Abstract
Tristetraprolin (TTP) and let-7 microRNA exhibit suppressive effects on cell growth through down-regulation of oncogenes. Both TTP and let-7 are often repressed in human cancers, thereby promoting oncogenesis by derepressing their target genes. However, the precise mechanism of this repression is unknown. We here demonstrate that p53 stimulated by the DNA-damaging agent doxorubicin (DOX) induced the expression of TTP in cancer cells. TTP in turn increased let-7 levels through down-regulation of Lin28a. Correspondingly, cancer cells with mutations or inhibition of p53 failed to induce the expression of both TTP and let-7 on treatment with DOX. Down-regulation of TTP by small interfering RNAs attenuated the inhibitory effect of DOX on let-7 expression and cell growth. Therefore, TTP provides an important link between p53 activation induced by DNA damage and let-7 biogenesis. These novel findings provide a mechanism for the widespread decrease in TTP and let-7 and chemoresistance observed in human cancers.
INTRODUCTION
The tumor suppressor p53 is a transcription factor that responds to various types of cellular stress, including DNA damage and oncogene activation. p53 regulates the expression of genes involved in a variety of cellular functions, including cell cycle arrest, DNA repair and apoptosis (1–3). Abnormalities in the p53 pathway are found in nearly all types of cancers, and p53 mutations are often associated with aggressiveness of tumor and poor patient prognosis (4). To date, ∼180 human genes have been experimentally confirmed as p53 targets (5).
MicroRNAs (miRNAs) are a class of short (21–25 nucleotides), single-stranded, non-coding RNAs. They bind to the 3′-untranslated regions (3′-UTRs) or protein-coding exons of specific mRNAs and inhibit translation or promote degradation of the transcript (6). Repression of miRNA biogenesis by suppression of the key components of miRNA processing machinery, such as Drosha and Dicer, promotes cellular transformation and tumorigenesis (7). This suggests that miRNAs might have intrinsic functions in tumor suppression and their down-regulation may accelerate oncogenesis. Recent studies have demonstrated a direct connection between p53 tumor suppressor networks and miRNA biogenesis machinery (8–12). This suggests that p53 mutations can down-regulate transcription and processing of miRNAs in human cancers. The miRNA let-7 functions as a tumor suppressor by targeting multiple oncogenes, and a reduction of let-7 levels is strongly associated with increased tumorigenicity and poor patient prognosis (13). Recently, Lin28 proteins have been identified as regulatory factors for let-7 biogenesis (14,15). In human tumors, Lin28 is up-regulated and reactivated to function as an oncogene promoting malignant transformation and tumor progression (16). Thus, down-regulation of Lin28 in tumors that overexpress this gene may provide a means to reactivate expression of the let-7 tumor suppressor (16).
Post-transcriptional regulation of gene expression can be mediated by AU-rich elements (AREs) located in the 3′-UTR of a variety of short-lived mRNAs such as cytokines and proto-oncogenes (17). The destabilizing function of AREs is regulated by ARE-binding proteins (18). One of the best-characterized ARE-binding proteins is tristetraprolin (TTP), which promotes degradation of ARE-containing transcripts (19,20). Previously, we have reported that Lin28a mRNA contains AREs within the 3′-UTR, and TTP acts as a positive regulator of let-7 biogenesis by down-regulating Lin28a expression in human cancer cells (21). However, TTP expression was shown to be significantly decreased in various cancers (22), which correlates with reduced expression of let-7 (13) and, as a result, may lead to abnormalities that contribute to cancer processes. Therefore, understanding the mechanisms that regulate TTP levels in cancer cells may provide new insights for controlling of the expression of oncogenes and the let-7 tumor suppressor.
We show here for the first time that p53 stimulated by the genotoxic agent doxorubicin (DOX) induced the expression of TTP in human cancer cells. This induction led to an increase in let-7 levels through down-regulation of Lin28a, a negative regulator of let-7. These studies thus represent a novel signaling pathway by which TTP complements p53 function through down-regulation of oncogenes and induction of let-7. These novel findings suggest that p53 serves as a positive regulator of both TTP and let-7 expression and provides a mechanism for the widespread decrease in TTP and let-7 observed in human cancers.
MATERIALS AND METHODS
Patients
Tissue samples were obtained from 45 patients with colonic adenocarcinoma who underwent surgical treatment at the Ulsan University Hospital during 2006 and 2007. Forty-eight samples of normal colonic mucosa were taken from tissues distant from the tumors. Tissue samples were fixed with 10% neutral formalin and embedded in paraffin. The Local Ethical Committee of Ulsan University Hospital approved this work.
Plasmids, small interfering RNAs, transfections and dual-luciferase assay
The pcDNA6/V5-TTP and the psiCHECK/Lin28a 3′-UTR (21) constructs were described previously. The pcDNA3/Flag-Lin28a construct was a gift from Dr. V. Narry Kim (Seoul National University, Seoul, Korea). pCMV-p53WT (containing human wild-type p53) and pCMV-p53MUT (human p53 mutant) constructs were purchased from Clontech (631922), and pGL3 basic and pRL-SV40 from Promega (E2231).
Small interfering RNAs (siRNAs) against human TTP (TTP-siRNA, sc-36761), human Lin28a (Lin28a-siRNA, sc-106829), human p53 (p53-siRNA, sc-29435) and control siRNA [scrambled siRNA (scRNA), sc-37007] were purchased from Santa Cruz Biotechnology (Santa Cruz). Cells were transfected at 24 h after plating using Lipofectamine™ RNAiMAX (Invitrogen) and were harvested at 48 h. The expression levels of TTP, p53 or Lin28 mRNA and protein were analyzed by RT-PCR and western blots. To monitor transfection efficiency, the GFP expression vector pEGFP-N1 (Clontech) was co-transfected with the plasmid construct or each oligonucleotide. After confirming transfection efficiency (>80%), cells were used for further study.
The sequence of the human TTP genomic locus at 19q13.1 (GenBank accession number NC_000019.9) was used to design PCR cloning primers. A 1411-bp genomic fragment containing the 5′-flanking region of the TTP gene was isolated by PCR amplification from human genomic DNA. Detailed procedures for generation of deletion and point mutants can be found in Supplemental Experimental Procedures.
For luciferase assays, cells were co-transfected with pGL3/TTPp-1343-luciferase reporter construct, pCMV-p53WT or pCMV-p53MUT (Clontech) and pRL-SV40 Renilla luciferase construct using TurboFect™ in vitro transfection reagent (Fermentas). Transfected cells were lysed with lysis buffer and mixed with luciferase assay reagent (Promega), and the chemiluminescent signal was measured in a SpectraMax L Microplate (Molecular Devices, Sunnyvale, CA). Firefly luciferase was normalized to Renilla luciferase in each sample. All luciferase assays reported here represent at least three independent experiments, each consisting of three wells per transfection.
