Abstract
Differentiation of skeletal muscle cells is accompanied by drastic changes in gene expression programs that depend on activation and repression of genes at defined time points. Here we identify the serine/threonine kinase homeodomain-interacting protein kinase 2 (HIPK2) as a corepressor that inhibits myocyte enhancer factor 2 (MEF2)-dependent gene expression in undifferentiated myoblasts. Downregulation of HIPK2 expression by shRNAs results in elevated expression of muscle-specific genes, whereas overexpression of the kinase dampens transcription of these genes. HIPK2 is constitutively associated with a multi-protein complex containing histone deacetylase (HDAC)3 and HDAC4 that serves to silence MEF2C-dependent transcription in undifferentiated myoblasts. HIPK2 interferes with gene expression on phosphorylation and HDAC3-dependent deacetylation of MEF2C. Ongoing muscle differentiation is accompanied by elevated caspase activity, which results in caspase-mediated cleavage of HIPK2 following aspartic acids 916 and 977 and the generation of a C-terminally truncated HIPK2 protein. The short form of the kinase loses its affinity to the repressive multi-protein complex and its ability to bind HDAC3 and HDAC4, thus alleviating its repressive function for expression of muscle genes. This study identifies HIPK2 as a further protein that determines the threshold and kinetics of gene expression in proliferating myoblasts and during the initial steps of myogenesis.
INTRODUCTION
Homeodomain-interacting protein kinase 2 (HIPK2) is a proline-directed kinase that shares strong sequence homology with the kinases HIPK1 and HIPK3. Members of the HIPK family display a certain degree of functional redundancy, as mice deficient for the Hipk1 or Hipk2 genes are viable, whereas double-deficient mice die early in embryogenesis (1,2). HIPK2 bears its kinase domain in the N-terminus, which is followed by an interaction domain for homeodomain transcription factors and a C-terminal end that is rich in short repeats of serine, glutamine and alanine. Recent evidence shows that HIPK2 is generated at the ribosome in a constitutively active form by cis-autophosphorylation of its activation loop (3). Accordingly the amount of HIPK2 proteins needs to be tightly controlled by the activity of at least four different ubiquitin E3 ligases (4–7). HIPK2 in apoptotic cells can be further controlled by caspase-mediated removal of the C-terminal region (8). HIPK2 localizes mainly to subnuclear speckles and functions either as a proapoptotic mediator of the DNA-damage response or alternatively as a regulator of differentiation processes. Drosophila HIPK2 (dHIPK2) regulates eye development by phosphorylation of the corepressor Groucho, thus altering gene expression (9). Inactivation of the Dhipk2 gene results in small rough eyes and pupal lethality with rare escaper adults (10). This role is evolutionary conserved in mammals, as HIPK2−/− and HIPK1+/− mice often show small eyes with lens deficiency and abnormally thickened and laminated retinas (11). A number of further studies have used loss-of-function approaches to reveal a role of HIPK2 in a variety of developmental processes. These include the postnatal development of enteric dopaminergic neurons (12), primitive–definitive hematopoiesis, vasculogenesis, angiogenesis, neural tube closure (2), erythroid differentiation (13) and aorta-gonad-mesonephros hematopoiesis (14). HIPK2 plays a dual role in transcriptional regulation, as it can either activate or repress mRNA production. The activating function of HIPK2 largely relies on its ability to phosphorylate transcription factors such as p53 (15,16), members of the T cell factor (TCF) family and activating transcription factor 1 (17,18). On the other hand, HIPK2 can repress gene transcription on binding to general regulators of gene expression including methyl-DNA–binding proteins (19,20), the acetyl transferases p300 and cAMP response element-binding protein (CREB)-binding protein (CBP) as well as the polycomb group protein Pc2 (2,21). In line with the concept of HIPK2 as a negative regulator of gene expression, shRNA-mediated downregulation of the kinase is sufficient to allow inducible expression of specific target genes (4). HIPK2 is also found in a complex with the NK-3 homeodomain protein and histone deacetylase (HDAC) activity (22). We recently identified HDAC3 as a further HIPK2-interacting protein (23). HDAC3 belongs to the class I family of HDACs with homology to the budding yeast counterparts Rpd3 (24) and is a catalytic subunit contained in Silencing mediator of retinoic acid and thyroid hormone receptor (SMRT)–Nuclear receptor Corepressor (NCoR) nuclear complexes (25,26). The class IIa family of HDACs (HDAC4, HDAC5, HDAC7 and HDAC9) show nucleocytoplasmic shuttling that is regulated by signal-induced phosphorylation (27–29). Class IIa HDACs also bind to the family of myocyte enhancer factor (MEF) transcription factors and repress their activity (30,31).
MEFs cooperate with the transcription factor MyoD for the expression of gene products mediating myogenic differentiation. Differentiation in adult skeletal muscle is triggered by specific stimuli such as repair or exercise, which leads to cell cycle arrest of precursor cells (myoblasts), followed by increased expression of muscle function genes. Differentiation is terminated after fusion of myoblasts into multinucleated myofibers (32,33). The differentiation process proceeds through highly coordinated unleashes of distinct serial transcriptional programs. The apical regulator of myogenesis is MyoD, a basic helix-loop-helix transcription factor that binds short DNA elements called E boxes. MyoD increases expression of further transcription factors such as myogenin and members of the MEF2 family (MEF2A-D), which in turn cooperate with MyoD in the expression of muscle-specifying genes such as myosin light chain (MyLC), myosin heavy chain and myogenin (33,34). The activity of MEF2 transcription factors is highly controlled by posttranslational modifications. These include stimulatory acetylation, inhibitory SUMO (small ubiquitin-like modifier) modification and regulatory phosphorylation by a variety of enzymes. This also ensures that MEF2 proteins, which are already detectable in myoblasts, are kept in a transcriptionally inactive state (34). MEF2 proteins bind to class IIa HDACs by 18 conserved amino acids in the amino-terminal extensions of HDAC4, −5 and −7. These HDACs suppress the myogenic transcriptional program and thus block the differentiation program of skeletal muscle cells (35). The association of MEFs with HDACs catalyzes local histone deacetylation but also attracts further chromatin-modifying enzymes such as methyltransferases that trigger histone H3 lys9 methylation (36). Silencing of MEF activity is also due to HDAC-mediated deacetylation of MEF2 proteins (37). Removal of the activating acetyl groups proceeds either by direct deacetylation or in the case of HDAC4, by two indirect mechanisms: one mechanism is based on the recruitment of HDAC3 (38), while the second mechanism uses a direct competition between acetylation and HDAC4-mediated SUMOylation of a specific lysine (39). But HDAC4 is also associated with further enzymes such as an incompletely characterized kinase activity that leads to an inhibitory MEF2D phosphorylation (40). The negative role of HDAC4-mediated phosphorylation is corroborated by the positive effect of Calcineurin on muscle differentiation, as this phosphatase dephosphorylates MEF2 proteins (41) and thus antagonizes its inhibitory SUMOylation (42). The complex network of accessory proteins and posttranslational modifications that shift MEF2 proteins from the repressed to the activated state is incompletely understood and new regulatory proteins that control the threshold and kinetics of myoblast differentiation need to be identified.
