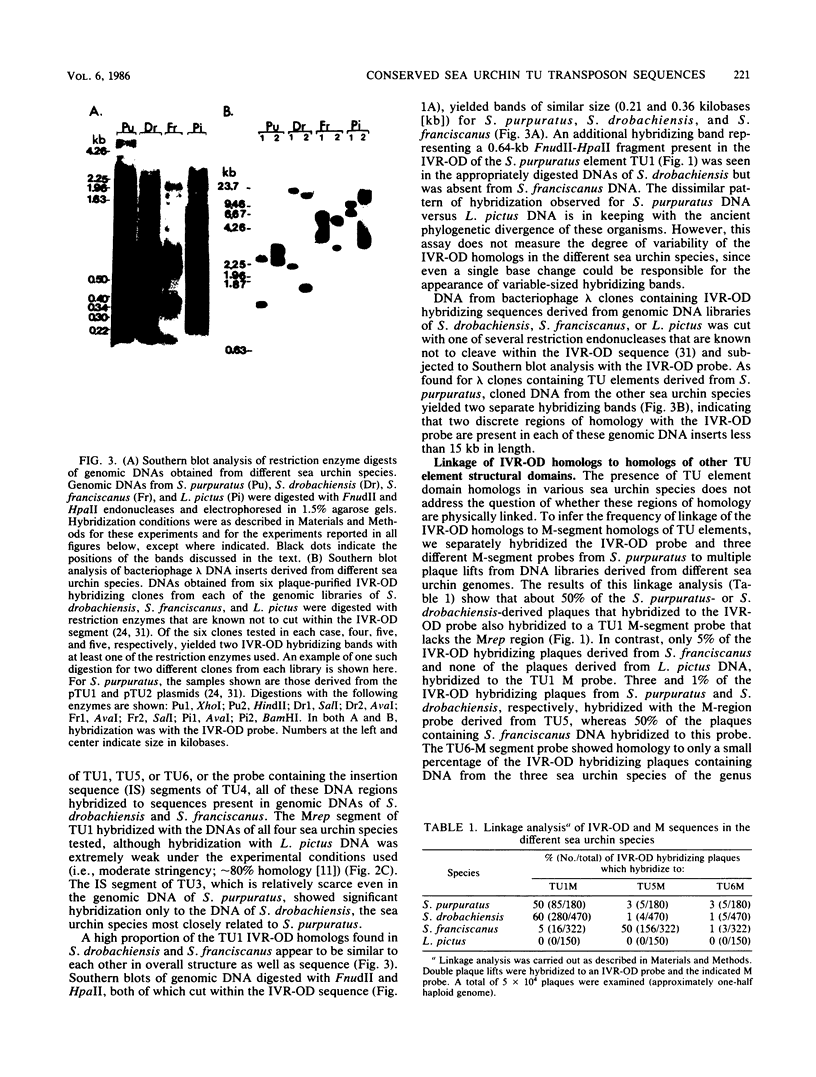
Sequences homologous to various structural domains of the Strongylocentrotus purpuratus TU family of transposons are present in sea urchin species closely related to S. purpuratus and were found in close proximity to each other in linkage patterns that differed for different species. Sequence homologs of the inverted repeat outer domain (IVR-OD) segment were, in addition, present in a sea urchin related only distantly to S. purpuratus and in all other eucaryotic organisms surveyed. In humans, a polymorphic hybridization pattern was seen for genomic DNA obtained from different individuals. Sequence comparisons revealed that repeated sequence motifs similar to those making up the 15-base-pair direct repeat unit of the IVR-OD domain of the TU elements exist in the IVRs of transposons identified in Drosophila melanogaster and maize and in the transcription control regions of certain eucaryotic viral and cellular genes. The remarkable evolutionary conservation of IVR-OD homologs may reflect a biological role for these sequences in DNA transposition, the regulation of gene expression, or both.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. M., Scheller R. H., Posakony J. W., McAllister L. B., Trabert S. G., Beall C., Britten R. J., Davidson E. H. Repetitive sequences of the sea urchin genome. Distribution of members of specific repetitive families. J Mol Biol. 1981 Jan 5;145(1):5–28. doi: 10.1016/0022-2836(81)90332-6. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Barker R. F., Thompson D. V., Talbot D. R., Swanson J., Bennetzen J. L. Nucleotide sequence of the maize transposable element Mul. Nucleic Acids Res. 1984 Aug 10;12(15):5955–5967. doi: 10.1093/nar/12.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Cairns J. The origin of human cancers. Nature. 1981 Jan 29;289(5796):353–357. doi: 10.1038/289353a0. [DOI] [PubMed] [Google Scholar]
- Conrad S. E., Botchan M. R. Isolation and characterization of human DNA fragments with nucleotide sequence homologies with the simian virus 40 regulatory region. Mol Cell Biol. 1982 Aug;2(8):949–965. doi: 10.1128/mcb.2.8.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Posakony J. W. Repetitive sequence transcripts in development. Nature. 1982 Jun 24;297(5868):633–635. doi: 10.1038/297633a0. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Saffer J. D., Lee W. S., Tjian R. Transcription factor Sp1 recognizes promoter sequences from the monkey genome that are simian virus 40 promoter. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4915–4919. doi: 10.1073/pnas.82.15.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Barbara McClintock's controlling elements: now at the DNA level. Cell. 1984 Dec;39(2 Pt 1):253–259. doi: 10.1016/0092-8674(84)90002-3. [DOI] [PubMed] [Google Scholar]
- Döring H. P., Tillmann E., Starlinger P. DNA sequence of the maize transposable element Dissociation. Nature. 1984 Jan 12;307(5947):127–130. doi: 10.1038/307127a0. [DOI] [PubMed] [Google Scholar]
