Abstract
The observation that long tracts of RNA are associated with replicating molecules of mitochondrial DNA (mtDNA) suggests that the mitochondrial genome of mammals is copied by an unorthodox mechanism. Here we show that these RNA-containing species are present in living cells and tissue, based on interstrand cross-linking. Using DNA synthesis in organello, we demonstrate that isolated mitochondria incorporate radiolabeled RNA precursors, as well as DNA precursors, into replicating DNA molecules. RNA-containing replication intermediates are chased into mature mtDNA, to which they are thus in precursor–product relationship. While a DNA chain terminator rapidly blocks the labeling of mitochondrial replication intermediates, an RNA chain terminator does not. Furthermore, processed L-strand transcripts can be recovered from gel-extracted mtDNA replication intermediates. Therefore, instead of concurrent DNA and RNA synthesis, respectively, on the leading and lagging strands, preformed processed RNA is incorporated as a provisional lagging strand during mtDNA replication. These findings indicate that RITOLS is a physiological mechanism of mtDNA replication, and that it involves a ‘bootlace' mechanism, in which processed transcripts are successively hybridized to the lagging-strand template, as the replication fork advances.
INTRODUCTION
Chromosomal DNA replication in both prokaryotes and eukaryotes occurs via coupled leading- and lagging-strand DNA synthesis, although many plasmids and viruses use alternative replication mechanisms (1). Vertebrate mitochondrial DNA (mtDNA), typically organized as supercoiled circular monomers of ∼16 kb (2), is comparable with many extrachromosomal elements of prokaryotes (1). The different nucleotide composition of the two strands of mtDNA has led them to be defined as heavy (H) and light (L). In the 1970s, replication of mtDNA was proposed to follow the strand-displacement model, based on electron microscopy (EM) (3) as well as mapping of mtDNA 5′ ends (4,5). The model envisioned replication as initiating via synthesis of an extended RNA primer mapping to a portion of the major non-coding region, which was designated as the origin of H-strand synthesis (OH) (2). After synthesis of approximately two-thirds of the nascent H-strand, a specific site (OL, the ‘origin’ of light-strand synthesis) is exposed in the parental H-strand, at which point synthesis of the L-strand was proposed to initiate. In this model, synthesis of each strand was envisaged to occur continuously from a single initiation site (2), with completion of the nascent L-strand beyond OH occurring after separation of the daughter circles. During the prolonged delay between initiation of first- and second-strand DNA synthesis, the lagging strand template was proposed to remain single stranded.
Although this model has many attractive features, our own studies, using both neutral 2D agarose gel electrophoresis (2D-AGE) and EM (6–8), found no evidence of the predicted partially single-stranded replication intermediates, when DNA prepared from highly purified mitochondria was analyzed. Instead, all replicating molecules detected were substantially duplex. However, we also showed that extensive single-stranded regions were generated in many replication intermediates when they were treated with RNase H, which degrades RNA strands when they are hybridized to DNA (8). The inference that mtDNA replication intermediates contain long stretches of RNA/DNA hybrid was also supported by immunopurification using an antibody specific for such polynucleotides (6). Extensive regions of RNA/DNA hybrid are not predicted by the strand-displacement model, but the observation that they were easily converted to partially single-stranded species by treatment with RNase H, or during nucleic-acid extraction from crude mitochondrial fractions, suggested that the molecular species from whose analysis the strand-displacement model was inferred were, in fact, a preparative artifact.
The extended RNA tracts found in replication intermediates were found to comprise uniquely the L-strand of mtDNA (7,8), which has recently been corroborated by another group (9). The identification of lengthy tracts of RNA hybridized to the parental H-strand, led us to propose that Ribonucleotides are Incorporated ThroughOut the Lagging Strand, RITOLS (8), although until now we were not able to infer the mechanism or significance of this process. In addition, we found evidence for a distinct population of mitochondrial replication intermediates (mtRIs) that were resistant to both RNase H and single-strand nuclease, and so were inferred to comprise fully double-stranded DNA (dsDNA) (10,11). Although these are usually in the minority (8), they can be readily detected under specific experimental conditions (10,11); however, their significance remains to be clarified. One obvious possibility is that two or more mtDNA replication mechanisms can operate simultaneously, on different template molecules.
The unidirectional nature of replication and the approximate location of the origin are shared between the RITOLS and strand-displacement models. The crucial difference is that while RITOLS explains the presence of extensive tracts of RNA/DNA hybrid in replication intermediates, the strand-displacement model predicts instead extensive regions of single strandedness. Although RITOLS intermediates have been characterized using a variety of techniques (6,8,10), two possible objections can be raised against the RITOLS concept. First, mtDNA replication intermediates containing tracts of RNA/DNA hybrid have not previously been shown to exist in vivo. Conceivably, they could arise as an artifact during nucleic acid extraction, by hybridization of a displaced H-strand, as predicted to exist by the strand-displacement model, with excess L-strand RNA. Second, even if molecules containing tracts of RNA/DNA hybrid could be shown to be present in vivo, it remains to be demonstrated that they are true intermediates in DNA replication, and not merely dead-end products of transcription and replication. If RNA-containing mtRIs are, indeed, present in vivo, then the most urgent questions concern the subsequent fate of these species and the source of the RNA, which could provide a clear indication of the mechanism of RITOLS mtDNA replication.
To address these issues, we implemented two procedures. First, we subjected intact cells, isolated organelles and tissue homogenates to nucleic acid cross-linking, and tested whether replication intermediates containing RNA/DNA hybrid were present in the cross-linked material. Second, using metabolic labeling of isolated mitochondria with both DNA and RNA precursors, we tested whether the species containing tracts of RNA/DNA hybrid were genuine replication intermediates in precursor–product relationship with fully replicated mtDNA. These experiments confirmed that RITOLS intermediates are present in vivo, and are processed further to fully replicated mtDNA. The source of the RNA incorporated into RITOLS intermediates was found to be preformed transcripts, rather than extended RNA primers generated as the replication fork progresses. On the basis of these findings, we propose a new model for mtDNA replication. Under this ‘bootlace model', preformed L-strand RNA is incorporated in vivo at the mtDNA replication fork, via 3′ to 5′ hybridization with the displaced H-strand, and is replaced by lagging-strand DNA in a subsequent maturation process.
MATERIALS AND METHODS
Mitochondrial DNA preparation from cells and tissue
mtDNA from sucrose-gradient purified mitochondria was prepared as described previously (12), except that isolated mitochondria were treated with Proteinase K at 4°C for 10–45 min before detergent lysis and successive phenol and chloroform/isoamylalcohol (24:1) extractions. In some cases, whole tissue DNA was extracted directly from homogenized rat liver using phenol and chloroform, after being solubilized with 1% sodium N-lauroylsarcosinate and incubated with 100 µg/ml Proteinase K on ice for 30 min. DNA was isolated from human cultured cells as previously described (11). Gel extraction and analysis of mtRIs from Balb C mouse livers was performed as described previously (8).
