Abstract
The past decade has produced an avalanche of experimental data on the structure and dynamics of the ribosome. Groundbreaking studies in structural biology and kinetics have placed important constraints on ribosome structural dynamics. However, a gulf remains between static structures and time dependent data. In particular, x-ray crystallography and cryo-EM studies produce static models of the ribosome in various states, but lack dynamic information. Single molecule studies produce information on the rates of transitions between these states but do not have high-resolution spatial information. Computational studies have aided in bridging this gap by providing atomic resolution simulations of structural fluctuations and transitions between configurations.
Introduction
Early computational efforts focused on structural models of the ribosome in the classical state, before high-resolution x-ray crystallography structures were available [1–6]. New RNA homology methods produced predictive models of the E. coli 30S ribosomal subunit and the T. thermophilus 70S ribosome that were highly consistent with cryo-EM data and used for phasing x-ray crystals [7,8]. After high-resolution x-ray structures of individual subunits were solved, it was then possible to perform short timescale molecular dynamics simulations of important functional regions [9,10] as well as normal mode analysis [11]* and Poisson-Boltzmann analysis on the intact ribosome[12]. As the speed of supercomputers increased, molecular dynamics simulations of the intact ribosome were possible [13]**, along with exhaustive sampling simulations of functional regions [14]. New reduced-model techniques and even faster supercomputers are paving the way to exhaustive-sampling molecular simulations of the intact ribosome [15]**. Ultimately, one would like to obtain the equilibrium free energy landscape of the ribosome complexed with its various ligands.
Due to the large size (Fig. 1) of the ribosome system (~3 million atoms for simulations with explicit water molecules) and formidable time scales involved (typical functional rates measured in vitro are hundreds of milliseconds), computational studies of the ribosome use various approximations, making tradeoffs between force field accuracy and the time scale sampled. As often occurs in molecular simulations, high levels of detail in the force field are typically attained at the cost of the time scale sampled, and in turn, accuracy of the entropic contribution to the free energy. Specifically, explicit calculations of the equilibrium free energy may be obtained by the expression -kT log P(Q), where P is the probability of sampling order parameter value Q for a suitable order parameter. Computing the free energy requires exhaustive sampling of configurational space. Thus, deficiencies in sampling produce inaccurate free energies, specifically in the entropic contribution to the free energy. In the case of the ribosome, it is difficult to obtain exhaustive sampling due to its large size. Enhanced sampling methods that attempt to produce exhaustive sampling must either (1) reduce the level of detail in the force field or (2) simulate portions of the ribosome.
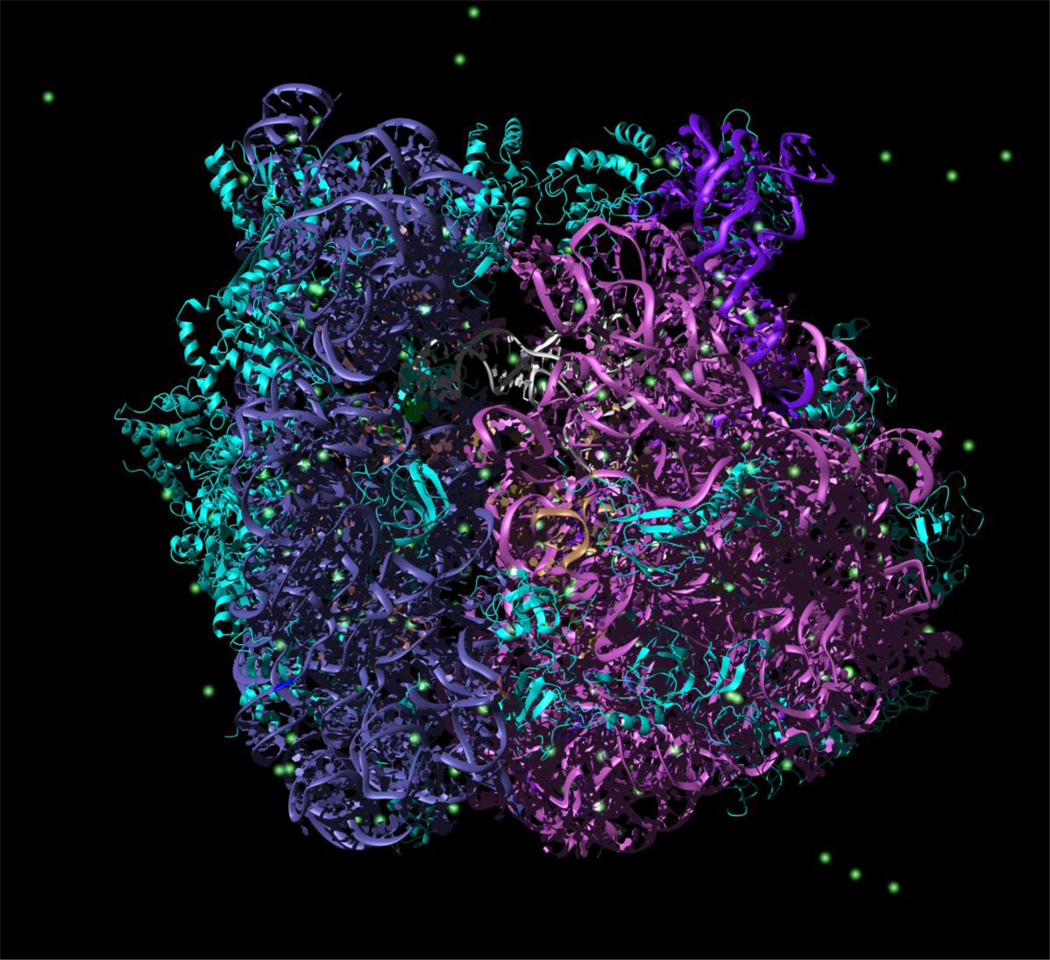
Figure 1.