Here we identify HIPK2 as a new component of the MEF2C–HDAC-associated multi-protein complex that serves to repress MEF2C activity in undifferentiated myoblasts. HIPK2 antagonizes gene expression in a kinase-dependent way by phosphorylation of MEF2C and indirectly by mediating HDAC3-dependent deacetylation of MEF2C. But HIPK2 only controls the transcription threshold in undifferentiated myoblasts and during the first days of muscle differentiation, as its caspase-mediated cleavage results in the generation of a truncated form that lacks a gene-repressive function.
MATERIALS AND METHODS
Antibodies, plasmids and reagents
All the information is given in the Supplementary Table S1.
Cell culture and transfections
Human osteosarcoma U2OS, HEK293T cells and murine C2C12 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FCS and 1% (w/v) penicillin/streptomycin at 37°C and 5% CO2. HEK293T cells were plated out 1 day before transfection, which was done using polyethylenimine as described (43) or with Rotifect (Roth) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Knockdown of HIPK2 in C2C12 cells was done by lentiviral delivery of a specific pLKO.1-puro vector expressing a HIPK2-specific shRNA (MISSION® shRNA vectors, Sigma-Aldrich). The production of lentiviruses was done as described (23). Overexpression of HIPK2 in C2C12 was done either with an adenoviral vector (a kind gift of Dr. Silvia Soddu, Regina Elena Cancer Institute, Rome, Italy) or a lentiviral vector encoding a Flag-tagged version of HIPK2 (23). Virally transduced cells were further selected with puromycin for 3 days and then shifted to differentiation medium (DMEM containing 2% horse serum). The differentiation medium was complemented after 2 days with 10 μM AraC to eliminate the non-differentiated myoblasts.
Monoclonal anti-HIPK2 antibodies
To develop monoclonal antibodies recognizing the N- or C-terminal parts of HIPK2, the regions encompassing sequences between 2–191 (HIP-N) and 781–1191 (HIP-C) were expressed as His-tagged proteins in Escherichia coli. The HIPK2 fragments were purified under denaturing conditions and dialyzed against phosphate buffered saline (PBS). The purified N-His-fusion proteins (HIP-N or HIP-C) (50 µg) were injected intraperitoneally (i.p.) and subcutaneously (s.c.) into LOU/C rats using incomplete Freund’s adjuvant supplemented with 5 nmol CpG 2006 (TIB MOLBIOL, Berlin, Germany). After a 6-week interval, a final boost with 50 µg HIP-C or HIP-N and CpG 2006 was given i.p. and s.c. 3 days before fusion. Fusions of the myeloma cell line P3X63-Ag8.653 with the rat immune spleen cells were performed according to standard procedures. Hybridoma supernatants were tested in a solid-phase immunoassay with HIP-C or HIP-N coated to ELISA plates. Antibodies from tissue culture supernatant bound to HIP-C or HIP-N were detected with horseradish peroxidase (HRP)-conjugated mAbs against the rat IgG isotypes (TIB173 IgG2a, TIB174 IgG2b, TIB170 IgG1 all from ATCC, R-2c IgG2c homemade), thus avoiding mAbs of IgM class. HRP was visualized with ready-to-use TMB (1-StepTM Ultra TMB-ELISA, Thermo). MAbs that reacted specifically with HIP-C or HIP-C were further analyzed in western blotting. N6A10 (rat IgG1) and C1B3 (rat IgG2b) were used in this study.
Cell lysis protocols
Harvested cells were washed in PBS and directly lysed in NP-40 buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM phenylmethylsulfonylfluoride, 10 mM NaF, 0.5 mM sodium orthovanadate, leupeptine (10 µg/ml), aprotinin (10 µg/ml), 1% (v/v) NP-40 and 10% (v/v) glycerol). The lysate was incubated for further 20 min on ice and centrifuged for 10 min at 4°C. Sodium dodecyl sulphate (SDS) sample buffer was added to the supernatant, followed by heating for 5 min at 95°C and separation of proteins by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Extracts that were tested for protein phosphorylation using λ phosphatase treatment were lysed in NP-40 buffer lacking all phosphatase inhibitors.
Immunoprecipitation experiments and western blotting
Cell lysates in NP-40 buffer were precleared for 1 h at 4°C by incubation with protein A/G sepharose. The immunoprecipitation (IP) was done on addition of 2 μg of precipitating antibodies or isotype-matched control antibodies together with 25 µl of protein A/G sepharose. The tubes were rotated in the cold room for 2 h on a spinning wheel. The immunoprecipitates were washed five times with NP-40 buffer and eluted by addition of 1× SDS sample buffer and boiling. Equal amounts of protein were separated by SDS-PAGE, followed by semidry blotting to a polyvinylidene difluoride membrane (Millipore) as previously described (44).
Indirect immunofluorescence
U2OS cells were grown on coverslips in 12-well dishes and transfected with various expression vectors. Fixation of cells was done with a cold methanol:acetone solution (1:1), followed by rehydration in PBS and blocking in PBS containing 10% (v/v) goat serum. The primary antibody (diluted in 1× PBS containing 1% (v/v) goat serum) was incubated overnight at 4°C. The next day, cells were washed several times in PBS and then incubated with the Cy3-coupled secondary antibody diluted in PBS containing 1% (v/v) goat serum. After washing the cells with PBS, nuclei were stained with Hoechst 33342 (1 μg/ml) and cells were mounted with Kaiser’s glycerol gelatine. Cells were analyzed with a Nikon eclipse TE2000-E microscope. Dying or mitotic cells and cells expressing aberrantly high levels of the proteins were not analyzed.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from lysed cells using the RNeasy mini kit (Qiagen). RNA quality was tested on ethidium bromide–stained agarose gels. cDNA was synthesized using Oligo (dT) 20 primers and the Superscript II first strand synthesis system (Invitrogen). Real-time polymerase chain reaction (PCR) was performed using specific primers (Supplementary Table S1) and Absolute SYBR Green ROX Mix (Thermo Scientific). Gene expression was determined using an Applied Biosystems 7300 real-time PCR system, all experiments were performed in triplicates and quantitation was done using the comparative ΔΔCT method. For that, data were normalized to the housekeeping gene β-actin, and the resulting ΔCT values were compared with a sample that was chosen as a calibrator. The relative expression level was then calculated according to the following formula: R = 2−ΔΔCT.