Electrophoretic mobility shift assay
Nuclear extracts of PA1 cells were prepared with the NE-PER nuclear extraction reagent (Pierce). Biotin end-labeled double-stranded oligonucleotides (hTTP-p53RE-134 WT, 5′-CGGAAGGGAACCAGTCCAGGGCCAGCCAGGCTGCGCCGG-3′; hTTP-p53RE-134 MUT, 5′-CGGAAGGGAATCGCTTCGCGGTCGCTCGCGCTGCGCCGG-3′) were generated using an oligonucleotide synthesizer (IDT DNA Technologies). For competition electrophoretic mobility shift assay (EMSA), 100-fold (4 pmol) excess of unlabeled double-stranded probe was added to the binding reaction. Detailed procedures for EMSA assay can be found in Supplemental Experimental Procedures.
RNA EMSA
TTP binding to Lin28 mRNA 3′-UTR was determined by RNA EMSA as described previously (21). Briefly, cytoplasmic extracts were prepared from DOX-treated PA1 cells using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Pierce Biotechnology Scientific, Rockford, IL). EMSAs were performed using biotinylated RNA probes (21) and the LightShift™ Chemiluminescent EMSA kit (Thermo Pierce Biotechnology Scientific). Anti-TTP antibody (T5327, Sigma) and control antibody (I-5381, Sigma) were used for RNA EMSA supershift.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-Magna ChIP™ G Kit (Millipore) according to the manufacturer’s instructions. The nuclei were isolated from cells. Isolated nuclei were sonicated to shear the DNA (average fragment size was 500 bp), and chromatin was immunoprecipitated with anti-p53 antibody or isotype control. The complexes were collected on protein-G-Magnetic beads and subsequently extracted from the beads. Bound DNA was purified and amplified by PCR with primers: TTPp primers (5′-GCCCAGGGCCGGGCGGAA-3′ and 5′-AGGAAACTGCAAGCACGG-3′), which amplified the 130-bp region (−157 to −28) of the TTP promoter containing target p53 motifs; p21p primers (5′-CTGGACTGGGCACTCTTGTC-3′ and 5′-CTCCTACCATCCCCTTCCTC-3′), which amplified the 214-bp region (−2376 to −2163) of the p21 promoter containing target p53 motif.
SDS–PAGE analysis and immunoblotting
Proteins were resolved by SDS–PAGE, transferred onto Hybond-P membranes (Amersham Biosciences Inc.) and probed with appropriate dilutions of the following antibodies: rabbit anti-human TTP (T5327, Sigma), anti-human Lin28a (ab46020, Abcam), anti-p53 (1026-1, Epitomics), anti-phospho-p53 (#9284, Cell Signaling), anti-p21 (2990-1, Epitomics), anti-caspase-3 (sc-7272, Santa Cruz) and anti-β-actin (A2228, Sigma). Immunoreactivity was detected using the ECL detection system (Amersham Biosciences Inc.). Films were exposed at multiple time points to ensure that the images were not saturated.
RNA kinetics, quantitative real-time PCR and semi-qRT-PCR
For RNA kinetic analysis, we used actinomycin D and assessed TTP mRNA expression by quantitative real-time PCR (qRT-PCR). DNase I-treated total RNA (3 µg) was reverse transcribed using oligo-dT and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed by monitoring in real-time the increase in fluorescence of SYBR Green dye (QIAGEN, Hilden, Germany) using StepOnePlus™ real-time PCR systems (Applied Biosystems). Semi-qRT-PCR was performed using Taq polymerase (Solgent, Daejeon, Korea) and PCR primer pairs (Table S1).
miRNA analysis
Total RNA was isolated with Trizol reagent (Ambion). Quantitative levels of mature let-7 b and let-7f were determined using TaqMan® MicroRNA Assay kits (let-7 b-002619 and let-7f-000382, Applied Biosystems) according to the manufacturer’s protocol. Mature miRNAs were detected using StepOne Plus by monitoring in real-time the increase in fluorescence of TaqMan probe-based detection (Applied Biosystem).
Statistical analysis
Differences in the expression of TTP and p53 among clinicopathological groups were evaluated by the Student’s t-test. A P value of <0.05 was considered statistically significant.
RESULTS
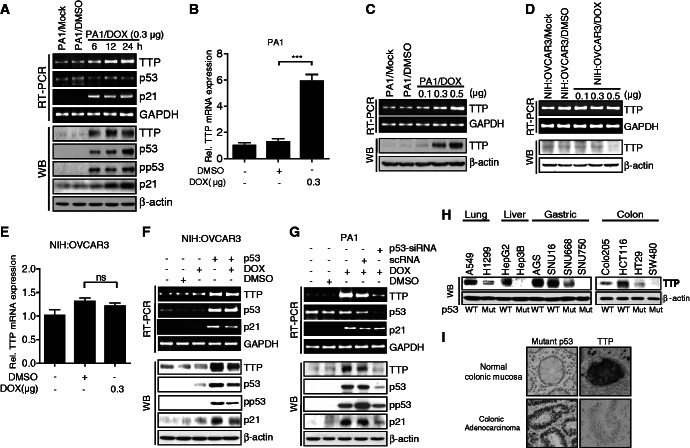
TTP expression is induced by DOX in a p53-dependent manner
A search for transcription factor–binding sites within the TTP promoter region using online software (MatInspector) revealed the presence of three potential p53 half-sites. We first determined the involvement of p53 in TTP expression by measuring TTP levels after treatment with the DNA-damaging agent DOX, a potent p53 inducer, in the p53 wild-type PA1 human ovarian cancer cell line. DOX treatment increased the levels of total and phosphorylated p53 proteins in a time-dependent manner (Figure 1A). We found that exposure to DOX increased not only a known p53 target, p21, but also both TTP protein and mRNA levels (Figure 1A and B). We also confirmed that DOX treatment increased TTP levels in a dose-dependent manner (Figure 1C). The levels of TTP transcript and protein were also increased in p53 wild-type MCF7 human breast cancer cells (Supplementary Figure S1A–C) and p53 wild-type Colo205 human colon cancer cells (Supplementary Figure S2A and B) after DOX treatment. To determine whether TTP can be induced by other DNA-damaging agents, MCF7 cells were treated with 0.1 μg/ml 5-FU or 50 μM etoposide for 24 h and analyzed for the levels of p53, p21 and TTP. They increased both TTP protein and mRNA levels (Supplementary Figure S3), indicating that induction of TTP is a global response to DNA damage.
Figure 1.
