Abstract
Basic helix–loop–helix/Per–Arnt–Sim (bHLH/PAS) transcription factors function broadly in development, homeostasis and stress response. Active bHLH/PAS heterodimers consist of a ubiquitous signal-regulated subunit (e.g., hypoxia-inducible factors, HIF-1α/2α/3α; the aryl hydrocarbon receptor, AhR) or tissue-restricted subunit (e.g., NPAS1/3/4, Single Minded 1/2), paired with a general partner protein, aryl hydrocarbon receptor nuclear translocator (Arnt or Arnt2). We have investigated regulation of the neuron-enriched Arnt paralogue, Arnt2. We find high Arnt/Arnt2 ratios in P19 embryonic carcinoma cells and ES cells are dramatically reversed to high Arnt2/Arnt on neuronal differentiation. mRNA half-lives of Arnt and Arnt2 remain similar in both parent and neuronal differentiated cells. The GC-rich Arnt2 promoter, while heavily methylated in Arnt only expressing hepatoma cells, is methylation free in P19 and ES cells, where it is bivalent with respect to active H3K4me3 and repressive H3K27me3 histone marks. Typical of a ‘transcription poised’ developmental gene, H3K27me3 repressive marks are removed from Arnt2 during neuronal differentiation. Our data are consistent with a switch to predominant Arnt2 expression in neurons to allow specific functions of neuronal bHLH/PAS factors and/or to avoid neuronal bHLH/PAS factors from interfering with AhR/Arnt signalling.
INTRODUCTION
The mammalian basic helix–loop–helix/Per–Arnt–Sim (bHLH/PAS) family of transcription factors consists of 19 structurally related proteins that are essential for a plethora of biological processes, including oxygen homeostasis, xenobiotic response, neurogenesis, appetite control and circadian rhythm (1,2). Prototypical signal-regulated members of this family include the aryl hydrocarbon receptor (AhR) and hypoxia-inducible factor-alphas (HIF-αs), which exert their activities by heterodimerizing with the common bHLH/PAS partner protein aryl hydrocarbon receptor nuclear translocator (Arnt), to form active DNA-binding complexes. In addition, Arnt has been demonstrated to homodimerize to regulate E-Box (CACGTG) harbouring adenovirus major late promoter-driven reporter gene expression (3–5). Some endogenous target genes of the homodimer have been proposed (6).
In addition to Arnt, mammals also express an Arnt paralogue known as Arnt2, which shares 80% amino acid identity to Arnt across the N-terminal bHLH and PAS regions (7), but is more divergent through the C-terminus. Some intriguing differences exist between the two Arnt paralogues. First of all, Arnt and Arnt2 show marked distinctions in their tissue-expression patterns. The Arnt transcripts and proteins are almost ubiquitously expressed both during fetal development and throughout adulthood, although the expression level is low in certain parts of the brain (8–10). In contrast, Arnt2 is much more tissue restricted, being expressed predominately in the central nervous system (CNS) and developing kidney (8–11). Elevated Arnt2 expression has also been detected in the tumour tissues of some breast cancer patients (12), where increased Arnt2 mRNA was strongly correlated with relapse-free survival and overall survival (12).
The reciprocal expression pattern of the two Arnt paralogues in regions of the CNS and the correlation between presence of Arnt2 and favourable outcome for patients with mammary tumours led to the proposal that there are unique functions of Arnt2 that cannot be performed by Arnt. Several studies following targeted disruption of the Arnt2 gene in mouse and zebrafish suggest that adequate expression of Arnt2 is required for specific areas of brain development (13–17). It has also been proposed that Arnt2 functions as the preferred binding partner of neuronal bHLH/PAS proteins such as Single Minded 1 (Sim1) and Neuronal PAS 4 (NPAS4), owing to their generally overlapping expression patterns (17,18). However, in transient transfection of cells with reporter genes, both Arnt and Arnt2 are able to heterodimerize with Sim1 and NPAS4 and regulate transcription (19,20 and unpublished observations).
The second major difference between Arnt and Arnt2 is the distinct phenotypes exhibited by Arnt- and Arnt2-knockout (KO) mice. Arnt-null mice die in utero between E9.5 and E10.5 owing to severe vascular defects (21,22), similar to those seen in HIF-1α-null mice (23,24). In contrast, Arnt2-null mice die perinatally as a result of impaired hypothalamus development (15,16), which phenocopies the defects seen in Sim1-null mice (25). The drastically different phenotypes between Arnt- and Arnt2-null mice again point to non-overlapping functions of these two proteins, especially during fetal development. On the other hand, genotypes of progeny from crossed compound heterozygous mutant Arnt+/− : Arnt2+/− mice did not follow expected Mendelian inheritance, with offspring null for either of Arnt or Arnt2 in combination with being heterozygote for the other form of Arnt not surviving beyond E8.5 (16). This early embryonic lethality suggests that partial redundancy may also exist between Arnt and Arnt2, at least at an early stage of fetal development.
Biochemical and reporter gene experiments indicate that Arnt2, in similar fashion to Arnt, can form functional heterodimers with AhR, HIF-αs, Sim1 & 2 and NPAS4 in vitro and in cell cultures (7,17,18,26). However, partnering of AhR with Arnt2 does not lead to expression of the endogenous AhR target gene Cyp1a1, potentially due to the presence of inhibitory proteins that weaken either AhR/Arnt2 heterodimerization and/or transactivation (10,27). Thus, the choice between partnering Arnt or Arnt2 may be a mechanism for providing variable outputs for bHLH/PAS proteins in vivo.
To better understand the differential roles of Arnt and Arnt2 in development, we analysed the expression of Arnt and Arnt2 in both embryonic carcinoma P19 cells and mouse embryonic stem (ES) cells following retinoic acid (RA)-induced neuronal differentiation. Both P19 and ES cells express intermediate levels of endogenous Arnt2, which could be induced several fold when differentiated towards neuronal lineage. In contrast, robust levels of Arnt rapidly decrease on differentiation, revealing a switch between Arnt and Arnt2. We explore the mechanism of this marked increase in Arnt2 expression and the mechanism by which Arnt2 is repressed in non-neural cells. Our data are consistent with current biological data supporting specific neuronal functions of Arnt2 and/or a need to avoid interference with mitigated AhR/Arnt function in neural tissue.
MATERIALS AND METHODS
Cell cultures
Murine embryonic carcinoma P19 cells and hepatoma Hepa1c1c7 cells were routinely grown in minimum essential medium alpha (MEM alpha) (Gibco/Cat No. 12571), supplemented with 10% fetal calf serum (FCS), 20 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin, and cultured at 37°C, with 5% CO2. Mouse embryonic stem cells D3 (ES D3) were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Gibco/Cat No. 11995), supplemented with 10% FCS, 20 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 1000 U/ml leukemia inhibitory factor (LIF) (Millipore/Cat. No. ESG1107) and 0.1 mM β-mercaptoethanol (Sigma/Cat No. 21985), and cultured at 37°C, with 5% CO2. Mouse V6.5 ES cells were maintained in ES D3 growth medium with irradiated (30 Grays) mouse embryonic fibroblast (MEF) feeder layer.