Snapshot from explicit solvent simulation of the 70S ribosomal complex. Left, small subunit (30S). Right, large subunit (50S). Cyan, ribosomal protein. Steel blue, small subunit ribosomal RNA (16S rRNA). Magenta, large subunit ribosomal RNA (23S rRNA). Dark purple, 5S rRNA. Green, magnesium ions. White/grey, P-site tRNA.
While approximations must be made, each computational technique used to study the ribosome is quite useful in its own way and provides a new window into ribosome dynamics that was previously unexplored experimentally. There are currently no experimental techniques available capable of studying ribosome dynamics in full atomic detail. In this review, we summarize many of the various computational techniques used to study ribosome dynamics over the past decade. In accordance with journal guidelines, we pay particular attention to publications within the previous 2 years. While the review is by no means completely comprehensive, it serves as a starting point for future treatments of ribosome simulation.
Computational studies of intact ribosomes
Normal mode analysis was used to determine the possible overall global motions of the ribosome at nucleotide resolution [11,16,17]. Specifically, these calculations produced ratchet-like pivoting of the small subunit relative to the large subunit, along with motion of the head and large subunit stalks. This technique has also been incorporated into the fitting of cryo-EM reconstructions with success (see below) [18,19]. Recent calculations have examined the coupled motions of tRNA and mobile domains of the ribosome showing that A- and P- site tRNAs, as well as the L1 protein, effect E-tRNA dynamics[20]. Electrostatic calculations have determined charged regions on the ribosome [12]. Detailed analysis has shown the surface of the ribosome to be mostly negative with scattered positively charged regions corresponding to ribosome proteins.
Explicit solvent molecular dynamics simulations have examined large-scale conformational changes of the ribosomal complex (Fig. 2). The most tractable conformational change during elongation is the movement (“accommodation”) of tRNA from its partially bound state (A/T) to its fully bound state (A/A) that occurs during the process of tRNA selection (also known as decoding). This conformational change is limited largely to the tRNA and the base of the L7/L12 stalk. Targeted simulations correctly predicted the regions of the large subunit that interact with the tRNA during this conformational change (“accommodation corridor”) [13]. These predictions have been verified by several recent experimental studies. Dinman and co-workers performed snoRNA deletions in yeast, producing ribosomes with unmodified nucleotides in the accommodation corridor[21,22]. The resulting ribosomes displayed fidelity phenotypes including defects in -1 frameshifting, peptide bond formation and misreading [22]. Recent mutational studies by the same group demonstrated that mutations of the predicted three-dimensional gate (A2556, 2492 and 2573) are viable for normal ribosomes, but dramatically effect growth when treated by antimicrobials [23]. These three nucleotides are considered “promising targets for antimicrobial therapy” [23]. Most recently, the Dontsova lab has also performed mutations on the same three nucleotides [24,25]. This group found that two of the nucleotides impair peptide release. The Dontsova and Rodnina labs found that mutations of the third nucleotide are lethal and act to block peptide bond formation, presumably by impeding tRNA accommodation into the fully bound (A/A) conformation. Recent molecular dynamics simulations of the intact ribosome have also examined short time scale dynamics of the L1 stalk (60 ns) and the ribosome exit tunnel in the large subunit of the ribosome [26–28]. The L1 stalk investigations examined interactions with initiator and elongator tRNAs in addition to producing all-atom fits of cryo-EM reconstructions. Interestingly, in studies of an isolated L1 stalk / tRNA subsystem, the simulations of the L1 stalk using the CHARMM 27 force field were found to be unstable while simulations using the AMBER99 force field were stable. Studies of the ribosome exit tunnel simulated 50 ns of physiological time and examined the ribosome-Sec Y channel interaction. A second study of the tunnel included approximately 5% (125,000 atoms) of the full ribosome system, achieved 2.1 microseconds of sampling and characterized interactions with nascent peptide chains.
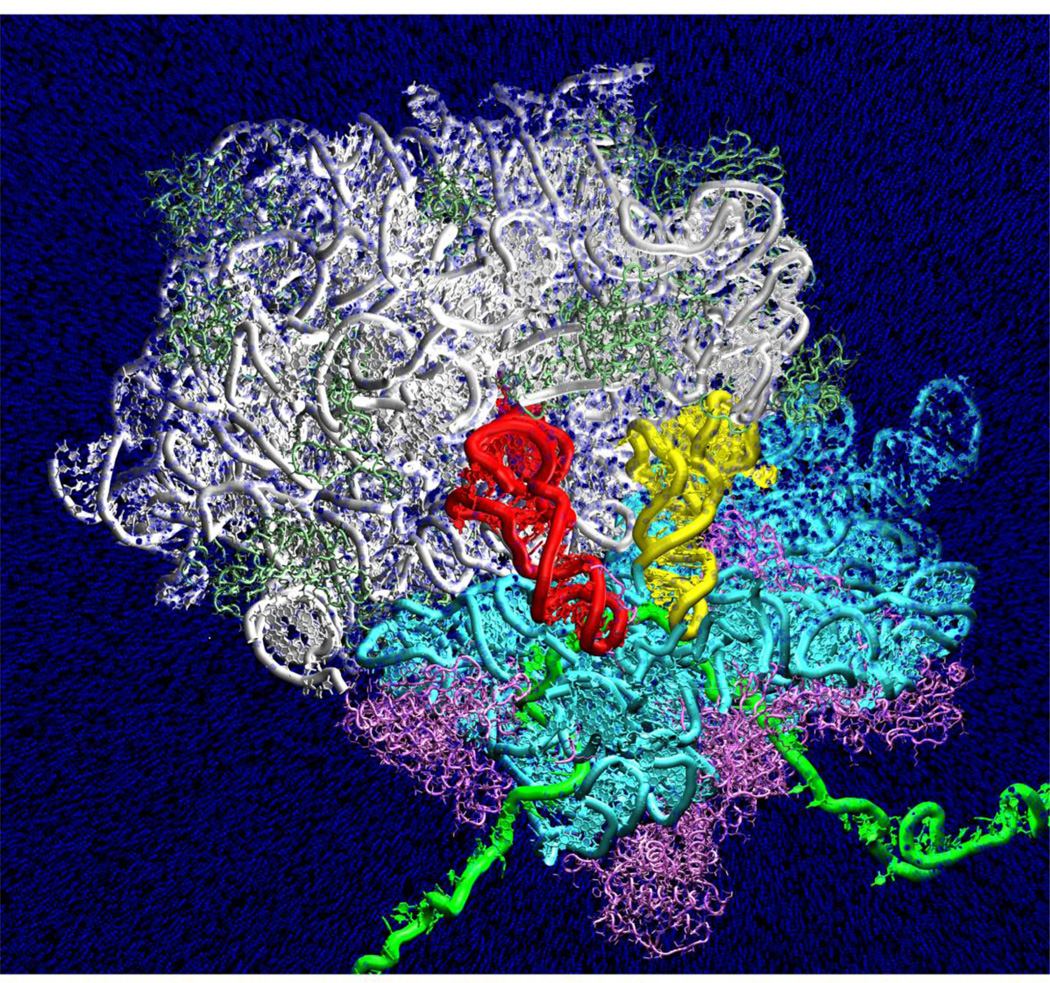
Figure 2.