DNA/protein pull-down experiments
A luciferase reporter gene controlled by the myogenin promoter was used as a template for a PCR reaction to amplify specific regions using one of the primers in the biotinylated form. One PCR product encompasses the MEF2 binding sites, and the control PCR product encompasses a part from the luciferase-coding region. After purification of the biotinylated PCR products, the binding assay was performed as follows: 8 µg of nuclear extract from C2C12 cells lysed at different differentiation points was incubated with 1 µg of the biotinylated DNA and 1 µg of poly-(dI-dC) in a final volume of 20 µl in binding buffer (20 mM HEPES, pH 7.9, 0.1 mM EDTA, 60 mM KCl, 8% (v/v) glycerol, 1 mM DTT, 500 µg/ml bovine serum albumin and 0.05% (v/v) NP-40). After 30-min incubation at room temperature, streptavidin-coupled magnetic beads pre-equilibrated in binding buffer were added to the reaction and incubated for another 30 min at room temperature. The DNA–protein complexes were washed four times with binding buffer using a magnetic stand. After boiling the mixture for 5 min in 1× SDS sample buffer, DNA-bound proteins were identified by western blotting using specific antibodies as indicated.
Chromatin immunoprecipitation experiments
Five flasks with C2C12 were cross-linked with 1% (v/v) formaldehyde for 10 min at room temperature. Cells were then incubated for 5 min in 0.1 M glycine to stop the cross-linking, followed by lysis in RIPA buffer as described (45). A Branson sonifier 250 was used to shear the genomic DNA by sonification. After removal of cellular debris by centrifugation and digestion of RNAs with RNAse A, equal amounts of DNA were incubated with 2 μg of N6A10 anti-HIPK2 antibodies or control IgG antibodies previously bound to protein G-coupled Dynabeads®. After extensive washing, the precipitated DNA fragments were eluted. Sequences of the primers used for chromatin immunoprecipitation (ChIP) experiments are given in the supplementary section; the PCR product was quantified using the Applied Biosystems 7300 real-time PCR system.
RESULTS
HIPK2 represses transcription of muscle-specific genes
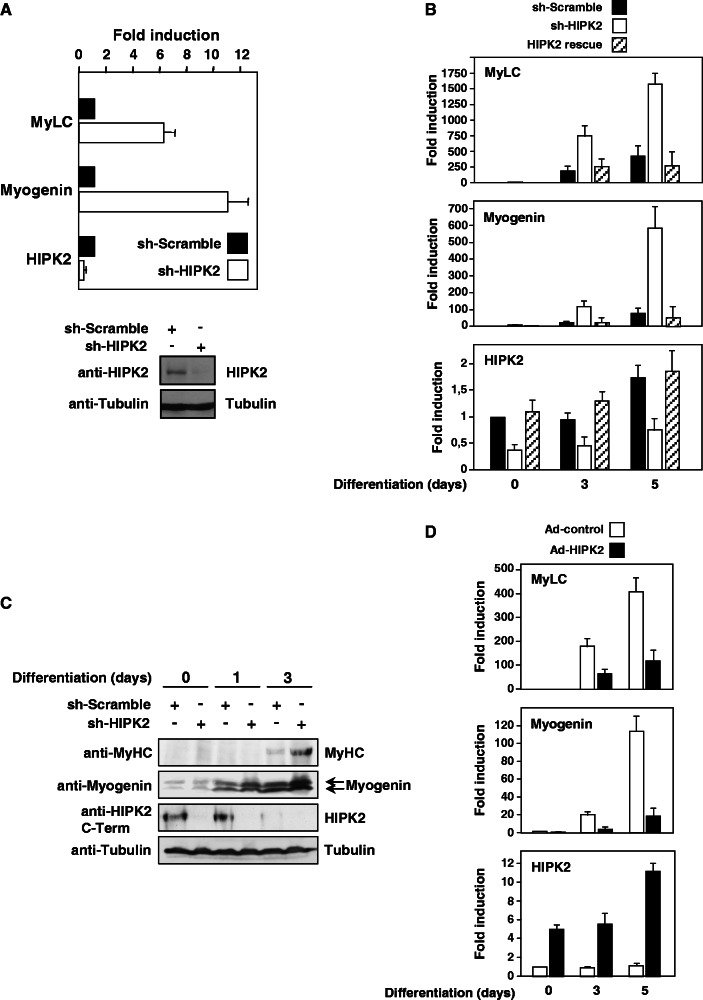
Given the importance of HIPK2 in a variety of differentiation processes and its expression in skeletal muscle (46), it was interesting to investigate its potential contribution to myogenesis. To address this question, we took advantage of the availability of the well-established C2C12 cell model that allows efficient conversion of myoblasts to myotubes and thus faithfully recapitulates muscle differentiation in vitro (47). To address the role of HIPK2 for basal expression of muscle-specific genes such as myogenin or MyLC in C2C12 cells, endogenous HIPK2 was knocked down on expression of a specific shRNA, followed by quantitative analysis of gene expression using qPCR. Knockdown of HIPK2 was paralleled by increased transcription of myogenin and MyLC (Figure 1A upper), suggesting that HIPK2 serves to repress basal expression of these genes in non-differentiated cells. To monitor HIPK2 protein levels in the murine C2C12 cells with high sensitivity and specificity, we developed monoclonal antibodies recognizing the N- or C-terminal parts of the kinase, which allowed us to confirm that the shRNA also resulted in reduced protein expression (Figure 1A lower). To reveal the contribution of HIPK2 for gene expression in differentiating myoblasts, the kinase was knocked down on expression of a specific shRNA, followed by induction of the differentiation process by the addition of differentiation medium. The differentiation process resulted in increased expression of myogenin and MyLC, and knockdown of HIPK2 further augmented transcription (Figure 1B). Off-target effects were excluded by rescue experiments where exaggerated gene expression after HIPK2 depletion was reverted on coexpression of an shRNA-resistant form of HIPK2 (Figure 1B), thus suggesting that HIPK2 serves to repress basal and also inducible transcription of these genes. The knockdown of HIPK2 also allowed an accelerated and enhanced expression of muscle function proteins such as myogenin and MyHC (Figure 1C), showing that the repressive effect of HIPK2 is also mirrored at the protein level. In these experiments, we also noted that differentiation was accompanied by a steady decrease of full-length HIPK2, which is in line with published data (48). To reveal the contribution of HIPK2 for gene expression by an independent experimental approach, C2C12 cells were virally transduced to overexpress the kinase, followed by the initiation of differentiation and the analysis of gene expression by qPCR (Figure 1D). The induced transcription of myogenin and MyLC genes was dampened on expression of HIPK2, corroborating the finding that HIPK2 restricts transcription of muscle-specific genes.