Neuronal differentiation of P19 and ES cells
P19 cells were harvested and resuspended in P19 Induction Medium (IM; MEM alpha, supplemented with 5% FCS, 20 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin), supplemented with 5 × 10−7 M RA. 1 × 106 cells in 10 ml P19 IM with RA were seeded onto 90-mm non-adherent petri dish (Techno Plas) and cultured at 37°C, with 5% CO2 to encourage embryoid body (EB) formation. After 4 days, EBs were harvested by gentle centrifugation and washed once in P19 IM without RA. To dissociate EBs and form a single-cell suspension, EBs were resuspended in 0.05% trypsin/EDTA in PBS supplemented with 20 µg/ml DNase I (Sigma/Cat No. DN-25) and incubated in 37°C water bath for 10 min, shaking every 2 min. P19 IM was added to EB/trypsin mixture at 1:1 ratio to inactivate trypsin, and cells were harvested by centrifugation. 2 × 106 cells were plated onto a gelatine-coated 6-cm dish in P19 IM without RA. One day after plating, P19 IM was changed to N2B27 medium as previously described (28) with medium changed every 2 days.
For ES D3 cells, following washing and resuspension in ES IM [high-glucose DMEM (Gibco/Cat No. 11995), supplemented with 5% FCS, 20 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin], 1 × 106 cells were seeded onto 90-mm non-adherent petri dish in ES IM without RA for 4 days, followed by 1 × 10−7 M RA treatment for 4 days, with medium changed every 2 days. EBs were dissociated and plated onto poly-d-lysine-coated dishes, and the neural-like cells were maintained as described above.
For chromatin immunoprecipitation (ChIP) experiments, V6.5 ES cells were subjected to direct neuronal differentiation protocol as previously described (29). Briefly, cells were passaged at 1:5 ratio on to a feeder-free gelatinized 10-cm dish. The following day, cells were harvested and washed once with serum-free DMEM to remove traces of LIF. Following resuspension in N2B27 (ES) medium (with 50 µg/ml bovine serum albumin fraction V and 0.1 mM β-mercaptoethanol), cells were counted, and 1 × 106 cells were seeded per 10-cm dish in N2B27 (ES) medium. One day after plating, medium was changed completely. Afterwards, 50% of the medium was refreshed every day.
Muscle cell differentiation of P19 cells
Similar to P19 neuronal differentiation, muscular differentiation of P19 cells was initiated by EB formation in P19 IM, supplemented with 1% DMSO. Following 4 days culturing, EBs were resuspended in DMSO-free P19 IM, re-seeded on 10-cm tissue culture dishes and cultured for 16 days, with medium change every 2 days.
Immunoblotting
Whole cell extracts were prepared as previously described (30). Fifty microgram of cell lysate was separated on 10% acrylamide SDS–PAGE gels and transferred onto nitrocellulose membranes using a wet transfer apparatus (Bio-Rad). Proteins were detected with antibodies against Arnt (MA-515, Affinity BioReagents), Arnt2 (M-165, Santa Cruz Biotechnology), β-III-tubulin (T2200, Sigma), MHC (MF20, Developmental Studies Hybridoma Bank), α-tubulin (MCA78G, Serotec) and horseradish peroxidase-conjugated secondary antibodies, and visualized with Immobilon Western Chemiluminescent Substrate (Millipore). For densitometry plot, band intensities were quantified by ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health).
RNA extraction, cDNA synthesis and quantitative real time PCR
Total RNA was extracted and reverse transcribed to cDNA as previously described (30). Intron spanning qRT-PCR primers were designed using online program Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Sequences for all qRT-PCR primers can be found at Supplementary Table S1. Target gene expression was normalized against house-keeping gene β-actin, which was largely unchanged during the course of differentiation. All experiments were performed in triplicates for three biological replicates.
Immunofluorescence
P19 cells were seeded onto coated round glass cover slips in a four-well multiwell tray (Nunc) at defined densities (see RA-induced neuronal differentiation protocols above). Cells were fixed using 4% paraformaldehyde for 10 min. The following antibodies were used for protein detection: mouse anti-Arnt (1A1, Origene), rabbit anti-Arnt2 (M-165, Santa Cruz Biotechnology), rabbit anti-β-III-tubulin (T2200, Sigma), mouse anti-β-III-tubulin (T8578, Sigma). Following incubation with fluorophore-conjugated secondary antibodies (Invitrogen), slides were mounted onto ProLong Gold with DAPI (P36935, Invitrogen).
Bisulfite conversion of genomic DNA and sequencing
Bisulfite sequencing was performed as previously described (31) with the following modifications. Briefly, genomic DNA was isolated using High Pure PCR Template Preparation Kit (Roche). One microgram of genomic DNA in 100 µl H2O was denatured at an alkaline pH by adding 300 µl of 3 M NaOH, and incubated at 55°C for 30 min. Immediately 900 µl of freshly made bisulfite conversion solution [4.7 M sodium bisulfite (Sigma), 2.4 mM hydroquinone (Sigma) and 361.5 mM NaOH] was added, overlayed with 120 µl of mineral oil (Bio-Rad) and incubated for a further 20 h at 55°C. The bisulfite-treated DNA was then purified using QIAquick Gel Extraction Kit (Qiagen), eluted in 2 × 40 µl of H2O and alkaline desulfonated with 8 µl of 3 M NaOH for 15 min at 37°C. DNA was precipitated with 50 µl of 7.5 M ammonium acetate, 700 µl of 100% EtOH and 1 µl of glycogen at −80°C for 30 min, followed by 1 × 700 µl 75% EtOH wash, and resuspended in 30 µl of H2O. Sequencing primers for amplifying Arnt2 promoter were designed by online program MethPrimer (http://www.urogene.org/methprimer/index1.html), and were mArnt2_meth_F: 5′ GAATGGTGTTTAATTTGGTTTTGA 3′ and mArnt2_meth_R: 5′ AAAAAATCCCAAAAATTCTACCCTAT 3′. Taq DNA polymerase (New England Biolabs) was used for PCR with the following conditions: 5 min at 95°C, followed by 36 cycles of 30 s at 95°C, 30 s at 60°C and 15 s at 72°C, and a final extension for 5 min at 72°C. Amplified products were subcloned directly into pGEM-T Easy vector (Promega) according to manufacturer’s instructions. For each cell line/condition, two biological replicates were analysed, and a total of 10 individual clones were selected and sequenced with M13 primers (5′ GTAAAACGACGGCCAGT 3′). Sequences were combined and analysed using BiQ Analyzer software (http://biq-analyzer.bioinf.mpi-inf.mpg.de/). Clones that were likely to come from the same chromosome of the same cell or with <95% C to T conversion rate of non-CpG cytosine residues were excluded from the analysis.