- Erwin D. H., Valentine J. W. "Hopeful monsters," transposons, and Metazoan radiation. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5482–5483. doi: 10.1073/pnas.81.17.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J. Transposable elements in eukaryotes. Int Rev Cytol. 1985;93:281–326. doi: 10.1016/s0074-7696(08)61376-5. [DOI] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Hartzell S. W., Byrne B. J., Subramanian K. N. Mapping of the late promoter of simian virus 40. Proc Natl Acad Sci U S A. 1984 Jan;81(1):23–27. doi: 10.1073/pnas.81.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell S. W., Yamaguchi J., Subramanian K. N. SV40 deletion mutants lacking the 21-bp repeated sequences are viable, but have noncomplementable deficiencies. Nucleic Acids Res. 1983 Mar 11;11(5):1601–1616. doi: 10.1093/nar/11.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Liebermann B., Liebermann D., Kedes L. H., Cohen S. N. TU elements: a heterogeneous family of modularly structured eucaryotic transposons. Mol Cell Biol. 1985 May;5(5):991–1001. doi: 10.1128/mcb.5.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. A., Davidson E. H., Britten R. J. Insertion of a short repetitive sequence (D88I) in a sea urchin gene: a typical interspersed repeat? J Mol Evol. 1984;20(3-4):195–201. doi: 10.1007/BF02104726. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Cohn R. H., Lowry J. C., Chang A. C., Cohen S. N. The organization of sea urchin histone genes. Cell. 1975 Nov;6(3):359–369. doi: 10.1016/0092-8674(75)90185-3. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Klein G. The role of gene dosage and genetic transpositions in carcinogenesis. Nature. 1981 Nov 26;294(5839):313–318. doi: 10.1038/294313a0. [DOI] [PubMed] [Google Scholar]
- Levis R., Collins M., Rubin G. M. FB elements are the common basis for the instability of the wDZL and wC Drosophila mutations. Cell. 1982 Sep;30(2):551–565. doi: 10.1016/0092-8674(82)90252-5. [DOI] [PubMed] [Google Scholar]
- Liebermann D., Hoffman-Liebermann B., Weinthal J., Childs G., Maxson R., Mauron A., Cohen S. N., Kedes L. An unusual transposon with long terminal inverted repeats in the sea urchin Strongylocentrotus purpuratus. Nature. 1983 Nov 24;306(5941):342–347. doi: 10.1038/306342a0. [DOI] [PubMed] [Google Scholar]
- MCCLINTOCK B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- McCutchan T. F., Singer M. F. DNA sequences similar to those around the simian virus 40 origin of replication are present in the monkey genome. Proc Natl Acad Sci U S A. 1981 Jan;78(1):95–99. doi: 10.1073/pnas.78.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. C., Spence A., Smith M. The distal transcription signals of the herpesvirus tk gene share a common hexanucleotide control sequence. Cell. 1984 May;37(1):253–262. doi: 10.1016/0092-8674(84)90321-0. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mishoe H., Brady J. N., Radonovich M., Salzman N. P. Simian virus 40 guanine-cytosine-rich sequences function as independent transcriptional control elements in vitro. Mol Cell Biol. 1984 Dec;4(12):2911–2920. doi: 10.1128/mcb.4.12.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982 May 20;297(5863):201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Potter S., Truett M., Phillips M., Maher A. Eucaryotic transposable genetic elements with inverted terminal repeats. Cell. 1980 Jul;20(3):639–647. doi: 10.1016/0092-8674(80)90310-4. [DOI] [PubMed] [Google Scholar]
- Queen C., Lord S. T., McCutchan T. F., Singer M. F. Three segments from the monkey genome that hybridize to simian virus 40 have common structural elements. Mol Cell Biol. 1981 Dec;1(12):1061–1068. doi: 10.1128/mcb.1.12.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Grula J. W., Posakony J. W., Hudspeth R., Davidson E. H., Britten R. J. Comparison of sea urchin and human mtDNA: evolutionary rearrangement. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4614–4618. doi: 10.1073/pnas.80.15.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N., Kress M., Gruss P., Khoury G. BK viral enhancer element and a human cellular homolog. Science. 1983 Nov 18;222(4625):749–755. doi: 10.1126/science.6314501. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B., Liao L. W., Hirsh D. Sequence of the C. elegans transposable element Tc1. Nucleic Acids Res. 1983 Jun 25;11(12):4201–4209. doi: 10.1093/nar/11.12.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer J. D., Singer M. F. Transcription from SV 40-like monkey DNA sequences. Nucleic Acids Res. 1984 Jun 11;12(11):4769–4788. doi: 10.1093/nar/12.11.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller R. H., Anderson D. M., Posakony J. W., McAllister L. B., Britten R. J., Davidson E. H. Repetitive sequences of the sea urchin genome. II. Subfamily structure and evolutionary conservation. J Mol Biol. 1981 Jun 15;149(1):15–39. doi: 10.1016/0022-2836(81)90258-8. [DOI] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. The genome of human papovavirus BKV. Cell. 1979 Dec;18(4):963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Syvanen M. The evolutionary implications of mobile genetic elements. Annu Rev Genet. 1984;18:271–293. doi: 10.1146/annurev.ge.18.120184.001415. [DOI] [PubMed] [Google Scholar]
- Truett M. A., Jones R. S., Potter S. S. Unusual structure of the FB family of transposable elements in Drosophila. Cell. 1981 Jun;24(3):753–763. doi: 10.1016/0092-8674(81)90101-x. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Zuker C., Cappello J., Lodish H. F., George P., Chung S. Dictyostelium transposable element DIRS-1 has 350-base-pair inverted terminal repeats that contain a heat shock promoter. Proc Natl Acad Sci U S A. 1984 May;81(9):2660–2664. doi: 10.1073/pnas.81.9.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]