In organello labeling of mitochondrial nucleic acid
Labeling of mitochondrial nucleic acids in isolated organelles was performed by a modification of a previously described method (13). Mitochondria from rat liver were isolated at 4°C using DNase and RNase free reagents. Isolated liver tissue from 3- to 5-week-old female Sprague–Dawley rats was minced and homogenized in 4 ml/g of tissue in sucrose tris edta (STE)-buffer [320 mM sucrose, 10 mM Tris–HCl (pH 7.4), 1 mM EDTA and 1 mg/mL essentially fatty acid–free bovine serum albumin (BSA)] using a motorized tight-fitting teflon pestle. Cellular debris was pelleted by differential centrifugation 1000gmax for 5 min. The resulting supernatant was centrifuged at 9000gmax for 5 min to pellet mitochondria. Mitochondria were washed once in STE-buffer, pelleted and equilibrated in incubation buffer [10 mM Tris–HCl (pH 8.0), sucrose and glucose 20 mM each, 65 mM d-sorbitol, 100 mM KCl, 10 mM K2HPO4, 05 μM EDTA, 1 mg/mL BSA, 1 mM ADP, MgCl2, glutamate and malate 5 mM each]. In organelle labeling was performed at 37°C with rotation, using 4 mg/mL mitochondria in incubation buffer supplemented with dCTP, dGTP and dTTP (50 µM each) and [α-32P]-dATP (Hartmann, 3000 Ci/mmol) at 6.6 nM. RNA was labeled using 6.6 nM [α-32P]-UTP (Perkin-Elmer, 12 000 Ci/mmol), ATP, CTP and GTP (50 µM each), and all four dNTPs (50 µM each). Mitochondria were recovered by centrifugation and the nucleic acids extracted as described previously (12).
Nucleic acid modification, fractionation and detection
Psoralen cross-linking: 4–5 mg aliquots of sucrose-gradient purified rat liver mitochondria, or 15–16 mg aliquots of rat liver homogenate were treated with 0 and 5 µM psoralen or 0 and 25 µM psoralen, respectively (4, 5′, 8-trimethylpsoralen, SIGMA) in 1 ml of 20 mM HEPES [pH 7.8], 50 mM EDTA. For cross-linking of mitochondrial nucleic acids in human osteosarcoma or embryonic kidney cells, the cells were grown to 80% confluence on 90 mm plates, washed with phosphate buffered saline (PBS) and treated with 25 µM psoralen in 1 ml of PBS. After incubation on ice for 2 min in the dark, samples were irradiated with 120 mJ/cm2 of UV light of wavelength 254 nm, for 15 min. After the cross-linking step, nucleic acids were isolated from rat liver homogenates, purified mitochondria or human cells, as described above. Restriction digestions were performed on 2–10 µg of purified nucleic acids according to the manufacturer’s (New England Biolabs) instructions. Nuclease treatments were performed before or after restriction digestion and ethanol precipitation as follows: RNase H (Promega) 1 U, 15 min at 37°C; Ribonuclease T1 (Roche) 100 U in 10 mM Tris–HCl (pH 7.4), 40 mM NaCl, 1 mM MgCl2 for 30 min at 37°C.
Neutral 2D-AGE was carried by the standard method (14). Briefly, first dimension separation was 30 V for 16 h in a 0.4% (w/v) agarose gel, at room temperature, followed by a second dimension electrophoresis step of 260 mA for 6 h in 1% agarose, at 4°C. After electrophoresis, gels were dried or alkaline transferred to nylon membranes (GE Osmonics Inc.), UV cross-linked and hybridized to either DNA or RNA radiolabeled probes, based on (15) for rat mtDNA, (16) for human mtDNA and (17) for mouse mtDNA (see supplementary data for probe details).
Riboprobes were generated from amplified mouse mtDNA containing a T7 promoter, using 32P-CTP or 32P-UTP (Hartmann) and an in vitro transcription kit (Ambion). L-strand specific riboprobes were 1, nt 14 903–15 339 (cyt b); 2, 13 874–14 280 (cyt b, c-tRNAGlu and RNA5); 3, 12 788–13 334 (RNA5); 4, 11 546–11 742 (tRNAHis, tRNASer(AGY) and tRNALeu(CUN)); 5, 5568–6044 (COXI); 6, 4951–5326 (tRNATrp, c-tRNAAla, c-tRNAAsn, OL, c-tRNACys and c-tRNATyr); 7, 4502–4920 (ND2) and 8, 3253–3682 (ND1). Nucleotide numbers were based on the revised mouse reference sequence (17), with target transcripts indicated in parentheses; c-tRNAs are the complementary sequence of the corresponding transfer RNA (tRNA). Primers are listed in supplementary Data. Hybridization of immobilized mitochondrial nucleic acid to probe was in 0.25 M sodium phosphate (pH 7.4), 7% SDS, 10 mM EDTA overnight, at 60°C for riboprobes, or 65°C for DNA. Post-hybridization washes were 1× SSC three times, followed by 1× SSC, 0.1% SDS twice, each for 20 min at 65°C. Filters were exposed to phosphorscreens and scanned using a Typhoon™ phosphorimager (GE Healthcare).
Quantitation of relative signals from Southern blotting and in organello labeling
After in organello labeling, restriction digestion and fractionation by 2D-AGE, mtDNA fragments were transferred to nylon membranes (GE Osmonics Inc.), UV cross-linked and exposed to phosphorscreens for 7–14 days. When the radioactive signal had decayed to the point where it was no longer discernible above the background signal, the membranes were hybridized to DNA radiolabeled probes and exposed for 1–2 days to determine the level of mtDNA species in the steady state. Signal was quantitated using a Typhoon™ phosphorimager (GE Healthcare). Equivalent portions of the arcs and spots on the 2D gel blots were selected for quantitation, always avoiding overlapping areas. These and other radioactive signals from in organello labeling were expressed as a percentage of the total signal from the panel and standardized to that of the steady state detected by Southern hybridization.
Quantitation of mtDNA and RNA levels
Transcript and mtDNA levels were determined by quantitative PCR, using GFP as external reference (18).
RESULTS
Nucleic acid cross-linking confirms the presence of RNA/DNA hybrid in mtRIs
Psoralen, a photo-activatable cross-linking reagent that introduces predominantly interstrand cross-links into nucleic acids, has been used previously to study RNA/DNA hybrids created by in vitro transcription of single-stranded bacteriophage fd DNA (19). Crucially, cross-linking of RNA/DNA hybrids renders them resistant to RNase H (an enzyme that specifically degrades the RNA component of RNA/DNA hybrids), owing to the accompanying change in conformation from B to Z-duplex (20,21), as demonstrated for a model substrate (Supplementary Figure S1A). Therefore, interstrand cross-linking of mtDNA was performed to distinguish whether RNA associates with replication intermediates in mitochondria in vivo, or, alternatively, whether this association could be an extraction artifact. The rationale for this experiment is as follows. If RNA is in hybrid with DNA in bona fide replication intermediates in vivo, cross-linking should protect it from digestion by RNase H. Conversely, if the RITOLS-type intermediates arise as an artifact during extraction, due to hybridization of mitochondrial RNA to a displaced parental DNA strand after protease treatment, the RNA will remain RNase H sensitive after in vivo cross-linking.