mRNA stability studies
The half-lives of Arnt and Arnt2 mRNAs were determined by Click-iT® Nascent RNA Capture Kit (Life Technologies) according to manufacturer’s instructions. Briefly, undifferentiated and 4-day RA-treated P19 cells were labelled with 0.2 mM ethynyl uridine (EU) and incubated at 37°C for 3 h. Cells were then allowed to recover in EU-free medium for 0, 1, 2, 4, 6 h, respectively. Total RNA was extracted (30), and 5 µg of total RNA was mixed with Click-iT reaction cocktail (25 μl Click-iT EU buffer, 4 μl 25 mM CuSO4 and 2.5 μl 10 mM Biotin azide). Immediately, the reaction buffer additive 1 was added, following by reaction buffer additive 2 exactly 3 min after adding of the first additive, and the reaction was carried out for 30 min at room temperature. Following incubation, the ‘clicked’ RNA was re-purified by ammonium acetate precipitation (see above), and 0.5 µg of purified RNA was bound to 25 μl of streptavidin magnetic beads with 80 units of RNAseOUT Recombinant Ribonuclease Inhibitors (Life Technologies) for 30 min. Beads were then washed 5 × 300 μl of Click-iT wash buffer 1, followed by 5 × 300 μl of wash buffer 2, and resuspended in a final volume of 25 μl wash buffer 2. The captured RNA was in-bead converted to cDNA as per manufacturer’s instructions using SuperScript III Reverse Transcriptase (Life Technologies). Relative levels of Arnt and Arnt2 mRNAs at each time point were measured by qRT-PCR. All experiments were performed in triplicates for three biological replicates.
Chromatin immunoprecipitation
ChIP was performed as previously described (32) using 2 μg of anti-trimethyl histone H3K4 (ab8580, Abcam) or 5 μg of anti-trimethyl histone H3K27 (07-449, Millipore) antibodies, or 5 μg non-specific IgG as control (Upstate). All real-time qPCR quantifications were performed in triplicates for three biological replicates. Primer sequences used in the ChIP experiments can be found in Supplementary Table S1.
RESULTS
A switch in the ratio of Arnt/Arnt2 during neuronal differentiation of P19 cells
A number of previous studies have shown that in contrast to ubiquitously expressed Arnt, the paralogue Arnt2 has expression restricted predominantly to the brain and developing kidneys (8,9,11). Consistent with this pattern, N39, an immortalized mouse embryonic hypothalamus-derived cell line (33), showed clear expression of Arnt2 mRNA, while conversely, hepatoma Hepa1c1c7 cells expressed little or no Arnt2 (Supplementary Figure S1). Furthermore, both embryonic carcinoma P19 cells and ES cells expressed Arnt2 at comparable levels with that found in the hypothalamic N39 cells (Supplementary Figure S1).
Arnt2-null mice show perinatal lethality owing to defective secretory neuron formation in the supraoptic nuclei (SON) and paraventricular nuclei (PVN) of hypothalamus (15), revealing a critical role for Arnt2 in certain areas of brain development. To further characterize the functions of Arnt2 during neurogenesis, we adapted a widely used P19 differentiation system with kinetics reminiscent of in vivo neuronal differentiation in murine embryos (28), to monitor the change in Arnt and Arnt2 expression during neuronal differentiation (Figure 1A). Both Arnt and Arnt2 transcripts can be clearly detected in undifferentiated P19 cells (Supplementary Figure S1). Given the relative amplification efficiencies of these two primer sets (93.1 and 95.7%, respectively), it was estimated that the amount of Arnt transcripts in undifferentiated P19 cells was approximately nine times higher than that of Arnt2.
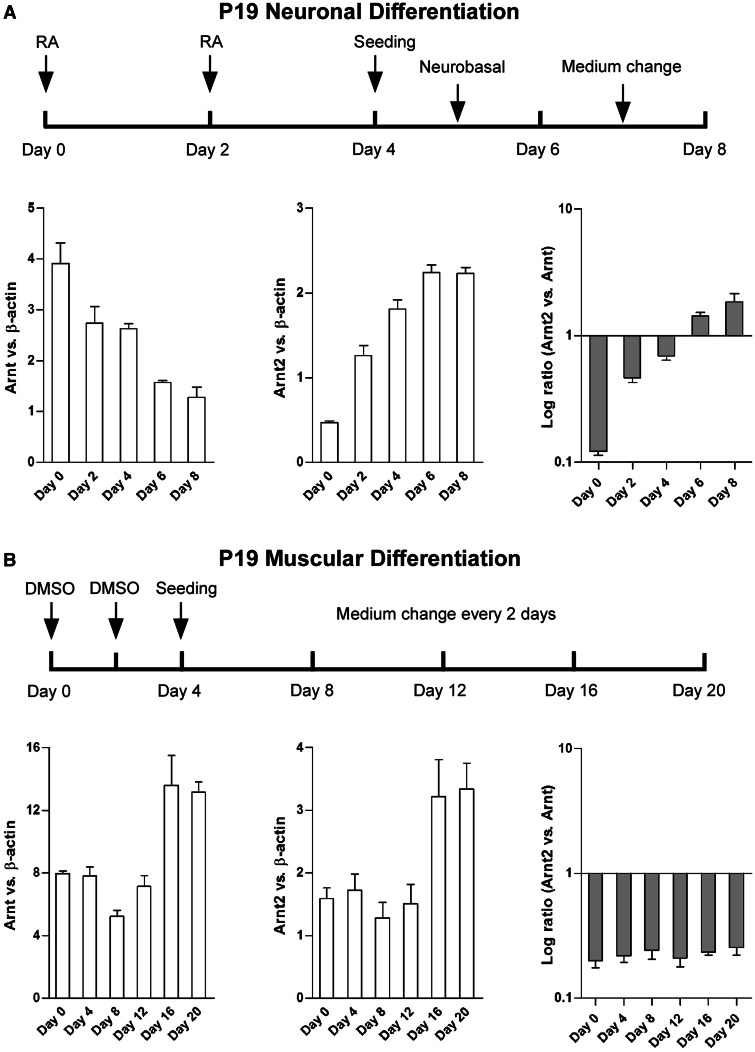
Figure 1.
Neuronal differentiation of P19 cells increased expression of Arnt2 mRNA concomitant with decreased expression of Arnt mRNA. (A) Scheduled protocol for neuronal differentiation of P19 cells via growth of EBs in the presence of 500 nM RA, followed by seeding and incubation in neurobasal N2B27 media. Differentiation of P19 cells into NPCs and neurons resulted in a switch between the Arnt and Arnt2 transcript ratios as assayed by real time qRT-PCR. (B) Scheduled protocol for P19 muscle cell differentiation by the addition of 1% DMSO. In contrast to neuronal differentiation, muscle cell differentiation did not alter the ratio of Arnt2 versus Arnt transcripts. All data are mean ± SEM, n = 3.
Differentiation of P19 cells led to rapid reduction of the pluripotent marker gene Oct4, as well as induction of the neuron progenitor cell (NPC) and postmitotic neuron marker genes Sox1 and β-III-tubulin, respectively, indicating a successful enrichment of neuronal cell populations in the culture (Supplementary Figure S2). Intriguingly, neuronal differentiation of P19 cells also led to marked increase in Arnt2 transcript levels, coincident with decreased Arnt mRNA expression (Figure 1A). Thus, differentiation of P19 cells into neurons resulted in a clear switch between the levels of Arnt and Arnt2 mRNAs, leading to predominant Arnt2 expression at the later stage of P19 differentiation.