Figure 1.
HIPK2 represses transcription of muscle-specific genes. (A) Mouse C2C12 myoblasts were lentivirally transduced to express a HIPK2-specific shRNA or a scrambled control. Transduced cells were further selected for 3 days in puromycin-containing medium. A fraction of the cells was lysed and tested by immunoblotting for efficient HIPK2 knockdown using the N6A10 antibody (lower), while the remaining cells were analyzed for expression of muscle-specific genes by quantitative real-time PCR (upper). To facilitate comparison, gene expression in the presence of a control shRNA was arbitrarily set as 1. Two independent experiments were performed in triplicates, error bars display standard errors of the mean (SEMs). (B) C2C12 cells were lentivirally transduced to express a scrambled control, a HIPK2-specific shRNA or a shRNA-resistant form of HIPK2. Transduced cells were selected for 3 days in the presence of puromycin. Cells remained untreated or differentiation was triggered by addition of differentiation medium for the indicated periods. Expression of the indicated genes was determined by qPCR, and transcription of undifferentiated cells expressing the control shRNA was set as 1. Error bars display SEMs derived from two independent experiments that were performed in triplicates. (C) The experiment was done as in (B) with the difference that cells were analyzed for protein expression of HIPK2 (using the C1B3 monoclonal antibody) and the muscle-specific proteins Myogenin and MyHC as shown. (D) C2C12 cells were transduced with an adenovirus to express HIPK2 as shown, followed by the induction of differentiation for 1 or 5 days and quantification of gene expression by qPCR. Gene expression in undifferentiated cells expressing the control shRNA was set as 1, error bars show SEMs derived from three independent experiments that were performed in triplicates.
HIPK2 associates with the MEF2C–HDAC4 complex and phosphorylates MEF2C
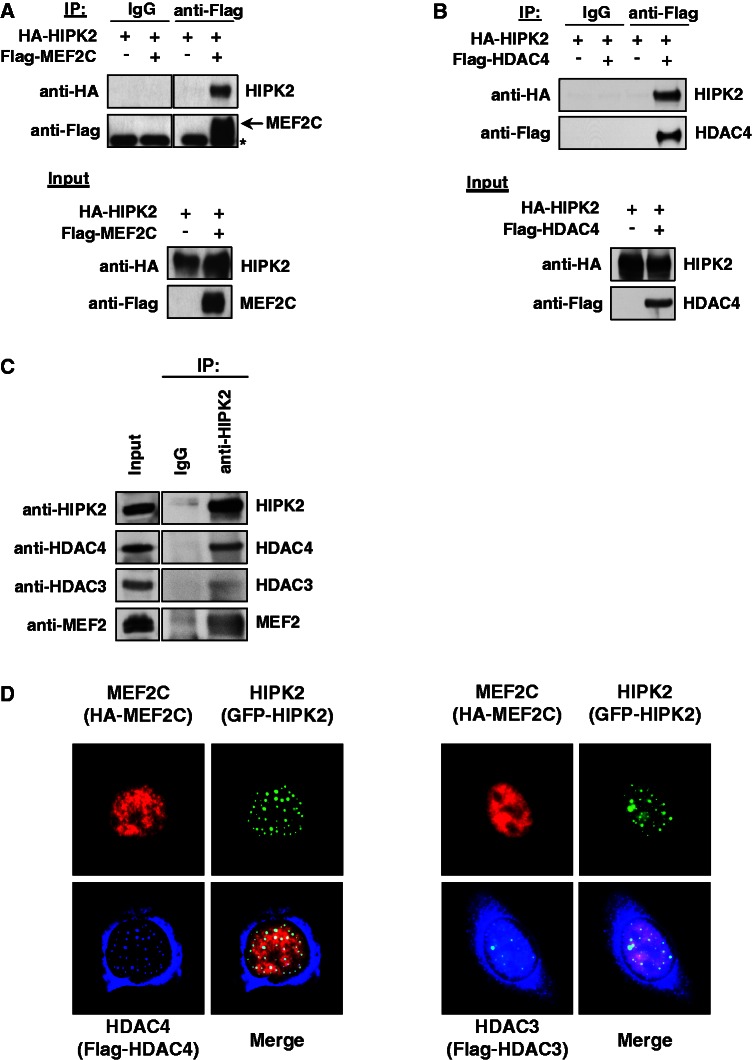
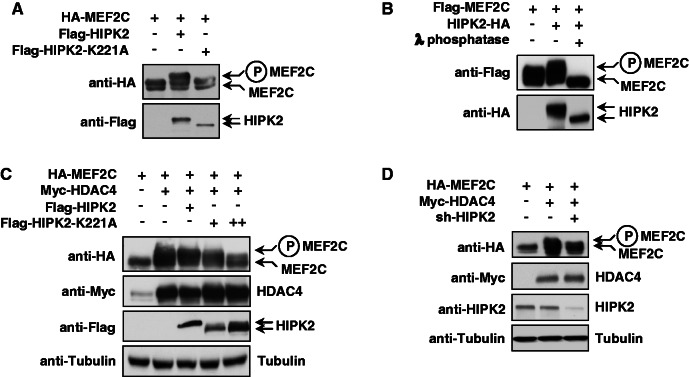
The gene-repressing effect of HIPK2 raises the question whether the kinase can bind to MEF2 and further components of the multi-protein complex controlling the expression of muscle-specific genes. To explore this issue, cells were transfected to express epitope-tagged versions of HIPK2 and MEF2C, followed by coimmunoprecipitation experiments. IP of Flag-tagged MEF2C allowed to detect binding of HIPK2 (Figure 2A). A similar approach was used to test the interaction between HIPK2 and HDAC4, which were also found to be associated (Figure 2B). Further coimmunoprecipitation experiments were performed with lysates from C2C12 cells to display the proteins that are constitutively associated with HIPK2 in undifferentiated myoblasts. Immunoprecipipitation of the endogenous kinase followed by immunoblotting revealed association with MEF2, HDAC3 and HDAC4 (Figure 2C). Indirect immunofluorescence showed colocalization between all proteins in the nucleus (Figure 2D). While HIPK2 and the nuclear fraction of HDAC4 show almost complete colocalization in nuclear speckles, only a fraction of HDAC3 binds to and colocalizes with HIPK2. This differential colocalization between HIPK2 and both HDACs is also reflected by the coimmunoprecipitation experiments that revealed preferential binding to HDAC4 (see Figure 2C), thus suggesting that the HIPK2-containing protein complex contains only substoichiometric amounts of HDAC3. In the course of these experiments, we noted that the coexpression of HIPK2 and MEF2C resulted in the occurrence of an upshifted MEF2C band in the presence of phosphatase inhibitors in the lysis buffer. To test the possible HIPK2-mediated phosphorylation of MEF2C, the transcription factor was coexpressed with the wild-type kinase or a kinase-inactive point mutant (HIPK2 K221A). Western blotting showed that the upshifted form of MEF2C occurred only in the presence of the wild-type kinase (Figure 3A). To investigate whether the slower electrophoretic migration of MEF2C is caused by phosphorylation, extracts from cells coexpressing HIPK2 and MEF2C were incubated with λ phosphatase. This treatment converted the slower migrating form of MEF2C into a band that migrated even slightly faster than the band that occurs on expression of MEF2C alone (Figure 3B), demonstrating that the upshifted band represents phosphorylated MEF2C. As published data report the association of HDAC4 with a kinase activity leading to MEF2 phosphorylation (42), it was interesting to investigate a possible contribution of HIPK2 to this enzymatic activity. Cells were transfected to express MEF2C along with HDAC4 and kinase-inactive HIPK2 that can associate with the endogenous kinase (3) and thus exert a trans-dominant negative function. Consistent with published data, the expression of HDAC4 was sufficient to trigger MEF2 phosphorylation (42). Expression of HIPK2 K221A dose-dependently diminished MEF2C phosphorylation (Figure 3C). Similarly, MEF2C phosphorylation by the HDAC4-associated kinase activity was significantly impaired on shRNA-mediated knockdown of HIPK2 (Figure 3D), thus revealing HIPK2 as another HDAC4-associated kinase.