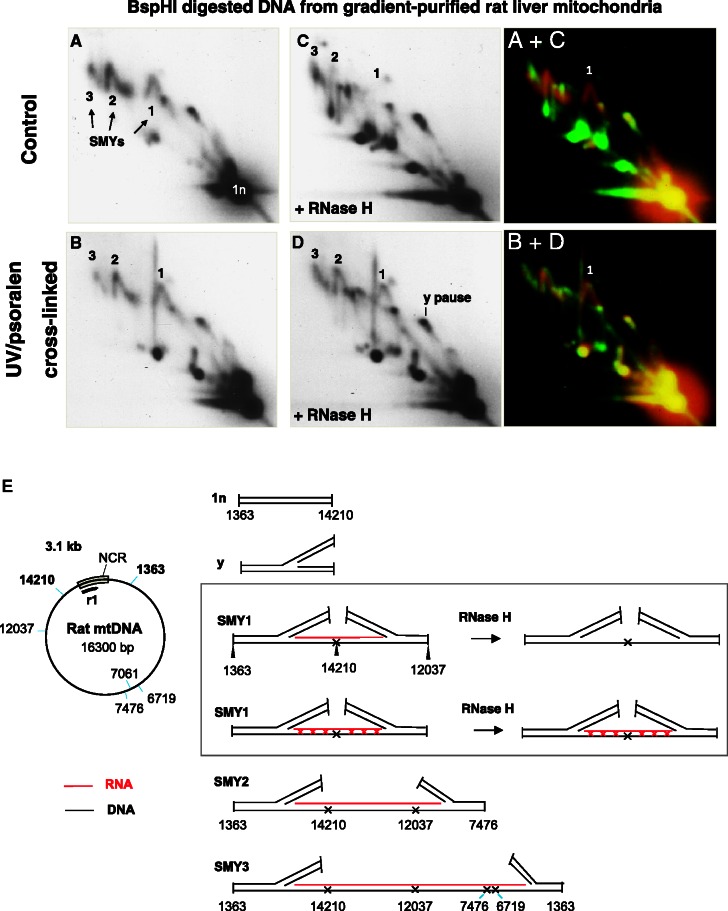
When intact rat liver mitochondria (Figure 1, Supplementary Figure S1B) or tissue homogenates (Supplementary Figure S1C) were treated with psoralen before mtDNA extraction, mtRIs proved markedly more resistant to RNase H than those of controls, when analyzed by 2D-AGE after digestion with BspHI. Cross-linking applied to intact human cultured cells also conferred RNase H resistance on mtRIs, and markedly improved the preservation of fragments containing the origin(s) of replication, which form so-called bubble arcs (Supplementary Figure S1D). The fact that the RNA associated with mtRIs can be cross-linked in isolated mitochondria, fresh tissue homogenates and intact cells, without the prior removal of protein, strongly indicates that the RNA is, indeed, already hybridized to replicating molecules of mtDNA in vivo.
Figure 1.
Psoralen cross-linking in vivo renders mtRIs resistant to RNase H. (A–D) DNA from cross-linked or control mitochondria were isolated, digested with BspHI and then, where indicated, digested further with RNase H, before separation by 2D-AGE. After transfer, the membrane was probed with probe r1 (see Supplementary Data for probe details). SMYs 1, 2 and 3 are SMY replication fork arcs, or their RNase H-modified counterparts. (E) schematic BspHI restriction map of rat mtDNA. To the right of the map are illustrated interpretations of species detected by probe r1, including typical intermediates of the three principle SMY arcs produced by BspHI digestion of rat mtDNA. In the case of SMY1 (inset), the effect of RNase H is interpreted according to whether the mtDNA was subjected to psoralen cross-linking in the organelle. Black lines represent DNA, red lines are RNA, red crosses indicate RNA/DNA psoralen-mediated interstrand cross-links, DNA/DNA cross-links are not shown for simplicity; a black cross is indicative of a blocked restriction site on one branch of the RI, owing to RNA/DNA hybrid. To the right of the gel images are merged false-colour images of A and C, and B and D, providing a direct comparison of the effect of RNase H on control and cross-linked samples: red—untreated (A and B), green—treated with RNase H (C and D). Species resistant to RNase H appear yellow; those modified or generated by RNase H appear red and green, respectively. BspHI was chosen because it produces a series of well-resolved SMY arcs covering almost the entire mitochondrial genome.
Radiolabeled dNTPs are incorporated into RNA-containing replication intermediates in organello
Establishing the presence of RNA hybridized in vivo to mtDNA does not of itself prove that the RITOLS species are true intermediates of mtDNA replication. To address this issue, we combined 2D-AGE analysis with a protocol for in organello labeling of newly synthesized DNA in isolated mitochondria (13). In this procedure, the mitochondria are not synchronized for DNA replication, and so molecules at all stages in the replication cycle should become labeled by the supply of a radioactive deoxynucleotide precursor. Both nascent DNA strands should become labeled, regardless of whether DNA synthesis is strand synchronous. This should include molecules forming a bubble arc, in an origin-containing restriction fragment, as well as molecules forming replication fork (Y) arcs or slow-moving Y-like (SMY) arcs derived from all restriction fragments of the genome. Importantly, if RITOLS is a physiological mechanism of mtDNA replication, many of the intermediates labeled in organello should be subject to modification by RNase H, unless psoralen-UV cross-linked before extraction. Furthermore, if RNA-containing species are generated at the replication fork, they should be detectable by the shortest incubation times required to incorporate label into replication intermediates.
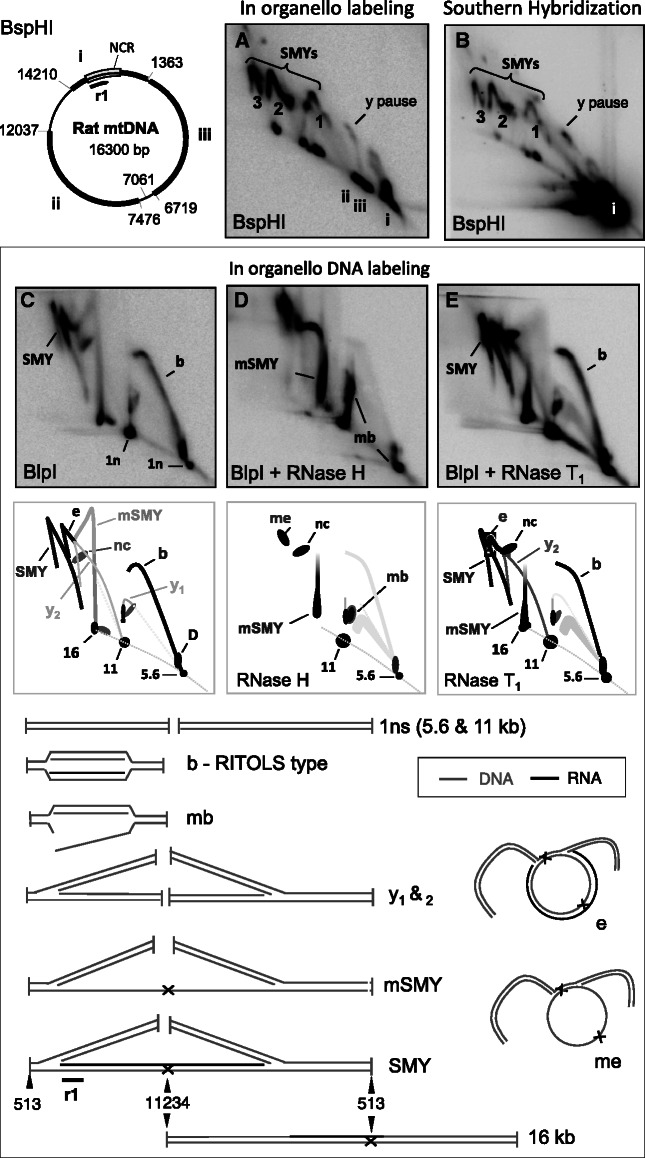
To test these predictions, isolated rat liver mitochondria were incubated in the presence of radiolabeled dATP, after which the extracted nucleic acids were digested with BspHl and analyzed by 2D-AGE (Figure 2A). The ‘1n’ linear fragments of non-replicating DNA seen prominently in the steady state (Southern hybridizations) (species i, Figure 2B) accounted for only a minor fraction of the material labeled in organello after a 5-min incubation (species i, Figure 2A). In most other respects, the major mtDNA species labeled in organello were similar to those detected by Southern hybridization (Figure 2A compared to B); i.e. [α32P]-dATP was incorporated chiefly into those species previously assigned as mtRIs (7,8,23). Next, the restriction enzyme BlpI was tested because it cuts rat mtDNA into two fragments of 5.6 and 10.7 kb, and the short fragment contains the putative origin region (OH), thus allowing the expected bubble arc to be well resolved, in a region of the gel distinct from the expected SMY arcs. Here again the majority of putative RITOLS intermediates that became labeled in organello had the same mobility as those detected in the steady state by Southern hybridization (Supplementary Figure S2A compared to D), and they displayed a pronounced sensitivity to RNase H (Figure 2C versus D). Specifically, RNase H digestion collapsed the bubble arc and grossly modified the SMY arc, similar changes as those observed in parallel (or previously) by Southern hybridization [Supplementary Figure S2B and E; and (7,8)]. The labeled replication intermediates of the bubble and SMY arcs were also largely resistant to single-strand specific RNase T1 (Figure 2E), consistent with the interpretation that much of their RNA content is in the form of RNA/DNA hybrid. Psoralen/UV cross-linking following labeling, but before nucleic acid extraction from mitochondria, conferred resistance to RNase H (compare Supplementary Figure S2A and B to G and H), providing further confirmation that the replication intermediates detected by direct labeling are essentially the same as those present in the steady state in vivo. The amount of DNA label incorporated into mtRIs increased linearly with time in both rat and mouse liver mitochondria, prepared in the same way (Supplementary Figure S3A–C). Labeling was 2 - to 4-fold more efficient with [α32P]-dATP than with [α32P]-dCTP (Supplementary Figure S3D), and pre-incubating mitochondria with the DNA chain terminator 2′, 3′- dideoxyadenosine-5′-triphiosphate (ddATP) inhibited the labeling of mtDNA (Supplementary Figure S3E and F).