In addition to neuronal differentiation, P19 cells can also be differentiated into cardiomyocytes and skeletal myocytes of mesoderm linage in the presence of 1% DMSO (34). To test whether the ratio of Arnt to Arnt2 was also altered by differentiation into an alternative lineage, we used this protocol and demonstrated, as expected, an increased expression of muscle precursor and skeleton muscle cell markers MyoD and MHC (myosin heavy chain), respectively (Supplementary Figure S3). However, in stark contrast to neuronal differentiation, muscular differentiation of P19 cells did not alter the relative ratios of Arnt2 and Arnt at transcript levels (Figure 1B), suggesting neuronal differentiation-specific mechanisms exist to invoke a switch in the ratio of Arnt/Arnt2 expression.
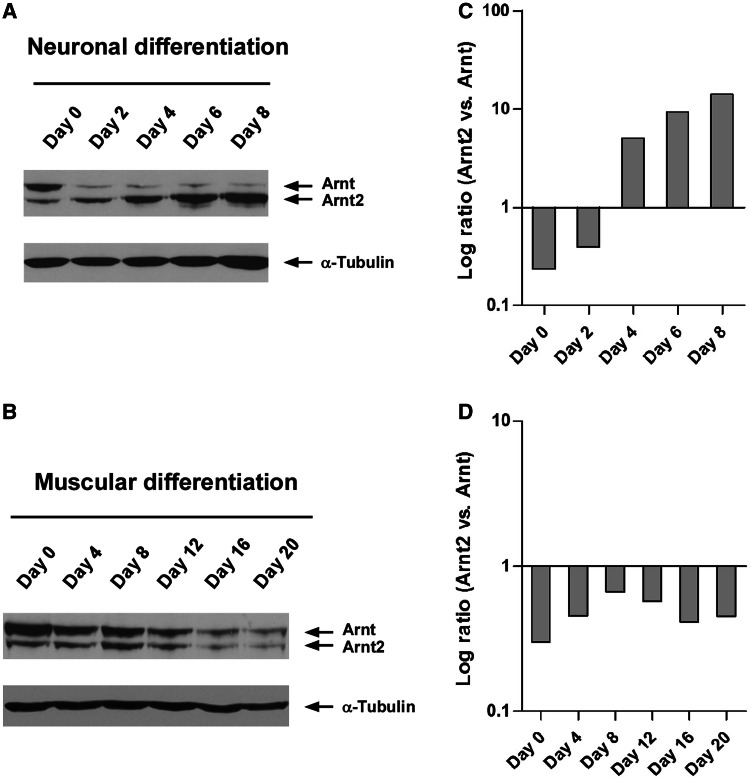
The Arnt and Arnt2 protein levels were also examined at different stages of P19 differentiation by immunoblotting. Similar to that found for mRNAs, the Arnt/Arnt2 switch could also be detected at the protein level following P19 neuronal differentiation, but was absent during muscular differentiation (Figure 2). The increased expression of Arnt2 protein during neuronal differentiation followed a similar kinetics as that of Arnt2 mRNA (Figure 1A). In contrast, reduction of Arnt protein appeared to be more rapid, with almost 10-fold reduction after the first 2 days of differentiation, while the Arnt mRNA declined gradually during the course of neuronal differentiation (Figure 1A).
Figure 2.
Neuronal differentiation of P19 cells increased Arnt2 and decreased Arnt protein expression. (A) Immunoblotting showing increased Arnt2 and decreased Arnt expression during neuronal differentiation of P19 cells as outlined in Figure 1A. (B) Immunoblotting of Arnt2 and Arnt proteins during muscle cell differentiation of P19 cells as outlined in Figure 1B. Relative levels of Arnt2 versus Arnt were largely unchanged during muscle cell differentiation. (C and D) Densitometry plots showing the log ratios of Arnt2 versus Arnt protein levels using the data presented in parts A and B, respectively. Data shown are representative from two independent experiments.
A switch in the ratio of Arnt/Arnt2 during neuronal differentiation of ES cells
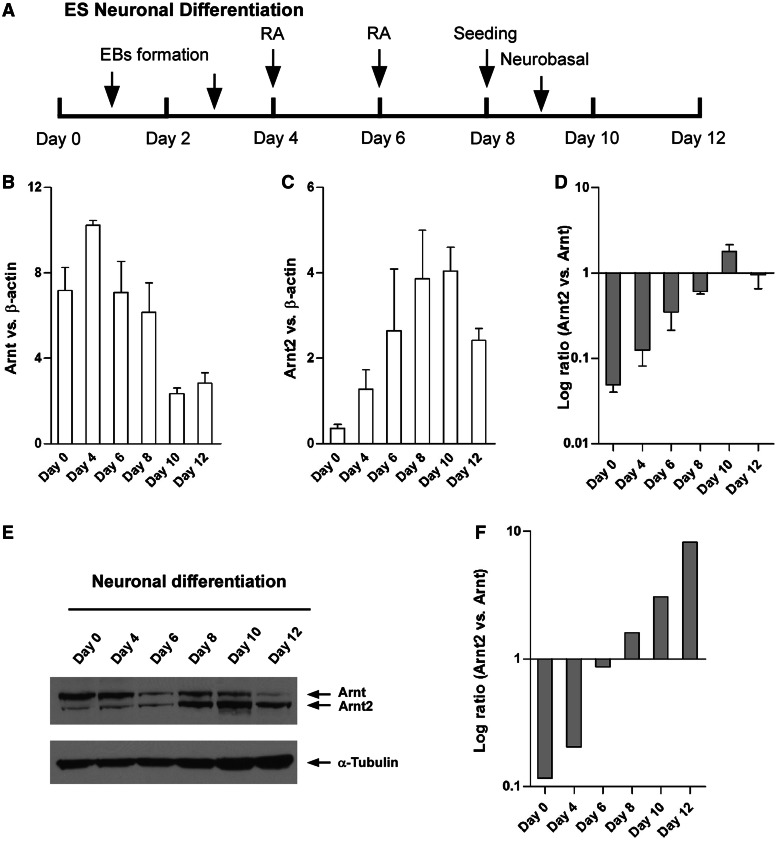
To confirm that increased Arnt2 and decreased Arnt expression during neuronal differentiation is a general phenomenon and not just peculiar to P19 cells, we repeated our neuronal differentiation experiments with ES cells. Neuronal differentiation of ES cells led to a marked reduction of Oct4 and increased expression of Sox1 and β-III-tubulin (Supplementary Figure S4), very similar to that seen in P19 differentiation, demonstrating a successful enrichment of NPC and neuron populations in the culture. This was also supported by microscopic analysis showing extensive neurite formation in differentiated ES cultures (Supplementary Figure S4). Furthermore, consistent with P19 neuronal differentiation, differentiation of ES cells led to increased Arnt2 and decreased Arnt expression at both mRNA and protein levels (Figure 3). Thus, the trend of an Arnt/Arnt2 switch was consistent in both P19 and ES differentiation models.
Figure 3.
