Abstract
Despite convincing evidence that 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)—a heterocyclic amine generated by cooking meats at high temperatures—is carcinogenic in animal models, it remains unclear whether PhIP exposure leads to increased cancer risk in humans. PhIP-DNA adduct levels were measured in specimens from 534 prostate cancer case-control pairs nested within a historical cohort of men with histopathologically benign prostate specimens. We estimated the overall and race-stratified risk of subsequent prostate cancer associated with higher adduct levels. PhIP-DNA adduct levels in benign prostate were significantly higher in Whites than African Americans (0.274 Optical Density Units (OD) ±0.059 vs. 0.256 OD ±0.054; p<0.0001). Prostate cancer risk for men in the highest quartile of PhIP-DNA adduct levels was modestly increased (Odds Ratio (OR) = 1.25; 95% confidence interval (CI) = 0.76-2.07). In subset analyses, the highest risk estimates were observed in White patients diagnosed more than 4 years after cohort entry (OR=2.74; 95% CI=1.01-7.42) or under age 65 (OR=2.80; 95% CI=0.87-8.97). In Whites, cancer risk associated with high grade prostatic intraepithelial neoplasia combined with elevated PhIP-DNA adduct levels (OR=3.89; 95% CI=1.56-9.73) was greater than risk associated with either factor alone. Overall, elevated levels of PhIP-DNA adducts do not significantly increase prostate cancer risk. However, our data show that White men have higher PhIP-DNA adduct levels in benign prostate tissue than African American men, and suggest that in certain subgroups of White men high PhIP-DNA adduct levels may predispose to an increased risk for prostate cancer.
Keywords: dna adducts; nested case-control study; immunohistochemistry; carcinogens; imidazoles; biopsy, needle
INTRODUCTION
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most abundant heterocyclic amine (HCA) formed during the cooking of meat 1, and a potential dietary risk factor for prostate and other cancers. A direct correlation between PhIP exposure, DNA adduct formation, and other indicators of prostate carcinogenesis is supported by animal models and in vitro studies of human tissues. Rats fed a PhIP-laden diet for 52 weeks had PhIP-DNA adducts in all prostate lobes and subsequently developed prostate cancer 2; PhIP exposure in rats is also associated with elevated mutation frequencies in prostate tissue 3 and increased prostate tumor incidence 4. Mice administered PhIP showed positive staining for PhIP-DNA adducts in human prostate xenografts 5. More recently, inflammation, atrophy of acini, and prostatic intraepithelial neoplasia were observed in the prostate glands of a CYP1A-humanized mouse model, following a single oral dose of PhIP 6.
Several in vitro studies of human prostate tissue incubated in PhIP-laden milieu have demonstrated detectable PhIP-DNA adducts in prostate cells 7-9. While one study found a low prevalence of detectable PhIP-DNA adducts in prostate tissue using the 32P-postlabeling method10, our own studies have demonstrated that PhIP-DNA adduct levels in prostate are related to dietary intake11;12 and tumor grade13. In vitro experiments using the comet assay and human prostate epithelial cells have shown that increased doses of PhIP result in increased DNA damage14. A study using a modified in vitro mutagen sensitivity assay, with activated PhIP (N-OH-PhIP) as the challenge mutagen and chromosomal aberrations as the endpoint, found that prostate cancer cases showed significantly higher numbers of breaks15, suggesting a greater susceptibility to PhIP-induced carcinogenesis in prostate cancer cases.
Despite the strong evidence for PhIP-induced prostate carcinogenesis from animal and in vitro studies, studies of dietary PhIP exposure and human prostate cancer risk are largely equivocal 16-21. One limitation of these studies is their reliance on food frequency questionnaires to estimate PhIP exposure. While dietary intake data is informative, it is ultimately a poor measure of biologically-effective dose, as it does not account for individual variation in PhIP metabolism or DNA repair capacity, which can influence DNA adduct formation 11. Cellular and molecular changes are likely to be more relevant to disease outcome than measurement of PhIP in the diet. As such, the detection and quantification of PhIP-DNA adducts within the tissue of interest is an important step toward understanding the connection between exposure and cancer development. Case-control studies of PhIP-DNA adducts and cancer risk are limited to two studies, one of breast 22 and the other of pancreas 23, both of which found elevated PhIP-DNA adducts in cancer patients. No studies have examined PhIP-DNA adducts levels in benign tissue and subsequent prostate cancer risk. Our own previous research on PhIP-DNA adduct levels was a cross-sectional study without a control group, using prostate tissue from cancer cases11-13.
In the current study, we advance the molecular epidemiologic study of DNA adducts and cancer risk by measuring PhIP-DNA adduct levels in histopathologically normal tissue specimens taken from the target organ, and assess the relationship between adduct levels, pre-neoplastic histological markers, and subsequent cancer risk using a case-control study nested within a large historical cohort. In addition to testing whether adduct levels in histopathologically benign target tissue were associated with incident prostate cancer and tumor aggressiveness, we also explored race-specific cancer associations.
METHODS
Study Sample and Medical Record Review
After obtaining approval from the Henry Ford Health System Institutional Review Board, we identified a historical cohort of 6,692 men with a benign prostate specimen collected by needle core biopsy or transurethral resection of the prostate (TURP) between January 1990 and December 2002. A nested case-control sample was drawn from this cohort based on eligibility criteria that included a recorded prostate specific antigen (PSA) level within a year of cohort entry and no history of a previous prostate cancer diagnosis. ‘Date of cohort entry’ was defined as the date that the initial benign prostate specimen was acquired; ‘date of case diagnosis’ was the date of first cancer-positive tissue specimen or the date a clinician first reported a clinical diagnosis of prostate cancer. Patients diagnosed with prostate cancer less than one year from date of cohort entry were ineligible for the study. We identified 808 potentially eligible cases diagnosed with prostate cancer prior to July 2007.
Incidence density sampling was used to select controls with replacement from all cohort members at risk at the time of case occurrence. Controls were randomly selected from among those cohort members who were free of prostate cancer at a follow-up duration greater than or equal to the time between cohort entry and diagnosis dates of the matched cases, with the end of follow-up denoted as the ‘reference date’ for controls. Matching criteria included age at entry into cohort (±2 years), date of entry into cohort (±2 years), race (African American or White), and type of specimen (biopsy or TURP). We were able to match 802 of 808 potentially eligible cases. Further review reduced the final analytic sample to 574 case-control pairs 24, of which we were able to analyze the PhIP-DNA adduct levels in 534 pairs; the remaining 40 pairs were excluded due to absence of sufficient numbers of epithelial cells in the tissue specimen.
Smoking status, and clinical, demographic, and co-morbidity data were abstracted from patients’ medical records, from five years before the date of cohort entry through the date of diagnosis (for prostate cancer cases) or reference date (for controls). All medical data used in study analyses are based on the date of cohort entry unless otherwise noted.
Immunohistochemistry
Consecutive sections (5 microns thick) were cut from each formalin-fixed, paraffin-embedded prostate specimen; one slide was used for PhIP-DNA adduct detection as described below and the other was hematoxylin and eosin (H&E) stained and examined by a single genitourinary pathologist (ONK) blinded to disease progression. The pathology examination included evaluation of the specimen for the presence of cancer, high-grade prostatic intraepithelial neoplasia (HGPIN), atrophy, and inflammation24.
Paraffin-embedded sections were heated to 50°C for one hour, deparaffinized in xylene, and rehydrated in serial alcohol. After treatment with RNase and Proteinase K, the sections were blocked using 3% BSA and normal goat serum. Sections were incubated in a humid 4°C chamber overnight with a 1:500 dilution of the primary anti-PhIP-DNA adduct polyclonal antibody 2;25; then incubated at room temperature for 30 minutes with a 1:200 dilution of the biotinylated secondary antibody. Specimens were bathed in 0.3% hydrogen peroxide in methanol for 20 minutes to block endogenous peroxidase activity.