Figure 2.
The majority of in organello synthesized mtRIs have the mobility and RNase sensitivity of those present in vivo. After in organello labeling of rat liver mitochondria (6.6 nM [α32P]-dATP, 5 min, 1 mg protein per reaction), mtDNA was extracted, digested with BspHI and subjected to 2D-AGE. Phosphorimager analysis revealed a series of SMY replication fork arcs (A), substantially the same as those detected by Southern hybridization after probing with r1 (B). Note that probe r1 detects a specific fragment of mtDNA (i), whereas incubation of mitochondria with [α32P]-dATP labels all fragments of the mtDNA during the reaction (i, ii, iii). (C–E) In organello labeled rat liver mtDNA was digested with BlpI, separated by 2D-AGE, transferred to filter-membrane and phosphorimaged. Where indicated, samples were treated additionally, after restriction digestion, with RNase H or RNase T1. Interpretations of the arcs and spots appear immediately below the 2D gel images. SMY—SMY replication fork arc; b—origin-containing bubble arc of the RITOLS type [see (8), y—standard replication (y) fork arcs, e—eyebrow arc (8), mSMY—modified SMY, mb—modified bubble (22), me—modified eyebrow, 16 kb—late maturation intermediate where RNA remains at the restriction site at nucleotide 513, but not at 11 234, representative examples are illustrated. Also illustrated are the 1 n linear restriction fragments of 5.6 and 11 kb. Not illustrated are nicked (uncut) circles (nc).
One other feature of the in organello–labeled material that distinguishes it from that detected by Southern hybridization was the near absence of the Y arcs, representing fully duplex DNA intermediates (i.e. fully duplex forms resistant to RNase H). Such arcs are seen routinely in Southern blots of 2D gels of material from solid tissues and cultured cells, and have been proposed to be products of conventional strand-coupled DNA synthesis (23). Standard Y arcs were detected by Southern hybridization in material from the isolated mitochondria used for in organello labeling (Supplementary Figure 2D–F), indicating that their integrity was not compromised during the incubations, although they did not label efficiently in organello (Figure 2A and C); nevertheless the standard Y arc was visible after 30 min of labeling (Supplementary Figure S3B).
RNA-containing replication intermediates labeled in organello are in precursor–product relationship with mature mtDNA
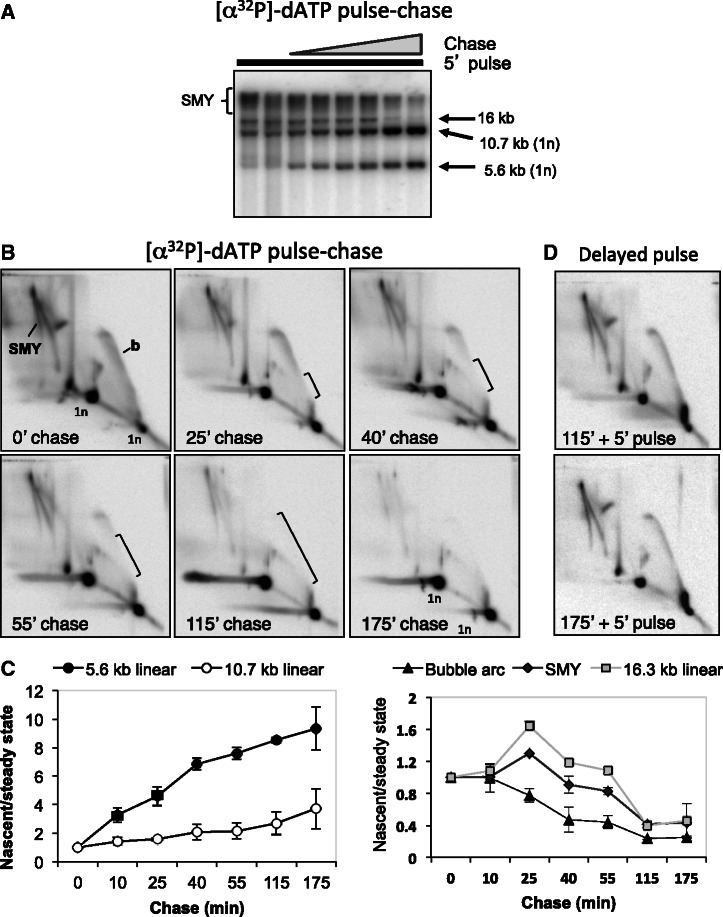
If the RNA-containing molecules pulse-labeled in organello are genuine intermediates in DNA replication, they should become converted to fully replicated mtDNA during a subsequent chase. To test this, we labeled rat liver mtRIs in organello using [α32P]-dATP for 5 min, conditions sufficient to incorporate substantial amounts of label (Figure 2A, Supplementary Figure S3), then subjected the mitochondria to cold chases of 25–175 min, using excess non-radioactive dATP. 1D- (Figure 3A) and 2D-AGE (Figure 3B) were used to identify the species into which label accumulated subsequently. Non-linear (fork-containing) species larger than the restriction fragments of mature mtDNA were labeled preferentially in a 5-min pulse, but over the course of a 3-h chase most of the label accumulated into the linear BlpI fragments of 5.6 and 10.7 kb (Figure 3A). 2D-AGE analysis confirmed that all the mtRIs diminished during the course of the chase (the bubble arc, the SMY arc and the final maturation intermediate (revealed in the digest as a 16.3 kb linear species), whereas label increased in linear fragments of 5.6 and 10.7 kb (Figure 3B), representing fully matured mtDNA. In the case of the arc of replication bubbles, the loss of signal advanced from its base to the tip of the arc (Figure 3B), consistent with new rounds of initiation incorporating chiefly cold dATP, during the chase, and arguing against the incorporation being the result of DNA repair. Quantitation of the different species indicated that the increase in signal for the unit length fragments was essentially linear (Figure 3C), implying that replication proceeded at a constant rate throughout the experiments. The decrease in signal of the mtRIs followed a logical pattern, with (origin-containing) bubble arcs being the first to decline, followed by SMY arcs, with the final maturation intermediate (16 kb linear species still containing RNA at one BlpI site) transiently increasing >1.5-fold during the chase, before declining to low levels (Figure 3C). Replication in the mitochondria remained active for the duration of the reactions, as mtRIs were efficiently labeled by a 5-min pulse of [α32P]-dATP after pre-incubations of 2 or 3 h (Figure 3D). Intact, unlabeled mtRIs were also preserved over such times, based on Southern hybridization (Supplementary Figure S4).