An Arnt/Arnt2 switch during neuronal differentiation of ES cells. (A) Scheduled protocol for neuronal differentiation of ES cells via formation of EBs and treatment with 500 nM RA, followed by seeding and incubation in neurobasal N2B27 media. (B and C) Real time qRT-PCR results showing increased Arnt2 and decreased Arnt expression during ES cell neuronal differentiation. Data are mean ± SEM, n = 3. (D) log ratios of Arnt2 versus Arnt derived from parts (B) and (C). (E) Immunoblotting showing increased Arnt2 and decreased Arnt protein expression during neuronal differentiation of ES cells. Data are representative from two independent experiments. (F) Relative band intensities for Arnt and Arnt2 proteins in part (E) were determined by densitometry and plotted as log ratio of Arnt2 versus Arnt.
Arnt2 is predominantly expressed in the neuronal cell population in differentiated P19 cells
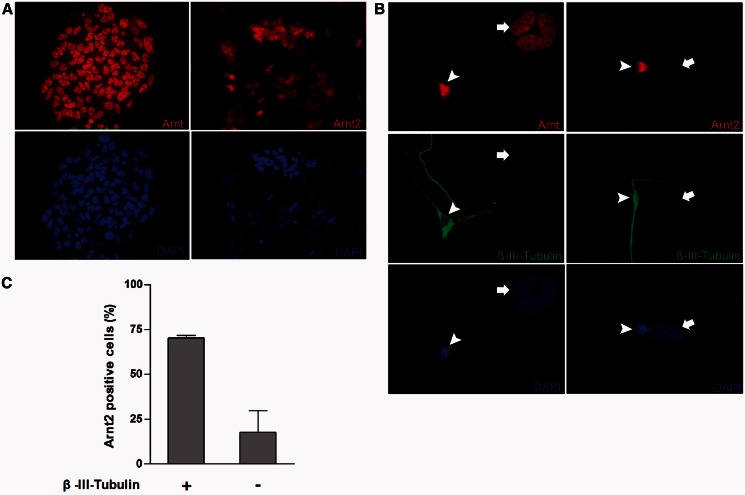
Although mRNA and protein expression analyses demonstrated a switch in the ratio of Arnt to Arnt2 levels during RA-induced neuronal differentiation, they did not show that the increase in Arnt2 levels occurred specifically in neurons. We therefore performed immunocytochemistry to determine whether Arnt2 was co-localized with the neuron marker β-III-tubulin. Consistent with mRNA and protein expression data in untreated P19 cells, both Arnt and Arnt2 were present in all non-differentiated P19 cells (Figure 4A). Upon differentiation towards neuronal lineages, Arnt expression is still observed in β-III-tubulin-negative and -positive cells. β-III-tubulin-negative cells, however, express barely detectable levels of Arnt2 compared with relatively high levels in β-III-tubulin-positive cells (Figure 4B). We counted immunostained cells from two independent differentiation experiments and found that the β-III-tubulin-positive cell population consistently contained a higher percentage of Arnt2-positive cells (average of 70.2 ± 1.4%) compared with the β-III-tubulin-negative cell population (average of 9.3 ± 17.8%) (Figure 4C). This indicates that, similar to the in vivo expression pattern (15), Arnt2 is enriched in cells of neuronal lineage. Put together, these observations demonstrate that the switch in ratio is due to enrichment of the neuronal cell population, which expresses Arnt2 at high levels.
Figure 4.
Arnt2 is predominantly expressed in neuronal cells following differentiation. (A) Arnt and Arnt2 proteins were detected in the nuclei of undifferentiated P19 cells by immunocytochemistry. (B) After an 8-day differentiation protocol as outlined in Figure 1A, Arnt was observed in both β-III-tubulin-positive (arrowhead) and -negative (arrow) cells, while Arnt2 was strong in β-III-tubulin-positive cells but barely detectable in β-III-tubulin-negative cells. Nuclei are visualized by blue DAPI staining. Data are representative of two independent experiments. (C) There are more Arnt2-positive cells within a population of randomly selected β-III-tubulin-positive cells than within a population of randomly selected β-III-tubulin-negative cells. Approximately 70 cells were counted for each β-III-tubulin status group per experiment. Data are mean ± SD, n = 2.
Increased expression of Arnt2 in differentiated P19 cells is a result of increased Arnt2 transcription
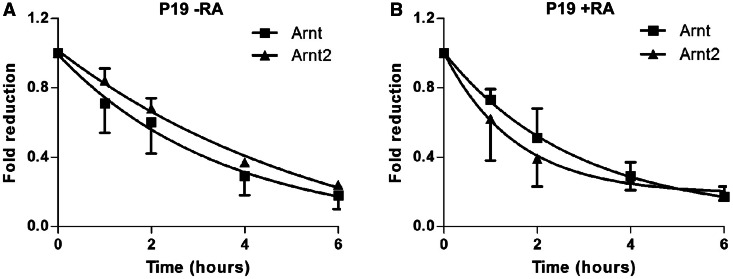
During neuronal differentiation, both P19 and ES cells exhibit a several fold increase in Arnt2 transcripts levels (Figures 1 and 3). However, whether this increase is the result of increased Arnt2 gene transcription or whether it is due to enhanced mRNA stability is unknown. To distinguish between these two possibilities, we used an RNA pulse labelling strategy followed by purification and quantitative PCR to measure the half-lives of Arnt and Arnt2 transcripts in both undifferentiated and 4-day RA-treated P19 cells (Figure 5). The half-lives of Arnt and Arnt2 mRNAs were relatively constant in both undifferentiated and differentiated cells. In addition, the Arnt and Arnt2 mRNAs displayed very similar half-lives, measured to be ∼2.5 h in P19 cells, although the Arnt2 transcript may have a slightly longer half-life than that of Arnt. These results indicate that increased Arnt2 expression during differentiation results from increased Arnt2 gene transcription, with only a minor component, if any, due to increased Arnt2 mRNA stability.
Figure 5.
Neuronal differentiation of P19 cells has little effect on the stability of Arnt or Arnt2 mRNAs. Half-lives of Arnt and Arnt2 mRNAs in undifferentiated (A) and 4-day RA-treated (B) P19 cells were measured by Click-iT Nascent RNA Capture Kit as described in Materials and Methods. The half-lives of Arnt and Arnt2 mRNAs were respectively calculated to be 2.3 ± 0.6 and 2.9 ± 0.1 h in undifferentiated P19 cells, and 2.3 ± 0.4 and 2.6 ± 0.2 h in RA-induced P19 cells. Data are mean ± SEM, n = 3.
Methylation status of the Arnt2 promoter in neuronal and non-neuronal cells
In an effort to further characterize the mechanism underlying enhanced Arnt2 expression during neuronal differentiation, transcription factor-binding site analysis on the proximal promoters of human, mouse and rat Arnt2 loci was performed. From this analysis, binding sites for transcriptional repressor protein REST (RE1 silencing transcription factor, also known as NRSF or neuron-restrictive silencer factor) were found to be overrepresented at the Arnt2 promoter of all three species. In addition, REST was shown to be recruited to the human Arnt2 promoter in a genome wide ChIP-seq study (35). As REST is a negative master regulator of neurogenesis that represses expression of neuronal genes in non-neuronal cells by recruiting histone deacetylase (HDAC) (36,37), we thought it logical to test the role of REST in regulating Arnt2 gene expression. Of note, a recent study in our laboratory showed that neuron-restricted expression of NPAS4, another bHLH/PAS protein, is REST dependent (D. Bersten, manuscript in preparation).