The PhIP-DNA polyclonal antibody recognizes DNA adducts at the C8 position of deoxyguanosine as the epitope. The PhlP-modified DNA antigen contains N2-(2′-deoxyguanosin-8-yl)-PhIP25, which is recognized as the major adduct formed between PhIP and DNA26. Specificity of the PhIP-DNA adduct antibody was evaluated in liver tissue of rats separately exposed for six weeks to heterocyclic amines as follows: 3-amino-1,4-dimethyl-5H-pyrido[4,3-b]indole (Trp-P-1, 150ppm); 2-aminodipyrido[1,2-a:3′,2′-d]imidazole (Glu-P-2, 500ppm); 2-amino-3-methylimidazo[4,5-f]quinoline (MeIQ, 300ppm); or 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx, 400ppm). Presence of DNA adducts was confirmed by the 32P-postlabeling method27. No cross-reactivity with any heterocyclic amine-DNA adducts was found (S. Takahashi, unpublished data).
The antibody complex was detected using an avidin-biotin-peroxidase complex solution and visualized using 3,3’-diaminobenzidine chromogen (Zymed Laboratories, San Francisco, CA). A negative control was included in each experiment by omitting the primary antibody. A cytospin sample of MCF-7 cells without PhIP treatment was included in each batch of staining. Staining was measured by absorbance image analysis using the Cell Analysis System 200 (Becton Dickinson, San Jose, CA). Absorbance of light at a wavelength of 500 nmol/L was measured in optical density units (OD). Previous calibration studies using N-hydroxy-PhIP-treated MCF-7 cells have demonstrated optical density sensitivity to N-hydroxy-PhIP adducts ranging from 1-100μM with a detection limit of about 1/107 for PhIP-DNA adducts22. For each specimen, a technician scored 50 epithelial cells (five fields, ten cells per field) selected to be representative, in terms of intensity, of the cells in the field.
Statistical Analysis
Conditional logistic regression analyses were used to estimate both unadjusted and adjusted odds ratios (ORs) for prostate cancer incidence during follow-up. Individual matching controlled for age, race, and specimen type (biopsy or TURP). Analyses were performed using adduct levels expressed as both continuous and categorical variables; for the latter, adduct distribution was segmented among control subjects into referent categories. Potential confounders were identified by first testing whether the variable was associated with case status or adduct levels; associated variables were then tested in multivariable models to determine whether their inclusion changed the effect estimate by 10% or more. Comparisons between stratified models were assessed using conditional logistic regression with interaction terms.
RESULTS
Study sample and PhIP-DNA adduct levels
In the analytic sample of 534 pairs, cases were an average of 65.4 years old at cohort entry and 40% were African American (Supplementary Table 1). The 40 pairs with unanalyzable adduct data were significantly older (2.5 years, p=0.05), had 0.8 more PSA tests between cohort entry and diagnosis (p=0.05), and entered the cohort earlier (median 21 months, p=0.03) than those that were analyzed. In the full sample, median time to diagnosis was 1-4 years after cohort entry, with the remaining diagnosed 4-15 years after cohort entry. Cases had significantly higher PSA levels at time of cohort entry (7.7 ± 7.3 ng/ml vs. 5.6 ± 5.3 ng/ml; p<0.0001) and averaged two more PSA tests between cohort entry and diagnosis. The majority of cases (52.6%) had Stage 2 tumors; 29% of cases had advanced tumor grade defined as either Gleason score 8 and above or Gleason score 7 with a primary grade 4. Mean PhIP-DNA adduct levels were slightly elevated in cases compared with controls, but the difference was not statistically significant (0.263 OD ±0.058 vs. 0.260 OD ±0.045; p=0.32).
Factors associated with PhIP-DNA adduct levels
Given the known association between race and differences in both exposure to and metabolism of PhIP 13;28-30, we tested whether PhIP-DNA adduct levels varied by race. Mean adduct levels were significantly higher in Whites than African Americans (0.276 ±0.059 vs. 0.257 ±0.053 OD; p<0.0001) and followed a normal distribution in both racial groups (Supplementary Figure 1).
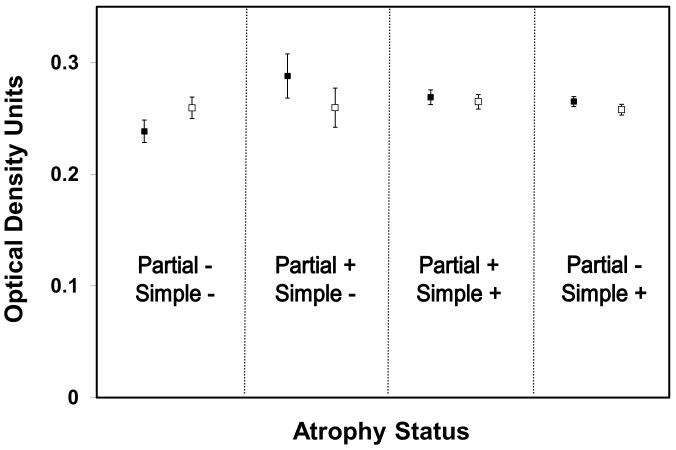
To better understand how the histological characteristics of prostate tissue might affect PhIP-DNA adduct levels, we estimated the mean adduct levels by histological variables that were previously described in this sample (Table 1) 24. In White patients with partial atrophy, we observed significantly higher levels of PhIP-DNA adducts than in those without partial atrophy (0.285 ±0.060 vs. 0.272 ±0.059 OD; p=0.01); the same trend in adduct levels was observed in the benign prostate specimens derived from African American patients, but differences between groups were smaller. Conversely, in African Americans with glandular inflammation, adduct levels were significantly higher than in those without glandular inflammation (0.239 ±0.057 vs. 0.260 ±0.051 OD; p=0.005), and a similar, but less significant inverse association was observed in Whites. In an effort to further tease apart the association of atrophy with adduct levels, we modeled PhIP-DNA adduct levels with covariates for both simple and partial atrophy adjusting for race and glandular inflammation. The least squares mean estimates of PAH-DNA adduct levels for the four possible combinations of simple and partial atrophy are shown in Figure 1. In cases, PhIP-DNA adduct levels were lowest when no atrophy was present, highest when only partial atrophy was present, and intermediate when simple atrophy was present (irrespective of whether partial atrophy was also present). In controls, there was no association between PhIP-DNA adduct levels and atrophy status; controls had higher levels of PhIP-DNA adducts than cases when no atrophy was observed, but much lower levels than cases when only partial atrophy was noted in the specimen. This same general pattern was observed when the data were stratified by race.
Table 1. Mean PhIP-DNA Adduct Levels in Optical Density Units Stratified by Race and Histological Variables.
