Abstract
The initiation of reverse transcription of the human immunodeficiency virus type 1 (HIV-1) requires the opening of the three-dimensional structure of the primer tRNALys 3 for its annealing to the viral RNA at the primer binding site (PBS). Despite the fact that the result of this rearrangement is thermodynamically more stable, there is a high-energy barrier that requires the chaperoning activity of the viral nucleocapsid protein. In addition to the nucleotide complementarity to the PBS, several regions of tRNALys 3 have been described as interacting with the viral genomic RNA. Among these sequences, a sequence of the viral genome called PAS for “primer activation signal” was proposed to interact with the T-arm of tRNALys 3, this interaction stimulating the initiation of reverse transcription. In this report, we investigate the formation of this additional interaction with NMR spectroscopy, using a simple system composed of the primer tRNALys 3, the 18 nucleotides of the PBS, the PAS (8 nucleotides) encompassed or not in a hairpin structure, and the nucleocapsid protein. Our NMR study provides molecular evidence of the existence of this interaction and highlights the role of the nucleocapsid protein in promoting this additional RNA-RNA annealing. This study presents the first direct observation at a single base-pair resolution of the PAS/anti-PAS association, which has been proposed to be involved in the chronological regulation of the reverse transcription.
Introduction
In retroviruses, the initiation of reverse transcription is primed by a cellular tRNA that is encapsidated in viral particles. tRNALys 3 is the natural primer of all immunodeficiency viruses, including the type 1 human immunodeficiency virus (HIV-1). Indeed, the primer tRNA is strongly bound to the genomic RNA through Watson–Crick base-pairing of its 18 3′-terminal nucleotides with the complementary viral primer binding site (PBS) (for reviews, see [1], [2]). The annealing of tRNALys 3 to the PBS requires the action of the nucleocapsid protein (NCp7) that acts as an RNA chaperone [3], [4], [5]. HIV-1 NCp7 is a short basic protein with two zinc-finger domains that destabilizes base-pairing in the primer tRNA without opening its structure [6]. Addition of a viral template containing the PBS results in RNA-RNA complex formation and significant structural changes in both RNAs. Both basic and zinc-finger domains of NCp7 are required for proper annealing of the tRNA/RNA complex. The basic domains help to destabilize the base-pairing in the four-way junction of the tRNA structure whereas the zinc-finger domains disrupt the ternary interactions within the tRNA molecule [6], [7], [8], [9].
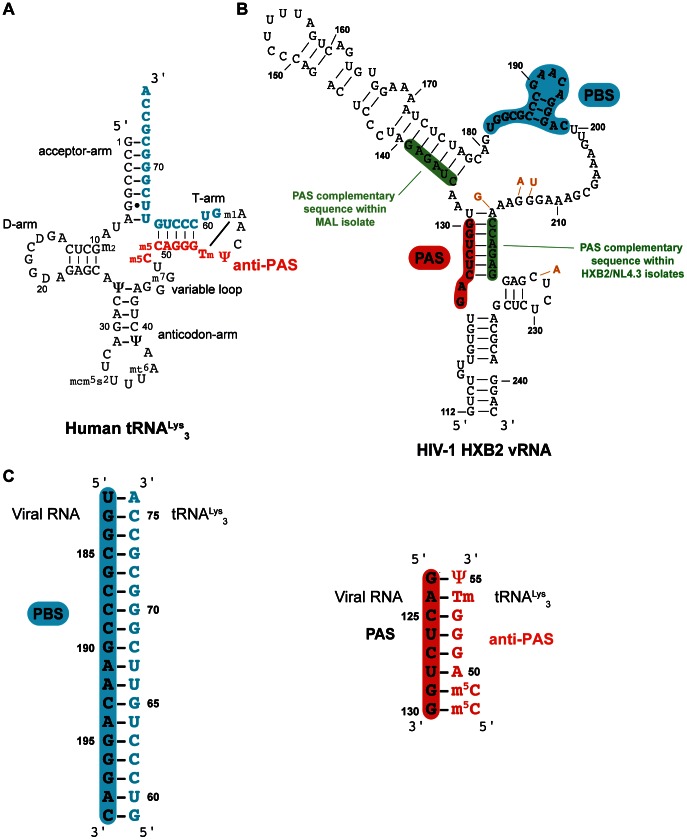
The specificity for tRNALys 3 as the primer of reverse transcription is strictly maintained in HIV-1 evolution. Earlier experiments from several groups have shown that altering the PBS sequence alone is not sufficient to stably switch tRNA usage [10], [11], [12]. To improve the understanding of the exclusive usage of tRNALys 3 as primer by HIV-1, the search for the determinants of specific tRNALys 3 selection, other than the PBS, was the subject of extensive studies [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27]. Additional contacts between tRNALys 3 and the viral RNA have been naturally considered as potential secondary determinants and were proposed to play a role in reverse transcription [20], [28], [29], [30]. The different interactions (reviewed in [31]) are located in the PBS domain of the viral RNA and include an A-rich loop, a C-rich region and the primer activation signal (PAS). The A-rich loop interaction was found to play a role in the initiation of reverse transcription of the HIV-1 MAL strain but not in the NL4.3 or HXB2 isolates. By adaptation of both the PBS and PAS motifs, the HIV-1 leader could be changed to accommodate tRNALys 1,2 as the primer of reverse transcription in vitro [16]. Additional studies mutating the PAS sequence in the viral RNA showed that the PAS is critically involved in stimulating tRNALys 3-primed reverse transcription initiation in the HIV-1 HXB2 and NL4.3 strains [17], [18]. Indeed, the PAS is an 8-nucleotide sequence in the U5 region (nucleotides 123–130, Figure 1) that was proposed to be involved in a base-pairing interaction with a region located in the T-arm of tRNALys 3 (the anti-PAS region: nucleotides 48–55, Figure 1). This interaction would thus lead to the formation of a higher order RNA structure that is suitable for stimulating initiation of reverse transcription. Indeed, the annealing of the tRNA primer to the PBS requires the opening of the acceptor and T stems of tRNALys 3 cloverleaf structure, thus releasing the anti-PAS motif (Figure 1). However, no direct evidence of the PAS/anti-PAS association can be obtained by probing experiments [20], [32], [33]. Nevertheless, one strong argument in favour of its existence during the viral replication cycle is the very strong conservation of the PAS-like vRNA–tRNA interactions among all HIV-1 isolates, but also in SIV isolates, and even possibly in all retroviruses [16], [17], [34]. An attractive model for the regulation of HIV reverse transcription has been proposed by the group of Ben Berkhout. The PAS sequence is initially masked in the viral RNA through base-pairing in the U5 leader stem (Figure 1B). Later, this sequence anneals with the anti-PAS sequence in the primer tRNA, which stimulates the reverse transcription initiation. Altogether, the presence of the PAS enhancer motif, initially masked and repressed by base-pairing, would provide a unique mechanism for positive and negative regulation of HIV-1 reverse transcription [18]. Very recently, using FRET spectroscopy, Beerens et al. [35] obtained new results that are consistent with a model for tRNA annealing that involves a secondary interaction between the tRNALys 3 molecule and the PAS sequence. In this study, the tRNALys 3/PAS interaction appears to be dynamic and stimulated by the nucleocapsid protein [35].