Figure 2.
HIPK2 interacts with MEF2C and HDAC4. (A) Epitope-tagged versions of MEF2C and HIPK2 were coexpressed in 293T cells. A fraction of the cell lysates was tested for the correct expression of the transfected proteins by immunoblotting (lower), while the remaining extracts were used for IP with anti-Flag antibodies or isotype-matched controls. After elution of bound proteins in 1× SDS sample buffer, coprecipitated HIPK2 was visualized by western blotting as shown. An asterisk indicates a non-specific band. (B) The experiment was done as in (A) with the exception that Flag-HDAC4 was expressed instead of Flag-MEF2C. (C) Lysates from C2C12 cells were used for IP with anti-HIPK2 antibodies or adequate controls. The coprecipitating endogenous MEF2 proteins, as well as HDAC3 and HDAC4, were revealed by immunoblotting as shown. (D) U2OS cells were transfected to express the indicated proteins and further analyzed by indirect immunofluorescence for the intracellular distribution of HIPK2 and its interaction partners. The merged pictures display colocalizing proteins in the white areas; representative pictures are displayed.
Figure 3.
HIPK2 phosphorylates MEF2C. (A) Epitope-tagged versions of HIPK2 or HIPK2 K221A were coexpressed with MEF2C in 293T cells. Cell lysates were tested by western blotting for the electrophoretic behavior of MEF2C, and the position of the phosphorylated form is indicated. (B) Cells were transfected to express HIPK2 and MEF2C as shown, and lysates were either left untreated or incubated with λ phosphatase as shown. Equal amounts of protein were separated by SDS-PAGE and analyzed by immunoblotting with the specified antibodies. (C) MEF2C was coexpressed with HDAC4, HIPK2 or increasing amounts of HIPK2 K221A. Extracts were further analyzed for MEF2C phosphorylation by western blotting, as revealed by the occurrence of the slower migrating phosphorylated form. (D) Cells transfected to express a HIPK2-specific shRNA were selected for 3 days in the presence of puromycin, followed by plating and retransfection to express MEF2C and HDAC4. Immunoblotting of cell extracts with specific antibodies ensured efficient HIPK2 knockdown, MEF2C phosphorylation was scored by the occurrence of the slower migrating form as shown.
HIPK2 governs the acetylation status of MEF2C via HDAC3
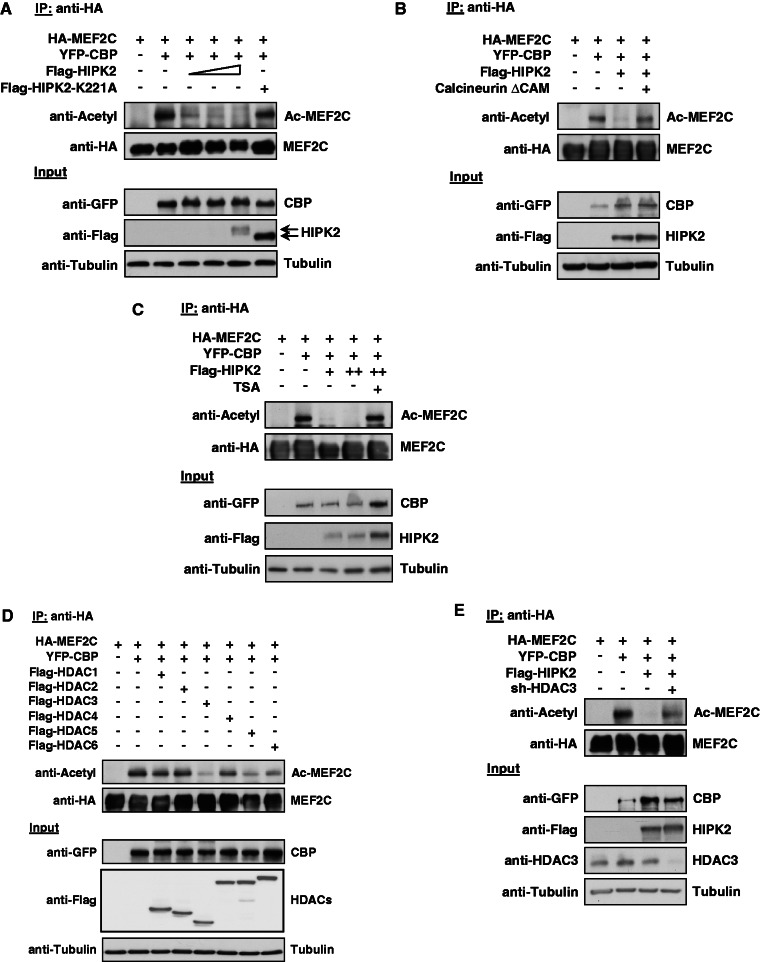
The known association of HIPK2 with acetyl transferases (2,16) and HDAC3 (23) raises the possibility that HIPK2 may also indirectly affect MEF2C acetylation. To address this question, cells were transfected to express MEF2C along with CBP, HIPK2 K221A or increasing amounts of HIPK2. Following IP of MEF2C, its acetylation status was analyzed by immunoblotting with an antibody recognizing acetylated lysines (Figure 4A). CBP caused robust acetylation of MEF2C, while coexpression of minute amounts of HIPK2 completely prevented this modification. The antagonizing function of HIPK2 for MEF2C acetylation was dependent on its intact kinase function, as acetylation of MEF2C was not affected by HIPK2 K221A. To investigate whether the effect of HIPK2 on MEF2C acetylation is phosphorylation dependent, we tested whether HIPK2-mediated deacetylation of MEF2C also occurs in cells expressing a constitutively active form of the calcineurin (Calcineurin ΔCam), an enzyme that removes inhibitory MEF2 phosphorylations (42). HIPK2-dependent deacetylation of MEF2C was completely restored in the presence of Calcineurin ΔCam (Figure 4B), suggesting that phosphorylation is required for the inhibitory effect of HIPK2 on MEF2C acetylation. HIPK2-mediated interference with the acetylation status could be due to decreased acetylation or alternatively to increased deacetylation. To reveal a possible role of HDACs for this process, the impact of HIPK2 for the acetylation status of MEF2C was also determined in cells where the activity of class I and II HDACs was inhibited by Trichostatin A (TSA). HIPK2-dependent deacetylation of MEF2C was completely lost in TSA-treated cells (Figure 4C), thus revealing the relevance of HDACs for this process. To identify the HDAC(s) responsible for the removal of CBP-triggered MEF2C acetylation, we performed a mini-screen on coexpression of MEF2C and CBP together with various different HDACs, followed by the determination of MEF2C acetylation by immunoblotting. These experiments showed largely deacetylated MEF2C in the presence of ectopically expressed HDAC3 and a minor deacetylation in the presence of HDAC5 and HDAC6 (Figure 4D). To substantiate a possible role of HDAC3 for HIPK2-regulated MEF2C acetylation by an independent experimental approach, HIPK2-dependent deacetylation of MEF2C was also determined in cells where HDAC3 expression was downregulated by a specific shRNA. Immunoblotting showed that HIPK2-mediated loss of MEF2C acetylation did not occur in HDAC3-depleted cells (Figure 4E), thus identifying HDAC3 as a relevant mediator of this process.
Figure 4.