| Variable | Mean Optical Density Units ± Standard Deviation |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Whole Sample | African Americans | Whites | |||||||
| Present | Absent | P value | Present | Absent | P value | Present | Absent | P value | |
| HGPIN | 0.272 ± .054 (n=74) |
0.268 ± .058 (n=838) |
0.60 | 0.253 ± .048 (n=31) |
0.257 ± .053 (n=323) |
0.69 | 0.285 ± .055 (n=43) |
0.275 ± .060 (n=515) |
0.28 |
| Atrophy | 0.269 ± .058 (n=736) |
0.266 ± .058 (n=176) |
0.45 | 0.257 ± .054 (n=285) |
0.256 ± .049 (n=69) |
0.86 | 0.277 ± .059 (n=451) |
0.272 ± .063 (n=107) |
0.43 |
| Simple atrophy | 0.269 ± .057 (n=656) |
0.269 ± .060 (n=256) |
0.95 | 0.256 ± .053 (n=253) |
0.260 ± .051 (n=101) |
0.51 | 0.277 ± .057 (n=403) |
0.274 ± .065 (n=155) |
0.62 |
| Post-atrophic hyperplasia | 0.282 ± .050 (n=18) |
0.268 ± .058 (n=894) |
0.34 | 0.308 ± .021 (n=3) |
0.257 ± .053 (n=351) |
0.09 | 0.276 ± .053 (n=15) |
0.279 ± .060 (n=543) |
0.99 |
| Simple atrophy - cyst formation |
0.264 ± .061 (n=175) |
0.270 ± .057 (n=737) |
0.28 | 0.258 ± .064 (n=54) |
0.257 ± .050 (n=300) |
0.88 | 0.267 ± .060 (n=121) |
0.279 ± .059 (n=437) |
0.07 |
| Partial atrophy | 0.277 ± .059 (n=267) |
0.265 ± .057 (n=645) |
0.006 | 0.260 ± .051 (n=91) |
0.256 ± .053 (n=263) |
0.45 | 0.285 ± .060 (n=176) |
0.272 ± .059 (n=382) |
0.01 |
| Inflammation | 0.267 ± .056 (n=551) |
0.272 ± .059 (n=361) |
0.18 | 0.254 ± .053 (n=219) |
0.261 ± .051 (n=135) |
0.24 | 0.275 ± .057 (n=332) |
0.278 ± .063 (n=226) |
0.48 |
| Glandular inflammation | 0.259 ± .059 (n=169) |
0.271 ± .057 (n=743) |
0.02 | 0.239 ± .057 (n=56) |
0.260 ± .051 (n=298) |
0.005 | 0.270 ± .058 (n=113) |
0.278 ± .060 (n=445) |
0.18 |
| Periglandular inflammation | 0.264 ± .058 (n=364) |
0.272 ± .057 (n=548) |
0.04 | 0.251 ± .055 (n=148) |
0.262 ± .051 (n=206) |
0.05 | 0.273 ± .058 (n=216) |
0.278 ± .060 (n=342) |
0.31 |
| Stromal inflammation | 0.266 ± .056 (n=430) |
0.271 ± .059 (n=482) |
0.19 | 0.254 ± .053 (n=171) |
0.259 ± .052 (n=183) |
0.39 | 0.274 ± .056 (n=259) |
0.278 ± .063 (n=299) |
0.37 |
Figure 1.
Least squares means estimates of PhIP-DNA adduct levels by case (shaded square)/control (open square) status and type of atrophy in the benign sample.
Prostate Cancer risk associated with higher levels of PhIP-DNA adducts
While mean PhIP-DNA adduct levels did not differ significantly between cases and controls, previous studies have shown that the effect of DNA adducts on cancer-related outcomes tends to be non-linear 31;32. Therefore, to determine whether prostate cancer risk was associated with elevated adduct levels, we tested two models, one in which adduct levels were categorized into quartiles, and another in which levels were dichotomized above and below the median (Table 2). Due to the differences of adduct levels by race, models for both the full sample as well as race-stratified models were tested. For the full sample, quartile risk estimates did not reach statistical significance nor was the trend statistically significant (p=0.36); similarly, the odds ratio for adduct levels greater than the median was also non-significant. When the sample was stratified by race, increased risk associated with elevated PhIP-DNA adduct levels trended upward in Whites: 12% for the 2nd quartile, 48% for the 3rd quartile, and 73% for the 4th quartile, but none were statistically significant, nor was the trend statistically significant (p=0.07). Models adjusting for inflammation, atrophy, and number of PSA tests were also tested, but changes in risk estimates were nominal (data not shown).
Table 2. Association of PhIP-DNA Adduct Levels with Prostate Cancer.
| Sample PhIP-DNA Adduct Level |
ORa | 95% CI | P value |
|---|---|---|---|
| Whole Sample (n=534 case-control pairs) | |||
| 2nd Quartile | 1.20 | 0.83-1.72 | 0.34 |
| 3rd Quartile | 1.29 | 0.87-1.92 | 0.20 |
| 4th Quartile | 1.25 | 0.76-2.07 | 0.38 |
| Trend Test | 0.36 | ||
| High PhIP levela | 1.16 | 0.84-1.59 | 0.37 |
| African Americans (n=213 case-control pairs) | |||
| 2nd Quartile | 1.32 | 0.75-2.32 | 0.33 |
| 3rd Quartile | 0.96 | 0.49-1.86 | 0.90 |
| 4th Quartile | 0.73 | 0.32-1.63 | 0.44 |
| Trend Test | 0.35 | ||
| High PhIP levela | 0.76 | 0.44-1.32 | 0.33 |
| Whites (n= 321 case-control pairs) | |||
| 2nd Quartile | 1.12 | 0.69-1.81 | 0.65 |
| 3rd Quartile | 1.48 | 0.90-2.44 | 0.13 |
| 4th Quartile | 1.73 | 0.89-2.34 | 0.10 |
| Trend Test | 0.07 | ||
| High PhIP levela | 1.44 | 0.97-2.14 | 0.07 |
all risk estimates use lowest level as referent group
above median level in controls
Table 3 reports risk estimates stratified by selected matching factors (including race) and tumor grade (for cases). In the full sample, little evidence for heterogeneity of high PhIP-DNA adduct levels by strata exists. In the White sub-sample—where elevated PhIP-DNA adduct levels had suggestive associations with prostate cancer risk—odds ratios were greater for cases with high tumor grade, longer follow-up, later cohort entry, and younger age at diagnosis. In White cases diagnosed before age 65, the risk of prostate cancer was elevated 70-80% in the 2nd and 3rd quartiles, and 180% in the 4th quartile, but neither risk estimates nor the linear trend test reached statistical significance.
Table 3. Association of PhIP-DNA Adduct Levels with Prostate Cancer, Stratified by Matching Factors and Case Characteristics.