Figure 1. Secondary structure of A) the human tRNALys 3, primer of the HIV-1 reverse transcription, in blue the sequence complementary to the PBS and in red the anti-PAS sequence, B) the PBS domain within the HIV-1 HXB2 isolate, in blue the PBS sequence, in red the PAS sequence and in green the sequences complementary to the PAS sequence, the nucleotides that are different between HXB2 and NL4.3 are indicated in orange, and C) the duplexes corresponding to the tRNALys 3/PBS and to the tRNALys 3/PAS complexes.
In the present report, the PAS/anti-PAS interaction and the role of the nucleocapsid protein have been directly investigated in vitro at a single base-pair resolution using NMR spectroscopy. We show here that the PAS/anti-PAS complex exists under certain conditions that dramatically depend on the action of the nucleocapsid protein.
Results
RNA sequences, labelling schemes, and NMR assignments of the free RNAs
Even if important progresses have been made over the last years, NMR analysis of large RNA complexes remains technically challenging. In order to simplify the interpretation of NMR spectra, we used a selective labelling strategy in which the 15N labelling of tRNALys 3 primer allowed us to only observe the NMR signals of its imino groups and not the ones of other RNA partners. In addition, the use of 1H-15N TROSY NMR experiments [36], [37] substantially improved the spectral resolution and sensitivity on these large RNA complexes. In RNA, imino protons are carried by G and U nucleotides. The intensity of imino proton NMR signals depends dramatically on their degree of protection towards exchange with the solvent. In practice, only protected imino protons that are forming stable hydrogen-bonds are visible in NMR spectra. Therefore, the NMR signal of an imino proton constitutes the signature of a base-pair within a folded RNA or between two different RNA molecules interacting to form a higher order structure. Structural information can therefore be obtained at a single base-pair resolution using NMR signals of iminos as reporter probes.
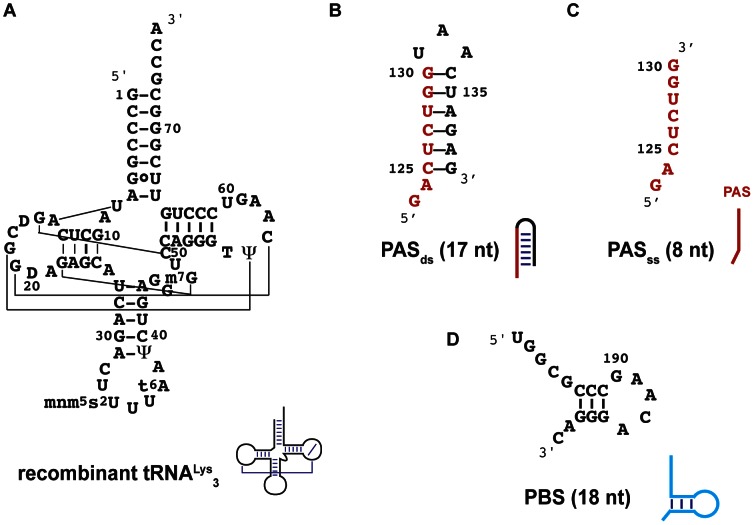
A similar labelling strategy using the NMR signals of the imino protons of tRNALys 3 as reporter signals has already been successfully employed to study the secondary structure of the HIV reverse transcription initiation complex and to reveal the different steps of annealing of tRNALys 3 to the PBS in the presence or in the absence of the nucleocapsid protein [7], [38]. In the present study, tRNALys 3 15N-labelling is also an appropriate tool to observe the annealing of the PAS and anti-PAS sequences (Figure 1C) as the largest number of sequential imino protons (carried by Gs and Us) belongs to tRNALys 3, and can therefore be observed in 2D 1H-15N correlation spectra. The human tRNALys 3 was thus produced as a recombinant tRNA in E. coli providing the incorporation of modified nucleotides by E. coli RNA modification enzymes as well as a uniform 15N labelling (see Materials and Methods). This recombinant tRNA (Figure 2A) bears the modified nucleotides crucial for the initiation of HIV-1 reverse transcription [21], [39]. In the case of the PAS activation, it was shown that reverse transcription primed by natural or synthetic tRNALys 3 is similarly activated, indicating that the PAS/anti-PAS interaction is not dependent on modified nucleotides within tRNALys 3 [18].
Figure 2. Secondary structures of RNA used in this study.
A) the recombinant tRNALys 3 expressed in E. coli, B) the hairpin encompassing PAS sequence, the double-stranded PAS (PASds) with the numbering of nucleotides in the HXB2 isolate, C) the single-stranded PAS sequence (PASss) and D) the PBS. A cartoon symbolizing the secondary structure of these RNAs is drawn next their names.
We performed our analysis on a model system involving critical fragments of the U5 region of HIV-1 viral RNA. The RNA molecules used to investigate the formation of the PAS/anti-PAS complex will be described below. Interestingly, the PAS sequence is involved in base-pairing within the viral RNA whatever the isolate. In the MAL isolate, the PAS sequence is part of a stem-loop structure [40] like shown in Figure 2B and S1. In the HXB2 isolate, the PAS sequence is involved in a duplex structure [41] (Figure 1B and S1). The reason for these differences comes from the fact that the PAS sequence has two potential complementary sequences within the viral RNA (see Figure 1B in green). For our NMR study, we thus chose to embed the PAS sequence in a minimal RNA hairpin (thereafter called PASds; Figure 2B) with the MAL isolate sequence and secondary structure context. This PASds RNA hairpin was chosen as a compromise between the size of the RNA (to be kept to a minimum to facilitate NMR spectra interpretation) and the ability to mimic the natural pairing occurring in both the MAL and HXB2 isolates. We also worked with a single-stranded sequence of PAS (PASss; Figure 1B and 2C) that only contains the 8 nt of the PAS sequence. Finally, our PBS RNA sequence (PBS; Figure 1B and 2D) contained the 18 nucleotides complementary to the nucleotides at the 3′-end of tRNALys 3. Importantly, the use of different RNA fragments to account for the different critical regions of the U5 leader RNA not only simplified the complexity of the NMR spectra for these large RNAs, but also allowed us to perform experiments with different combinations of the fragments to study the influence of the PBS sequence on the PAS/anti-PAS association (see paragraphs below).
