Abstract
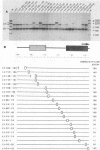
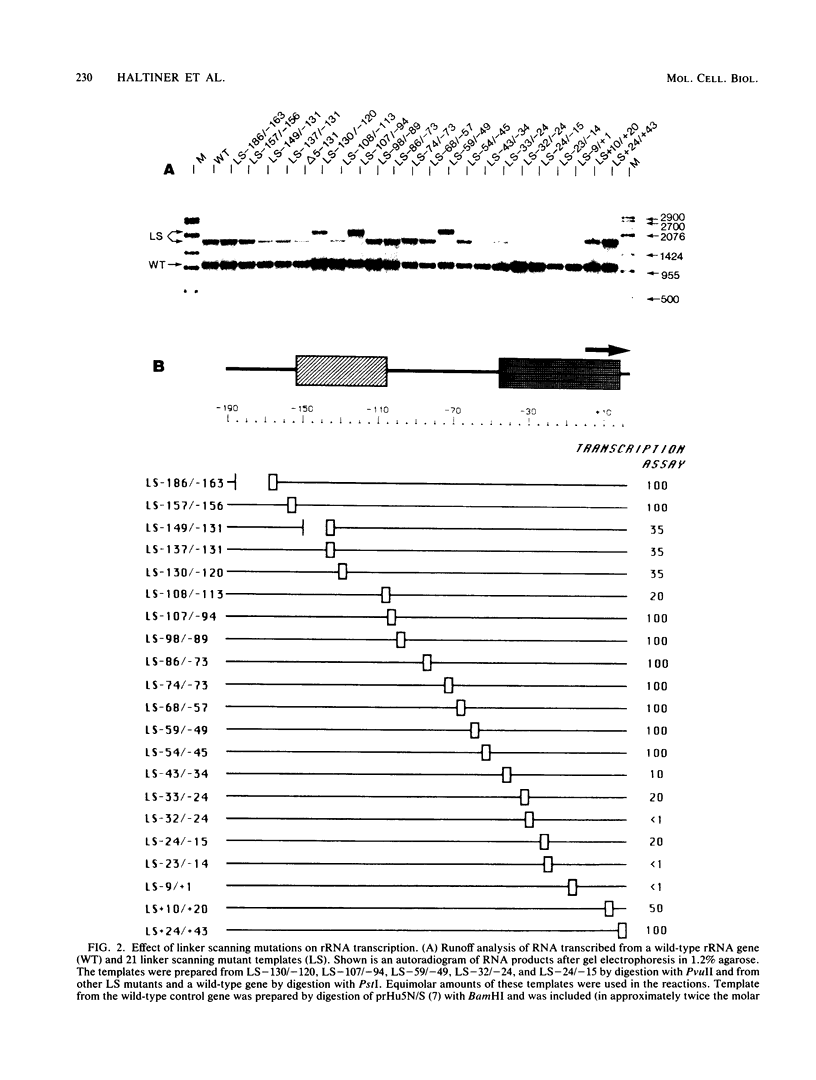
A cell-free RNA polymerase I transcription system was used to evaluate the transcription efficiency of 21 linker scanning mutations that span the human rRNA gene promoter. Our analysis revealed the presence of two major control elements, designated the core and upstream elements, that affect the level of transcription initiation. The core element extends from -45 to +18 relative to the RNA start site, and transcription is severely affected (up to 100-fold) by linker scanning mutations in this region. Linker scanning and deletion mutations in the upstream element, located between nucleotides -156 and -107, cause a three- to fivefold reduction in transcription. Under certain reaction conditions, such as the presence of a high ratio of protein to template or supplementation of the reaction with partially purified protein fractions, sequences upstream of the core element can have an even greater effect (20- to 50-fold) on RNA polymerase I transcription. Primer extension analysis showed that RNA synthesized from all of these mutant templates is initiated at the correct in vivo start site. To examine the functional relationship between the core and the upstream region, mutant promoters were constructed that alter the orientation, distance, or multiplicity of these control elements relative to each other. The upstream control element appears to function in only one orientation, and its position relative to the core is constrained within a fairly narrow region. Moreover, multiple core elements in close proximity to each other have an inhibitory effect on transcription.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Nucleotide sequence requirements for specific initiation of transcription by RNA polymerase I. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6908–6911. doi: 10.1073/pnas.79.22.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I., Smith V. A., Grummt F. Amino acid starvation affects the initiation frequency of nucleolar RNA polymerase. Cell. 1976 Mar;7(3):439–445. doi: 10.1016/0092-8674(76)90174-4. [DOI] [PubMed] [Google Scholar]
- Haltiner M., Kempe T., Tjian R. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 1985 Feb 11;13(3):1015–1025. doi: 10.1093/nar/13.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U., England B., Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985 Jul;41(3):965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- Hershko A., Mamont P., Shields R., Tomkins G. M. "Pleiotypic response". Nat New Biol. 1971 Aug;232(33):206–211. [PubMed] [Google Scholar]
- Jones K. A., Yamamoto K. R., Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985 Sep;42(2):559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- Kishimoto T., Nagamine M., Sasaki T., Takakusa N., Miwa T., Kominami R., Muramatsu M. Presence of a limited number of essential nucleotides in the promoter region of mouse ribosomal RNA gene. Nucleic Acids Res. 1985 May 24;13(10):3515–3532. doi: 10.1093/nar/13.10.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Cordes S., Tjian R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol. 1985 Jun;5(6):1358–1369. doi: 10.1128/mcb.5.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Smale S. T., Haltiner M. M., Tjian R. Regulation of human ribosomal RNA transcription. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3558–3562. doi: 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1(6):575–584. [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- May P., May E., Bordé J. Stimulation of cellular RNA synthesis in mouse-kidney cell cultures infected with SV40 virus. Exp Cell Res. 1976 Jul;100(2):433–436. doi: 10.1016/0014-4827(76)90175-0. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. C., Spence A., Smith M. The distal transcription signals of the herpesvirus tk gene share a common hexanucleotide control sequence. Cell. 1984 May;37(1):253–262. doi: 10.1016/0092-8674(84)90321-0. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Species-specific rDNA transcription is due to promoter-specific binding factors. Mol Cell Biol. 1984 Feb;4(2):221–227. doi: 10.1128/mcb.4.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. J., Dev V. G., Miller D. A., Tantravahi R., Eliceiri G. L. Transcription and processing of both mouse and Syrian hamster ribosomal RNA genes in individual somatic hybrid cells. Exp Cell Res. 1978 Sep;115(2):457–460. doi: 10.1016/0014-4827(78)90309-9. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOWELL P. C. Phytohemagglutinin: an initiator of mitosis in cultures of normal human leukocytes. Cancer Res. 1960 May;20:462–466. [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Raskas H. J., Thomas D. C., Green M. Biochemical studies on adenovirus multiplication. XVII. Ribosome synthesis in uninfected and infected KB cells. Virology. 1970 Apr;40(4):893–902. doi: 10.1016/0042-6822(70)90135-2. [DOI] [PubMed] [Google Scholar]
- Salomon C., Türler H., Weil R. Polyoma-induced stimulation of cellular RNA synthesis is paralleled by changed expression of the viral genome. Nucleic Acids Res. 1977;4(5):1483–1503. doi: 10.1093/nar/4.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J. A., Ohrlein A., Grummt I. In vitro mutagenesis and transcriptional analysis of a mouse ribosomal promoter element. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2137–2141. doi: 10.1073/pnas.81.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S. T., Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol Cell Biol. 1985 Feb;5(2):352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Wilkinson J. A., Roan J., Reeder R. H. Nested control regions promote Xenopus ribosomal RNA synthesis by RNA polymerase I. Cell. 1983 Nov;35(1):199–206. doi: 10.1016/0092-8674(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Soprano K. J., Dev V. G., Croce C. M., Baserga R. Reactivation of silent rRNA genes by simian virus 40 in human-mouse hybrid cells. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3885–3889. doi: 10.1073/pnas.76.8.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantravahi R., Miller D. A., D'Ancona G., Croce C. M., Miller O. J. Location of rRNA genes in three inbred strains of rat and suppression of rat rRNA activity in rat-human somatic cell hybrids. Exp Cell Res. 1979 Mar 15;119(2):387–392. doi: 10.1016/0014-4827(79)90368-9. [DOI] [PubMed] [Google Scholar]
- Yamamoto O., Takakusa N., Mishima Y., Kominami R., Muramatsu M. Determination of the promoter region of mouse ribosomal RNA gene by an in vitro transcription system. Proc Natl Acad Sci U S A. 1984 Jan;81(2):299–303. doi: 10.1073/pnas.81.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]