DOX induces expression of TTP mRNA and protein by a p53-dependent mechanism in human cancer cells. (A–C) DOX increases TTP levels in p53 wild-type human ovarian cancer PA1 cells. PA1 cells were treated with the indicated concentration of DOX for (A) the indicated length of time or (B, C) 24 h. The levels of TTP, p53 and p21 were measured by semi-qRT-PCR (A and C, top), qRT-PCR (B) and western blotting (A and C, bottom). The level of phospho-p53 (pp53) was measured by western blotting (A, bottom). Values are means ± SD (n = 3). ***P < 0.001. (D–G) p53 is required for DOX-induced expression of TTP. (D, E) p53 mutant human ovarian cancer NIH:OVCAR3 cells were treated with the indicated concentration of DOX for 24 h. The level of TTP was measured by semi-qRT-PCR (D, top), qRT-PCR (E) and western blotting (D, bottom). Vaules are means ± SD (n = 3). ns, not significant. (F) NIH:OVCAR3 cells were transfected with pCMV-p53WT. After treatment with 0.3 μg/ml of DOX for 24 h, the levels of TTP, p53 and p21 were measured by semi-qRT PCR (top) and western blotting (bottom). (G) PA1 cells were transfected with siRNA against p53 (p53-siRNA) or scRNA. After treatment with DOX for 24 h, the levels of TTP, p53, and p21 were measured by semi-qRT PCR (top) and western blotting (bottom). (H) The level of TTP protein is inversely correlated with that of p53 protein in several human cell lines. Levels of TTP and p53 proteins were determined by western blot analysis in lung adenocarcinoma (A549 and H1299), hepatocellular carcinoma (HepG2 and Hep3B), gastric adenocarcinoma (AGS, SNU16, SNU668 and SNU750), colon adenocarcinoma (Colo205, HCT116, HT29 and SW480). WT, p53 wild type; Mut, p53 mutant. (I) TTP expression levels are inversely correlated with that of mutant p53 in human colon tissues. Representative immunohistochemical staining of TTP and mutant p53 in normal and colon adenocarcinoma tissues.
To investigate whether DOX-induced expression of TTP is functionally dependent on p53, we measured TTP levels in p53 mutant human cancer cells such as NIH:OVCAR3 human ovarian cancer cells and MDA-MB231 human breast cancer cells after DOX treatment. In both NIH:OVCAR3 (Figure 1D–F) and MDA-MB231 cells (Supplementary Figure S1D–F), DOX treatment did not significantly increase the levels of p53, p21 or TTP. To further confirm the involvement of p53 in DOX-induced expression of TTP, we examined the effect of silencing endogenous p53 using siRNA on TTP expression induced by DOX in PA1, MCF7 and Colo205 cells. Down-regulation of p53 significantly attenuated the expression of TTP transcript and protein induced by DOX (Figure 1G, Supplementary Figure S1G and S2C). We next transfected NIH:OVCAR3 and MDA-MB231 cells with pCMV-p53WT to overexpress wild-type p53. DOX treatment of NIH:OVCAR3 and MDA-MB231 cells together with ectopic expression of wild-type p53 increased the levels of TTP transcript and protein (Figure 1F and Supplementary Figure S1F). These data indicate that the human TTP promoter is under positive regulation by p53 in cancer cells.
Expression of TTP is inversely correlated with p53 mutations in human cancer cell lines and colonic adenocarcinoma tissues
To determine the relationship between p53 mutations and TTP expression, we analyzed the expression of TTP in six p53 wild-type (A549, HepG2, AGS, SNU16, Colo205 and HCT116) and six p53 mutant (H1299, Hep3B, SNU668, SNU750, HT29 and SW480) human cancer cell lines. While cell lines with wild-type p53 showed high expression levels of TTP, those with mutant p53 showed a low level of TTP (Figure 1H), suggesting a negative correlation between p53 mutations and TTP expression in human cancer cell lines. To evaluate the contribution of p53 in TTP expression in p53 wild-type cancer cells, we determine the effect of p53 down-regulation on the TTP expression level in p53 wild-type MCF7 cells. Knock-down of p53 by siRNA treatment decreased TTP level (Supplementary Figure S4A), indicating that TTP expression in p53 wild-type cancer cells dependents on p53. TTP expression has been reported to be decreased in many tumor types (22). To compare the TTP expression levels in Colo205 and HCT116 colon cancer cells with those in human normal colonic mucosa, we conducted western blotting. The TTP levels in both Colo205 and HCT116 were lower than those in normal tissues (Supplementary Figure S4B). We next determined whether TTP protein expressed in cancer cells is active in promoting mRNA decay. A549 (with high TTP level) and H1299 (with low TTP level) cells were transfected with a luciferase reporter gene linked to 3′-UTR fragments of Lin28a, a TTP target gene (21). The luciferase activity in A549 cells was significantly lower than that in H1299 cells (Supplementary Figure S5). TTP knock-down by siRNA treatment significantly increased the luciferase activity in A549 cells (Supplementary Figure S5), indicating that TTP in p53 wild-type cancer cells is active.
It has been reported that missense mutations of p53 increase its stability and high levels of p53 correlate with p53 mutations in tumor samples (23). To determine the correlation between p53 mutations and TTP expression, we examined the expression of p53 and TTP proteins in 45 colonic adenocarcinoma tissues by immunohistochemical staining. Forty-eight normal colonic mucosa tissues distant from the cancer of the surgically resected specimens were used as normal controls. p53 expression was extremely low in normal mucosa (mean staining score, 0.65 ± 0.14) and high in colon cancer (mean staining score, 9.00 ± 0.64) (P < 0.0001) (Figure 1I, Table 1). In contrast, expression of TTP was significantly low in colon cancer (mean staining score, 2.29 ± 0.31) but high in non-malignant tissues (mean staining score, 8.35 ± 0.37) (P < 0.0001; Figure 1I and Table 1). These results suggest that the expression level of TTP is inversely correlated with expression of p53 in colon carcinoma.
Table 1.
TTP and p53 expression and clinicopathologic features of patients with colon cancers
| Disease index | n | p53 |
TTP |
||
|---|---|---|---|---|---|
| Staining scorea | P value | Staining scorea | P value | ||
| Tumor | 45 | 9.00 ± 0.64 | <0.0001 | 2.29 ± 0.31 | <0.0001 |
| Normal mucosa | 48 | 0.65 ± 0.14 | 8.35 ± 0.37 | ||
aMean ± s.e.m.