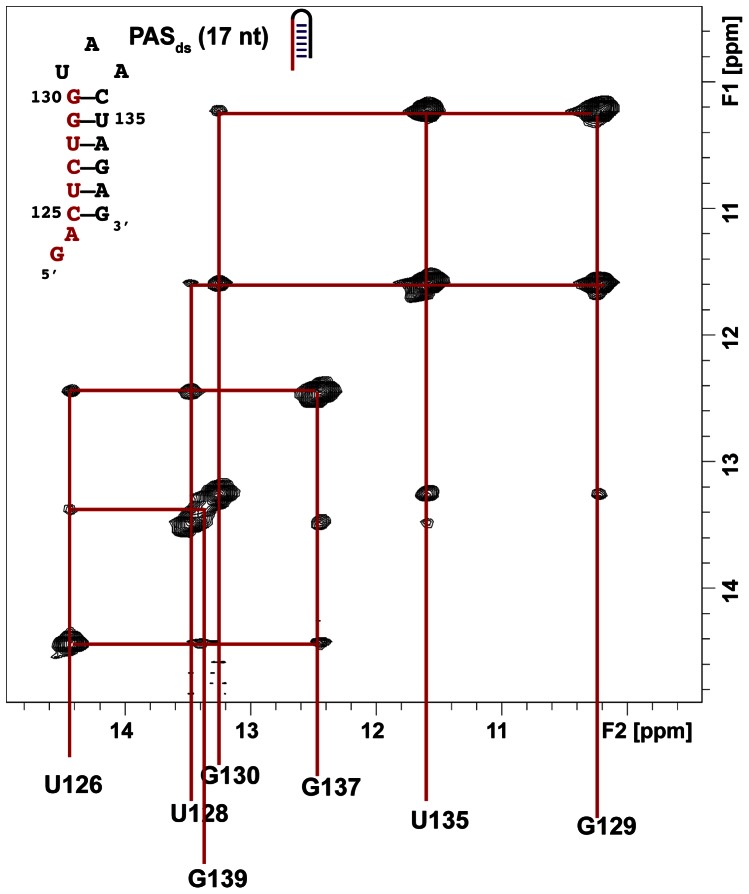
Assignments of tRNALys 3 imino groups were previously performed [6] and are indicated in Figure 3. To monitor the imino signals of PASds in NOESY experiments, we first assigned their resonances. The PASds hairpin contains six base-pairs and seven imino protons. Figure 4 shows the imino region of a NOESY experiment carried out at 15°C on PASds. Briefly, a GU base-pair gives rise to an intense NOE as the imino protons of the guanine and the uridine are facing each other in such a base-pairing at around 2.5 Å. In addition, in a GU pair, the imino proton of the guanine is the most shielded. Therefore, the imino proton of G129 was assigned to 10.21 ppm and that of U135 to 11.60 ppm. From this starting point, the assignment of all imino protons was straightforward and is summarized in Figure 4.
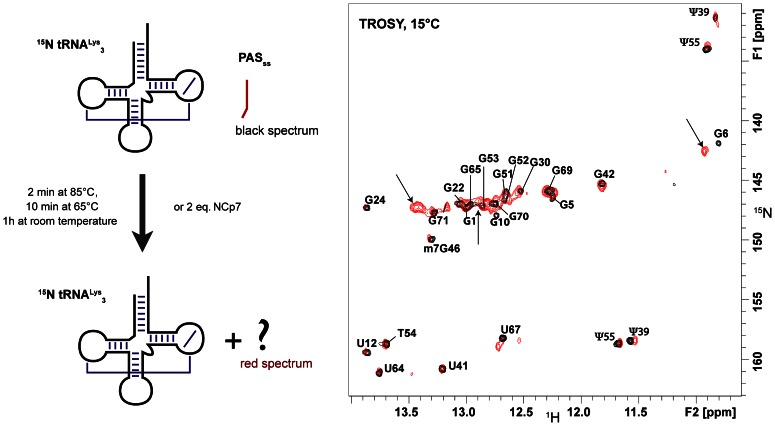
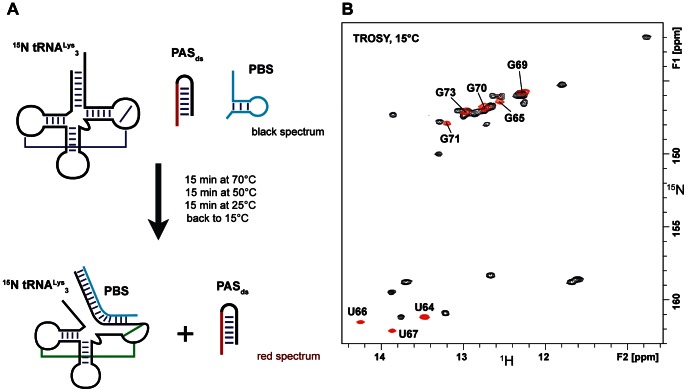
Figure 3. Tests of annealing of PASss to tRNALys 3.
A) Schematic drawing of the experimental procedure, B) Superimposition of two TROSY experiments recorded at 15°C showing the imino groups of tRNALys 3 (0.1 mM) alone (in black, reference spectrum) and after the heat-annealed procedure with 1 equivalent of PASss (red spectrum). The arrows indicate peaks corresponding to the PAS/anti-PAS complex described in Figure 7.
Figure 4. Assignment of the imino proton resonances of PASds.
NOESY experiment recorded at 15°C with a mixing time of 150 ms with a sample of PASds at 1.6 mM in a phosphate buffer (10 mM, pH 6.5) with 50 mM KCl.
The involvement of the PAS sequence in a doubled-stranded helix prevents it from annealing to tRNALys 3
To investigate the annealing of the PAS signal to tRNALys 3, we first devised two different strategies described in the materials and methods section. Briefly, the first procedure is a heat annealing-based experiment whereas the second one uses the chaperone activity of the NCp7 protein. The 1H-15N correlation NMR spectra recorded after performing these procedures on a 15N-labelled tRNALys 3/PASds mixture are identical to that of the free tRNALys 3 (Figure S2). These experiments therefore demonstrated that tRNALys 3 remained in the same state, i.e. folded on its own. Consequently, the annealing of PASds to tRNALys 3 could not be promoted neither by the heat-annealed nor by the NCp7-mediated procedures.
We subsequently used the single-stranded PAS sequence to conduct the two procedures in parallel. All the signals of the free tRNALys 3 remain after these treatments (Figure 3) but the appearance of new peaks indicates that an other entity has been formed (Figure 3). We attribute the appearance of these new signals to the formation of a binary complex between tRNALys 3 and PASss. The peaks indicated by an arrow were assigned to PAS/anti-PAS complex with the help of subsequent experiments (see paragraph below). Noticeably, the presence or the absence of NCp7 did not change the ratio between these two species (data not shown). Moreover, the comparison of the results obtained with PASss or with PASds strongly suggests that the embedment of the PAS sequence in base-paired region prevents it from annealing to the anti-PAS sequence of tRNALys 3. In addition, the feature that NCp7 did not affect the ratio of the different RNA entities as compared with the heat annealing procedure is in complete accordance with NCp7 being an RNA chaperone [4], [42], [43], [44]. Taking together, these observations support the idea that the accessibility of the PAS sequence can modulate its association with the tRNA primer and thus be a regulator element of the initiation of reverse transcription [45].