HIPK2 leads to HDAC3-dependent deacetylation of MEF2C. (A) 293T cells were transfected to express MEF2C along with the acetyl transferase CBP, HIPK2 K221A and increasing amounts of HIPK2. A fraction of the cell lysates was tested for the correct expression of the transfected proteins by western blotting (lower). Equal amounts of protein in the remaining extracts were used for IP with anti-HA antibodies, followed by determination of acetylation using an antibody recognizing acetylated lysines (upper). (B) The experiment was performed similar to the one in (A), but with the exception that cells were transfected with the indicated constructs along with a constitutively active form of the phosphatase Calcineurin (Calcineurin ΔCAM) (C) The experiment was done as in (A), but cells coexpressing MEF2C, CBP and HIPK2 were also incubated for 6 h in the presence of 1 μM TSA as shown. (D) The indicated combinations of MEF2C, CBP and the different HDAC proteins were coexpressed in 293T cells, followed by IP of MEF2C with anti-HA antibodies and the analysis of MEF2C acetylation by immunoblotting (upper). The lower part shows correct expression of the transfected proteins. (E) 293T cells were transfected with vectors for a HDAC3-specific shRNA or the empty vector as a control, followed by selection of transfected cells with puromycin for 3 days. Cells were replated and then transfected to express the indicated combinations of MEF2C, CBP and HIPK2 together with the shRNA-producing plasmids as shown. Cell lysates were analyzed for acetylation of immunoprecipitated MEF2C (upper) and for correct protein expression (lower) as shown.
Caspases activated by myoblast differentiation cleave HIPK2 at aspartic acids 977 and 916
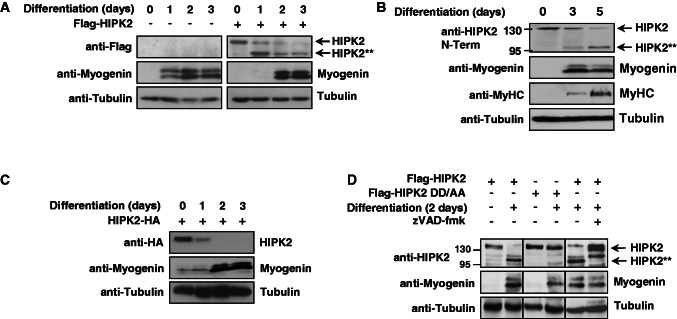
The loss-of-function approaches displayed in Figure 1 had shown a repressive role of HIPK2 for the expression of muscle-specific proteins. It was then interesting to determine muscle differentiation in the presence of the overexpressed kinase. C2C12 cells were virally transduced with a vector encoding Flag-HIPK2 or an adequate control, followed by the induction of muscle differentiation. Cell extracts were prepared at different time points, and differentiation was scored by the determination of myogenin levels. These experiments showed a delayed onset of myogenin expression in cells overexpressing HIPK2 (Figure 5A), consistent with the ability to downregulate MEF2-dependent transcription. In these experiments, immunoblotting with anti-Flag antibodies allowed to observe that differentiation was accompanied by a steady decrease of full-length HIPK2 and a concomitant appearance of a shorter form. A follow-up experiment performed in differentiating C2C12 cells showed that also the endogenous HIPK2 protein showed a time-dependent disappearance and the parallel induction of a shorter isoform (Figure 5B). Of note, this pattern could be only observed with the monoclonal antibody N6A10 recognizing the N-terminal part, while the C1B3 monoclonal antibody that only recognizes the C-terminus failed to detect the shorter form (see also Figure1C). Cleavage of the endogenous kinase occurs later than cleavage of overexpressed HIPK2. This discrepancy is attributable to the fact that overexpression of HIPK2 as such is sufficient to induce caspase activity (49), a phenomenon also displayed in Supplementary Figure S1. The possibility that muscle differentiation generates a HIPK2 form lacking the C-terminal part was further investigated on expression of a C-terminally tagged HIPK2 variant in C2C12 cells. The induction of differentiation resulted in the disappearance of the full-length form without a concomitant generation of the short form (Figure 5C), suggesting that muscle differentiation activates a proteolytic activity removing the C-terminal part of HIPK2. As muscle differentiation is accompanied by a strong increase in caspase activity (50) and HIPK2 can be cleaved by caspases following aspartic acids 916 and 977 (8), it was then self-evident to investigate a possible cleavage of HIPK2 by caspases. To address this question, we tested the effects of the irreversible broad-spectrum caspase inhibitor zVAD-fmk on HIPK2 cleavage in differentiating myoblasts. While controls showed the efficient conversion of HIPK2 to the smaller fragment, the induction of differentiation followed by addition of zVAD-fmk largely blocked HIPK2 processing and only allowed the generation of a HIPK2 form that was slightly larger than the small fragment (Figure 5D). Further experiments showed that this slightly larger band exactly comigrated with the incompletely cleaved HIPK2 form encompassing the residues 1–977 (data not shown). As zVAD does not only impair the cleavage of HIPK2, but also of further identified (51,52) and unidentified caspase substrates, this experimental setting does not allow conclusions on the specific role of HIPK2 for myogenin expression. Next, it was investigated whether mutation of both known caspase cleavage sites at Asp977 and Asp916 to alanine (HIPK2 DD/AA) renders the kinase resistant to caspase-mediated cleavage in differentiating myoblasts. The analysis of differentiating C2C12 cells transiently transfected to express HIPK2 with mutations in both cleavage sites showed the complete protection of the mutant protein from processing (Figure 5D). In summary, these data show that the transcriptional repressor HIPK2 is cleaved by caspases in differentiating myoblasts.