Figure 3.
Pulse-chase experiments establish a precursor–product relationship between RNA-containing mtRIs and mature mtDNA. (A, B and D) Five-minute pulse-labeling of rat liver mitochondria with 6.6 nM [α32P]-dATP, (A, B) followed by chases of 10, 25, 40, 55, 115 or 175 min, with 660 µM unlabeled dATP or (D) following pre-incubation without [α32P]-dATP for 115 or 175 min. Extracted DNA was BlpI digested, separated by 1D (A) or 2D-AGE (B, D) and phosphorimaged after membrane transfer. Brackets demarcate the loss of signal on the replication bubble arc. (C) Quantitation of label in mtRIs, during chase, based on phosphorimaging. Values on vertical axes are based on signal from in organello labeling (nascent) normalized to Southern blot (steady state) signal for the different species analyzed. The resulting ratios for the chase samples were expressed as relative to no chase (0’ chase), set as 1.
Labeling of mtRIs with an RNA precursor
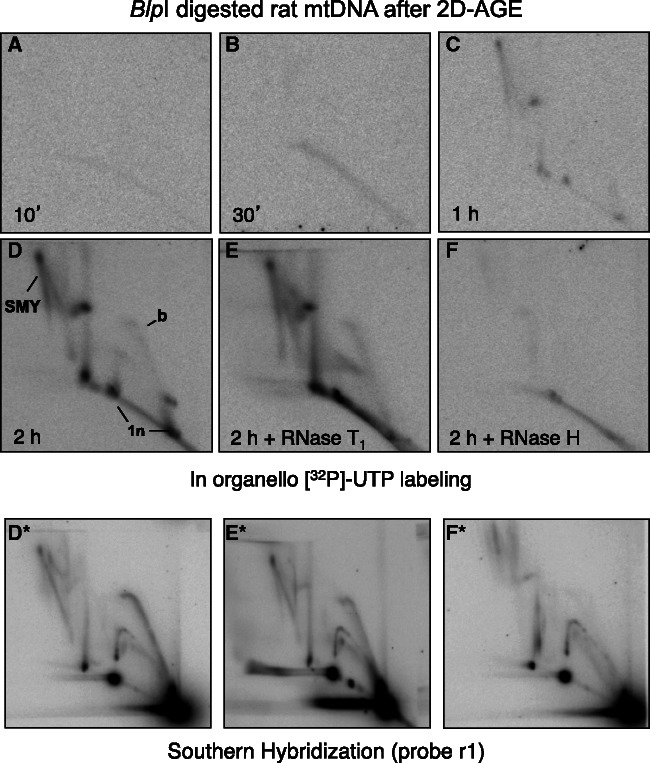
Because the RITOLS model of replication predicts that the lagging strand template is hybridized to RNA (8), ribonucleotide incorporation into mtRIs was tested, in organello, by using [α32P]-UTP in place of [α32P]-dATP. Although, there was no detectable labeling of mtRIs after 10- or 30-min incubations with [α32P]-UTP (Figure 4A and B), a faint pattern, similar to that seen with [α32P]-dATP, was discernible after 1 h of labeling (Figure 4C), and by 2 h of incubation with [α32P]-UTP, the characteristic pattern of mtRIs was evident (Figure 4D). During their synthesis, mitochondrial transcripts are expected to be sensitive to RNases, and yet the UTP-labeled mtRIs were largely resistant to RNase T1 (Figure 4E). In contrast, almost all of the mtRIs that became labeled with [α32P]-UTP in organello were degraded by RNase H (Figure 4F). Because RNase H-treated mtRIs labeled with [α32P]-dATP were modified, but still detectable (Figure 2D), and the same was true of unlabeled mtRIs in the [α32P]-UTP labeling experiments, as detected by Southern hybridization (Figure 4F*), the ribonucleotide precursor cannot have been converted to DNA once inside mitochondria. As a control, the efficient synthesis of bulk mitochondrial RNA was verified, during 20 min of [α32P]-UTP labeling in organello (Supplementary Figure S5A).
Figure 4.
In organello labeling of mtRIs using an RNA precursor. (A–F) Isolated rat liver mitochondria were incubated for 10 min, 30 min, 1 h or 2 h with [α32P]-UTP. Extracted mtDNA was digested with BlpI, and treated with RNase H or RNase T1, as indicated, before 2D-AGE. (D*, E* and F*) Southern hybridizations, performed on duplicate samples incubated for 2 h prepared in parallel with the [α32P]-UTP labeling reactions. Longer exposures of panels A and B (the 10 and 30 min incubations) are shown to demonstrate that the gels were not blank.