The nucleocapsid protein can promote the annealing of PAS to the anti-PAS part of tRNALys 3 if the PBS is also annealed to tRNALys 3
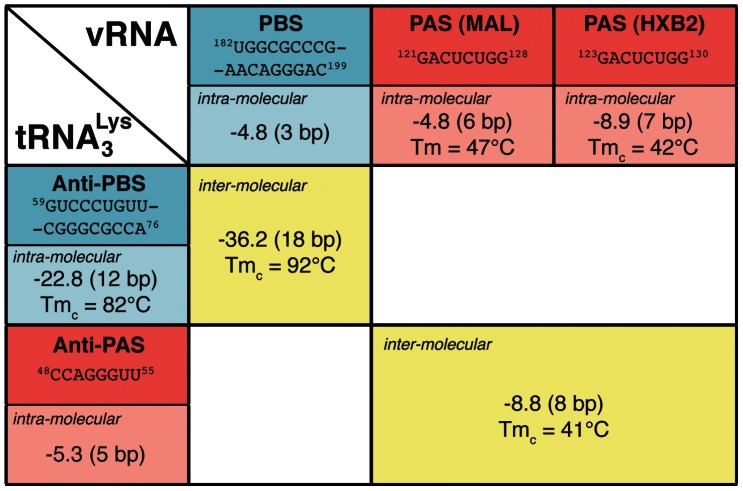
From a thermodynamic point of view, the annealing of tRNALys 3 to the PBS is strongly favoured (Figure 5, ΔG = −36.2 kcal/mol and Tmc = 92°C). In addition, the annealing of tRNALys 3 to the PBS will partially open the tRNA structure and thus increase the accessibility to the anti-PAS sequence. Therefore, in order to investigate whether the PBS could influence the annealing of the PAS sequence with tRNALys 3, we decided to conduct the heat-annealed procedure in the presence of the PBS. Figure 6 shows the NMR footprint experiment resulting from this procedure. The assignment of tRNALys 3/PBS imino groups was previously published [7]. The imino groups of tRNALys 3 (black spectrum – Figure 6) have disappeared to give rise to imino groups of tRNALys 3 involved in base-pairing with the PBS (red spectrum – Figure 6). A NOESY experiment (data not shown) substantiates that the PASds is still free as its imino groups are still observable at the chemical shifts of the imino groups of the free PASds like in Figure 4. In conclusion, when we mixed 15N-tRNALys 3 to the PBS and to the PASds at the same time, the heat-annealed complex is composed of the PBS bound to tRNALys 3 and the PAS remains free in solution (Figure 6). This complex is stable and does not change with time.
Figure 5. Thermodynamic stability of the interaction involving the PBS (in blue) or the PAS (in red) sequences.
ΔG values calculated with UNAFold [57] are indicated in kcal/mol and the number of base pairs are indicated in parenthesis. The melting temperature Tmc was calculated by UNAFold and the melting temperature Tm was measured by UV spectroscopy (Figure S5). See also Figure S1 for details on the different sequence and secondary structures used to model the intra-molecular interaction within the MAL and HXB2 isolates.
Figure 6. Test of annealing of PASds to tRNALys 3 in presence of the PBS using the heat-annealed procedure.
A) Schematic drawing of the annealing procedure, B) Superimposition of two TROSY experiments recorded at 15°C showing the imino groups of tRNALys 3 alone (in black, reference spectrum) and after the heat-annealed procedure with 1 equivalent of PBS and PASds (red spectrum).
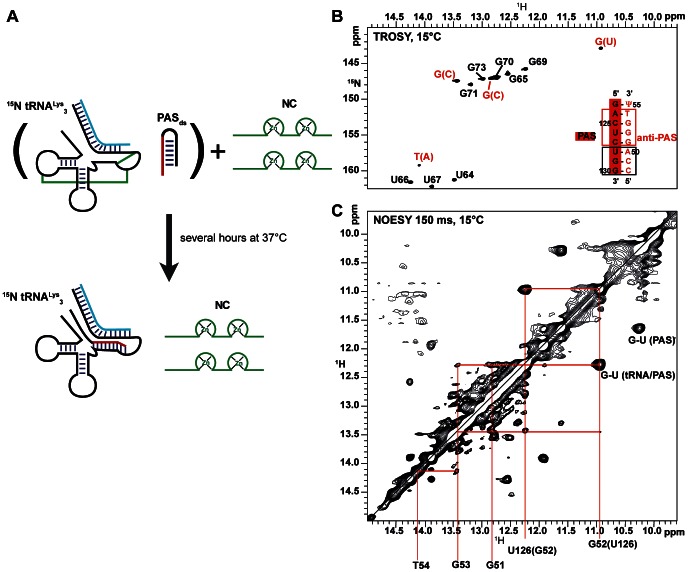
Subsequently, we added two equivalents of NCp7 to this mixture and left it for several hours at 37°C. Interestingly, new signals in addition to those originating from the tRNALys 3/PBS complex further appeared in the NMR footprint experiment (Figure 7) indicating the formation of new base-pairs. Because the PBS and the PAS sequences are not labelled, only the imino groups of tRNALys 3 are observable in 1H-15N TROSY experiments whereas all the imino signals from tRNALys 3, PBS and PAS are observable in 1H-1H NOESY experiments. Moreover, N1 of Gs and N3 of Us and Ts can be easily distinguished by their 15N chemical shift. Therefore, we could easily assign the four new signals to three 15N-labelled guanines and one 15N-labelled pyrimidine involved in new base-pairings and belonging to tRNALys 3. A NOESY experiment enabled us to assign the new signals to the imino groups of the PAS/anti-PAS complex as highlighted on Figure 7C. In short, the G52-U126 pair is readily assigned since the 15N-labelled G at 10.93 ppm in the proton dimension is connected through an intense NOE with a non-labelled U at 12.27 ppm as expected for a G-U pair. Both nucleotides are connected with a 15N-labelled guanine at 13.44 ppm that is further connected to the T resonating at 14.10 ppm. In addition, the 15N chemical shift of T54 at 159.16 ppm and the sequence of the PAS/anti-PAS complex further supports that this peak corresponds to the T54 of tRNALys 3 involved in the complex. The last signal corresponding to a guanine in the TROSY spectrum (12.85 ppm-147.04 ppm) was assigned to G51 since its imino proton gives an NOE to both imino protons of the G52-U126 pair. The imino groups carried by the PAS sequence (U128, G129, G130) were more difficult to assign since they are not labelled and therefore do not give rise to peaks in the TROSY experiment. We proceeded using a stepwise strategy: first we identified in the NOESY experiment of the mixture (Figure 7) the cross-peaks originated from the PASds alone and that from the tRNALys 3/PBS complex using previously recorded NOESY experiments (Figure S3). Then, the comparison of these three NOESY experiments revealed imino cross-peaks originating from the tRNA/PAS complex (black peaks alone). In short, we assigned U128 to 13.63 ppm, G129 to 13.26 ppm and G130 to 12.37 ppm by superimposing and comparing these three NOESY experiments (Figure S3). Only the G-Ψ base-pair, located at one extremity of the PAS/anti-PAS interaction region, was not observed in our experiments. These data clearly demonstrate for the first time the existence of a PAS/anti-PAS association at a single base-pair resolution.