Figure 5.
HIPK2 is cleaved by caspases during myogenic differentiation. (A) C2C12 cells were infected with lentiviruses to express Flag-HIPK2 as shown, followed by the induction of differentiation for the indicated periods. Protein extracts were analyzed by western blotting for the expression of myogenin and HIPK2 with specific antibodies. The shorter HIPK2 form is indicated as HIPK2**. (B) C2C12 myoblasts were stimulated with differentiation medium to enter the differentiation program and harvested at different time points. Cell extracts were analyzed by immunoblotting to reveal the differentiation state, as revealed by increased expression of myogenin and MyHC. HIPK2 processing was analyzed with N6A10 antibodies and the full-length and cleaved forms are shown. The positions of molecular weight marker proteins are displayed. (C) C2C12 cells were transfected to express HIPK2 fused to a C-terminal HA-tag, followed by the induction of differentiation and western blot analysis as shown. (D) C2C12 cells were transfected to express Flag-HIPK2 or Flag-HIPK2 DD/AA as shown. Muscle differentiation was induced for 2 days in the absence or presence of the caspase inhibitor zVAD-fmk (20 μM added for the last 12 h before harvest). Cell lysates were tested for HIPK2 cleavage and expression of myogenin with specific antibodies.
HIPK2 cleavage impedes its repressive function for MEF2
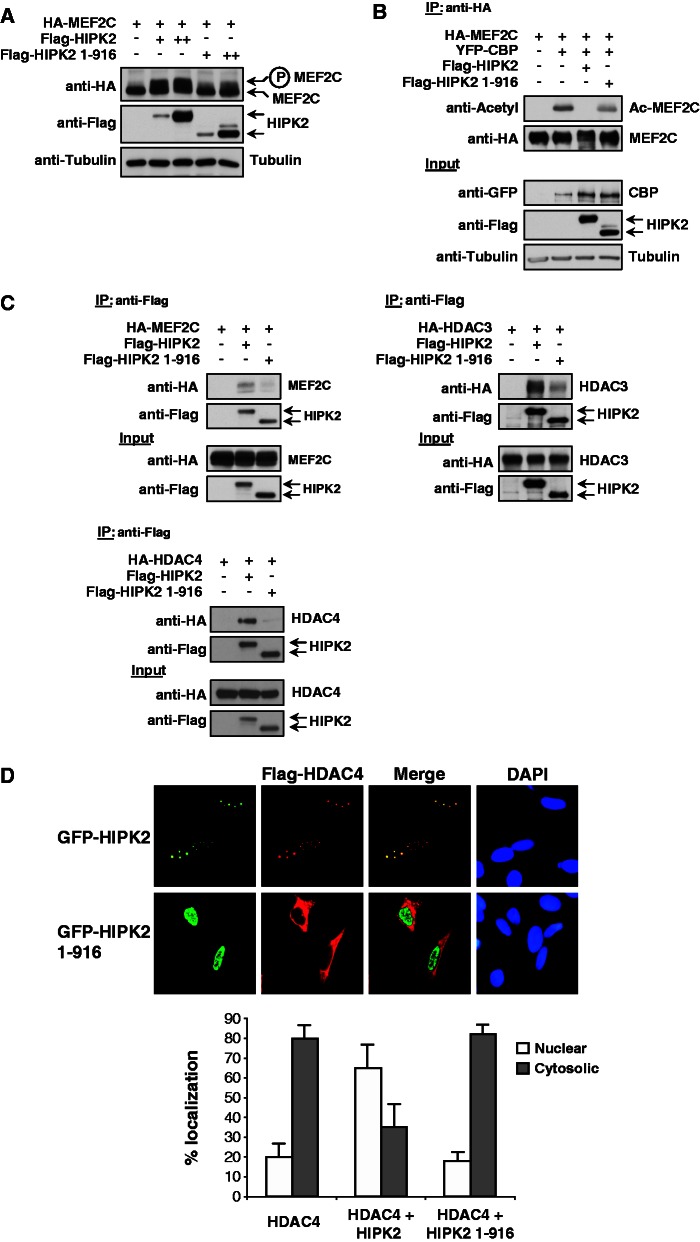
To address the functional consequences of HIPK2 cleavage on its ability to modify MEF2C, cells were transfected to coexpress MEF2C along with the wild-type form of HIPK2 or a HIPK2 variant representing the C-terminally truncated form (HIPK2 1-916). Subsequent immunoblotting showed impaired MEF2C phosphorylation by HIPK2 1-916 (Figure 6A). In the next step, both HIPK2 forms were compared for their ability to influence the acetylation state of MEF2C. CBP-triggered acetylation of MEF2C was completely reversed on coexpression of HIPK2, while the C-terminally truncated form reduced MEF2C acetylation only mildly (Figure 6B). It was then interesting to test the impact of C-terminal HIPK2 truncation on its ability to bind MEF2C and both HDACs. Coimmunoprecipitation experiments revealed that the truncated form of HIPK2 (1-916) has a strongly impaired ability to bind MEF2C and HDAC3/4 (Figure 6C), thus revealing that removal of the C-terminal part largely precludes these protein–protein interactions.
Figure 6.