| Sample PhIP Level |
Whole Sample | African Americans | Whites | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ORa | 95% CI | P value | OR | 95% CI | P value | OR | 95% CI | P value | |
| Low Tumor Grade | (n=367 case-control pairs) | (n = 137 case-control pairs) | (n = 230 case-control pairs) | ||||||
| 2nd Quartile | 1.40 | 0.84-2.31 | 0.19 | 1.94 | 0.83-4.55 | 0.13 | 1.17 | 0.62-2.19 | 0.63 |
| 3rd Quartile | 1.37 | 0.81-2.33 | 0.25 | 1.42 | 0.53-3.85 | 0.49 | 1.30 | 0.69-2.46 | 0.41 |
| 4th Quartile | 1.29 | 0.65-2.54 | 0.47 | 1.09 | 0.30-4.01 | 0.90 | 1.36 | 0.61-3.04 | 0.46 |
| Trend Test | 0.51 | 0.98 | 0.43 | ||||||
| High Tumor Grade | (n= 138 case-control pairs) | (n = 56 case-control pairs) | (n = 82 case-control pairs) | ||||||
| 2nd Quartile | 0.92 | 0.45-1.86 | 0.81 | 1.14 | 0.38-3.44 | 0.82 | 0.87 | 0.33-2.28 | 0.77 |
| 3rd Quartile | 1.57 | 0.68-3.65 | 0.29 | 0.94 | 0.22-4.08 | 0.93 | 2.04 | 0.69-6.07 | 0.20 |
| 4th Quartile | 1.03 | 0.31-3.39 | 0.97 | 0.45 | 0.07-2.96 | 0.41 | 1.84 | 0.34-10.00 | 0.48 |
| Trend Test | 0.76 | 0.43 | 0.33 | ||||||
| 1-4 years of Follow-up | (n= 266 case-control pairs) | (n = 103 case-control pairs) | (n = 163 case-control pairs) | ||||||
| 2nd Quartile | 1.63 | 0.91-2.92 | 0.10 | 1.83 | 0.76-4.44 | 0.18 | 1.48 | 0.68-3.24 | 0.32 |
| 3rd Quartile | 1.46 | 0.75-2.84 | 0.27 | 0.99 | 0.31-3.16 | 0.99 | 1.68 | 0.74-3.84 | 0.22 |
| 4th Quartile | 1.15 | 0.49-2.69 | 0.74 | 0.95 | 0.23-4.02 | 0.95 | 1.23 | 0.42-3.55 | 0.71 |
| Trend Test | 0.82 | 0.75 | 0.67 | ||||||
| 4-15 years of follow-up | (n= 267 case-control pairs) | (n = 110 case-control pairs) | (n = 157 case-control pairs) | ||||||
| 2nd Quartile | 0.90 | 0.51-1.58 | 0.72 | 0.82 | 0.32-2.11 | 0.67 | 0.99 | 0.48-2.05 | 0.99 |
| 3rd Quartile | 1.34 | 0.75-2.37 | 0.32 | 0.90 | 0.32-2.55 | 0.84 | 1.60 | 0.78-3.27 | 0.20 |
| 4th Quartile | 1.46 | 0.70-3.03 | 0.31 | 0.62 | 0.17-2.18 | 0.45 | 2.74 | 1.01-7.42 | 0.05 |
| Trend Test | 0.21 | 0.53 | 0.03 | ||||||
| Early Cohort entry | (n = 263 case-control pairs) | (n = 88 case-control pairs) | (n = 175 case-control pairs) | ||||||
| 2nd Quartile | 1.28 | 0.69-2.40 | 0.44 | 2.46 | 0.69-8.76 | 0.17 | 1.02 | 0.49-2.12 | 0.96 |
| 3rd Quartile | 1.28 | 0.68-2.40 | 0.44 | 1.92 | 0.56-6.53 | 0.30 | 1.09 | 0.52-2.29 | 0.83 |
| 4th Quartile | 1.73 | 0.78-3.86 | 0.18 | 2.76 | 0.60-12.75 | 0.19 | 1.42 | 0.54-3.72 | 0.48 |
| Trend Test | 0.21 | 0.27 | 0.47 | ||||||
| Late Cohort entry | (n=270 case-control pairs) | (n = 125 case-control pairs) | (n = 145 case-control pairs) | ||||||
| 2nd Quartile | 1.10 | 0.67-1.84 | 0.69 | 1.00 | 0.48-2.10 | 1.00 | 1.14 | 0.55-2.36 | 0.72 |
| 3rd Quartile | 1.40 | 0.76-2.57 | 0.28 | 0.60 | 0.20-1.77 | 0.35 | 2.18 | 0.99-4.78 | 0.05 |
| 4th Quartile | 0.98 | 0.46-2.13 | 0.97 | 0.29 | 0.08-1.08 | 0.06 | 2.14 | 0.73-6.24 | 0.16 |
| Trend Test | 0.89 | 0.06 | 0.09 | ||||||
| Age <65 | (n=274 case-control pairs) | (n = 110 case-control pairs) | (n = 164 case-control pairs) | ||||||
| 2nd Quartile | 1.30 | 0.72-2.35 | 0.39 | 0.92 | 0.38-2.24 | 0.85 | 1.79 | 0.79-4.09 | 0.16 |
| 3rd Quartile | 1.35 | 0.72-2.53 | 0.35 | 0.95 | 0.35-2.60 | 0.92 | 1.71 | 0.74-3.95 | 0.21 |
| 4th Quartile | 1.08 | 0.47-2.47 | 0.85 | 0.38 | 0.10-1.42 | 0.15 | 2.80 | 0.87-8.97 | 0.08 |
| Trend Test | 0.83 | 0.18 | 0.09 | ||||||
| Age 65+ | (n=259 case-control pairs) | (n = 103 case-control pairs) | (n = 156 case-control pairs) | ||||||
| 2nd Quartile | 1.10 | 0.64-1.89 | 0.72 | 1.71 | 0.65-4.50 | 0.27 | 0.82 | 0.41-1.62 | 0.56 |
| 3rd Quartile | 1.45 | 0.79-2.67 | 0.23 | 1.02 | 0.29-3.60 | 0.97 | 1.55 | 0.76-3.16 | 0.22 |
| 4th Quartile | 1.54 | 0.72-3.29 | 0.26 | 1.52 | 0.33-7.03 | 0.59 | 1.46 | 0.57-3.45 | 0.47 |
| Trend Test | 0.23 | 0.78 | 0.28 | ||||||
all risk estimates use lowest quartile as referent group
The other stratum with markedly higher risk estimates was White cases diagnosed four years or more after cohort entry, where risk of prostate cancer was unchanged for the 2nd quartile, but increased 60% for the 3rd quartile, and 174% for the 4th quartile. The risk estimate in the 4th quartile was marginally statistically significant (OR=2.74; 95% Confidence Interval (CI)=1.01-7.42) and the linear trend of risk estimates across quartiles was also statistically significant (p=0.03). To further investigate whether this suggested a temporal relationship between adduct levels and prostate cancer, we analyzed the association between adduct quartile and time to diagnosis among White cases (Supplementary Figure 2). While cases with elevated adduct levels were diagnosed more rapidly within 2-3 years of follow-up, the greatest difference in risk between the lowest and highest quartiles was observed 5-7 years after cohort entry. Overall, the four quartile curves for time to diagnosis were significantly different (log rank p-value=0.05).
We next tested whether known clinical or histological prostate cancer risk factors modified the relationship between elevated PhIP-DNA adduct levels and prostate cancer (Table 4). Overall, no factors significantly modified the risk associated with elevated adduct levels and prostate cancer, although some interesting trends emerged. Partial atrophy and glandular inflammation were both associated with PhIP-DNA adducts in this study population in a race-specific manner, but neither modified the risk of prostate cancer associated with high adduct levels. Glandular inflammation had the highest interaction odds ratio (OR=2.19 in African Americans; 1.97 in Whites), and although neither odds ratio was statistically significant, it appeared that elevated PhIP-DNA adduct levels increased risk for prostate cancer only in the presence of glandular inflammation. HGPIN was associated with prostate cancer in this study population 24, and it modestly enhanced the association between elevated adduct levels and prostate cancer in Whites. In the absence of HGPIN, elevated PhIP-DNA adducts increased the risk for prostate cancer by 38%, but in the presence of both HGPIN and high PhIP-DNA adducts level, the risk increased almost 4-fold (OR=3.89; CI=1.56-9.73). This is in contrast to what was observed in African Americans where the combination of HGPIN and elevated PhIP-DNA adduct levels actually decreased prostate cancer risk.
Table 4. Effect Modification of PhIP-DNA Adduct Levela Associations with Prostate Cancer.