Figure 7. Assignment of the NCp7-mediated tRNALys 3/PBS/PASds complex (0.5.
mM of each component). A) Schematic drawing of the experimental procedure, B) TROSY experiment recorded at 15°C showing the imino groups of tRNALys 3 (0.5 mM) after the NCp7-mediated procedure described in A), C) NOESY experiment recorded on the same sample as in A at 15°C with a mixing time of 150 ms showing the region of the imino groups. The red square indicates base-pairings assigned thanks to the use of the TROSY (A) and the NOESY (B) experiments whereas the black square indicates base-pairings assigned from the comparison of NOESY experiments.
It is interesting to notice that in addition to the signals of the PAS annealed to tRNALys 3, the signals of the free PASds are still observable in the NOESY experiment, and this even though tRNALys 3 and PAS have been mixed at a 1∶1 ratio. It is easily observable on the GU pairs. The free and the bound PAS contain each a GU pair that gives rise to an intense peak appearing at a different chemical shift for the free and the bound PAS (Figure 7C). These characteristic GU peaks represent therefore a clear signature that not all the PASds has annealed to tRNALys 3. The comparison of the intensity of these two GU cross-peaks in the NOESY experiment indicates that approximately 50% of the PASds is annealed to tRNALys 3. Whatever the salt concentration, the amount of NCp7 (not more than 4 equivalents to avoid aggregation), we did not succeed in obtaining more than 50% of PAS/anti-PAS complex. We could not rule out that the amount of NCp7 added in our samples is not sufficient to reach higher amount of the PAS/anti-PAS complex. Briefly, we want here to mention that no exchange cross-peaks between individual imino protons in the free and annealed form of the PAS RNA are observed in the different NOESY spectra (Figure 7 and S3). This absence is most probably attributable to a rapid exchange of the unprotected imino protons with the solvent during the inter-conversion between the free and bound forms of the PAS. In conclusion, the PASds signal can anneal to the anti-PAS segment of tRNALys 3 when the PBS is present under the action of the nucleocapsid protein. In contrast with the annealing of the PBS to tRNALys 3, only a fraction of the PAS (∼50%) is annealed to tRNALys 3.
For the single-stranded PAS sequence, we cannot quantify the amount of PASss/anti-PAS complex that is formed, since the free single-stranded PASss has no observable imino proton. However, we investigated the possibility to make this complex with the heat-annealing or the NCp7-mediated procedures. Interestingly, the heat-annealed complex is present in sufficient amount to give observable NMR signals when 1 equivalent of PASss is mixed with equimolar quantity of tRNALys 3 and PBS (Figure S4). The addition of PASss at 5 equivalents did not change the resulting NMR footprint. The addition of two equivalents of NCp7 protein led to the same results suggesting that NCp7 would be crucial for destabilizing the base-pairings within PASds, but would be dispensable in the case of an unpaired PAS sequence, as seen here with the PASss RNA. Indeed, the destabilization of RNA helices is a well-known feature of the NCp7 chaperone activity [46], [47], [48], [49]. For this reason, we ultimately investigated the interaction between NCp7 and PASds by 1D 1H NMR. Interestingly, we observed an important broadening of all imino proton NMR signals indicating that NCp7 binds non-specifically to PASds and is likely to induce a destabilization of the hairpin (Figure S6).
Discussion
In this report, we provide direct evidence at a molecular level and with a single base pair resolution of base-pairings between the PAS in the viral RNA and the anti-PAS in tRNALys 3. In addition, we showed that NCp7 promotes the annealing of the PAS sequence to its complementary anti-PAS sequence within the primer tRNALys 3. Importantly, we were unable to simulate the action of the nucleocapsid protein with a simple heat-annealing procedure when the PAS sequence is engaged in a double-stranded region, accentuating the importance of NCp7 in this RNA structural rearrangement. Our results are in agreement with a recent study [35] showing a two-fold increase of annealed tRNALys 3 molecules to the template viral RNA containing the PAS sequence in the presence of NCp7 compared to a heat-annealing procedure. Moreover, differences between viral RNA/tRNALys 3 complex promoted by heat or by NCp7 have been noticed, in particular, the conformational heterogeneity of this complex becomes smaller in the presence of NCp7 [35]. This study thus suggests that NCp7 facilitates tRNA annealing to the PAS motif. Our work provides further and complementary evidences at the level of single base-pairs, regarding the formation of the PAS/tRNALys 3 complex using small fragments of viral RNA.
Thermodynamic analysis
Thermodynamic considerations are important to properly describe and understand these large structural rearrangements involving three different RNA molecules. The formation of the complex between tRNALys 3 and the PBS is largely thermodynamically favoured (Figure 5, ΔG = −36.2 kcal/mol, Tmc = 92°C). As a result, the tRNA/PBS complex can easily be promoted by both the heat-annealed and the NCp7 procedures [7].
The situation concerning the formation of the binary complex between tRNALys 3 and the PAS is very different. tRNALys 3 on its own is more stable than paired with the PAS sequence. Indeed, the free energy (ΔG) reaches −22 kcal/mol with a melting temperature Tmc calculated by UNAFold above 80°C whereas for the PAS/anti-PAS complex, ΔG is only −8.8 kcal/mol with a Tmc around 40°C (Figure 5). In addition, the free PAS is already involved within the viral RNA in quite stable secondary structures (ΔG = −4.8 kcal/mol, Tm = 47°C for the MAL isolate and ΔG = −8.9 kcal/mol, Tmc = 42°C for the HXB2 strain; Figure 5). As a result, the tRNA/PAS binary complex is clearly disfavoured and we showed that it cannot be promoted neither by the heat-annealed nor the NCp7-mediated procedures (Figure S2). The situation is different in the case of the binary complex between tRNALys 3 and the single-stranded PAS molecule (PASss). Indeed, in this case, the thermodynamic parameters of the tRNA/PAS complex have not changed, but the PAS sequence cannot fold on its own and only 6 bp (Figure 5 and 1A) at the level of the tRNALys 3 T-arm need to be disrupted. However, the T-arm is stacked on the acceptor arm forming a long helix (12 bp) that further stabilizes this region. This could explain why the single-stranded PAS can only be partially annealed to the primer tRNALys 3 (Figure 3).
Finally, the situation governing the formation of the ternary complex between tRNALys 3, the PBS, and the PAS is of interest. The tRNALys 3/PBS complex is thermodynamically greatly favoured thereby freeing the tRNALys 3 anti-PAS sequence. The PAS sequence within the HXB2 viral RNA or annealed with the anti-PAS region of tRNALys 3 are of similar thermodynamic stability. Altogether, these thermodynamic considerations can help explaining why the association of the PAS sequence with the anti-PAS region of tRNALys 3 can only be observed in the presence of the PBS (Figure 6), unless one uses the single-stranded PAS molecule (Figure S4). Indeed, as NCp7 acts as an RNA chaperone, it helps remodelling RNA structures towards the most stable assembly. Here, the strand exchange property of NCp7 is definitely crucial to promote the formation of the ternary complex (tRNALys 3/PBS/PAS) that was shown to stimulates the initiation of reverse transcription [17], [18]. For the PAS/anti-PAS interaction, the fact that the intra-molecular PAS association within the viral RNA and the inter-molecular association with tRNALys 3 are of comparable stability (see Figure 5), can very likely explain why we did observed a 50/50 mixture of the PAS sequence annealed with tRNALys 3 or with “itself” in the PASds hairpin.