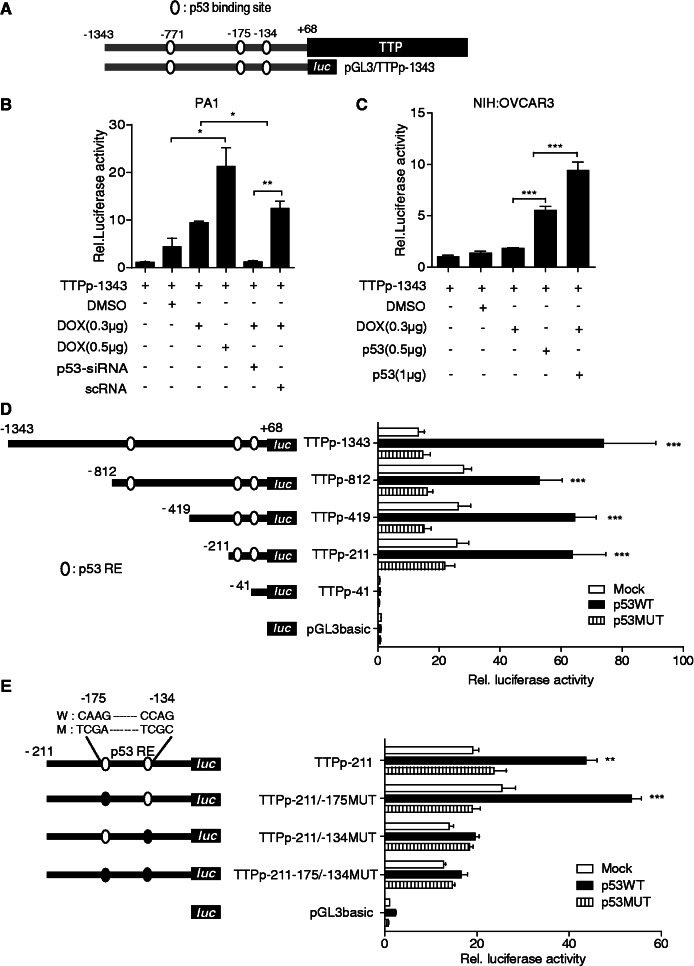
p53 stimulated by DOX enhances TTP promoter activity
The experiments above indicated that DOX was able to induce TTP transcription and its induction was p53-dependent. We first determined whether DOX treatment affects the TTP mRNA stability. As shown in Supplementary Figure S6, TTP mRNA stability was not affected by DOX treatment, suggesting that DOX-induced expression of TTP does not result from the increase in the TTP mRNA stability. We next tested whether DOX treatment transactivates the TTP promoter in a reporter assay. The p53 wild-type PA1 cells were transiently transfected with pGL3/TTPp-1343 construct (−1343 to +68) containing three potential p53 half-sites of the human TTP gene (Figure 2A) followed by treatment with DOX. As shown in Figure 2B, DOX treatment significantly increased TTP promoter activity in a dose-dependent manner in PA1 cells at 6 h after the treatment. To confirm the involvement of p53 in DOX-induced activation of the TTP promoter, we inhibited p53 expression in PA1 cells using siRNA. Down-regulation of p53 by siRNA (Figure 1G) abrogated DOX induction of the TTP promoter in PA1 cells (Figure 2B). Coinciding with this result, DOX treatment did not stimulate the TTP promoter in p53 mutant NIH:OVCAR3 cells (Figure 2C). We next transfected NIH:OVCAR3 cells with pCMV-p53WT to overexpress wild-type p53. DOX treatment of NIH:OVCAR3 cells together with ectopic expression of wild-type p53 increased TTP promoter activity (Figure 2C). These data indicate that the human TTP promoter is under positive regulation by p53 in human cancer cells.
Figure 2.
DOX induces TTP promoter activity in a p53-dependent manner. (A) Schematic structure of the reporter construct pGL3/TTPp-1343. The TTP promoter contains three potential p53 half-sites at positions −771, −175 and −134 relative to the transcription start site. (B, C) DOX induces TTP promoter activity in a p53-dependent manner. (B) PA1 cells were co-transfected with pGL3/TTPp-1343 containing the TTP promoter (−1343 to +68) and p53-siRNA or scRNA. (C) NIH:OVCAR3 cells were co-transfected with pGL3/TTPp-1343 and pCMV-p53WT (p53 wild type) or pCMV-p53MUT (p53 mutant). After treatment with DOX for 24 h, luciferase activity was measured. The expression levels obtained from pGL3/TTPp-1343-transfected cells without DOX treatment were set to 1. Values are means ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (D, E) p53 half-site at −134 relative to the transcription start site is required for the transcriptional activation of the TTP promoter. (D) Deletion and (E) mutation analysis of the TTP promoter in NIH:OVCAR3 cells. Schematic structures of the deletion and point mutation reporter constructs are shown on the left. Three potential p53 half-sites are indicated with open circles. The point mutations introduced into the binding sites are represented as closed circles. NIH:OVACAR3 cells were co-transfected with the TTP promoter construct and pCMV-p53WT or pCMV-p53MUT. After treatment with DOX for 24 h, cells were lysed and assayed for luciferase activity. The levels of firefly luciferase activity were normalized to Renilla luciferase activity. The expression levels obtained from pGL3 basic-transfected cells were set to 1. Values are means ± SD (n = 3). **P < 0.01; ***P < 0.001.
p53 half-site at −134 is responsible for DOX-inducible promoter activity
To evaluate the contribution of three potential p53 half-sites (5′-RRRCWWGYYY-3′; positions −771, −175 and −134 relative to the transcription start site) (Figure 2A) in the regulation of TTP promoter activity by DOX, a series of deletion constructs were prepared by inserting different fragments of the human TTP promoter with a fixed 3′-end at the +68 position (relative to the transcription start site) but a different 5′-end into the pGL3-Basic vector. NIH:OVCAR3 cells were co-transfected with the TTP promoter construct and pCMV-p53WT or pCMV-p53MUT, and promoter activity was assessed by measuring luciferase activity after DOX treatment. While DOX induced luciferase levels in cells transfected with pCMV-p53WT, it did not in cells transfected with pCMV-p53MUT (Figure 2D). As shown in Figure 2D, deletion from −1343 to −211 (pGL3/TTPp-211) did not reduce luciferase levels. However, further deletion from −211 to −41 (pGL3/TTPp-41) dramatically reduced luciferase levels to <5% of those of the parental construct pGL3/TTPp-1343, suggesting the presence of a positive regulatory element in this region. This region contains two potential p53 half-sites (positions −175 and −134 relative to the transcription start site) (Figure 2D). To evaluate the contribution of these two potential p53 half-sites in the regulation of TTP promoter activity by DOX, we made point mutations of them in the context of the reporter construct pGL3/TTPp-211, and mutant promoter activity was compared with that of the wild-type promoter. Mutation of the first p53 half-site (−175) (pGL3/TTPp-211/-175MUT) did not reduce the ability of DOX to activate transcription through this promoter (Figure 2E). However, mutation of the second p53 half-site (−134) (pGL3/TTPp-211/-134MUT) completely abrogated DOX-mediated promoter induction in PA1 null cells (Figuer 2E). These results indicate that the p53 half-site at −134 plays important role in DOX induction of TTP promoter activity.