| Whole Sample (n=534 case-control pairs) |
African Americans (n=213 case-control pairs) |
Whites (n=321 case-control pairs) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect Modifier | OR | 95% CI | P value | OR | 95% CI | P value | OR | 95% CI | P value |
| PSA at cohort entry | |||||||||
| PSA<4 ng ml/Low PhIP | 1 | 1 | 1 | ||||||
| PSA<4 ng ml/High PhIP | 1.35 | 0.79-2.32 | 0.27 | 0.89 | 0.35-2.27 | 0.81 | 1.63 | 0.84-3.18 | 0.15 |
| PSA≥4 ng ml/Low PhIP | 3.39 | 2.14-5.36 | <0.0001 | 3.73 | 1.83-7.62 | 0.0003 | 3.08 | 1.69-5.63 | 0.0002 |
| PSA≥4 ng ml/High PhIP | 3.21 | 2.00-5.15 | <0.0001 | 2.37 | 1.08-5.19 | 0.03 | 3.62 | 1.99-6.58 | <0.0001 |
| PSA≥4 ng ml × High PhIP Interaction |
0.70 | 0.39-1.27 | 0.24 | 0.71 | 0.26-1.93 | 0.51 | 0.72 | 0.34-1.52 | 0.39 |
| Glandular Inflammation | |||||||||
| No Inflammation/Low PhIP | 1 | 1 | 1 | ||||||
| No Inflammation/High PhIP | 1.06 | 0.75-1.48 | 0.75 | 0.70 | 0.40-1.25 | 0.23 | 1.29 | 0.84-1.96 | 0.25 |
| Inflammation/Low PhIP | 0.78 | 0.48-1.27 | 0.32 | 1.01 | 0.49-2.07 | 0.98 | 0.64 | 0.32-1.26 | 0.20 |
| Inflammation/High PhIP | 1.54 | 0.91-2.61 | 0.11 | 1.56 | 0.54-4.52 | 0.42 | 1.61 | 0.87-2.99 | 0.13 |
| Inflammation × High PhIP Interaction |
1.87 | 0.95-3.68 | 0.07 | 2.19 | 0.65-7.43 | 0.21 | 1.97 | 0.83-4.69 | 0.13 |
| HGPIN | |||||||||
| No HGPIN/Low PhIP | 1 | 1 | 1 | ||||||
| No HGPIN/High PhIP | 1.16 | 0.84-1.61 | 0.37 | 0.82 | 0.47-1.44 | 0.48 | 1.38 | 0.92-2.07 | 0.12 |
| HGPIN/Low PhIP | 2.16 | 1.07-4.37 | 0.03 | 2.88 | 1.04-7.98 | 0.04 | 1.52 | 0.55-4.16 | 0.42 |
| HGPIN/High PhIP | 2.25 | 1.12-4.51 | 0.02 | 0.79 | 0.24-2.59 | 0.69 | 3.89 | 1.56-9.73 | 0.004 |
| HGPIN × High PhIP Interaction | 0.90 | 0.35-2.28 | 0.82 | 0.33 | 0.08-1.46 | 0.15 | 1.86 | 0.50-6.88 | 0.35 |
| Partial Atrophy | |||||||||
| No Atrophy/Low PhIP | 1 | 1 | 1 | ||||||
| No Atrophy/High PhIP | 1.23 | 0.87-1.74 | 0.25 | 0.85 | 0.47-1.53 | 0.58 | 1.48 | 0.95-2.30 | 0.08 |
| Atrophy/Low PhIP | 0.98 | 0.66-1.46 | 0.92 | 1.12 | 0.62-2.02 | 0.72 | 0.89 | 0.51-1.54 | 0.67 |
| Atrophy/High PhIP | 1.00 | 0.66-1.74 | 1.00 | 0.59 | 0.27-1.28 | 0.18 | 1.25 | 0.76-2.08 | 0.38 |
| Atrophy × High PhIP Interaction | 0.83 | 0.49-1.41 | 0.49 | 0.63 | 0.26-1.52 | 0.30 | 0.95 | 0.48-1.89 | 0.89 |
High PhIP was considered above median level in controls
DISCUSSION
We report for the first time a prospective analysis of PhIP-DNA adduct levels—a marker of biologically effective exposure to PhIP—measured in histopathologically benign tissue, and subsequent cancer risk for the same organ. Prior prospective studies of adduct levels and cancer risk have used surrogate tissues, such as white blood cells, but the correlation between adduct levels in these surrogate tissues versus the target organ is unclear 33;34. Here we find that higher levels of PhIP-DNA adducts in benign prostate specimens were associated with a modestly increased risk for prostate cancer in White men. When the analysis was restricted to White cases diagnosed more than four years after tissue collection, however, patients with the highest PhIP-DNA adduct levels had almost 3-fold increased risk of prostate cancer. In African Americans, we did not detect any observable increased risk associated with high PhIP-DNA adduct levels, in either the full sample or subgroups.
Until now, the question of whether PhIP increases risk for prostate cancer has largely been addressed using estimated dietary consumption of PhIP35. The first such effort found no increased risk of prostate cancer with increasing PhIP consumption36. A subsequent prospective study found a modest 22% increased prostate cancer risk for the highest PhIP consumption, with a statistically significant trend21. Since that study, multiple cohort 17;18;20 and case-control 16;19 questionnaire-based studies have been performed, collectively providing equivocal results concerning dietary PhIP consumption and prostate cancer risk. Given the high potential for measurement error using questionnaire data, biomarker studies are needed to address the potential carcinogenicity of this compound.
Despite availability of an anti-PhIP-DNA adduct antibody for immunohistochemistry studies 12;22;25, only two case-control studies of cancer have employed this method previously; both finding higher PhIP-DNA adduct levels in benign tissue of the cancer-affected organ of cases compared with the corresponding tissue of controls22;23. Adduct levels were four times more likely to be high in benign tissue of breast cancer cases than controls22 and 3.5 times more likely to be high in benign tissue of pancreatic cancer cases than controls23. To date, ours is the first such study to be performed in human prostate and the only study of PhIP-DNA adduct levels in pre-disease tissue.
Our previous study of men with prostate cancer found no racial differences in PhIP adduct levels in prostate tissue13, but in the present study, we found that Whites had significantly higher levels of PhIP adducts in benign prostate than African Americans. Dietary data strongly suggest African Americans have higher exposure to PhIP through food preparation methods that differ by race 28 and African Americans have been shown to excrete more PhIP in urine30. Both activation and detoxification of PhIP play a role in adduct formation. African American men have slightly higher activity levels of the PhIP-metabolizing enzyme SULT1A1 than Whites29. However, both African Americans and Whites show an association between prostate cancer risk, SULT1A1 levels, and the Arg213His functional polymorphism in the SULT1A1 gene29. African Americans also have higher enzymatic activity levels of CYP1A2 and N-acetyltransferase37, two enzymes important in the O-acetylation and N-oxidation of PhIP, respectively. PhIP and its carcinogenic metabolite N-hydroxy-PhIP (N-OH-PhIP) are extensively conjugated by UDP-glucuronosyltransferases (UGTs)9, and UGT1A1 is the predominant UGT involved in PhIP metabolism38; notably, the most prevalent UGT1A1 genotype in African Americans is associated with a lower capacity to detoxify PhIP39. While PhIP detoxification by UGT1A1 appears to be less efficient in African Americans, a study of UGT1A1 genetic variation and colon cancer risk found an elevated risk associated with intermediate- to low-activity UGT1A1 genotypes in Whites but not in African Americans40. This is consistent with our recent study that found non-specific genetic variation related to African ancestry to be a stronger predictor of PhIP-DNA adduct levels in prostate tissue than genetic variation in either UGT1A1 or SULT1A111. While the literature reviewed above would suggest that African Americans should have higher PhIP-DNA adduct levels and subsequently a greater risk of cancer associated with this marker than Whites, our results suggest the opposite. Clearly the carcinogenic potential of PhIP in human prostate is more complex than the sum of what is currently known about race-specific dietary exposure and genetic variation in PhIP metabolism41;42.