Relevance for HIV-1 initiation of reverse transcription
Obviously, the use of small viral RNA fragments and the absence of reverse transcriptase cannot reproduce all the events occurring during the initiation of HIV-1 reverse transcription. But, replacing our new findings in the context of previous works showing that the PAS sequence stimulates the initiation of HIV-1 reverse transcription [17], [18] is, however, very informative. First, we confirmed that the PAS/anti-PAS interaction requires the annealing of tRNALys 3 to the PBS thus, these new base-pairings will not drive the annealing of tRNALys 3 to the PBS. Consequently, our results are in agreement with works that demonstrated that the PAS sequence had no effect on the hybridization of the tRNA primer on the PBS [18]. Secondly, the interaction between tRNALys 3 and the PAS motif is dynamic [35], and probably short-lived during the reverse transcription, since the reverse transcriptase must penetrate the PAS/anti-PAS helix during the early elongation phase [50]. Our observation that only a fraction of the PAS sequence is annealed to tRNALys 3 is definitely related to its low relative stability, its dynamic behaviour and might certainly be put in relation with its transient nature in vivo. It also probably explains why chemical probing did not succeed to uncover this interaction [32], [33]. Interestingly, in the case of the A-rich loop of the viral RNA of MAL isolates that interacts with the anticodon loop of tRNALys 3, NMR did not succeed in observing it [7], [38] whereas a number of chemical probing studies did [20], [21], [22], [51]. Thirdly, the PAS sequence in the HIV-1 genome is occluded by base-pairing in the U5 leader stem, and thus, needs to be free for the interaction with the tRNA primer. We showed that the nucleocapsid, as expected, plays a major role to promote its annealing to the anti-PAS sequence, most probably by destabilizing the PAS duplex and promoting strand exchange between the viral RNA and tRNALys 3.
Finally, the presence of the PAS motif that is temporarily repressed by base-pairing provides a unique mechanism for regulation of HIV-1 reverse transcription. It was speculated that this mechanism may preclude premature reverse transcription in virus-producing cells such that the vRNA genome is copied only after it is appropriately packaged into virions [18], [52], [53]. Because NCp7 is released from the Gag precursor protein during maturation of virion particles, this mechanism will ensure the precise timing for activation of reverse transcription. tRNA packaging and annealing to the PBS in viral particles is not dependent on Gag processing [54], whereas efficient tRNA primer extension is. PAS accessibility may therefore regulate reverse transcription in the viral life cycle.
Conclusions
We showed in this report that base-pairings between the PAS in the viral RNA and the anti-PAS in tRNALys 3 can be observed by NMR under certain conditions. In our hands, the PAS/anti-PAS interaction is positively influenced by several factors: (i) the annealing of tRNALys 3 to the PBS, which results in the partial opening of the tRNA structure and therefore increases the accessibility to the anti-PAS sequence; (ii) the presence of the nucleocapsid chaperone protein, which might destabilize the initial PAS base-pairing within the viral RNA and favour RNA strand exchange. On the other hand, it is disfavoured by the molecular inaccessibility of the PAS sequence as a result of its embedment in a base-paired region within the viral RNA. Altogether, the PAS/anti-PAS interaction appears to be a new illustration of the chaperone activity of the NCp7 protein. In addition, our data highlight the power of NMR experiments to map global RNA folds and demonstrate that NMR is a powerful tool to characterize annealing processes in detail.
Materials and Methods
Sample Preparation
15N-labelled tRNALys 3 was expressed in E. coli (JM101TR strain) from a recombinant plasmid and purified as previously described [39]. Small fragments of HIV viral RNA namely the PBS (18 nucleotides), PASds (17 nucleotides) and PASss (8 nucleotides) were purchased from Dharmacon research with 2′-O-bis (acetoxyethoxy)-methyl (ACE) protection. The samples were deprotected following manufacturer recommendations and lyophylised. They were then dissolved in a phosphate buffer (10 mM, pH 6.5) with 50 mM KCl. The PBS and the PASds hairpin were folded by heating at 90°C for 5 min and snap-cooling on ice. The sequences of RNA samples used in this study are indicated in Figure 2. The sequence of the PASds hairpin corresponds to the sequence and secondary structure context of the MAL isolate, and was chosen as a compromise between the size of the RNA (to be kept to a minimum for the NMR study) and the ability to mimic the natural pairing occurring in both the MAL and HXB2 isolates (See also Figure S1).
The recombinant NCp7 protein (HIV-1 strain NL4.3, 55 residues) was overexpressed from the bacterial expression vector pRD2, which contains the NCp7 coding region from HIV-1 strain NL4-3 subcloned into pET-3a (Novagen, WI). This plasmid was kindly provided by M.F. Summers [55], [56]. pRD2 was transformed into E. coli strain BL21(DE3)pLysE and the overexpressed protein was purified as previously described [55], [56].
Heat-annealed and NCp7-mediated procedures
In the standard heat-annealed procedure, RNA samples were heated at 85°C for 2 minutes, next incubated at 65°C for 10 minutes before a slow cooling to room temperature and putting them in the NMR tube. In the experiment of Figure 6, as we acquired NMR spectra at each temperature, the sample containing tRNALys 3 (0.44 mM), PASds and PBS at ratio 1∶1∶1 was first heated at 70°C in the NMR spectrometer for the time necessary to acquire 1D 1H NMR spectrum (roughly 15 minutes in total). Then the temperature of the sample was progressively decreased to 50°C, 40°C, 25°C and 15°C in the NMR spectrometer and a 1D 1H NMR spectrum was acquired at each temperature. At 15°C, a NOESY and a TROSY NMR experiments were recorded to analyse the RNA complex in the tube.
In the NCp7-mediated procedure, two equivalents of NCp7 were added to RNA samples and the mixture was then heated at 37°C for 30 min to several hours. Both procedures were performed in the NMR buffer (10 mM KPO4 pH 6.5) containing 50 mM KCl or 125 mM KCl. The same results were obtained at both salt concentration.
Thermodynamic stabilities and melting temperatures reported in Figure 5 were calculated using the UNAFold web server (http://mfold.rna.albany.edu/) [57]. The melting temperature of PASds was measured by UV spectroscopy measuring the absorbance at 260 nm from 15°C to 80°C (Figure S5).