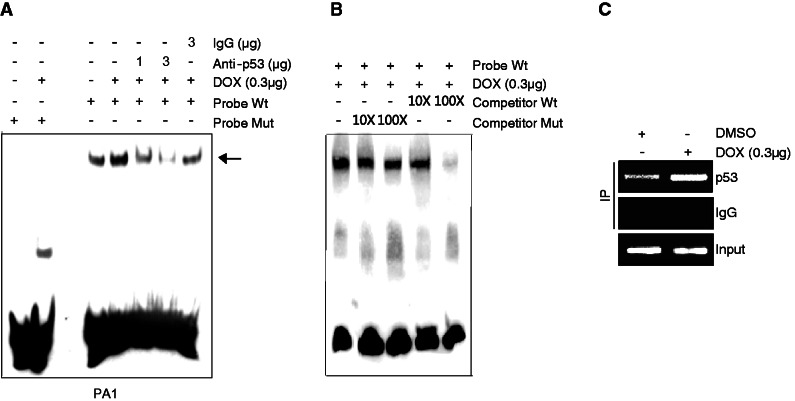
DOX enhances binding of p53 to the TTP promoter
To determine the nature of the interaction of p53 with the p53 half-site at −134 in the TTP promoter, we performed an EMSA in PA1 cells using a 23-bp oligonucleotide bearing the wild-type or mutant p53 half-site at −134 as a probe. When EMSA was conducted using the wild-type p53 probe, a dominant DNA–protein complex was observed, and DOX significantly increased the intensity of the DNA–protein complexes (Figure 3A, lanes 3 and 4). However, the mutant p53 probe prevented formation of this complex (Figure 3A, lanes 1 and 2). The complex competed with an excess of the unlabeled wild-type p53 probe but did not with unlabeled mutant p53 probe (Figure 3B), demonstrating that these DNA–protein complexes are p53 site specific. The DNA–protein complexes were dramatically reduced in a dose-dependent manner by preincubation of the reaction mixture with an anti-p53 antibody (Figure 3A, lanes 5 and 6). However, preincubation of the reaction mixture with an isotype control did not reduce the complexes (Figure 3A, lane 7). These results indicate that p53 can specifically interact with the p53 half-site at −134, and DOX increases this interaction. To confirm the direct binding of p53 to the TTP promoter in vivo, we performed a ChIP assay in PA1 cells treated with DOX. As a positive control, we determined the binding of p53 to the promoter of p21, a well-known p53 target gene. Chromatin was sonicated into fragments and precipitated using an anti-p53 or isotype control antibody. The precipitated DNA was subjected to PCR using primers designed to amplify a 130-bp fragment of the TTP promoter flanking the p53 half-site at −134 or a 214-bp fragment of the p21 promoter. In the absence of DOX, a significant level of p53 was found to be bound to the promoters of both p21 and TTP in the absence of DOX. DOX treatment significantly increased p53 binding to the promoter of both genes (Figure 3C, Supplementary Figure S7). This result indicates that DOX treatment increases recruitment of p53 to the TTP promoter in living cells.
Figure 3.
DOX enhances binding of p53 to the TTP promoter. (A, B) EMSA and (C) ChIP assay. PA1 cells were treated with DOX for 24 h. (A) EMSA was performed as described in Experimental Procedures. Biotinylated oligonucleotide containing wild-type (WT) or mutant (MUT) p53 half-site at −134 of the TTP promoter was used as a probe. To identify p53 immunoreactivity in the DNA–protein complexes, nuclear extracts were incubated with anti-p53 antibody or non-specific mouse IgG before addition of the biotinylated probe. An arrow indicates the position of the DNA–protein complexes containing p53. (B) Competition experiments were performed using a 100-fold excess of unlabeled WT and MUT p53 probes. (C) ChIP analysis was performed to confirm the interaction of p53 with the TTP promoter in vivo in PA1 cells. Formaldehyde cross-linked chromatin from PA1 cells was incubated with anti-p53 antibody or with non-specific IgG. Total input DNA at a 1:10 dilution was used as positive control for the PCR reaction. Immunoprecipitated DNA was analyzed by PCR with primers specific for the TTP promoter.
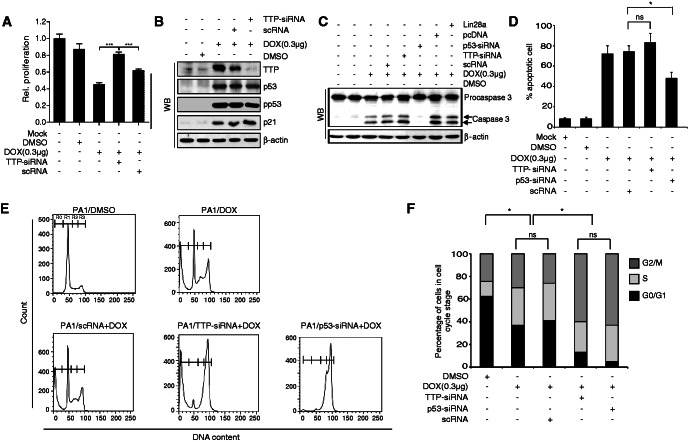
TTP mediates the inhibitory effect of DOX on cell growth
DOX treatment led to substantial inhibition (58% inhibition) of PA1 cell growth (Figure 4A). Our next goal was to determine whether TTP could mediate the growth inhibitory effect of DOX. We examined the effect of a TTP knock-down on DOX-induced inhibition of growth in PA1 cells. The treatment of siRNA against TTP (TTP-siRNA) significantly inhibited the expression of TTP in PA1 cells but showed a negligible effect on p53 and its target p21 (Figure 4B). Interestingly, TTP-siRNA but not scRNA attenuated the inhibitory effect of DOX on the growth of PA1 cells (Figure 4A). These results suggest that TTP is required for DOX-induced inhibition of PA1 cells.
Figure 4.
TTP mediates the inhibitory effect of DOX on cell growth. (A, B) Down-regulation of TTP by siRNA attenuates the inhibitory effect of DOX on cell growth. PA1 cells were transfected with TTP-specific (TTP-siRNA) or scRNA. After treatment with DOX for 24 h, (A) cell viability was assessed by measuring absorbance at 490 nm using a MTS cell proliferation assay and (B) the levels of TTP, p53, pp53 and p21 were measured by western blotting. The values obtained with mock-treated PA1 cells were set to 1. Values are means ± SD (n = 3). ***P < 0.001. (C, D) Down-regulation of TTP by siRNA does not affect DOX-induced apoptosis. PA1 cells were transfected with TTP-siRNA, p53-siRNA, scRNA), pcDNA3/Flag-Lin28a or pcDNA3/Flag. After treatment with DOX for 24 h, (C) the levels of procaspase-3 and caspase-3 were measured by western blotting. (D) Cells were labeled with an Annexin-V-FLUOS staining kit for quantitating cells undergoing apoptosis. Values are means ± SD (n = 3). *P < 0.05. ns, not significant. (E, F) Down-regulation of TTP by siRNA alters the effect of DOX on the cell cycle. PA1 cells were transfected with TTP-siRNA, p53-siRNA or scRNA. (E) After treatment with DOX for 24 h, cells were stained with propidium iodide and cell cycle distribution and apoptotic DNA fragmentation (Sub-G1) were analyzed by flow cytometry. Data are representative of three independent experiments. R0, sub-G1; R1, G0/G1; R2, S; R3, G2/M. (F) Percentage of cells at each phase in different groups. (n = 3). *P < 0.05. ns, not significant.