An RNA chain terminator inhibits the incorporation of labeled RNA into mtRIs, without blocking mtDNA replication
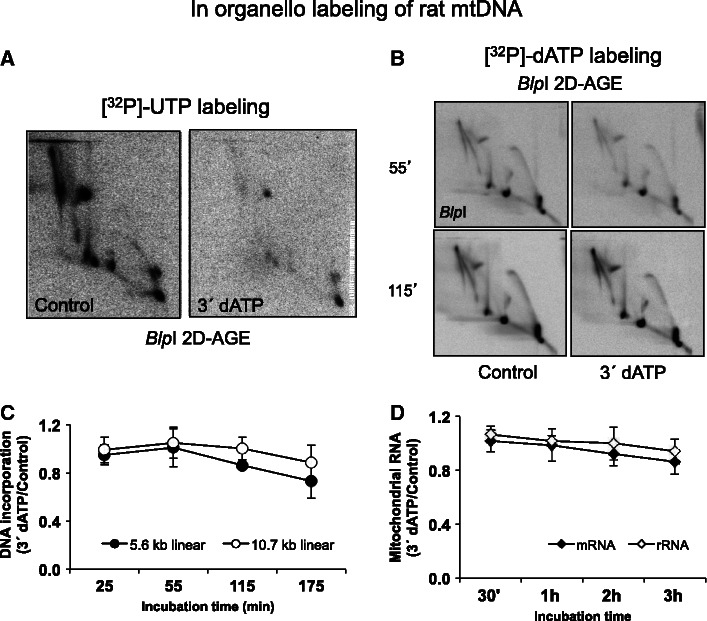
Two possible models for RNA incorporation during mtDNA replication have been proposed (8): one is based on the successive hybridization of mature transcripts to the lagging-strand template (the bootlace model); the other depends on a primase that generates long primers. A third possibility would be that ongoing transcription supplies the RNA that hybridize to replicating DNA molecules as the fork proceeds, yet this could only avoid substantial single-stranded regions forming if multiple active transcription complexes were arrayed on replicating mtDNAs ahead of the fork (Supplementary Figure S6). To discriminate between these different possibilities, we added the RNA chain terminator cordycepin triphosphate (3′-deoxyadenosine-5′-triphosphate or 3′ dATP) to in organello labeling reactions. Although the RNA chain terminator can also block synthesis of the primers needed for the initiation of mtDNA synthesis, the probability of it doing so will be much lower than for termination of the RNAs required for expression of mtDNA, which are synthesized as near genome–length polycistronic transcripts, or the synthesis of the RNA equivalent of Okazaki fragments. Hence, there should be a large dynamic range in which an RNA chain terminator would effectively inhibit synthesis of the long tracts of RNA for the lagging strand (whatever their unit size), while having little impact on primer synthesis at the origin. Incubation of mitochondria with cordycepin triphosphate, before the addition of [α32P]-UTP, produced a marked inhibition of free transcripts labeled in organello at a drug concentration of 20 μM (Supplementary Figure S5A). Cordycepin triphosphate also inhibited [α32P]-UTP labeling of mtRIs (Figure 5A Supplementary Figure S4B), yet the effect of the inhibitor on DNA labeling (with [α32P]-dATP) was much slower and less marked (Figure 5C, compare with Figures 2C and 5B) and produced only minor changes in the relative labeling of different mtRIs. Note that, in contrast to its marked effect on ongoing transcription (Supplementary Figure S5A), incubation with cordycepin triphosphate over 2 h did not substantially affect the steady state level of mitochondrial RNAs (Figure 5D).
Figure 5.
An RNA chain terminator rapidly inhibits nascent RNA formation in isolated mitochondria but affects mtDNA replication much more slowly. (A) 2D-AGE analysis of rat mtDNA labeled for 120 min with [α32P]-dUTP after pre-incubation without (control) or with 20 µM cordycepin triphosphate for 20 min. The intactness of the mtRIs is confirmed in Supplementary Figure S5C. (B) 2D-AGE analysis of rat mtDNA labeled for 5 min with [α32P]-dATP after pre-incubation without (control) or with 20 µM cordycepin triphosphate for 55 or 115 min. (C) Quantitation of the signals from the linear BlpI fragments of mtDNA (panel B and equivalent samples), during 3′ dATP treatment. Note the slow but gradual decrease in [α32P]-dATP incorporation into mtDNA. [α32P]-dATP incorporation was standardized to the level of steady-state mtRIs, based on Southern hybridization. (D) Steady-state levels, relative to zero time point, of mitochondrial rRNA (12S and 16S) and mRNA (COX2, ND1, ND3, ND5 and CYTB), after the indicated times of incubation with 20 µM cordycepin triphosphate. Measurements were based on Q-PCR values normalized to those for mtDNA.
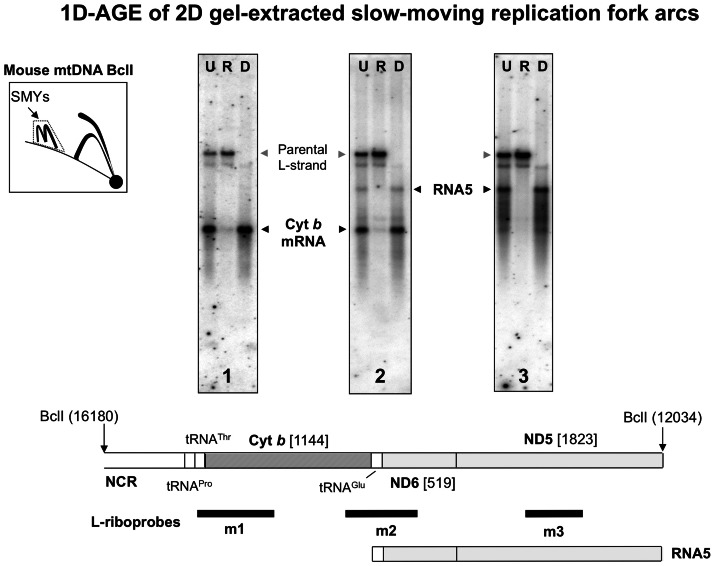
These results strongly suggest that preformed transcripts are the source of the RNA incorporated into replication intermediates, rather than de novo synthesis via a primase, or ongoing transcription. To analyze directly the nature of the RNA incorporated into replicating molecules, we eluted mouse mtRIs found in the SMY region of 2D gels, and analyzed the structure of the RNA therein. We chose the SMY region because, in the BclI digest, it is well separated on 2D gels from all other material. As a control, we first confirmed that there was no detectable trace of even the largest processed mitochondrial RNA species resolving in the vicinity of the SMY arcs when purified RNA was independently fractionated by 2D-AGE (Supplementary Figure S7). The RNA and DNA species found in the SMY arc–derived mtRIs were then analyzed by blot hybridization, following different enzymatic treatments, heat denaturation and fractionation by 1D-AGE (Figure 6, Supplementary Figure S8). A series of L-strand specific riboprobes revealed the SMY-derived mtRIs to contain mainly discrete RNA species, the most abundant of which correspond in size to previously characterized fully processed transcripts (24–27) from around the mitochondrial genome (Figure 6, Supplementary Figure S8). Thus, these processed mitochondrial transcripts are proposed to be the source of the lagging strand RNA that underpins RITOLS replication. It is noteworthy that they included antisense tRNAs (Supplementary Figure S8) that have no other predicted function than to cover the lagging-strand template, during the interval between the initiation of first- and second-strand mtDNA synthesis.
Figure 6.
mtRIs are associated with processed transcripts. Gel-excised material representing the SMY arcs from the indicated portion of a 2D gel of BclI-digested mouse liver mtDNA, blot-hybridized to L-strand specific riboprobes: 1 (nt 14 903–15 339), 2, nt 13 874–15 339 or 3, nt 13 584–14 014 (panels 1–3, respectively). Black arrowheads indicate species co-migrating with previously characterized L-strand mRNAs. Gray arrowheads indicate the parental L-strand DNA from this restriction fragment (nt 12 034–16 180). Gel-excised material was left untreated (U), treated with RNase H (R) or DNase (D), and then separated by 1D-AGE following heat denaturation. A map of the relevant BclI fragment of mouse mtDNA is shown below the gel panels. NCR—major non-coding region; Cyt b—cytochrome b gene; ND5, ND6—NADH dehydrogenase 5, 6 genes. RNA5 comprises antisense tRNAGlu and ND6 conjoined with the sense strand of ND5, representing the mRNA for ND5. For a full description of these and other transcripts of mammalian mtDNA, see (24–27) and Supplementary Figure S8.