Explicit solvent molecular dynamics simulation of aminoacyl-tRNA (yellow) accommodation into the ribosome (from Sanbonmatsu et al. J Struct. Biol. 2007). Every tenth water represented by blue sphere. Red, peptidyl-tRNA. Green, mRNA. Cyan, 16S rRNA. White, 23S rRNA. Magenta, small subunit ribosomal protein. Light green, large subunit ribosomal protein. Upper region of the simulation box (including 30S head) is clipped for the purposes of visualization to reveal positions of the tRNAs.
Most recently, explicit solvent simulations of the full 70S ribosome have connected measured kinetic rates of accommodation to free energy barriers[29]*. These simulations of the intact ribosome (3.2 million atoms) achieved aggregate sampling of 2.1 microseconds and measured the diffusion of tRNA inside the ribosome. The study produced an estimate of the pre-factor in the Arrhenius equation for this process. The end result of these calculations is a table converting measured rates to estimated free energy barriers.
Coarse-grained and reduced description molecular dynamics simulations have explored larger regions of configurational space and longer time scales. The first coarse-grained molecular dynamics study of the entire ribosome on a half a microsecond time scale accounted for large-scale motions by implementing a Morse potential function, allowing for breaking of interactions in the MD simulations [30]**. Here, large-scale collective motions such as the rotation of the subunits were produced. In addition, the anti-correlated movement of the ribosomal stalks positioned on the opposite sides of the large subunit was observed. Studies performed by Tryskla et al. also enabled the investigation of large-scale conformational changes coupled with electrostatic effects[31]. Tryskla and coworkers have also applied the Poisson-Boltzmann electrostatics model to the ribosome to determine the computational assembly map of the 30S subunit[32]. Recent studies have also used new coarse graining schemes designed for even longer sampling [33].
Finally, recent all-atom reduced model simulations based on a structure-based potential have enabled systematic studies of spontaneous large-scale conformational changes with atomistic detail (Fig. 3) [15]. Here, reversible excursions of tRNAs into and out of ribosomal binding sites were observed, consistent with recent single molecule FRET experiments [69]. These simulations produced sampling on the order of ~200 milliseconds for the ribosomal complex in atomistic detail. The simulations suggest that helix 89 plays a critical role in monitoring tRNA accommodation. In addition, the high entropy of the tRNA 3’-CCA end appears to present an entropic barrier for accommodation.