NMR spectroscopy
NMR data were measured on a Bruker Avance DRX600 spectrometer equipped with a TCI cryoprobe. 1D 1H NMR spectra were recorded using a watergate sequence for solvent suppression [58]. For samples at a concentration of 0.1 mM, 512 scans were usually acquired. 1H-15N TROSY experiments [36], [37] were carried out with 128 t1 increments and 2048 t2 data points per increment. The t1 dimension was acquired with the echo-antiecho method. The spectral widths were 3600 Hz and 1900 Hz in the proton and the nitrogen dimensions, respectively. NOESY experiments with a mixing time of 150 ms were recorded at 15°C using a watergate sequence for solvent suppression [58]. 256 t1 increments and 2048 t2 data points per increment were recorded. The t1 dimension was acquired with the States-TPPI method. The spectral widths were 12600 Hz in both dimension. Spectra were processed using TOPSPIN (Bruker) and analysed with TOPSPIN or CcpNMR (http://www.ccpn.ac.uk/).
The interaction between 15N-tRNALys 3 and others partners were carried out at a concentration of 0.45 mM when we needed to acquire a NOESY experiment and at a concentration of 0.1 mM when only a TROSY experiment was necessary to analyze the RNA complex in the NMR sample.
Supporting Information
RNA sequences used to model the secondary structure context of the PAS sequence within A) the MAL isolate and B) the HXB2 isolate. These RNA fragments were used to derive the thermodynamic parameters for the PAS sequence within the two different viral RNA isolates (See Figure 5).
(TIF)
Tests of annealing of PASds to tRNALys3. A) Schematic drawing of the experiment, B) Superimposition of two TROSY experiments recorded at 15°C showing the imino groups of tRNALys 3 (0.1 mM) alone (in black, reference spectrum) and after the heat-annealed procedure with 1 equivalent of PASds (red spectrum).
(TIF)
Superimposition of three NOESY experiments recorded at 15°C with a mixing time of 150 ms showing the imino-imino region, in blue for PASds, in red for a mixture of tRNALys3, PASds and PBS for which the annealing between the tRNALys3/PBS was promoted, in black for a mixture of tRNALys3, PASds, PBS, NCp7 for which the annealing tRNALys3/PBS/PASds was promoted (same experiment as Figure 7 ). The NOESY cross-peaks for GU base-pairs within PASds, and tRNALys 3/PAS complex are indicated. The assigment of the imino groups within the PAS/anti-PAS complex is indicated in purple for 15N-signals and the assignment for black cross-peaks corresponding to non-labelled imino groups are in orange.
(TIF)
Annealing of PASss to tRNALys3 in the presence of PBS. Superimposition of two TROSY experiments recorded at 15°C showing the imino groups of tRNALys 3 (0.1 mM), in black: in complex with PBS (1 equivalent) and PASss (1 equivalent) after the heat-annealed procedure; and in red: in complex with PBS (1 equivalent) and PASss (1 equivalent) after the NCp7-mediated procedure. Only the GC regions are shown since the T54 imino group was not observed in these experiments.
(TIF)
UV melting curve of the PASds.
(TIF)
Region of the imino protons of the 1H NMR spectra of the PASds alone (0.16 mM) and of the PASds (0.16 mM) mixed with NCp7 at 1∶1 ratio. The spectra were recorded at 15°C using a watergate sequence to [58] to achieve water signal suppression.
(TIF)
Funding Statement
D.S. was the recipient of a studentship from the “Ministère de la Recherche”. This work was supported by grants from the French National Agency for Research on AIDS and Viral Hepatitis (ANRS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Marquet R, Isel C, Ehresmann C, Ehresmann B (1995) tRNAs as primer of reverse transcriptases. Biochimie 77: 113–124. [DOI] [PubMed] [Google Scholar]
- 2. Mak J, Kleiman L (1997) Primer tRNAs for reverse transcription. J Virol 71: 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Darlix JL, Cristofari G, Rau M, Pechoux C, Berthoux L, et al. (2000) Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Advances in pharmacology 48: 345–372. [DOI] [PubMed] [Google Scholar]
- 4. Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K (2010) Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol 7: 754–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tisne C (2005) Structural bases of the annealing of primer tRNA(3Lys) to the HIV-1 viral RNA. Current HIV research 3: 147–156. [DOI] [PubMed] [Google Scholar]
- 6. Tisne C, Roques BP, Dardel F (2001) Heteronuclear NMR studies of the interaction of tRNA(Lys)3 with HIV-1 nucleocapsid protein. J Mol Biol 306: 443–454. [DOI] [PubMed] [Google Scholar]
- 7. Tisne C, Roques BP, Dardel F (2004) The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J Biol Chem 279: 3588–3595. [DOI] [PubMed] [Google Scholar]
- 8. Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K (2004) Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol 337: 951–968. [DOI] [PubMed] [Google Scholar]
- 9. Barraud P, Gaudin C, Dardel F, Tisne C (2007) New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie 89: 1204–1210. [DOI] [PubMed] [Google Scholar]
- 10. Li X, Mak J, Arts EJ, Gu Z, Kleiman L, et al. (1994) Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J Virol 68: 6198–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, et al. (2004) Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol 78: 2601–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wakefield JK, Wolf AG, Morrow CD (1995) Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA(3Lys). J Virol 69: 6021–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, et al. (1989) HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. The EMBO journal 8: 3279–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abbink TE, Beerens N, Berkhout B (2004) Forced selection of a human immunodeficiency virus type 1 variant that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme. J Virol 78: 10706–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abbink TE, Berkhout B (2007) HIV-1 reverse transcription: close encounters between the viral genome and a cellular tRNA. Advances in pharmacology 55: 99–135. [DOI] [PubMed] [Google Scholar]
- 16. Beerens N, Berkhout B (2002) Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA 8: 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beerens N, Berkhout B (2002) The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol 76: 2329–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beerens N, Groot F, Berkhout B (2001) Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem 276: 31247–31256. [DOI] [PubMed] [Google Scholar]
- 19. Dupuy LC, Kelly NJ, Elgavish TE, Harvey SC, Morrow CD (2003) Probing the importance of tRNA anticodon: human immunodeficiency virus type 1 (HIV-1) RNA genome complementarity with an HIV-1 that selects tRNA(Glu) for replication. J Virol 77: 8756–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R (1995) Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer). J Mol Biol 247: 236–250. [DOI] [PubMed] [Google Scholar]
- 21. Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B (1993) Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem 268: 25269–25272. [PubMed] [Google Scholar]
- 22. Isel C, Westhof E, Massire C, Le Grice SF, Ehresmann B, et al. (1999) Structural basis for the specificity of the initiation of HIV-1 reverse transcription. Embo J 18: 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG (2003) Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J Biol Chem 278: 14185–14195. [DOI] [PubMed] [Google Scholar]
- 24. Kang SM, Zhang Z, Morrow CD (1997) Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNA(Met). J Virol 71: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang SM, Zhang Z, Morrow CD (1999) Identification of a human immunodeficiency virus type 1 that stably uses tRNALys1,2 rather than tRNALys,3 for initiation of reverse transcription. Virology 257: 95–105. [DOI] [PubMed] [Google Scholar]