DISCUSSION
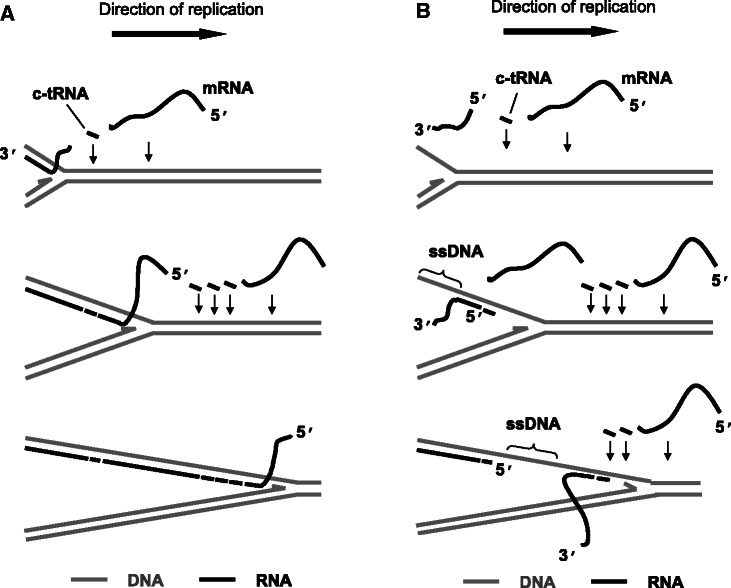
The application of psoralen/UV cross-linking and in organello labeling provide compelling evidence that RNA/DNA hybrids form naturally during the process of mammalian mtDNA replication, and are then replaced with DNA, via a maturation process. Furthermore, combined with the analysis of RNA species found in mtDNA replication intermediates by strand-specific probing, they support an unorthodox mechanism for the incorporation of RNA into replication intermediates, via 3′ to 5′ hybridization of preformed L-strand RNAs with the parental H-strand, as the replication fork progresses. This ‘bootlace mechanism’, illustrated in Figure 7, proposes the successive threading of processed transcripts onto the lagging-strand template, to create RNA/DNA hybrid tracts in which the RNA strand is discontinuous. At most points during replication there will be a portion of the RNA that has yet to be hybridized (Figure 7), and this might explain why RNase T1 enhanced the signal of the SMY arc (Figure 2E).
Figure 7.
The bootlace model of mtDNA Replication. Preformed (L-strand) transcripts, including complementary tRNA and mRNA hybridize to the template lagging strand of mammalian mtDNA as leading strand DNA synthesis proceeds. RNA recruitment is an ongoing process throughout the replication cycle, possibly mediated by specialized components of the mitochondrial (RITOLS) replisome. Incorporation of the transcripts 3′–5′ (A) avoids the formation of single-stranded (ss) sections of DNA, as indicated by the data reported here, whereas such segments would be unavoidable if the transcripts were laid down 5′–3′ (B).
Previous EM data showed that the mtDNA replication intermediates detectable in steady state are essentially duplex over their entire length (6). Furthermore, 2D-AGE, enzymatic digestion and immunopurification showed that a prevalent class of these duplex intermediates contained tracts of RNA/DNA hybrid, implying the incorporation of lagging-strand RNA across the whole genome, RITOLS (6–8). While we could not exclude that these molecules might contain one or more short singe-stranded gaps, their overall structure was fundamentally inconsistent with the predictions of the strand-displacement model. Moreover, species predicted by the strand-displacement model were completely undetectable in mtDNA prepared from highly purified mitochondria. Nevertheless, these earlier studies left open the possibility that the RNA-containing intermediates arose from products of strand-displacement replication hybridizing with excess mitochondrial RNA during extraction. They also did not exclude the possibility that the RNA-containing species might be dead-end products. The data presented here refute these ideas. Cross-linking in vivo (Figure 1, Supplementary Figure S1) protected the RNA/DNA hybrid-containing species from degradation by RNase H, indicating that they are already present before extraction. Metabolic labeling in organello confirmed that they are genuine intermediates in DNA replication, as they are in precursor–product relation to mature mtDNA (Figures 2 and 3). The RNA found hybridized to DNA in replication intermediates corresponds to the L-strand of mtDNA (Figure 6, Supplementary Figure S8). Hence, we conclude that RNA is laid down on the lagging strand as leading-strand DNA synthesis progresses around the circular genome. The predicted intermediates of the strand-displacement model were not synthesized in organello, even under the shortest labeling times required for detectable amounts of radiolabel to be incorporated into mtDNA. Thus, we can exclude the possibility that replication proceeds initially via the strand-displacement model, with RNA incorporated into replication intermediates in a subsequent step.
Comparison of the effects of DNA and RNA chain terminators (Figure 5, Supplementary Figures S3 and S5) showed that concomitant RNA synthesis is not required for RITOLS replication. The fact that over 1 h of exposure to cordycepin triphosphate failed substantially to modify mtDNA replication (Figure 5B), while effectively inhibiting transcription within 10 min, refutes the idea that the lengthy tracts of RNA incorporated on replicating molecules of mtDNA are the products of ongoing transcription. Equally, the lack of any effect of the RNA chain terminator on mtDNA replication excludes the possibility of a primase being responsible for the synthesis of the lagging-strand RNA. If such a primase were sensitive to inhibition by cordycepin triphosphate, replication would arrest, or the lagging-strand template would be left bare, profoundly altering the structure, mobility and enzymatic sensitivity of the mtRIs [see (6,8) for details]. Conversely, a primase insensitive to cordycepin triphosphate (28) should incorporate [α32P]-UTP into mtRIs irrespective of the presence of the RNA chain terminator, which was not the case (Figure 5A, Supplementary Figure S5B). Thus, the RNA species found in RITOLS replication intermediates must logically be synthesized before the onset of DNA replication. The obvious source of such RNAs is processed (mature) transcripts, which was confirmed by our analysis of the species found in mtRIs isolated from the SMY-arc region of 2D gels (Figure 6, Supplementary Figure S8). Although these RNA species display some heterogeneity, they are of substantial length, and the largest and most abundant correspond in size with previously characterized mature transcripts from the various regions of the genome interrogated. This finding provides strong support for a modified version of the bootlace model (8), as illustrated in Figure 7, in which processed lagging-strand transcripts are successively incorporated into replication intermediates as the replication fork advances, and are subsequently fragmented or recovered during the maturation process by an as yet unknown mechanism. Accordingly, the maturation process might account for some of the aforementioned length heterogeneity of the RNAs associated with mtRIs.
One curious observation is that while replication intermediates composed fully of dsDNA are readily detectable by Southern hybridization (10,11,23,29), they were barely detectable in the material labeled in organello using DNA precursors (Figure 2). As previously suggested, they might reflect a parallel mechanism of conventional strand-coupled replication occurring alongside RITOLS in a separate population of molecules. The fact that they do not become labeled in organello, as readily as RITOLS intermediates, implies that conventional strand-coupled replication is slow in mitochondria, or the isolated organelles are unable to sustain this mode of replication at the in vivo level, perhaps because of the disruption of the mitochondrial network. Alternatively, the intermediates composed fully of dsDNA could represent molecules in which replication has stalled, and in which, although the RNA has been replaced by DNA, their further processing occurs only slowly, causing them to accumulate over a period of many hours, which is barely detected in labeling reactions of maximally 3 h (Supplementary Figure S3B and C).