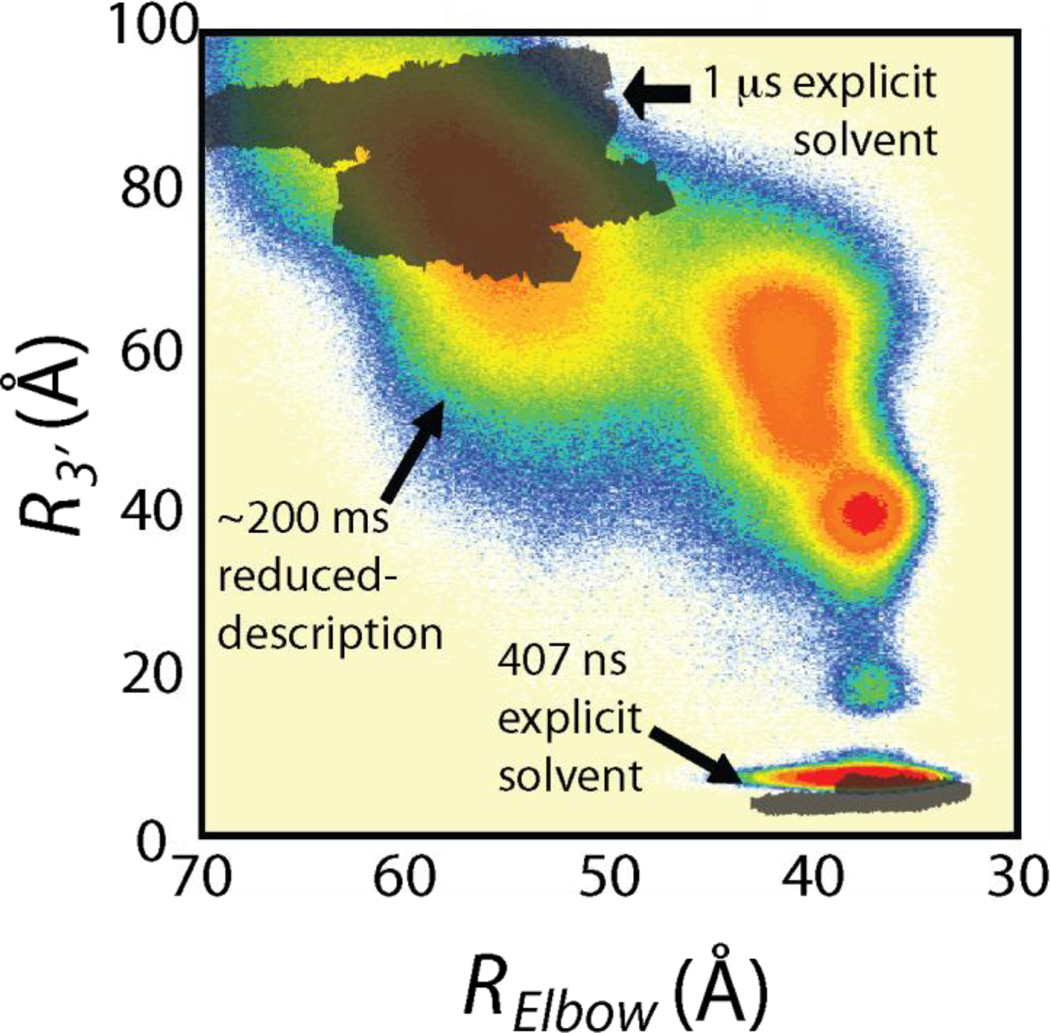
Figure 3.
Comparison of explicit solvent and all-atom structure-based simulations for tRNA accommodation into the ribosome. a) Comparison of spontaneous fluctuations in explicit solvent molecular dynamics simulation (grey, ~1.4 microsecond total sampling) and structure-based simulations (contours in color, ~200 milliseconds total sampling) displays overlap in sampling of A/T basin (upper left) and A/A basin (lower right).
Computational studies of functional regions of the ribosome
A second class of ribosome simulations focuses on localized regions of the ribosome that are critical for protein synthesis. Several quantum mechanical studies of the peptidyl transferase center have explored reaction mechanisms [34,35]. Warshel and co-workers found that solvation effects are significant and that while the 2’- hydroxyl group of tRNA A76 significantly accelerates the reaction rate, it does not directly catalyze the reaction. They concluded that substrate-assisted catalysis was unlikely[35]*. More recently, this group has examined the mechanism of GTP hydrolysis by EF-Tu during tRNA selection. They conclude that rather than behaving as a general base, the critical residue H84 contributes to an allosteric effect [36]. We note that Wieden and co-workers have performed extensive studies on the mechanism of EF-Tu, including one very interesting investigation that combines molecular dynamics simulations with rapid kinetics studies [37].
Aqvist and co-workers have also examined the mechanism of the peptidyl transferase reaction, combing an empirical valence bond description with molecular dynamics simulations. They also conclude that an acid-base catalysis method is unlikely. They favor a proton-shuttling mechanism that does include the P-site adenine O2' oxygen [34]. More recently the group has employed high-level quantum calculations to reveal a double proton shuttling mechanism, where water molecules play a key role[38]. Aqvist and co-workers have applied quantum chemical methods to the mechanism of termination[39]. They conclude that a methylated glutamine residue in the ribosome release factor makes an entropic contribution to lowering the free energy barrier.
Early (2003) short time scale simulations (20 ns sampling) of the decoding center complexed with mRNA and tRNA anticodon stem loops have examined the stability of the decoding center hydrogen bond network for a wide range of cognate, near-cognate and non-cognate mRNA-tRNA combinations [9]. In the simulations, ribosomal RNA had a stabilizing effect on cognate codon-anticodon interactions and a destabilizing effect on certain non-cognate combinations. Recent studies by Aqvist and co-workers have revisited this study with a slightly longer aggregate sampling of the A-site subsystem. Here, they attempt to calculate binding free energies using a set of nine simulations per system, with 1 ns of production time for each simulation. The total amount of sampling for the study was 54 ns of sampling for bound states and 27 ns of sampling for free states[40]. On this time scale, they observe that the Leu2 anticodon stemloop is more stable that the Ser stem loop when complexed with the Phe UUU codon. Aqvist and co-workers have applied a similar method to study stop-codon recognition, where they use 51 ns of sampling per system to calculate binding free energies[38].
A variety of studies have investigated the dynamics of the decoding center helix itself [14,41–43]. Explicit solvent simulations of the decoding center complexed with antibiotics have examined the stability of the drug-ribosome interactions (Fig. 4) [14]. Exhaustive sampling simulations (replica exchange molecular dynamics simulations) of the decoding center show a high degree of convergence after 0.5 microseconds in the absence of antibiotics and 12 microseconds (12,000 ns) in the presence of gentamicin. These simulations show the decoding center bases (A1492 and A1493) flipping in and out of helix 44 in absence of ligand [14,41]. In the presence of gentamicin, this equilibrium is shifted towards the flipped out state. Flipping time scales are consistent with 2-aminopurine fluorescence studies of the decoding helix[44]. We note that these studies do not include interactions with the large subunit, which could act to stabilize A1492 and A1493. The studies produced the first energy landscapes of the ribosome, and suggest that an entropy shuttling mechanism may move the drug from local minimum to local minimum until the binding site is reached.