Several studies have shown that caspase-3 activation is required for induction of apoptosis in response to DOX (24). Similar to previous reports, we also found that DOX activated caspase-3 in PA1 cells (Figure 4C). Next, we inhibited TTP expression by using siRNA against TTP and tested whether TTP is involved in DOX-induced caspase-3 activation. As shown in Figure 4C, while knock-down of p53 blocked DOX-induced caspase-3 activity, the TTP knock-down did not affect caspase-3 activity. Consistently, while knock-down of p53 attenuated DOX-induced apoptosis, inhibition of TTP did not reduce the apoptosis (Figure 4D and E). This suggests that, while p53 mediates DOX-induced caspase-3 activation and apoptosis, TTP is not involved in caspase-3 activation and apoptosis.
DOX-induced p53 can inhibit cell proliferation through cell cycle arrest (25,26). To determine the role of TTP in DOX-induced cell cycle arrest, we inhibited TTP expression in PA1 cells by using siRNA and analyzed the effect of TTP inhibition on DOX-induced cell cycle arrest. As a control, we used siRNA to also analyze the effect of p53 inhibition on DOX-induced cell cycle arrest. DOX treatment increased the percentage of cells in S and G2/M phases (Figure 4E and F), indicating that DOX induces S and/or G2/M phase cell cycle arrest. Interestingly, a further increase of cells in S and G2/M phases was observed in cells treated with either p53-siRNA or TTP-siRNA (Figure 4E and F). The increase of cells in S and/or G2/M phase did not seem to result in an increase in cell cycle arrest because inhibition of TTP attenuated the growth inhibitory effect of DOX on cell proliferation (Figure 4A). This result suggests that TTP plays some role in regulating the cell cycle in DOX-treated cells.
DOX-induced expression of TTP down-regulates the expression of Lin28
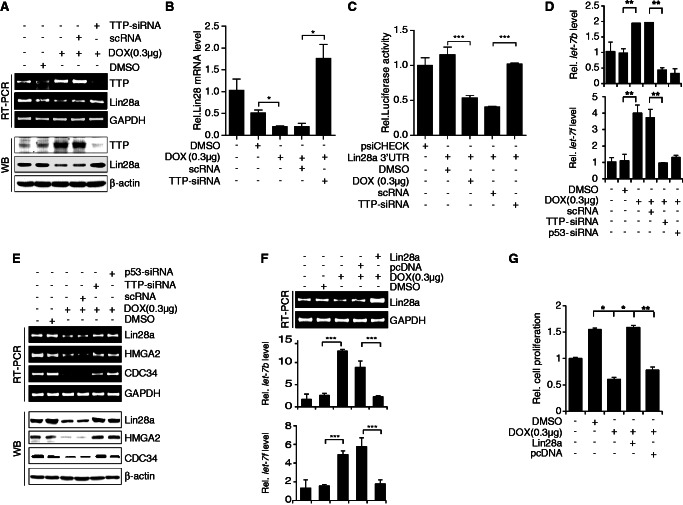
Previously, we reported that ectopic expression of TTP down-regulates the expression of Lin28a in PA1 cells (21). Our next goal was to determine whether TTP induced by DOX can inhibit the expression of Lin28a. As shown in Figure 5A and B, DOX treatment decreased the expression of Lin28a confirmed by semi-qRT-PCR (Figure 5A, top), qRT-PCR (Figure 5B) and western blot (Figure 5A, bottom). To confirm the involvement of TTP in DOX-induced down-regulation of Lin28a, we inhibited TTP expression using siRNA and analyzed Lin28a expression in DOX-treated PA1 cells. Down-regulation of TTP (Figure 5A) significantly attenuated the inhibitory effect of DOX on the expression of Lin28a transcript (Figure 5A and B) and protein (Figure 5A). In addition, treatment of siRNA against p53 (p53-siRNA) abrogated the inhibitory effect of DOX on the expression of Lin28a transcript and protein (Figure 5D). This result suggests that TTP induced by DOX down-regulates Lin28a expression in PA1 cells. TTP has been reported to down-regulate Lin28a expression by binding to the 3′-UTR of Lin28a mRNA (21). To determine whether down-regulation of Lin28a expression by DOX-induced TTP is mediated through the 3′-UTR of Lin28a mRNA, we used a luciferase reporter gene linked to Lin28a 3′-UTR fragments containing pentameric AUUUA in the plasmid psiCHECK2 (21). When PA1 cells were treated with DOX to induce TTP expression (Figure 5A), luciferase activity was inhibited (Figure 5C). The inhibitory effect of DOX on luciferase activity was attenuated by knock-down of TTP (Figure 5C). To test whether DOX increases TTP binding to Lin28a mRNA 3′-UTR, we performed RNA EMSA using the RNA probes as described previously (21). As shown in Supplementary Figure S8, wild-type Lin28a 3′-UTR probe (Lin28 WT probe) formed probe–protein complexes and DOX treatment increased the level of the complexes. The formation of the complexes was reduced by preincubation of the reaction mixture with anti-TTP antibody (Supplementary Figure S8). Taken together, these data indicate that the 3′-UTR of Lin28a mRNA is responsible for down-regulation of Lin28a by DOX-induced TTP.
Figure 5.