To test the role of REST in regulating Arnt2 expression, we transiently transfected P19 cells with a dominant negative mutant of REST, or treated cells with the HDAC inhibitor trichostatin A, and measured the expression level of Arnt2 mRNA following each treatment. Unexpectedly, neither of these treatments increased the expression level of Arnt2 (data not shown), consistent with a REST-independent pathway of Arnt2 regulation in differentiated P19 cells.
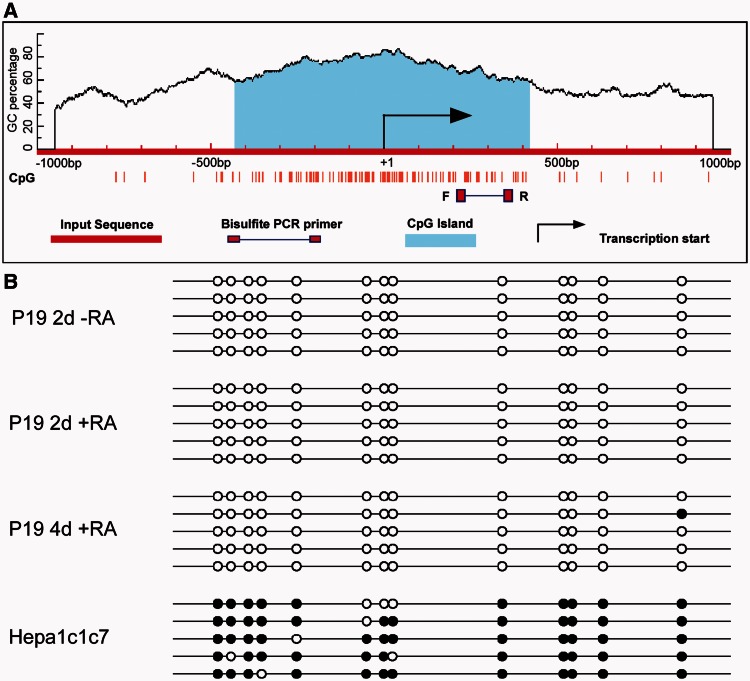
Inspection of the mouse Arnt2 locus also showed that the Arnt2 promoter is highly GC rich and contains a CpG island (Figure 6A) that is highly conserved among mammals (Supplementary Figure S5). As global DNA demethylation has been reported to be a permanent feature of P19 cells during neuronal differentiation (38), it is plausible that the change in expression of Arnt2 is a result of decreased methylation status of the Arnt2 promoter during neuronal differentiation. To test this hypothesis, genomic DNA was extracted from undifferentiated and RA-treated (2- and 4-day) P19 cells and subjected to bisulfite sequencing analysis. Unexpectedly, no DNA methylation was found at the promoter region of the Arnt2 locus in either undifferentiated or differentiated P19 cells, suggesting that the increased expression of Arnt2 in differentiated P19 cells is not a result of attenuated DNA methylation (Figure 6B). Conversely, the Arnt2 promoter in hepatoma Hepa1c1c7 cells, which do not express Arnt2, was highly methylated (Figure 6B). Thus, although the methylation status of the Arnt2 promoter did not contribute to the regulation of Arnt2 gene in P19 cells, hypermethylation of Arnt2 promoter is likely to occur in other tissue types where Arnt2 is not detectable, which is probably the mechanism for silencing the Arnt2 gene in tissues such as liver (Supplementary Figure S1).
Figure 6.
Mapping the DNA methylation status of the Arnt2 promoter in P19 and Hepa1c1c7 cells. (A) CpG island prediction in the murine Arnt2 promoter using MethPrimer software. Criteria used for prediction were island size >100 bp, GC percentage >30% and observed/expected CpG ratio >0.6. A single CpG island of 851 bp was identified, which spread across the transcriptional start site of the murine Arnt2 gene. (B) Bisulfite sequencing revealed the Arnt2 promoter was not methylated in either undifferentiated P19 cells (2d -RA) or 2 day and 4 day differentiated (i.e. 2d and 4d +RA treated) P19 cells, but was hypermethylated in hepatoma Hepa1c1c7 cells. The black circles indicate methylated CpG sites, and white circles indicate unmethylated CpG sites. PCR fragment analysed indicated by F and R boxes in (A).
The Arnt2 promoter displays the epigenetic hallmarks of a lineage-specific gene
As the Arnt2 promoter is not methylated in undifferentiated P19 cells, we hypothesized that epigenetic modifications might confer a level of repression that could be reversed in neuronal cells on differentiation. We therefore performed ChIP experiments to test for the presence of trimethylated histone 3 lysine 27 (H3K27me3), a common repressive marker, and H3K4me3, a common active marker, in Arnt and Arnt2 promoter regions. As expected, the Arnt promoter did not contain the repressive mark in undifferentiated ES cells, neuronal differentiated ES cells or Hepa1c1c7 cells, but was marked with H3K4me3 in all three of these cell populations (Figure 7A). These observations were consistent with the ubiquitous expression of Arnt across tissue types.
Figure 7.
Active and repressive histone methylation marks on the Arnt and Arnt2 promoters in undifferentiated ES cells, 7-day neuronal differentiated ES cells and Hepa1c1c7 cells. ChIP-qPCR data showed the Arnt promoter in all three cell types to contain the active marker H3K4me3, but no repressive marker H3K27me3 (A). The Arnt2 promoter was bivalently marked in undifferentiated and differentiated ES cells, but the repressive marker decreased in differentiated cells. Only the repressive marker was detected in Hepa1c1c7 cells (B). Data are mean ± SEM, n = 3.
The Arnt2 promoter in undifferentiated ES cells contained both H3K27me3 and H3K4me3, two opposing epigenetic markers (Figure 7B). This phenomenon, previously described as a bivalent marker, has been typically found in the promoters of genes that are poised for expression during early development (39–41). It has been proposed that H3K27me3 and the associated polycomb repressor complex tends to override the active marker H3K4me3 at bivalent promoters (39,41), consistent with our finding of the high Arnt/Arnt2 ratio in untreated ES cells.
On differentiation, bivalently marked genes can be activated or repressed in a tissue-specific manner via conversion into monovalency (41). In keeping with this model, we found a decrease in the presence of H3K27me3 at the Arnt2 promoter following 7-day neuronal differentiation of ES cells (Figure 7B), which correlated with the increase in Arnt2 expression. Notably, in confirmation of our results, global ChIP-sequencing analyses of H3K4me3 and H3K27me3-marked chromatin reported that the Arnt2 promoter contained both of these modifications in undifferentiated ES cells, but only H3K4me3 in neural progenitor cells (41).
In Hepa1c1c7, we found the Arnt2 promoter contained no H3K4me3 and some H3K27me3 (Figure 7B). This is again consistent with the lack of Arnt2 expression in non-neuronal differentiated cells.