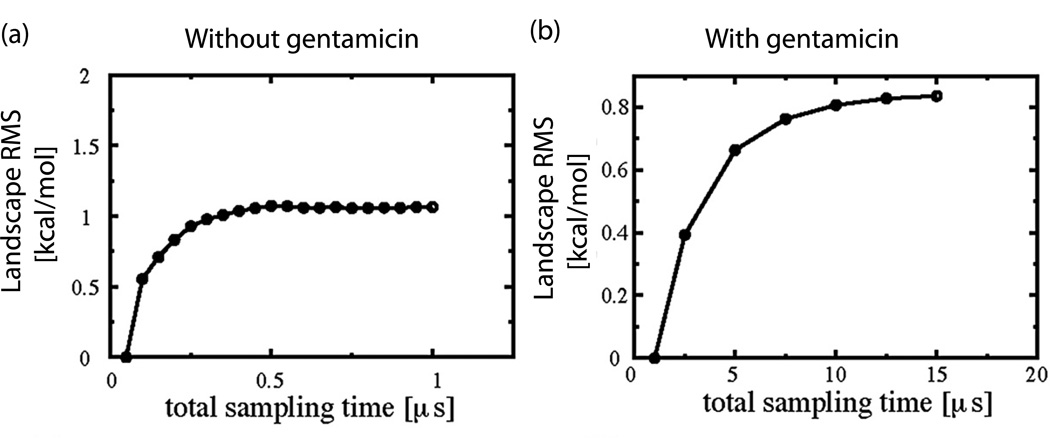
Figure 4.
Convergence studies of explicit solvent replica exchange molecular dynamics simulations (REMD) of the ribosomal decoding center, with and without the antibiotic Gentamicin. (a)-(b) Deviation (“Landscape RMS”) of two-dimensional potential of mean force landscape obtained after time t from those obtained at time t0. Deviation represents the point-wise RMSD between two free energy landscapes. Values of the deviation approach a plateau, suggesting convergence after ~0.5 microseconds without Gentamicin (a) and ~12 microseconds with Gentamicin (b).
Tryskla and co-workers performed explicit solvent molecular dynamics simulations of the decoding center, introducing various mutations to analyze how nucleotide substitutions change its physicochemical properties. They found differences in the internal mobility of the A-site, as well as in ion and water density distributions inside the binding cleft, between the prokaryotic and mutated RNA [42].
In a very interesting study, Tryskla and co-workers used Brownian dynamics simulations to examine antibiotic binding to the 30S subunit, showing that electrostatic steering is not the sole factor directing the aminoglycoside antibiotic toward the binding site on the 30S ribosomal subunit. The simulations explained helped elucidate the mechanism by which paromomycin overstabilizes the 70S ribosome complex. This study unveiled other possible binding clefts in the ribosomal RNA of the 30S subunit and explained the physical mechanisms of aminoglycoside diffusion and binding to the ribosomal RNA [45]*.
Sponer and co-workers have performed explicit solvent simulations of three-way junctions present in the ribosome, demonstrating that the junctions can act as anisotropic flexible elements contributing to functional fluctuations of the ribosome, including coupled dynamics between 5S rRNA, the A-site finger, and L7/L12 stalk region [46]*. Sponer, Frank and co-workers have also performed highly detailed studies of the A-site finger [47]. The team examines dynamics of H38 for several species and suggest that the kink-turn is responsible for large-scale motions of H38. Several groups have used reduced-description molecular simulations as well as explicit solvent molecular dynamics simulations to study peptide folding and dynamics within the exit tunnel of the large subunit of the ribosome [48–53]. Finally, a number of studies of localized ribosomal regions have focused on drug-ribosome interactions [14,42,43,54].
Cryo-EM fitting
Tama and co-workers have pioneered the use of simulation in fitting cryo-EM reconstructions of the ribosome. At the core of their method is the use of a correlation term that determines the quality of the fit. Here, a simulated EM map is produced from a structural model and correlated with the experimentally determined cryo-EM map. The correlation term is used in the potential itself to iteratively converge on a solution. They first included this in normal mode analysis, achieving close fits to the cryo-EM data [18]. Next, they used explicit solvent molecular dynamics simulations with a high degree of success [55]. Schulten and coworkers have also applied molecular dynamics flexible fitting to the ribosome with great success [56,57]. Recently, a technique using a reduced representation force field, MDfit, allowed molecular dynamics fitting to be performed on a desktop computer, which is available at mdfit.lanl.gov [58,59]. This so-called ‘1-step’ fitting uses a potential based on the x-ray structure and finds a conformation with a high correlation to cryo-EM density while maintaining the stereochemistry present in the x-ray structure. In a very interesting use of explicit solvent molecular dynamics simulation, Frank and co-workers performed simulations of the tRNA in solution and selected conformations with very close correlations to their tRNA cryo-EM density inside the ribosome [60]*. Caulfield and co-workers used a combination of biased and unbiased simulations to achieve atomistic fits of cryo-EM data. Their method uses a conformational selection technique akin to Monte Carlo called Maxwell’s Demon molecular dynamics[61]. Tama and co-workers have recently compared the various methods, demonstrating that a consensus between several fits is a better measure of the accuracy of the method[62].* Although not cryo-EM fitting, we note that Noller and co-workers have performed a very interesting computational study on ribosome dynamics, where they use translation-libration-screw (TLS) constraints to refine atomistic models based on x-ray diffraction data [63]. Here, they examine the mobilities of the peptidyl transferase center and the Shine-Dalgarno helix, as well as the 30S rotation relative to the 50S subunit. The group concludes that regions of the ribosome near the Shine-Dalgarno helix are stabilized by the Shine-Dalgarno helix, suggesting that the Shine-Dalgarno helix may help stabilize the 30S subunit during initiation.