DOX increases let-7 levels through the p53-TTP-Lin28 pathway. (A–C) DOX decreases Lin28 levels in a TTP-dependent manner. (A, B) PA1 cells were transfected with TTP-siRNA or scRNA. After treatment with DOX for 24 h, the levels of TTP and Lin28a were measured by semi-qRT-PCR (A, top), western blotting (A, bottom) and qRT-PCR (B). Values are means ± SD (n = 3). *P < 0.05. (C) PA1 cells were co-transfected with psiCHECK2 luciferase reporter constructs containing fragments derived from the 3′-UTR of Lin28a mRNA and TTP-siRNA or scRNA. After treatment with DOX for 24 h, cells were lysed and assayed for luciferase activity. The levels of firefly luciferase activity were normalized to Renilla luciferase activity. Luciferase activity obtained from PA1 cells transfected with psiCHECK2 alone was set to 1.0. Values are means ± SD (n = 3). ***P < 0.001. (D–G) DOX increases let-7 levels through the p53-TTP-Lin28 pathway. (D, E) PA1 cells were transfected with TTP-siRNA, p53-siRNA, or scRNA. After treatment with DOX for 24 h, (D) the levels of TTP, p53, Lin28, HMGA2, and CDC34 were measured by semi-qRT-PCR (top) and western blotting (bottom) and (E) the levels of mature-miRNA for let-7 b and let-7f were determined by qRT-PCR. The levels obtained from untreated PA1 cells were set to 1. Values are means ± SD (n = 3). **P < 0.01. (F, G) PA1 cells were transfected with pcDNA3/Flag-Lin28a or pcDNA3/Flag. After treatment with DOX for 24 h, (F) the level of Lin-28 a was measured by semi-qRT-PCR (top panel) and western blotting (bottom panel) and the levels of mature-miRNA for let-7 b and let-7f were determined by qRT-PCR and (G) cell viability was assessed by measuring absorbance at 490 nm using a MTS cell proliferation assay. Values are means ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
TTP mediates DOX-induced expression of let-7
Overexpression of TTP has been reported to increase let-7 levels through down-regulation of Lin28a (21). We here examined the effect of DOX-induced TTP on let-7 levels in PA1 cells. As shown in Figure 5E, DOX treatment increased the levels of let-7 b and let-7f and decreased the levels of let-7 targets, HMGA2 and CDC34 (Figure 5D). To determine whether TTP mediates the increase of let-7 levels in DOX-treated cells, we inhibited TTP expression using siRNA and analyzed let-7 levels in DOX-treated PA1 cells. Inhibition of TTP significantly attenuated the effect of DOX on the expression of let-7 b and let-7f (Figure 5E). Inhibition of p53 by siRNA also blocked the increase of let-7 in DOX-treated PA1 cells (Figure 5E). We next determined whether ectopic overexpression of p53 in p53 null mutant cells shows the same effects on the expression of Lin28 and let-7 as DOX treatment. We transiently transfected SKOV3 cells (p53 null mutant) with pCMV-p53WT and, after 24 h incubation, analyzed expression levels of TTP, Lin28a and let-7. As shown in Supplementary Figure S9, similar to DOX treatment, overexpression of p53 significantly increased the levels of TTP and let-7 but decreased Lin28a. To determine if ectopic expression of Lin28a could block the DOX-induced increase in let-7 b levels, we transfected PA1 cells with pcDNA3/Flag-Lin28a, which does not contain the Lin28a 3′-UTR, and analyzed let-7 b levels and cell proliferation. Overexpression of Lin28a (Figure 5F) abrogated the effect of DOX on the expression of let-7 b (Figure 5F). In addition, overexpression of Lin28a blocked the inhibitory effect of DOX on PA1 cell growth (Figure 5G). Collectively, these results suggest that DOX increases let-7 levels through the p53-TTP-Lin28 pathway.
DISCUSSION
Previously we have reported that TTP increases let-7 levels through down-regulation of the negative regulator Lin28 in cancer cells (21). Both TTP and let-7 can inhibit the growth of cancer cells through down-regulation of oncogenes (13,19,20) However, a significant decrease in the expression of both TTP and let-7 has been found in many cancer cells (13,22,27). Until now, the mechanisms for the reduced expression of both TTP and let-7 in cancer cells have not been clarified. Here, we demonstrated that p53 and p53 inducers such as DOX can act as key regulators for the expression of TTP and let-7 in cancer cells. We first provided evidence that TTP is a target gene of p53: the TTP promoter region contains p53 responsible elements (REs); levels of TTP transcript and protein were increased by p53; p53 increased the expression of a luciferase reporter gene linked to a p53 RE from the TTP promoter; EMSA and ChIP analyses demonstrated that p53 binds to the p53 RE from the TTP promoter. We found that the DNA-damaging agent DOX also enhanced TTP levels. DOX is a widely used anticancer drug (28) and has been reported to exert its inhibitory functions through p53-dependent and -independent pathways (29). The induction of TTP by DOX seems to be mediated by the p53-dependent pathway because DOX did not enhance TTP levels in cells with p53 mutations and inhibition of p53 by using siRNA blocked the effect of DOX on TTP levels.
If p53 is the inducer of TTP expression in cancer cells, it is possible to speculate that TTP levels might be low in cells with p53 mutations. In accordance with our expectations, TTP levels were low in cells with p53 mutations compared with wild-type p53 cells. We also determined the relationship between p53 and TTP levels in human colonic adenocarcinoma tissues. Approximately half of human cancers have inactivating mutations of p53, and most of the remaining tumors deactivate the p53 pathway (30). Most mutant p53 proteins produced by missense mutations are resistant to degradation and thus have prolonged half-lives, allowing their detection by immunohistochemical staining (23). In the present study, immunohistochemical staining demonstrated a negative relationship between p53 and TTP expression. Thus, our results suggest that TTP levels are low in human tumor tissues with p53 mutations, perhaps explaining the low TTP levels observed in most tumor tissues (22).
Previously, we reported that TTP overexpression increases let-7 levels through down-regulation of the negative regulator Lin28 (21). In this study, we demonstrated that the DNA-damaging agent DOX induces the expression of TTP through p53 activation, suggesting the possibility that DOX may enhance let-7 levels through the p53/TTP/Lin28 pathway in cancer cells. Here, we provided evidence supporting this hypothesis: DOX treatment decreased Lin28a levels; DOX treatment increased let-7 levels and decreased let-7 target genes, HMGA2 and CDC34; inhibition of p53 or TTP by siRNAs blocked the effect of DOX on the expression of Lin28a, let-7, HMGA2 and CDC34; overexpression of Lin28a cDNA without the 3′-UTR blocked the effect of DOX on cell growth and expression of let-7, HMGA2 and CDC34. It has been reported that p53 can increase the levels of miRNAs by enhancing transcription and/or processing of miRNAs: p53 activates transcription of the miR-34 gene family, leading to production of miRNAs that promote apoptosis or senescence (8); p53 interacts with the Drosha complex, facilitating the processing of a subset of pri-miRNAs to pre-miRNAs (11). Our results uncover the existence of a novel p53/TTP/Lin28/let-7 pathway that links p53 activation with the expression of let-7. Mutational inactivation of p53 is frequently observed in various human cancers (30), and our results provide an explanation for low levels of let-7 in cancer cells (13). Besides let-7, Lin28a can negatively regulate the expression of several other miRNAs such as miR-107, miR-143 and MiR-200C (31), raising the possibility that concerted up-regulation of several Lin28 target miRNAs by p53 may occur under certain physiological conditions. Further investigation will provide insights into how miRNAs are regulated by the p53-TTP-Lin28 pathway.