In summary, Arnt2 expression in undifferentiated ES cells was dampened through bivalent epigenetic modification, and loss of the repressive marker on neuronal differentiation led to a substantial increase in expression. Arnt2 was fully repressed in non-neuronal Hepa1c1c7 cells, through hypermethylation of its promoter and persistence of the H3K27me3-repressive epigenetic modification. Ubiquitous Arnt expression across various cell types was in agreement with presence of the active H3K4me3 epigenetic marker and absence of repressive H3K27me3 in its promoter.
DISCUSSION
In this study, we provide evidence that neuronal differentiation of P19 and ES cells alters the expression levels of Arnt and Arnt2, leading to increased Arnt2 and decreased Arnt at both mRNA and protein levels (Figures 1–4). Increased Arnt2 expression has also been reported in NGF (nerve growth factor)-treated rat pheochromocytoma PC12 cells, a rat model for neuronal differentiation (42), providing a third example of this phenomenon and demonstrating that the mode of Arnt/Arnt2 switch following neuronal differentiation is likely to be general and independent of cell lines studied. Furthermore, the expression pattern of Arnt/Arnt2 in our in vitro models also matches well with the in vivo expression pattern of these two proteins (8–11).
Alternation of Arnt/Arnt2 expression following neuronal differentiation is a distinctly regulated process, as muscular differentiation of P19 cells did not reproduce this phenomenon (Figures 1 and 2). It appears that at least two independent mechanisms exist in regulating Arnt2 expression in non-neuronal cells. First of all, the expression of Arnt2 in non-neuronal non-pluripotent cells can be silenced by DNA hypermethylation, resulting in a complete repression of Arnt2 gene expression. This was demonstrated by studies in hepatoma Hepa1c1c7 cells, where the expression of Arnt2 was fully repressed (Figure 6 and Supplementary Figure S1). In addition, expression of Arnt2 in pluripotent ES and P19 cells could also be subject to polycomb-dependent regulation, which only partially represses Arnt2 expression. In this scenario, repression can be readily reversed by alteration of polycomb-associated epigenetic marks such as H3K27me3. Alternatively, a bivalent Arnt2 promoter may receive signals to remove positive epigenetic marks such as H3K4me3 during differentiation to non-neural cell types. It will now be interesting to test this model by tracing epigenetic marks on the Arnt2 promoter while subjecting ES cells to some of the emerging in vitro differentiation protocols shown capable of providing a range of cell lineages.
In attempts to understand the mechanism behind reduced Arnt expression, we examined the effects of P19 neuronal differentiation on miR-107 expression, as this was recently shown to reduce Arnt levels in response to p53 activity in human colon cancer specimens (43), and analysis of mouse embryos suggest that miR-107 is expressed predominately in the brain and neural tube, but not in the other developing organs (44). However, qRT-PCR analysis showed only minor increases in the expression levels of both pre-miR-107 and the miR-107-harbouring Pank1 gene at different stages of P19 neuronal differentiation (data not shown), making it unlikely miR-107 exerts a major influence on attenuated Arnt expression. As Arnt protein loss is more rapid than the reduction in Arnt mRNA levels, we suspect that Arnt protein loss most likely results from a signal-induced ubiquitylation/proteasome pathway or protease activity.
Although it is important to explore the mechanism of the switch between Arnt species during neuronal differentiation, the question of why this switch occurs now needs to be addressed. We have observed that as Arnt2 becomes expressed during the differentiation process, the neural bHLH/PAS transcription factors Sim1 and NPAS4 also begin to be expressed (Hao, unpublished results). The data imply that a switch may be needed for function of Sim1 and/or NPAS4, or perhaps other neuronal transcription factors such as NPAS1 and NPAS3. Mouse knock out studies have revealed Arnt2 to be the in vivo partner of Sim1 in the hypothalamus (17), and Arnt2 has also been reported to be the predominant partner of NPAS4 in the brain (19). Ooe et al. suggest that NPAS4 has higher affinity for Arnt2 than Arnt, consistent with the idea that Arnt2 may be needed for neuronal functions unable to be performed by Arnt. Interestingly, a reciprocal scenario exists for the AhR, which activates its classic endogenous target gene Cyp1a1 when co-expressed with Arnt, but not with Arnt2 (10,27). Thus, the species of Arnt partner protein may have a previously unappreciated role in defining output of varying bHLH/PAS complexes. Although in vitro experiments and transfected reporter gene assays have generally failed to detect specificity between Arnt and Arnt2 when paired with a range of bHLH/PAS proteins, activities on endogenous target genes, which are largely ill-defined for Sim1 and NPAS4, have yet to be analysed.
Ancestral genes of the AhR play roles in neuron development in both Caenorhabditis elegans (45) and Drosophila (46), both of which express a single equivalent of Arnt. Mammalian AhR is ubiquitous and has recently been reported to function in proliferation, survival and differentiation of neurons in the dentate gyrus of the hippocampus (47), a region of the brain which shows high NPAS4 expression (18). A possible reason for the altered ratio of Arnt/Arnt2 expression may be to allow AhR and NPAS4 to function in separate pathways without competing for a common partner factor, thus avoiding cross interference. Analysis of cell extracts has revealed the AhR to have a higher affinity for Arnt than Arnt2 within the cell (10,27,48); thus, increasing Arnt2 expression in neurons could provide a second Arnt species with dampened interaction with the AhR, clearing the way for dimerization with NPAS4 in the hippocampus, or likewise Sim1 in the hypothalamus, or perhaps other bHLH/PAS proteins elsewhere in the brain. In higher organisms, the extra form of Arnt may well have arisen to allow increased complexity and specialization of neuronal bHLH/PAS factors. In keeping with this model, BMAL (Brain and Muscle ARNT-like) is a further bHLH/PAS protein with homology to Arnt in the bHLH and PAS domains, yet in vivo BMAL seems to exclusively dimerize with the circadian rhythm factor, Clock (1). Clock/BMAL complexes drive the circadian pacemaker in the suprachiasmatic nucleus of the hypothalamus, providing a prototypical example of specific output from an Arnt-like protein. In conclusion, although it is apparent that both Arnt and Arnt2 have the capacity to interact with a wide range of bHLH/PAS proteins (e.g. Sim1, Sim2, NPAS4, HIF-1α, HIF-2α, AhR), specific complexes may be favoured in a given cell type, determined by factors such as the relative expression level of each Arnt together with possible cell-specific modifying factors or signal-induced posttranslational modifications.
Finally, the AhR has recently been shown to function as a ubiquitin ligase for certain nuclear receptors, such as the Estrogen Receptor (ER), with Arnt being found in the AhR ubiquitin ligase complex (49). An unexplored alternative possibility is that decreased Arnt in neurons may be needed to dampen detrimental AhR-induced ubiquitylation, and associated proteasomal degradation, of proteins important for neuron function, e.g. the ER. A switch to predominantly Arnt2 in neurons would thus allow neuronal bHLH/PAS factors to perform essential functions free of possible side effects that may occur in the presence of high levels of Arnt.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
ACKNOWLEDGEMENTS
The mouse monoclonal antibody against myosin heavy chain (MF-20) developed by Dr D.A. Fischman was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, USA.