Conclusions
Outstanding issues and future outlook
Significant progress has been made over the past decade in using computational methods to bridge the gap between detailed static structures[64–67] of the ribosome and measurements of ribosome dynamics [68–71] with low spatial resolution. Explicit solvent simulations of ribosomal conformal changes have been performed that have been validated experimentally [13,23–25]. Reduced-model simulations of the entire ribosome in atomic detail have reached time scales on the order of ~200 milliseconds [15]. In addition, enhanced sampling simulations in explicit solvent of localized regions have revealed the energy landscape for local conformational changes [14,41]. The next step is to produce free energy landscapes of global conformational changes of the ribosomal complex[72]. We estimate that this will require approximately one second of sampling for each barrier. This level of sampling may be reached with all-atom reduced-model simulations using supercomputers in the near future. However, achieving this level of sampling in explicit solvent will require significant advances in compute power. While initial attempts at determining the free energy landscape will certainly be useful, we caution the reader that aggregate sampling time scales of 100 ms – 1 s may be necessary to produce accurate entropic components of the free energy for the entire ribosome. When merged together with kinetic experiments, however, one can estimate free energy barriers with relatively short simulations (~1 microsecond) [29]. Another frontier lies at the level of systems biology, where computer simulations are used to examine the overall kinetics and energy balance for the various substeps of protein synthesis. While only a handful of studies of this nature have been performed, this area may grow quite rapidly in the coming years [73,74].
Highlights.
It is not possible to study ribosome dynamics at atomic resolution experimentally
Computation provides a window into dynamics that is unexplored experimentally
Computational studies of the ribosome have been tested and validated experimentally
Supercomputers produced microsecond explicit solvent simulations (3 million atoms)
All-atom reduced simulations of large, spontaneous conformational changes have been performed with 200 ms sampling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brimacombe R, Atmadja J, Stiege W, Schuler D. A detailed model of the three-dimensional structure of escherichia coli 16 s ribosomal rna in situ in the 30 s subunit. Journal of molecular biology. 1988;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra A, Tan RK, Harvey SC. Prediction of the three-dimensional structure of escherichia coli 30s ribosomal subunit: A molecular mechanics approach. Proc Natl Acad Sci U S A. 1990;87(5):1950–1954. doi: 10.1073/pnas.87.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easterwood TR, Major F, Malhotra A, Harvey SC. Orientations of transfer rna in the ribosomal a and p sites. Nucleic Acids Res. 1994;22(18):3779–3786. doi: 10.1093/nar/22.18.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malhotra A, Harvey SC. A quantitative model of the escherichia coli 16 s rna in the 30 s ribosomal subunit. J Mol Biol. 1994;240(4):308–340. doi: 10.1006/jmbi.1994.1448. [DOI] [PubMed] [Google Scholar]
- 5.VanLoock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC. Movement of the decoding region of the 16 s ribosomal rna accompanies trna translocation. J Mol Biol. 2000;304(4):507–515. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- 6.Fink DL, Chen RO, Noller HF, Altman RB. Computational methods for defining the allowed conformational space of 16s rrna based on chemical footprinting data. RNA. 1996;2(9):851–866. [PMC free article] [PubMed] [Google Scholar]
- 7.Tung CS, Joseph S, Sanbonmatsu KY. All-atom homology model of the escherichia coli 30s ribosomal subunit. Nat Struct Biol. 2002;9(10):750–755. doi: 10.1038/nsb841. [DOI] [PubMed] [Google Scholar]
- 8.Tung CS, Sanbonmatsu KY. Atomic model of the thermus thermophilus 70s ribosome developed in silico. Biophys J. 2004;87(4):2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanbonmatsu KY, Joseph S. Understanding discrimination by the ribosome: Stability testing and groove measurement of codonanticodon pairs. J Mol Biol. 2003;328(1):33–47. doi: 10.1016/s0022-2836(03)00236-5. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Ma B, Shapiro B. Binding interactions between the core central domain of 16s rrna and the ribosomal protein s15 determined by molecular dynamics simulations. NUCLEIC ACIDS RESEARCH. 2003;31(2):629–638. doi: 10.1093/nar/gkg149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chacon P, Tama F, Wriggers W. Mega-dalton biomolecular motion captured from electron microscopy reconstructions. J Mol Biol. 2003;326(2):485–492. doi: 10.1016/s0022-2836(02)01426-2. This first treatment of the ribosome using normal mode analysis displayed experimentally observed overall global motions of the ribosomal complex including 30S rotation relative to 50S subunit and stalk movement.
- 12.Baker N, Sept D, Joseph S, Holst M, McCammon J. Electrostatics of nanosystems: Application to microtubules and the ribosome. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of trna into the ribosome during decoding. Proc Natl Acad Sci U S A. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. Proc Natl Acad Sci U S A (2005) 102 (44):15854–15859. The first explicit solvent molecular dynamics simulation of the ribosome remains one of the largest systems simulated to date in computational biology. The study defined the accommodation corridor and has been validated experimentally by two independent groups.
- 14.Vaiana AC, Sanbonmatsu KY. Stochastic gating and drug-ribosome interactions. J Mol Biol. 2009;386(3):648–661. doi: 10.1016/j.jmb.2008.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY. Accommodation of aminoacyl-trna into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16(6):1196–1204. doi: 10.1261/rna.2035410. RNA (2010) 16:1196–1204. The study simulated spontaneous largescale conformational changes of the ribosome (accommodation events) with ~200 milliseconds total sampling using a reduced-representation potential. Reversible accommodation events were consistent with single molecule FRET experiments.