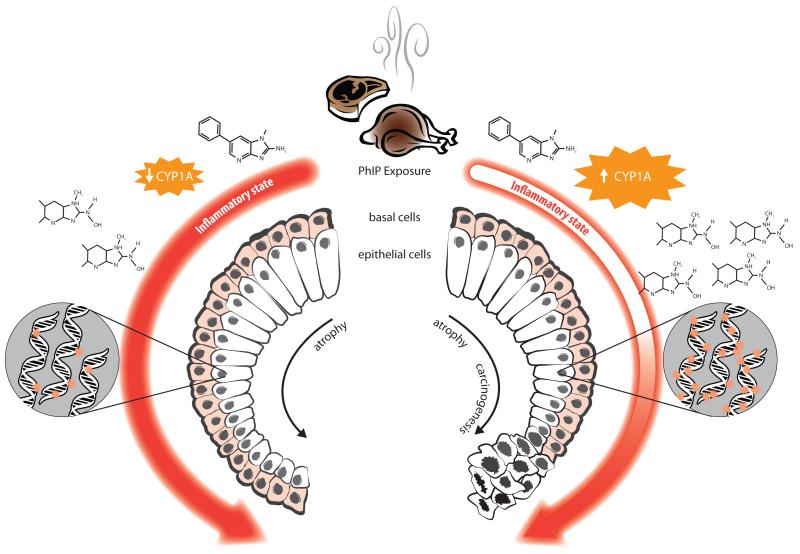
Our findings may be better understood by considering the histological cofactors we found associated with adduct levels. Inflammation and atrophy are hypothesized to be precursors of prostate cancer 43;44. Inflammatory cytokines are known to suppress the activity of CYP1A enzymes45 which is expressed in human prostate tumor and normal cells 46;47; experimental evidence has shown that human prostate cells can activate PhIP and incur subsequent downstream effects such as DNA adduct formation and damage 8;48. We found that adduct levels were higher when partial atrophy was present, and that PhIP-DNA adducts and HGPIN may act synergistically to increase prostate cancer risk in White men. In both races, adduct levels were highest in cases with partial but no simple atrophy and prostate cancer risk was increased only in the presence of both high PhIP-DNA adduct levels and glandular inflammation. Based on these findings we propose the model described in Figure 2 as to how PhIP might increase prostate cancer risk in humans. This model assumes variation in inherited susceptibility to metabolize PhIP and other dietary cofactors. The state of the inflammatory environment within the PhIP-exposed prostate is potentially related to both the amount of PhIP-DNA adducts created and prostate cancer risk.
Figure 2. Proposed model of PhIP metabolism and DNA adduct formation in human prostate in relation to inflammation, atrophy, and cancer risk.
Depending on inherited susceptibility and level of dietary intake, PhIP exposure elicits a variable inflammatory response. A strong response (left side of figure) may dampen the expression of CYP1A enzymes, leading to accumulation of fewer active PhIP metabolites (N2-hydroxy-PhIP), and subsequently lower levels of DNA adducts; a strong inflammatory response would also accelerate atrophy of prostate glandular cells, but not necessarily lead to carcinogenesis. Alternatively, a weak inflammatory response to PhIP exposure (right side of figure) may result in higher CYP1A activity levels, generation of more active PhIP metabolites, and subsequently higher levels of DNA adducts, but the progression of cellular atrophy would not be as rapid. A late elevated inflammatory response coupled with high levels of DNA adducts could incite prostate carcinogenesis.
Our study was observational, and while the duration of at-risk follow-up was equal for cases and controls, cohort members differed in their medical follow-up and screening behavior. Cases had significantly more PSA tests between cohort entry and diagnosis than controls, and the frequency of PSA tests in our study sample was greater than current screening recommendations, even in controls. However, there is no a priori reason why screening behavior should differ by adduct levels and, indeed, adjustment for number of PSA tests during follow-up did not substantively change our results. While we were able to analyze 93% of eligible pairs, included pairs were slightly over-represented by younger cases and newer tissue samples, which tended to show stronger associations between PhIP-DNA adduct levels and prostate cancer risk in Whites. Based on the age range of our cohort and the high prevalence of undiagnosed prostate cancer in older men49, some men in our cohort likely had synchronous prostate cancer that was missed on initial biopsy. One would expect these cohort members misclassified as “disease-free” to be diagnosed sooner50 and bias risk estimates towards the null. Hence risk estimates in men with longer follow-up may be less biased—suggesting that the greater prostate cancer risk associated with high PhIP-DNA adduct levels we observed in White men with four or more years of follow-up is closer to the true risk estimate.
In using a semi-quantitative immunohistochemical assay to measure PhIP-DNA adduct levels, our results are subject to several limitations. The specificity of the antibody we used to detect PhIP-DNA adducts has not been validated against the full range of possible mutagenic heterocyclic amines generated in well-done meats; most notably, we were unable to test whether the antibody could distinguish between PhIP-DNA and 2-amino-3,4,8-dimethylimidazo[4,5-f]quinoxaline (DiMeIQx)-DNA adducts. Despite this, we believe our results remain valid for several reasons. Although several studies using food preparation questionnaire data suggest an increased prostate cancer risk associated with DiMeIQx intake16;17, to our knowledge, laboratory studies have not detected DiMeIQx-DNA adducts in prostate tissue. Furthermore, we have previously reported a dose-response relationship between grilled red meat intake and prostate tissue levels of the same PhIP-DNA adduct antibody used in this study 12 even though DiMeIQx is present only in trace amounts in beef products53. Levels of PhIP in other cooked meats are also generally an order of magnitude greater than levels of DiMeIQx; as a result, DiMeIQx intake is unlikely to confound PhIP exposure measurements 51;52.More sensitive methods of PhIP-DNA adduct detection have been attempted in only two human studies; neither studied prostate tissue, and the percentage of “undetectable” samples varied significantly between them 54;55. Based on previously-reported calibration studies of the PhIP-DNA adduct antibody22, the absorbency measure we used can detect roughly a 100-fold range difference in PhIP-DNA adduct concentration, with a lower limit of detection around 1/107 nucleotides. While such a level might seem unrealistic in humans, a study using mass spectrometry detected PhIP-DNA adducts in lymphocytes at a level of 3 × 108 nucleotides55 Finally, because our study categorized the adduct data, it can be assumed that specimens with adduct levels undetectable by more sensitive methods would be in the lowest category, and serve as the reference group in statistical analyses. In addition, analysis of adduct data as categorical variables defined around the median ensured that measurement inaccuracies at the extremes would not over influence the results.
In summary, the increased risk of prostate cancer conferred by elevated levels of PhIP-DNA adducts in benign prostate tissue appears to be modest and confined to White men. However, the apparent dose-response nature of this relationship and its amplification in men with longer follow-up lends credence to this result. Based on animal models of PhIP-induced carcinogenesis, synergies between elevated adduct levels and pre-neoplastic changes are biologically plausible. Clearly, if PhIP is acting as a prostate carcinogen in humans, significant risk of cancer from PhIP exposure is likely confined to the subset of men with greater capacity to activate PhIP. Further study of determinates of PhIP metabolism in humans, and the role of inflammation in this process, is needed to identify men that may be at greatest risk for PhIP-induced carcinogenesis.
Supplementary Material
Distribution of PhIP-DNA adduct levels in benign prostate by race. Data for African American and White men are in open and shaded bars, respectively.
Time to diagnosis for White cases based on PhIP-DNA adduct quartile level.
Acknowledgements
The authors wish to thank the medical record abstractors and other study personnel that helped with data collection for this study. The authors especially thank Travis Wheeler and Nancy Lemke who processed all prostate specimens used in this study.
Funding: This work was supported by the National Institutes of Health (grant # 5R01-ES011126).
Footnotes
Conflict of interest: The authors have no conflict of interest to disclose.