FUNDING
Australian Research Council. Funding for open access charge: The University of Adelaide, Adelaide, South Australia, Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1.McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 2.Furness SG, Lees MJ, Whitelaw ML. The dioxin (aryl hydrocarbon) receptor as a model for adaptive responses of bHLH/PAS transcription factors. FEBS Lett. 2007;581:3616–3625. doi: 10.1016/j.febslet.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Antonsson C, Arulampalam V, Whitelaw ML, Pettersson S, Poellinger L. Constitutive function of the basic helix-loop-helix/PAS factor Arnt. Regulation of target promoters via the E box motif. J. Biol. Chem. 1995;270:13968–13972. doi: 10.1074/jbc.270.23.13968. [DOI] [PubMed] [Google Scholar]
- 4.Sogawa K, Nakano R, Kobayashi A, Kikuchi Y, Ohe N, Matsushita N, Fujii-Kuriyama Y. Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc. Natl Acad. Sci. USA. 1995;92:1936–1940. doi: 10.1073/pnas.92.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Shi S, Zhang R, Hankinson O. Identifying target genes of the aryl hydrocarbon receptor nuclear translocator (Arnt) using DNA microarray analysis. Biol. Chem. 2006;387:1215–1218. doi: 10.1515/BC.2006.150. [DOI] [PubMed] [Google Scholar]
- 7.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt) Mol. Cell. Biol. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain S, Maltepe E, Lu MM, Simon C, Bradfield CA. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech. Dev. 1998;73:117–123. doi: 10.1016/s0925-4773(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 9.Aitola MH, Pelto-Huikko MT. Expression of Arnt and Arnt2 mRNA in developing murine tissues. J. Histochem. Cytochem. 2003;51:41–54. doi: 10.1177/002215540305100106. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty EJ, Pollenz RS. Analysis of Ah receptor-ARNT and Ah receptor-ARNT2 complexes in vitro and in cell culture. Toxicol. Sci. 2008;103:191–206. doi: 10.1093/toxsci/kfm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeburg PB, Abrahamson DR. Divergent expression patterns for hypoxia-inducible factor-1beta and aryl hydrocarbon receptor nuclear transporter-2 in developing kidney. J. Am. Soc. Nephrol. 2004;15:2569–2578. doi: 10.1097/01.ASN.0000141464.02967.29. [DOI] [PubMed] [Google Scholar]
- 12.Martinez V, Kennedy S, Doolan P, Gammell P, Joyce H, Kenny E, Prakash Mehta J, Ryan E, O'Connor R, Crown J, et al. Drug metabolism-related genes as potential biomarkers: analysis of expression in normal and tumour breast tissue. Breast Cancer Res. Treat. 2008;110:521–530. doi: 10.1007/s10549-007-9739-9. [DOI] [PubMed] [Google Scholar]
- 13.Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 14.Hill AJ, Heiden TC, Heideman W, Peterson RE. Potential roles of Arnt2 in zebrafish larval development. Zebrafish. 2009;6:79–91. doi: 10.1089/zeb.2008.0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosoya T, Oda Y, Takahashi S, Morita M, Kawauchi S, Ema M, Yamamoto M, Fujii-Kuriyama Y. Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes Cells. 2001;6:361–374. doi: 10.1046/j.1365-2443.2001.00421.x. [DOI] [PubMed] [Google Scholar]
- 16.Keith B, Adelman DM, Simon MC. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc. Natl Acad. Sci. USA. 2001;98:6692–6697. doi: 10.1073/pnas.121494298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 2000;90:253–261. doi: 10.1016/s0925-4773(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 18.Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ooe N, Saito K, Kaneko H. Characterization of functional heterodimer partners in brain for a bHLH-PAS factor NXF. Biochim. Biophys. Acta. 2009;1789:192–197. doi: 10.1016/j.bbagrm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Woods SL, Whitelaw ML. Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J. Biol. Chem. 2002;277:10236–10243. doi: 10.1074/jbc.M110752200. [DOI] [PubMed] [Google Scholar]
- 21.Maltepe E, Schmidt JV, Baunoch D, Bradfield CA, Simon MC. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 22.Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev. Biol. 1997;191:297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- 23.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maltepe E, Keith B, Arsham AM, Brorson JR, Simon MC. The role of ARNT2 in tumor angiogenesis and the neural response to hypoxia. Biochem. Biophys. Res. Commun. 2000;273:231–238. doi: 10.1006/bbrc.2000.2928. [DOI] [PubMed] [Google Scholar]
- 27.Sekine H, Mimura J, Yamamoto M, Fujii-Kuriyama Y. Unique and overlapping transcriptional roles of arylhydrocarbon receptor nuclear translocator (Arnt) and Arnt2 in xenobiotic and hypoxic responses. J. Biol. Chem. 2006;281:37507–37516. doi: 10.1074/jbc.M606910200. [DOI] [PubMed] [Google Scholar]
- 28.Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J. Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 30.Hao N, Whitelaw ML, Shearwin KE, Dodd IB, Chapman-Smith A. Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Res. 2011;39:3695–3709. doi: 10.1093/nar/gkq1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao N, Lee KL, Furness SG, Bosdotter C, Poellinger L, Whitelaw ML. Xenobiotics and loss of cell adhesion drive distinct transcriptional outcomes by aryl hydrocarbon receptor signaling. Molecular pharmacology. 2012;82:1082–1093. doi: 10.1124/mol.112.078873. [DOI] [PubMed] [Google Scholar]
- 33.Belsham DD, Cai F, Cui H, Smukler SR, Salapatek AM, Shkreta L. Generation of a phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology. 2004;145:393–400. doi: 10.1210/en.2003-0946. [DOI] [PubMed] [Google Scholar]
- 34.McBurney MW, Jones-Villeneuve EM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- 35.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 37.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 38.Hatada I, Morita S, Kimura M, Horii T, Yamashita R, Nakai K. Genome-wide demethylation during neural differentiation of P19 embryonal carcinoma cells. J. Hum. Genet. 2008;53:185–191. doi: 10.1007/s10038-007-0228-0. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 40.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 41.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drutel G, Heron A, Kathmann M, Gros C, Mace S, Plotkine M, Schwartz JC, Arrang JM. ARNT2, a transcription factor for brain neuron survival? Eur. J. Neurosci. 1999;11:1545–1553. doi: 10.1046/j.1460-9568.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- 43.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl Acad. Sci. USA. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoesel B, Bhujabal Z, Przemeck GK, Kurz-Drexler A, Weisenhorn DM, Angelis MH, Beckers J. Combination of in silico and in situ hybridisation approaches to identify potential Dll1 associated miRNAs during mouse embryogenesis. Gene Expr. Patterns. 2010;10:265–273. doi: 10.1016/j.gep.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, Powell-Coffman JA, Jin Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development. 2004;131:819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- 46.Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latchney SE, Hein AM, Kerry O'Banion M, Dicicco-Bloom E, Opanashuk LA. Deletion or activation of the Aryl hydrocarbon receptor (AhR) alters adult hippocampal neurogenesis and contextual fear memory. J Neurochem. 2013;125:430–445. doi: 10.1111/jnc.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hankinson O. Why does ARNT2 behave differently from ARNT? Toxicol. Sci. 2008;103:1–3. doi: 10.1093/toxsci/kfn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc. Natl Acad. Sci. USA. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.