- 16.Tama F, Valle M, Frank J, Brooks CL., 3rd Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci U S A. 2003;100(16):9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Rader AJ, Bahar I, Jernigan RL. Global ribosome motions revealed with elastic network model. J Struct Biol. 2004;147(3):302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Gorba C, Miyashita O, Tama F. Normal-mode flexible fitting of highresolution structure of biological molecules toward one-dimensional low-resolution data. Biophys J. 2008;94(5):1589–1599. doi: 10.1529/biophysj.107.122218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, 3rd, Ban N, Frank J. Structure of the e. Coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438(7066):318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurkcuoglu O, Doruker P, Sen TZ, Kloczkowski A, Jernigan RL. The ribosome structure controls and directs mrna entry, translocation and exit dynamics. Phys Biol. 2008;5(4):046005. doi: 10.1088/1478-3975/5/4/046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rrna base modification. PLoS One. 2007;2(1):e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meskauskas A, Dinman JD. Ribosomal protein l3: Gatekeeper to the a site. Mol Cell. 2007;25(6):877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakauskaite R, Dinman JD. Mutations of highly conserved bases in the peptidyltransferase center induce compensatory rearrangements in yeast ribosomes. RNA. 2011;17(5):855–864. doi: 10.1261/rna.2593211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burakovsky DE, Sergiev PV, Steblyanko MA, Konevega AL, Bogdanov AA, Dontsova OA. The structure of helix 89 of 23s rrna is important for peptidyl transferase function of escherichia coli ribosome. FEBS letters. 2011;585(19):3073–3078. doi: 10.1016/j.febslet.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 25.Burakovsky DE, Sergiev PV, Steblyanko MA, Kubarenko AV, Konevega AL, Bogdanov AA, Rodnina MV, Dontsova OA. Mutations at the accommodation gate of the ribosome impair rf2-dependent translation termination. RNA. 2010;16(9):1848–1853. doi: 10.1261/rna.2185710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. The role of l1 stalk-trna interaction in the ribosome elongation cycle. Journal of molecular biology. 2010;402(4):741–760. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trabuco LG, Harrison CB, Schreiner E, Schulten K. Recognition of the regulatory nascent chain tnac by the ribosome. Structure. 2010;18(5):627–637. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17(11):1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitford PC, Onuchic JN, Sanbonmatsu KY. Connecting energy landscapes with experimental rates for aminoacyl-trna accommodation in the ribosome. J Am Chem Soc. 2010;132(38):13170–13171. doi: 10.1021/ja1061399. J Am Chem Soc (2010) 132:13170–13171. Microsecond sampling of the ribosome in explicit solvent allowed the calculation of tRNA diffusion within the ribosome. These diffusion coefficients were used to construct a table connecting experimentally measured kinetic rates to free energy barrier heights.
- 30. Trylska J, Tozzini V, McCammon JA. Exploring global motions and correlations in the ribosome. Biophys J. 2005;89(3):1455–1463. doi: 10.1529/biophysj.104.058495. This first coarse-grained simulation of the ribosome revealed large-scale motions of the ribosome.
- 31.Trylska J, Konecny R, Tama F, Brooks CL, 3rd, McCammon JA. Ribosome motions modulate electrostatic properties. Biopolymers. 2004;74(6):423–431. doi: 10.1002/bip.20093. [DOI] [PubMed] [Google Scholar]
- 32.Trylska J, McCammon JA, Brooks Iii CL. Exploring assembly energetics of the 30s ribosomal subunit using an implicit solvent approach. J Am Chem Soc. 2005;127(31):11125–11133. doi: 10.1021/ja052639e. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Sanbonmatsu KY, Voth GA. Key intermolecular interactions in the e Coli 70s ribosome revealed by coarse-grained analysis. Journal of the American Chemical Society. 2011;133(42):16828–16838. doi: 10.1021/ja2028487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(35):12395–12400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma PK, Xiang Y, Kato M, Warshel A. What are the roles of substrateassisted catalysis and proximity effects in peptide bond formation by the ribosome? Biochemistry. 2005;44(34):11307–11314. doi: 10.1021/bi0509806. One of the first quantum chemical studies of the ribosome, this calculation suggested that peptide bond formation does not process by acid-base catalysis.
- 36.Adamczyk AJ, Warshel A. Converting structural information into an allosteric-energy-based picture for elongation factor tu activation by the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(24):9827–9832. doi: 10.1073/pnas.1105714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wieden HJ, Mercier E, Gray J, Steed B, Yawney D. A combined molecular dynamics and rapid kinetics approach to identify conserved threedimensional communication networks in elongation factor tu. Biophysical journal. 2010;99(11):3735–3743. doi: 10.1016/j.bpj.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallin G, Aqvist J. The transition state for peptide bond formation reveals the ribosome as a water trap. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(5):1888–1893. doi: 10.1073/pnas.0914192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trobro S, Aqvist J. Mechanism of the translation termination reaction on the ribosome. Biochemistry. 2009;48(47):11296–11303. doi: 10.1021/bi9017297. [DOI] [PubMed] [Google Scholar]
- 40.Almlof M, Ander M, Aqvist J. Energetics of codon-anticodon recognition on the small ribosomal subunit. Biochemistry. 2007;46(1):200–209. doi: 10.1021/bi061713i. [DOI] [PubMed] [Google Scholar]
- 41.Sanbonmatsu KY. Energy landscape of the ribosomal decoding center. Biochimie. 2006;88(8):1053–1059. doi: 10.1016/j.biochi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Romanowska J, Setny P, Trylska J. Molecular dynamics study of the ribosomal a-site. J Phys Chem B. 2008;112(47):15227–15243. doi: 10.1021/jp806814s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaiana AC, Westhof E, Auffinger P. A molecular dynamics simulation study of an aminoglycoside/a-site rna complex: Conformational and hydration patterns. Biochimie. 2006;88(8):1061–1073. doi: 10.1016/j.biochi.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Kaul M, Barbieri CM, Pilch DS. Aminoglycoside-induced reduction in nucleotide mobility at the ribosomal rna a-site as a potentially key determinant of antibacterial activity. J Am Chem Soc. 2006;128(4):1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]
- 45. Dlugosz M, Trylska J. Aminoglycoside association pathways with the 30s ribosomal subunit. J Phys Chem B. 2009;113(20):7322–7330. doi: 10.1021/jp8112914. This investigation probed the pathways of drug binding to the ribosome using Brownian dynamics simulations of aminoglycosides binding to the ribosome.