References
- 1.Felton JS, Knize MG, Shen NH, Lewis PR, Andresen BD, Happe J, Hatch FT. The isolation and identification of a new mutagen from fried ground beef: 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Carcinogenesis. 1986;7:1081–6. doi: 10.1093/carcin/7.7.1081. [DOI] [PubMed] [Google Scholar]
- 2.Shirai T, Sano M, Tamano S, Takahashi S, Hirose M, Futakuchi M, Hasegawa R, Imaida K, Matsumoto K, Wakabayashi K, Sugimura T, Ito N. The prostate: a target for carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) derived from cooked foods. Cancer Res. 1997;57:195–8. [PubMed] [Google Scholar]
- 3.Nakai Y, Nelson WG, De Marzo AM. The Dietary Charred Meat Carcinogen 2-Amino-1-Methyl-6-Phenylimidazo[4,5-b]Pyridine Acts as Both a Tumor Initiator and Promoter in the Rat Ventral Prostate. Cancer Res. 2007;67:1378–84. doi: 10.1158/0008-5472.CAN-06-1336. [DOI] [PubMed] [Google Scholar]
- 4.Shirai T, Kato K, Futakuchi M, Takahashi S, Suzuki S, Imaida K, Asamoto M. Organ differences in the enhancing potential of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine on carcinogenicity in the prostate, colon and pancreas. Mutat Res. 2002;506-507:129–36. doi: 10.1016/s0027-5107(02)00159-8. [DOI] [PubMed] [Google Scholar]
- 5.Cui L, Takahashi S, Tada M, Kato K, Yamada Y, Kohri K, Shirai T. Immunohistochemical detection of carcinogen-DNA adducts in normal human prostate tissues transplanted into the subcutis of athymic nude mice: results with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and 3,2′-dimethyl-4-aminobiphenyl (DMAB) and relation to cytochrome P450s and N-acetyltransferase activity. Jpn J Cancer Res. 2000;91:52–8. doi: 10.1111/j.1349-7006.2000.tb00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G, Wang H, Liu AB, Cheung C, Reuhl KR, Bosland MC, Yang CS. Dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-induced prostate carcinogenesis in CYP1A-humanized mice. Cancer Prev Res. 2012;5:963–72. doi: 10.1158/1940-6207.CAPR-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang CY, Debiec-Rychter M, Schut HA, Morse P, Jones RF, Archer C, King CM, Haas GP. N-Acetyltransferase expression and DNA binding of N-hydroxyheterocyclic amines in human prostate epithelium. Carcinogenesis. 1999;20:1591–5. doi: 10.1093/carcin/20.8.1591. [DOI] [PubMed] [Google Scholar]
- 8.Williams JA, Martin FL, Muir GH, Hewer A, Grover PL, Phillips DH. Metabolic activation of carcinogens and expression of various cytochromes P450 in human prostate tissue. Carcinogenesis. 2000;21:1683–9. doi: 10.1093/carcin/21.9.1683. [DOI] [PubMed] [Google Scholar]
- 9.Malfatti MA, Felton JS. N-glucuronidation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and N_hydroxy-PhIP by specific human UDP-glucuronosyltransferases. Carcinogenesis. 2001;22:1087–93. doi: 10.1093/carcin/22.7.1087. [DOI] [PubMed] [Google Scholar]
- 10.Di Paolo OA, Teitel CH, Nowell S, Coles BF, Kadlubar FF. Expression of cytochromes P450 and glutathione S-transferases in human prostate, and the potential for activation of heterocyclic amine carcinogens via acetyl-coA- Int J Cancer. 2005;117:8–13. doi: 10.1002/ijc.21152. [DOI] [PubMed] [Google Scholar]
- 11.Rybicki BA, Neslund-Dudas C, Bock CH, Nock NL, Rundle A, Jankowski M, Levin AM, Beebe-Dimmer J, Savera AT, Takahashi S, Shirai T, Tang D. Red wine consumption is inversely associated with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-DNA adduct levels in prostate. Cancer Prev Res (Phila) 2011;4:1636–44. doi: 10.1158/1940-6207.CAPR-11-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang D, Liu JJ, Rundle A, Neslund-Dudas C, Savera AT, Bock CH, Nock NL, Yang JJ, Rybicki BA. Grilled meat consumption and PhIP-DNA adducts in prostate carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2007;16:803–8. doi: 10.1158/1055-9965.EPI-06-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang D, Liu JJ, Bock CH, Neslund-Dudas C, Rundle A, Savera AT, Yang JJ, Nock NL, Rybicki BA. Racial differences in clinical and pathological associations with PhIP-DNA adducts in prostate. Int J Cancer. 2007;121:1319–24. doi: 10.1002/ijc.22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kooiman GG, Martin FL, Williams JA, Grover PL, Phillips DH, Muir GH. The influence of dietary and environmental factors on prostate cancer risk. Prostate Cancer Prostatic Dis. 2000;3:256–8. doi: 10.1038/sj.pcan.4500489. [DOI] [PubMed] [Google Scholar]
- 15.El-Zein R, Etzel CJ, Lopez MS, Gu Y, Spitz MR, Strom SS. Human sensitivity to PhIP: A novel marker for prostate cancer risk. Mutat Res. 2006;601:1–10. doi: 10.1016/j.mrfmmm.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Punnen S, Hardin J, Cheng I, Klein EA, Witte JS. Impact of meat consumption, preparation, and mutagens on aggressive prostate cancer. PLoS One. 2011;6:e27711. doi: 10.1371/journal.pone.0027711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Major JM, Cross AJ, Watters JL, Hollenbeck AR, Graubard BI, Sinha R. Patterns of meat intake and risk of prostate cancer among African-Americans in a large prospective study. Cancer Causes Control. 2011;22:1691–8. doi: 10.1007/s10552-011-9845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koutros S, Cross AJ, Sandler DP, Hoppin JA, Ma X, Zheng T, Alavanja MC, Sinha R. Meat and meat mutagens and risk of prostate cancer in the Agricultural Health Study. Cancer Epidemiol Biomarkers Prev. 2008;17:80–7. doi: 10.1158/1055-9965.EPI-07-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John EM, Stern MC, Sinha R, Koo J. Meat consumption, cooking practices, meat mutagens, and risk of prostate cancer. Nutr Cancer. 2011;63:525–37. doi: 10.1080/01635581.2011.539311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander A, Linseisen J, Rohrmann S. Intake of heterocyclic aromatic amines and the risk of prostate cancer in the EPIC-Heidelberg cohort. Cancer Causes Control. 2011;22:109–14. doi: 10.1007/s10552-010-9680-9. [DOI] [PubMed] [Google Scholar]
- 21.Cross AJ, Peters U, Kirsh VA, Andriole GL, Reding D, Hayes RB, Sinha R. A prospective study of meat and meat mutagens and prostate cancer risk. Cancer Res. 2005;65:11779–84. doi: 10.1158/0008-5472.CAN-05-2191. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–7. [PubMed] [Google Scholar]
- 23.Zhu J, Rashid A, Cleary K, Abbruzzese JL, Friess H, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo [4,5-b]-pyridine (PhIP)-DNA adducts in human pancreatic tissues. Biomarkers. 2006;11:319–28. doi: 10.1080/13547500600667911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kryvenko ON, Jankowski M, Chitale DA, Tang D, Rundle A, Trudeau S, Rybicki BA. Inflammation and preneoplastic lesions in benign prostate as risk factors for prostate cancer. Mod Pathol. 2012;25:1023–32. doi: 10.1038/modpathol.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi S, Tamano S, Hirose M, Kimoto N, Ikeda Y, Sakakibara M, Tada M, Kadlubar FF, Ito N, Shirai T. Immunohistochemical demonstration of carcinogen-DNA adducts in tissues of rats given 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP): detection in paraffin-embedded sections and tissue distribution. Cancer Res. 1998;58:4307–13. [PubMed] [Google Scholar]
- 26.Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr., Kadlubar FF. Identification of N-(Deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem Res Toxicol. 1992;5:691–7. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi S, Hasegawa R, Mutai M, Ito N, Ochiai M, Nagao M, Sugimura T. Additive Action of Five Heterocyclic Amines in Terms of Induction of GST-P Positive Single Cells and Foci In Rat Liver-Correlation With DNA Adduct Formation. Journal of Toxicologic Pathology. 1994;7:423–8. [Google Scholar]