- 26. Wakefield JK, Kang SM, Morrow CD (1996) Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNA(His). J Virol 70: 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Z, Kang SM, LeBlanc A, Hajduk SL, Morrow CD (1996) Nucleotide sequences within the U5 region of the viral RNA genome are the major determinants for an human immunodeficiency virus type 1 to maintain a primer binding site complementary to tRNA(His). Virology 226: 306–317. [DOI] [PubMed] [Google Scholar]
- 28. Aiyar A, Cobrinik D, Ge Z, Kung HJ, Leis J (1992) Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J Virol 66: 2464–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang C, Li X, Rong L, Inouye P, Quan Y, et al. (1997) The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J Virol 71: 5750–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller JT, Ehresmann B, Hubscher U, Le Grice SF (2001) A novel interaction of tRNA(Lys,3) with the feline immunodeficiency virus RNA genome governs initiation of minus strand DNA synthesis. J Biol Chem 276: 27721–27730. [DOI] [PubMed] [Google Scholar]
- 31. Sleiman D, Goldschmidt V, Barraud P, Marquet R, Paillart JC, et al. (2012) Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus research 169: 324–339. [DOI] [PubMed] [Google Scholar]
- 32. Goldschmidt V, Ehresmann C, Ehresmann B, Marquet R (2003) Does the HIV-1 primer activation signal interact with tRNA3(Lys) during the initiation of reverse transcription? Nucleic Acids Res 31: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldschmidt V, Paillart JC, Rigourd M, Ehresmann B, Aubertin AM, et al. (2004) Structural variability of the initiation complex of HIV-1 reverse transcription. J Biol Chem 279: 35923–35931. [DOI] [PubMed] [Google Scholar]
- 34. Freund F, Boulme F, Litvak S, Tarrago-Litvak L (2001) Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex. Nucleic Acids Res 29: 2757–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beerens N, Jepsen MD, Nechyporuk-Zloy V, Kruger AC, Darlix JL, et al.. (2013) Role of the primer activation signal in tRNA annealing onto the HIV-1 genome studied by single-molecule FRET microscopy. RNA. [DOI] [PMC free article] [PubMed]
- 36. Pervushin K, Riek R, Wider G, Wuthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A 94: 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weigelt J (1998) Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc 120: 10778–10779. [Google Scholar]
- 38. Puglisi EV, Puglisi JD (2011) Secondary structure of the HIV reverse transcription initiation complex by NMR. J Mol Biol 410: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tisne C, Rigourd M, Marquet R, Ehresmann C, Dardel F (2000) NMR and biochemical characterization of recombinant human tRNA(Lys)3 expressed in Escherichia coli: identification of posttranscriptional nucleotide modifications required for efficient initiation of HIV-1 reverse transcription. RNA 6: 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, et al. (1993) Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol 229: 382–397. [DOI] [PubMed] [Google Scholar]
- 41. Berkhout B (1996) Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol 54: 1–34. [DOI] [PubMed] [Google Scholar]
- 42. Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, et al. (2006) Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res 34: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Levin JG, Guo J, Rouzina I, Musier-Forsyth K (2005) Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80: 217–286. [DOI] [PubMed] [Google Scholar]
- 44. Rein A, Henderson LE, Levin JG (1998) Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci 23: 297–301. [DOI] [PubMed] [Google Scholar]
- 45. Abbink TE, Berkhout B (2008) HIV-1 reverse transcription initiation: a potential target for novel antivirals? Virus research 134: 4–18. [DOI] [PubMed] [Google Scholar]
- 46. Azoulay J, Clamme JP, Darlix JL, Roques BP, Mely Y (2003) Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J Mol Biol 326: 691–700. [DOI] [PubMed] [Google Scholar]
- 47. Beltz H, Piemont E, Schaub E, Ficheux D, Roques B, et al. (2004) Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol 338: 711–723. [DOI] [PubMed] [Google Scholar]
- 48. Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC (2006) Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J Mol Biol 363: 867–877. [DOI] [PubMed] [Google Scholar]
- 49. Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, et al. (2001) Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci U S A 98: 6121–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ooms M, Cupac D, Abbink TE, Huthoff H, Berkhout B (2007) The availability of the primer activation signal (PAS) affects the efficiency of HIV-1 reverse transcription initiation. Nucleic Acids Res 35: 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Isel C, Keith G, Ehresmann B, Ehresmann C, Marquet R (1998) Mutational analysis of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucleic Acids Res 26: 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, et al. (2008) Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic acids research 36: 2311–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saadatmand J, Niu M, Kleiman L, Guo F (2009) The contribution of the primer activation signal to differences between Gag- and NCp7-facilitated tRNA(Lys3) annealing in HIV-1. Virology 391: 334–341. [DOI] [PubMed] [Google Scholar]
- 54. Cen S, Khorchid A, Gabor J, Rong L, Wainberg MA, et al. (2000) Roles of Pr55(gag) and NCp7 in tRNA(3)(Lys) genomic placement and the initiation step of reverse transcription in human immunodeficiency virus type 1. J Virol 74: 10796–10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF (1998) Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol 279: 633–649. [DOI] [PubMed] [Google Scholar]
- 56. De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, et al. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279: 384–388. [DOI] [PubMed] [Google Scholar]
- 57. Markham NR, Zuker M (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33: W577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2: 661–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA sequences used to model the secondary structure context of the PAS sequence within A) the MAL isolate and B) the HXB2 isolate. These RNA fragments were used to derive the thermodynamic parameters for the PAS sequence within the two different viral RNA isolates (See Figure 5).
(TIF)
Tests of annealing of PASds to tRNALys3. A) Schematic drawing of the experiment, B) Superimposition of two TROSY experiments recorded at 15°C showing the imino groups of tRNALys 3 (0.1 mM) alone (in black, reference spectrum) and after the heat-annealed procedure with 1 equivalent of PASds (red spectrum).
(TIF)
Superimposition of three NOESY experiments recorded at 15°C with a mixing time of 150 ms showing the imino-imino region, in blue for PASds, in red for a mixture of tRNALys3, PASds and PBS for which the annealing between the tRNALys3/PBS was promoted, in black for a mixture of tRNALys3, PASds, PBS, NCp7 for which the annealing tRNALys3/PBS/PASds was promoted (same experiment as Figure 7 ). The NOESY cross-peaks for GU base-pairs within PASds, and tRNALys 3/PAS complex are indicated. The assigment of the imino groups within the PAS/anti-PAS complex is indicated in purple for 15N-signals and the assignment for black cross-peaks corresponding to non-labelled imino groups are in orange.
(TIF)
Annealing of PASss to tRNALys3 in the presence of PBS. Superimposition of two TROSY experiments recorded at 15°C showing the imino groups of tRNALys 3 (0.1 mM), in black: in complex with PBS (1 equivalent) and PASss (1 equivalent) after the heat-annealed procedure; and in red: in complex with PBS (1 equivalent) and PASss (1 equivalent) after the NCp7-mediated procedure. Only the GC regions are shown since the T54 imino group was not observed in these experiments.
(TIF)
UV melting curve of the PASds.
(TIF)
Region of the imino protons of the 1H NMR spectra of the PASds alone (0.16 mM) and of the PASds (0.16 mM) mixed with NCp7 at 1∶1 ratio. The spectra were recorded at 15°C using a watergate sequence to [58] to achieve water signal suppression.
(TIF)