- 46. Besseova I, Reblova K, Leontis NB, Sponer J. Molecular dynamics simulations suggest that rna three-way junctions can act as flexible rna structural elements in the ribosome. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq414. This explicit solvent molecular dynamics simulation study showed that three-way junctions play a key role in ribosome dynamics.
- 47.Reblova K, Razga F, Li W, Gao H, Frank J, Sponer J. Dynamics of the base of ribosomal a-site finger revealed by molecular dynamics simulations and cryo-em. Nucleic acids research. 2010;38(4):1325–1340. doi: 10.1093/nar/gkp1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirmizialtin S, Ganesan V, Makarov DE. Translocation of a beta-hairpinforming peptide through a cylindrical tunnel. The Journal of chemical physics. 2004;121(20):10268–10277. doi: 10.1063/1.1807832. [DOI] [PubMed] [Google Scholar]
- 49.Ziv G, Haran G, Thirumalai D. Ribosome exit tunnel can entropically stabilize alpha-helices. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):18956–18961. doi: 10.1073/pnas.0508234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Contreras Martinez LM, Martinez-Veracoechea FJ, Pohkarel P, Stroock AD, Escobedo FA, DeLisa MP. Protein translocation through a tunnel induces changes in folding kinetics: A lattice model study. Biotechnology and bioengineering. 2006;94(1):105–117. doi: 10.1002/bit.20832. [DOI] [PubMed] [Google Scholar]
- 51.Ishida H, Hayward S. Path of nascent polypeptide in exit tunnel revealed by molecular dynamics simulation of ribosome. Biophysical journal. 2008;95(12):5962–5973. doi: 10.1529/biophysj.108.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrone PM, Snow CD, Lucent D, Pande VS. Side-chain recognition and gating in the ribosome exit tunnel. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16549–16554. doi: 10.1073/pnas.0801795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucent D, Snow CD, Aitken CE, Pande VS. Non-bulk-like solvent behavior in the ribosome exit tunnel. PLoS computational biology. 2010;6(10):e1000963. doi: 10.1371/journal.pcbi.1000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge X, Roux B. Absolute binding free energy calculations of sparsomycin analogs to the bacterial ribosome. J Phys Chem B. 2010;114(29):9525–9539. doi: 10.1021/jp100579y. [DOI] [PubMed] [Google Scholar]
- 55.Orzechowski M, Tama F. Flexible fitting of high-resolution x-ray structures into cryoelectron microscopy maps using biased molecular dynamics simulations. Biophys J. 2008;95(12):5692–5705. doi: 10.1529/biophysj.108.139451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16(5):673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor tu conformation control gtp hydrolysis. Proc Natl Acad Sci U S A. 2009;106(4):1063–1068. doi: 10.1073/pnas.0811370106. This ground-breaking study applied molecular dynamics fitting to EF-Tu bound ribosomes, producing key insights into tRNA selection.
- 58.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit trna hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CMT, Onuchic JN, Sanbonmatsu KY. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1108363108. Epub (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, Frank J. Transfer rna in the hybrid p/e state: Correlating molecular dynamics simulations with cryo-em data. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(42):16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caulfield TR, Devkota B, Rollins GC. Examinations of trna range of motion using simulations of cryo-em microscopy and x-ray data. J Biophys. 2011 doi: 10.1155/2011/219515. 2011(219515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ahmed A, Whitford PC, Sanbonmatsu KY, Tama F. Consensus among flexible fitting approaches improves the interpretation of cryo-em data. Journal of structural biology. 2011 doi: 10.1016/j.jsb.2011.10.002. Tama presents a systematic comparison of a large number of cryo-EM fitting techniques.
- 63.Korostelev A, Noller HF. Analysis of structural dynamics in the ribosome by tls crystallographic refinement. Journal of molecular biology. 2007;373(4):1058–1070. doi: 10.1016/j.jmb.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Demeshkina N, Jenner L, Yusupova G, Yusupov M. Interactions of the ribosome with mrna and trna. Curr Opin Struct Biol. 2010;20(3):325–332. doi: 10.1016/j.sbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 65.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 a resolution. Science. 2011 doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 66.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of trna binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annual review of biophysics. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 68.Blanchard SC. Single-molecule observations of ribosome function. Curr Opin Struct Biol. 2009;19(1):103–109. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. Conformational sampling of aminoacyl-trna during selection on the bacterial ribosome. J Mol Biol. 2010;399(4):576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the ef-g-bound ribosome. EMBO J. 2010;29(4):770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2010;107(2):709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: Mapping the free energy landscape of the ribosome using single-molecule fret. Biopolymers. 2008;89(7):565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sharma AK, Chowdhury D. Stochastic theory of protein synthesis and polysome: Ribosome profile on a single mrna transcript. J Theor Biol. 2011;289(36) doi: 10.1016/j.jtbi.2011.08.023. This study uses systems biology techniques to explore the overall ribosome profiles on the message.
- 74.Sharma AK, Chowdhury D. Distribution of dwell times of a ribosome: Effects of infidelity, kinetic proofreading and ribosome crowding. Phys Biol. 2011;8(2):026005. doi: 10.1088/1478-3975/8/2/026005. [DOI] [PubMed] [Google Scholar]