Recently it has been reported that p21 mRNA contains ARE and TTP promotes its decay (32). However, in this study, TTP down-regulated Lin28a expression without affecting the expression levels of p21. Our finding is consistent with other studies demonstrating the gene- and cell type-specific activity of TTP. In glioblastoma U87 cells, TTP was found to bind to the c-myc 3′-UTR and decay its mRNA (33). However, in HeLa cells, although TTP enhanced the mRNA decay of granulocyte/macrophage colony-stimulating factor, interleukin-3, tumor necrosis factor-α, interleukin-2 and c-fos, it did not regulate the stability of c-myc mRNA (34). In A549 cells, while TTP destabilized LAT2 mRNA, it did not affect the stability of c-fos and c-myc (27). It is not clear how TTP selectively regulates the stability of target mRNAs. Previously we have reported that the binding of TTP to the 3′-UTR of cIAP2 mRNA is not enough for degradation of the target gene. Besides the sequences for TTP binding, additional nucleotide sequences are required for degradation of the target gene (35). It is possible to speculate that TTP-mediated decay of mRNAs requires additional RNA-binding proteins with sequence- and cell type-specific activities. The possible candidates for the RNA-binding proteins are heterogeneous nuclear RNPs (hnRNPs). Approximately 30 hnRNPs have been identified and they have been reported to influence diverse cellular events, including transcription, splicing, polyadenylation, mRNA localization, translation and decay in sequence- and cell type–specific manner (36–39). One recent study suggested the possibility of association between TTP and hnRNP by demonstrating that transportin, which recognizes hnRNP A1, interacts with TTP and modulates TTP-mediated decay of ARE-containing mRNAs (40). Further characterizing the other factors (especially hnRNPs) involved in TTP-mediated mediated mRNA decay will elucidate the molecular mechanisms of gene and cell type specificity of mRNA decay.
DOX induces G2/M phase cell cycle arrest (25,26), caspase-3 activation (24) and apoptosis (41) in a p53-dependent manner. In this study, we found that DOX treatment induced caspase-3 activation, apoptosis and G2/M phase cell cycle arrest in PA1 cells. Inhibition of p53 by siRNA attenuated the effects of DOX on both cell cycle arrest and apoptosis. Similar to previous reports, this finding suggests that activation of p53 is a key mechanism of action of DOX. Interestingly, inhibition of TTP by siRNA did not affect caspase-3 activation or apoptosis induced by DOX treatment but altered the inhibitory effect of DOX on cell cycle arrest. Our results indicate that induction of TTP is a key mechanism of action of p53 in mediating DOX-induced cell cycle arrest. Activation of p53 by various types of cellular stresses induces the expression of different subsets of p53 target genes, which leads to a range of cellular outcomes from cell recovery to cell death (42). Besides DOX, other DNA-damaging agents may induce the expression of TTP in human cancer cells. However, it is not clear whether TTP induced by DNA-damaging agents other than DOX affects only the cell cycle and not apoptosis in cancer cells. Further study on the interaction between TTP, TTP target genes and p53 target genes will reveal the role of TTP in various types of cellular stresses and the mechanisms underlying how TTP mediates p53-induced cell cycle arrest.
In summary, multiple levels of evidence confirm that TTP is a direct target of p53 and mediates the inhibitory effect of DOX on cell proliferation through down-regulation of oncogenes and induction of the tumor suppressor let-7. This study provides a molecular mechanism for the reduced levels of both TTP and let-7 in cancer cells reported to have various mutations of p53. In addition, the novel p53-TTP-Lin28-let-7 pathway expands our understanding of the cooperative network of tumor suppressor genes in cancer cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–9 and Supplementary Methods.
FUNDING
Korea Research Foundation Grant funded by the Korean government (MOEHRD) [KRF-2008-C00249, BRL-2009-0087350, 2009-0094050, 2010-0023905 and 2009-0070260]; Korea Health 21 R&D Project, Ministry of Health & Welfare [A101086]. Funding for open access charge: Grants from the Korea Research Foundation Grant funded by the Korean government (MOEHRD) [BRL-2009-0087350].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 5.Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D'Erchia AM, et al. p53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinformatics. 2007;8(Suppl.1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 8.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 12.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 18.Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- 19.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 20.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim CW, Vo MT, Kim HK, Lee HH, Yoon NA, Lee BJ, Min YJ, Joo WD, Cha HJ, Park JW, et al. Ectopic over-expression of tristetraprolin in human cancer cells promotes biogenesis of let-7 by down-regulation of Lin28. Nucleic Acids Res. 2012;40:3856–3869. doi: 10.1093/nar/gkr1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris CC, Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N. Engl. J. Med. 1993;329:1318–1327. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 24.Yang XH, Sladek TL, Liu X, Butler BR, Froelich CJ, Thor AD. Reconstitution of caspase 3 sensitizes MCF-7 breast cancer cells to doxorubicin- and etoposide-induced apoptosis. Cancer Res. 2001;61:348–354. [PubMed] [Google Scholar]
- 25.Ling YH, el-Naggar AK, Priebe W, Perez-Soler R. Cell cycle-dependent cytotoxicity, G2/M phase arrest, and disruption of p34cdc2/cyclin B1 activity induced by doxorubicin in synchronized P388 cells. Mol. Pharmacol. 1996;49:832–841. [PubMed] [Google Scholar]
- 26.Zimmermann M, Arachchige-Don AS, Donaldson MS, Dallapiazza RF, Cowan CE, Horne MC. Elevated cyclin G2 expression intersects with DNA damage checkpoint signaling and is required for a potent G2/M checkpoint arrest response to doxorubicin. J. Biol. Chem. 2012;287:22838–22853. doi: 10.1074/jbc.M112.376855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HH, Vo MT, Kim HJ, Lee UH, Kim CW, Kim HK, Ko MS, Lee WH, Cha SJ, Min YJ, et al. Stability of the LATS2 tumor suppressor gene is regulated by tristetraprolin. J. Biol. Chem. 2010;285:17329–17337. doi: 10.1074/jbc.M109.094235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 1998;339:900–905. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Konorev EA, Kotamraju S, Joseph J, Kalivendi S, Kalyanaraman B. Doxorubicin induces apoptosis in normal and tumor cells via distinctly different mechanisms. intermediacy of H(2)O(2)- and p53-dependent pathways. J. Biol. Chem. 2004;279:25535–25543. doi: 10.1074/jbc.M400944200. [DOI] [PubMed] [Google Scholar]
- 30.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 31.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Al-Haj L, Blackshear PJ, Khabar KSA. Regulation of p21/CIP1/WAF-1 mediated cell-cycle arrest by RNase L and tristetraprolin, and involvement of AU-rich elements. Nucleic Acids Res. 2012;40:7739–7752. doi: 10.1093/nar/gks545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marderosian M, Sharma A, Funk AP, Vartanian R, Masri J, Jo OD, Gera JF. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- 34.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J. Cell Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 35.Kim CW, Kim HK, Vo MT, Lee HH, Kim HJ, Min YJ, Cho WJ, Park JW. Tristetraprolin controls the stability of cIAP2 mRNA through binding to the 3′UTR of cIAP2 mRNA. Biochem. Biophys. Res. Commun. 2010;400:46–52. doi: 10.1016/j.bbrc.2010.07.136. [DOI] [PubMed] [Google Scholar]
- 36.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 37.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 38.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 39.Singh R, Valcárcel J. Building specificity with nonspecific RNA- binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 40.Chang WL, Tarn WY. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 2009;37:6600–6612. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, Housman DE, Jacks T. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 42.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.