Many new questions arise from this work. The identity of the various enzymes and factors that execute and regulate the process of RNA incorporation during mtDNA replication, as well as the pathway by which the RNA containing intermediates are processed subsequently during maturation to dsDNA, remain to be elucidated, including the extent to which the incorporated RNA is used for priming. Poly(A) tails present on processed transcripts would remain unhybridized to the DNA template, but should not constitute a problem for incorporation, and may even facilitate the delivery and threading of successive RNA segments as the replication fork progresses, or may be used to initiate the maturation process. RNA incorporation is likely to require one or more RNA helicases to unwind the extensive secondary structure of RNAs, especially ribosomal and transfer RNAs. Extensive modifications to tRNAs might impair incorporation, and so it will be interesting to determine whether RITOLS intermediates contain fully or partially unmodified tRNAs.
The evolutionary rationale for the emergence of RITOLS, operating via the bootlace mechanism, is not obvious. However, for coating of the displaced parental strand during DNA synthesis, RNA has a major theoretical advantage over the mitochondrial single-stranded DNA-binding protein, which is the alternative according to the strand-displacement model. Although the fidelity of RNA synthesis is lower than DNA synthesis, RNA and DNA contain the same genetic information. Hybridized RNA thus provides an informational back up that could be used to repair damage that occurs to the lagging-strand template during mtDNA replication, in an environment rich in DNA-damaging oxygen radicals. In fact, RNA incorporation affords the possibility of having even less single-strandedness at the replication fork than conventional coupled leading and lagging strand DNA synthesis, which requires looping out of the lagging-strand template to permit Okazaki fragment synthesis. Alternatively, RNA incorporation may reflect a mechanism that minimizes the deleterious consequences of collisions between transcription and replication complexes (30), by inhibiting transcription on replicating molecules (31), or restraining the replisome.
The in organello mtDNA replication system provides a test-bed to study the role of proteins implicated in mtDNA replication, such as the mitochondrial RNA polymerase POLRMT (32), mitochondrial transcription factors (33), members of the mTERF family (34) and the DNA helicase Twinkle (C10orf2). The in organello system can augment in vitro systems based solely on purified proteins acting on artificial templates (35,36). In addition, for replication factors known or suspected to be shared between mitochondria and the nucleus, use of the in organello system allows physiological effects in mitochondria to be studied specifically, which would be considerably more difficult to accomplish in whole cells.
The in organello labeling procedure also has the potential to answer many outstanding questions about mtDNA replication in pathological states. For example, by studying the relative rate of incorporation of labeled nucleotide precursors into newly synthesized mtDNA, it should be possible to confirm or refute the idea that pathological Twinkle helicase mutants cause replication stalling (37). Nucleotide balance is also recognized as a critical parameter influencing mtDNA replication, as imbalances precipitate mtDNA depletion and disease (38,39). This too may lend itself to analysis in organello, although differences in the relative rate of uptake between different nucleotides will need to be taken into account.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8.
ACKNOWLEDGEMENTS
We thank our colleagues for support and advice, in particular Drs Michal Minczuk, Jaakko Pohjoismaki, Priit Jõers and Hans Spelbrink. The study was supported by the UK Medical Research Council, the Academy of Finland, Sigrid Juselius Foundation and Tampere University Hospital Medical Research Fund.
FUNDING
Cambridge University Commonwealth Trust, Fellowship (to L.K.). Medical Research Council, the Academy of Finland, Sigrid Juselius Foundation and Tampere University Hospital Medical Research Fund. Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kornberg A, Baker TA. DNA Replication. 2nd edn. New York: W. H. Freeman & Co.; 1992. [Google Scholar]
- 2.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 3.Kasamatsu H, Vinograd J. Unidirectionality of replication in mouse mitochondrial DNA. Nat. New Biol. 1973;241:103–105. doi: 10.1038/newbio241103a0. [DOI] [PubMed] [Google Scholar]
- 4.Crews S, Ojala D, Posakony J, Nishiguchi J, Attardi G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979;277:192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- 5.Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J. Biol. Chem. 1981;256:5109–5115. [PubMed] [Google Scholar]
- 6.Pohjoismaki JL, Holmes JB, Wood SR, Yang MY, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 8.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolesar JE, Wang CY, Taguchi YV, Chou SH, Kaufman BA. Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res. 2013;41:e58. doi: 10.1093/nar/gks1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes A, Yang MY, Bowmaker M, Holt IJ. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J. Biol. Chem. 2005;280:3242–3250. doi: 10.1074/jbc.M411916200. [DOI] [PubMed] [Google Scholar]
- 11.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Reyes A, Yasukawa T, Cluett TJ, Holt IJ. Analysis of mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol. Biol. 2009;554:15–35. doi: 10.1007/978-1-59745-521-3_2. [DOI] [PubMed] [Google Scholar]
- 13.Enriquez JA, Ramos J, Perez-Martos A, Lopez-Perez MJ, Montoya J. Highly efficient DNA synthesis in isolated mitochondria from rat liver. Nucleic Acids Res. 1994;22:1861–1865. doi: 10.1093/nar/22.10.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 15.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J. Mol. Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 16.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 17.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, et al. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen CK, Hsieh TS, Wang JC, Hearst JE. Photochemical cross-linking of DNA-RNA helices by psoralen derivatives. J. Mol. Biol. 1977;116:661–679. doi: 10.1016/0022-2836(77)90265-0. [DOI] [PubMed] [Google Scholar]
- 20.Arjumand S, Arif Z, Ali A, Ali R. Native DNA fragments photocrosslinked to psoralen binds to anti-B and anti-Z DNA antibodies. Immunol. Lett. 1995;48:215–219. doi: 10.1016/0165-2478(95)02474-3. [DOI] [PubMed] [Google Scholar]
- 21.Hasan R, Ali A, Ali R. Antibodies against DNA-psoralen crosslink recognize unique conformation. Biochim. Biophys. Acta. 1991;1073:509–513. doi: 10.1016/0304-4165(91)90223-4. [DOI] [PubMed] [Google Scholar]
- 22.Kalejta RF, Lin HB, Dijkwel PA, Hamlin JL. Characterizing replication intermediates in the amplified CHO dihydrofolate reductase domain by two novel gel electrophoretic techniques. Mol. Cell. Biol. 1996;16:4923–4931. doi: 10.1128/mcb.16.9.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 24.Clayton DA. Transcription of the mammalian mitochondrial genome. Annu. Rev. Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- 25.Bhat KS, Bhat NK, Kulkarni GR, Iyengar A, Avadhani NG. Expression of the cytochrome b-URF6-URF5 region of the mouse mitochondrial genome. Biochemistry. 1985;24:5818–5825. doi: 10.1021/bi00342a020. [DOI] [PubMed] [Google Scholar]
- 26.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 27.Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int. Rev. Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 28.Frick DN, Richardson CC. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 29.Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 30.Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentin T, Cherny D, Larsen HJ, Nielsen PE. Transcription arrest caused by long nascent RNA chains. Biochim. Biophys. Acta. 2005;1727:97–105. doi: 10.1016/j.bbaexp.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Matsushima Y, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells. J. Biol. Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 34.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 39.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.