- 28.Bogen KT, Keating GA. U.S. dietary exposures to heterocyclic amines. J Expo Anal Environ Epidemiol. 2001;11:155–68. doi: 10.1038/sj.jea.7500158. [DOI] [PubMed] [Google Scholar]
- 29.Nowell S, Ratnasinghe DL, Ambrosone CB, Williams S, Teague-Ross T, Trimble L, Runnels G, Carrol A, Green B, Stone A, Johnson D, Greene G, et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2004;13:270–6. doi: 10.1158/1055-9965.epi-03-0047. [DOI] [PubMed] [Google Scholar]
- 30.Kidd LC, Stillwell WG, Yu MC, Wishnok JS, Skipper PL, Ross RK, Henderson BE, Tannenbaum SR. Urinary excretion of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in White, African-American, and Asian-American men in Los Angeles County. Cancer Epidemiol Biomarkers Prev. 1999;8:439–45. [PubMed] [Google Scholar]
- 31.Kriek E, Rojas M, Alexandrov K, Bartsch H. Polycyclic aromatic hydrocarbon-DNA adducts in humans: relevance as biomarkers for exposure and cancer risk. Mutat Res. 1998;400:215–31. doi: 10.1016/s0027-5107(98)00065-7. [DOI] [PubMed] [Google Scholar]
- 32.Gammon MD, Santella RM, Neugut AI, Eng SM, Teitelbaum SL, Paykin A, Levin B, Terry MB, Young TL, Wang LW, Wang Q, Britton JA, et al. Environmental toxins and breast cancer on Long Island. I. Polycyclic aromatic hydrocarbon DNA adducts. Cancer Epidemiol Biomarkers Prev. 2002;11:677–85. [PubMed] [Google Scholar]
- 33.van Schooten FJ, Hillebrand MJ, van Leeuwen FE, Van ZN, Jansen HM, den Engelse L, Kriek E. Polycyclic aromatic hydrocarbon--DNA adducts in white blood cells from lung cancer patients: no correlation with adduct levels in lung. Carcinogenesis. 1992;13:987–93. doi: 10.1093/carcin/13.6.987. [DOI] [PubMed] [Google Scholar]
- 34.Wiencke JK, Kelsey KT, Varkonyi A, Semey K, Wain JC, Mark E, Christiani DC. Correlation of DNA adducts in blood mononuclear cells with tobacco carcinogen-induced damage in human lung. Cancer Res. 1995;55:4910–4. [PubMed] [Google Scholar]
- 35.Sinha R. An epidemiologic approach to studying heterocyclic amines. Mutat Res. 2002;506-507:197–204. doi: 10.1016/s0027-5107(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 36.Norrish AE, Ferguson LR, Knize MG, Felton JS, Sharpe SJ, Jackson RT. Heterocyclic amine content of cooked meat and risk of prostate cancer. J Natl Cancer Inst. 1999;91:2038–44. doi: 10.1093/jnci/91.23.2038. [DOI] [PubMed] [Google Scholar]
- 37.Relling MV, Lin JS, Ayers GD, Evans WE. Racial and gender differences in N-acetyltransferase, xanthine oxidase, and CYP1A2 activities. Clin Pharmacol Ther. 1992;52:643–58. doi: 10.1038/clpt.1992.203. [DOI] [PubMed] [Google Scholar]
- 38.Malfatti MA, Felton JS. Human UDP-glucuronosyltransferase 1A1 is the primary enzyme responsible for the N-glucuronidation of N-hydroxy-PhIP in vitro. Chem Res Toxicol. 2004;17:1137–44. doi: 10.1021/tx049898m. [DOI] [PubMed] [Google Scholar]
- 39.Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology. 2005;42:448–57. doi: 10.1002/hep.20770. [DOI] [PubMed] [Google Scholar]
- 40.Girard H, Butler LM, Villeneuve L, Millikan RC, Sinha R, Sandler RS, Guillemette C. UGT1A1 and UGT1A9 functional variants, meat intake, and colon cancer, among Caucasians and African-Americans. Mutat Res. 2008;644:56–63. doi: 10.1016/j.mrfmmm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knize MG, Kulp KS, Salmon CP, Keating GA, Felton JS. Factors affecting human heterocyclic amine intake and the metabolism of PhIP. Mutat Res. 2002;506:507–153. doi: 10.1016/s0027-5107(02)00162-8. [DOI] [PubMed] [Google Scholar]
- 42.Malfatti MA, Dingley KH, Nowell-Kadlubar S, Ubick EA, Mulakken N, Nelson D, Lang NP, Felton JS, Turteltaub KW. The urinary metabolite profile of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine is predictive of colon DNA adducts after a low-dose exposure in humans. Cancer Res. 2006;66:10541–7. doi: 10.1158/0008-5472.CAN-06-1573. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Bergh A, Damber JE. Morphological transition of proliferative inflammatory atrophy to high-grade intraepithelial neoplasia and cancer in human prostate. Prostate. 2009;69:1378–86. doi: 10.1002/pros.20992. [DOI] [PubMed] [Google Scholar]
- 44.Sfanos KS, De Marzo AM. Prostate cancer and inflammation: the evidence. Histopathology. 2012;60:199–215. doi: 10.1111/j.1365-2559.2011.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdel-Razzak Z, Loyer P, Fautrel A, Gautier JC, Corcos L, Turlin B, Beaune P, Guillouzo A. Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Mol Pharmacol. 1993;44:707–15. [PubMed] [Google Scholar]
- 46.Sterling KM, Jr., Cutroneo KR. Constitutive and inducible expression of cytochromes P4501A (CYP1A1 and CYP1A2) in normal prostate and prostate cancer cells. J Cell Biochem. 2004;91:423–9. doi: 10.1002/jcb.10753. [DOI] [PubMed] [Google Scholar]
- 47.Murray GI, Taylor VE, McKay JA, Weaver RJ, Ewen SW, Melvin WT, Burke MD. The immunohistochemical localization of drug-metabolizing enzymes in prostate cancer. J Pathol. 1995;177:147–52. doi: 10.1002/path.1711770208. [DOI] [PubMed] [Google Scholar]
- 48.Martin FL, Cole KJ, Muir GH, Kooiman GG, Williams JA, Sherwood RA, Grover PL, Phillips DH. Primary cultures of prostate cells and their ability to activate carcinogens. Prostate Cancer Prostatic Dis. 2002;5:96–104. doi: 10.1038/sj.pcan.4500579. [DOI] [PubMed] [Google Scholar]
- 49.Sakr WA, Grignon DJ, Crissman JD, Heilbrun LK, Cassin BJ, Pontes JJ, Haas GP. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20-69: an autopsy study of 249 cases. In Vivo. 1994;8:439–43. [PubMed] [Google Scholar]
- 50.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. 2009;69:1470–9. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keating GA, Bogen KT. Estimates of heterocyclic amine intake in the US population. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:127–33. doi: 10.1016/j.jchromb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 52.Keating GA, Bogen KT. Methods for estimating heterocyclic amine concentrations in cooked meats in the US diet. Food Chem Toxicol. 2001;39:29–43. doi: 10.1016/s0278-6915(00)00115-0. [DOI] [PubMed] [Google Scholar]
- 53.Sinha R, Rothman N, Salmon CP, Knize MG, Brown ED, Swanson CA, Rhodes D, Rossi S, Felton JS, Levander OA. Heterocyclic amine content in beef cooked by different methods to varying degrees of doneness and gravy made from meat drippings. Food Chem Toxicol. 1998;36:279–87. doi: 10.1016/s0278-6915(97)00162-2. [DOI] [PubMed] [Google Scholar]
- 54.Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 4-aminobiphenyl are infrequently detected in human mammary tissue by liquid chromatography/tandem mass spectrometry. Carcinogenesis. 2012;33:124–30. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magagnotti C, Pastorelli R, Pozzi S, Andreoni B, Fanelli R, Airoldi L. Genetic polymorphisms and modulation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-DNA adducts in human lymphocytes. Int J Cancer. 2003;107:878–84. doi: 10.1002/ijc.11492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of PhIP-DNA adduct levels in benign prostate by race. Data for African American and White men are in open and shaded bars, respectively.
Time to diagnosis for White cases based on PhIP-DNA adduct quartile level.