Functional characterization of the C-terminally truncated HIPK2 form. (A) 293T cells were transiently transfected to express MEF2C along with increasing amounts of HIPK2 or HIPK2 1-916. Cell extracts were analyzed by western blotting for MEF2C phosphorylation as revealed by the occurrence of the upshifted band as shown. (B) MEF2C and CBP were coexpressed with HIPK2 or HIPK2 1-916, followed by IP of MEF2C and the analysis of its acetylation status by immunoblotting (upper). The input material is displayed in the lower part. (C) The Flag-tagged full-length forms of HIPK2 or its cleavage product (HIPK2 1-916) were expressed alone or together with MEF2C, HDAC3 and HDAC4 as shown. Following IP with anti-Flag antibodies the coprecipitating proteins were revealed by immunoblotting. (D) U2OS cells were transfected to express Flag-HDAC4 together with GFP-HIPK2 or GFP-HIPK2 1-916, followed by indirect immunofluorescence to reveal the localization HIPK2 and HDAC4. Nuclear DNA was stained by Hoechst and the merge shows areas of colocalization in yellow (upper). The lower part shows a statistical analysis of the immunofluorescence data that were derived from the analysis of 100 interphase cells, error bars show SEMs.
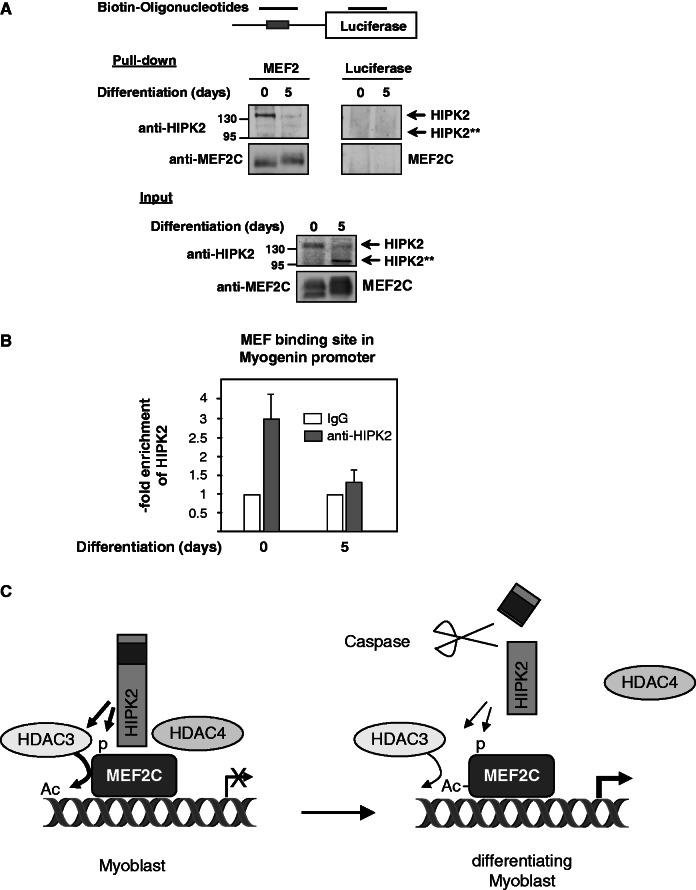
As the intracellular localization of HDAC4 can be regulated by protein kinases (53,54), it was interesting to compare the intracellular localization of HDAC4 with that of HIPK2 and HIPK2 1-916. Immunofluorescence experiments showed a complete colocalization between HIPK2 and HDAC4 in nuclear bodies (Figure 6D). In contrast, the C-terminally truncated form of HIPK2 was located in the nucleoplasm, while HDAC4 was mainly found in the cytosol in most of the cells, suggesting that only the full-length kinase can recruit HDAC4 to subnuclear structures. To explore whether HIPK2 is associated with the DNA-bound MEF2 complex, extracts from control and differentiated C2C12 cells were incubated with a biotinylated DNA oligonucleotide encompassing the MEF2-binding sites contained on the Myogenin promoter. The DNA–protein coprecipitation assay showed association of HIPK2 in extracts from undifferentiated myoblasts, while no binding was seen in cell extracts from cells that have been differentiated for 5 days (Figure 7A). To substantiate these findings in a complementary experimental approach, ChIP experiments were performed in undifferentiated and differentiated C2C12 cells. Primers spanning the MEF2 binding site in the murine myogenin promoter allowed the detection of endogenous HIPK2, while association of HIPK2 to this site was lost on induction of differentiation (Figure 7B). In summary, these data show that the transcriptional repressor HIPK2 is found in association with the MEF2 complex only in undifferentiated myoblasts, while it is removed during the process of ongoing differentiation.
Figure 7.
Promoter association of HIPK2. (A) Nuclear extracts from undifferentiated C2C12 or differentiated C2C12 cells were incubated with the indicated biotinylated PCR products spanning either the MEF2 binding sites contained in the Myogenin promoter or an unrelated region of the luciferase gene, as schematically shown at the top of the figure. After pulling down the DNA–protein complexes with streptavidin-coated magnetic beads, the interacting proteins were eluted and identified by western blotting with the indicated antibodies. (B) C2C12 cells were left untreated or induced to trigger the differentiation program for 5 days. Following cross-linking of protein–DNA complexes with formaldehyde, cells were further used for ChIP analysis using the indicated HIPK2-specific or control antibodies. HIPK2 association with the MEF2 binding site in the myogenin promoter was detected by real-time PCR using specific primers. The amount of HIPK2-associated DNA is presented as fold enrichment over the IgG control, and error bars display standard deviations obtained from two independent experiments performed in triplicates. (C) Schematic summary illustrating the repressive role of HIPK2 in undifferentiated myoblasts (left) and the loss of HIPK2-mediated repression during ongoing differentiation (right). For further details, see text.
DISCUSSION
HIPK2 as a regulator of signaling thresholds and amplitudes during myogenic differentiation
Differentiation of muscle cells involves several steps that require an exact control of cell cycle checkpoints, as proliferating myoblasts encountering an environment lacking mitogens react with the induction of differentiation and exit the cell cycle (32,33). A controlled attenuation of the cell cycle at specific stages is also characteristic for cells that have experienced mild DNA damage to allow subsequent induction of the DNA repair program. Accordingly, exposure to genotoxic agents causes a reversible inhibition of myogenic differentiation (55) and both related processes also use a set of overlapping mediators such as p53 (56) and HIPK2, as identified in this study. Consistent with the concept of HIPK2 as a signal integrator that receives input from developmental processes and the DNA damage response, we noted that more than one half of the known HIPK2 interactors (57) have a documented role for muscle cell differentiation and function, as displayed in Supplementary Table S2. But HIPK2 does not function as an ‘on–off switch’ of myogenesis, a function exemplified by MyoD (58). The kinase rather works as an accessory regulator that serves to control the signaling threshold and amplitude of gene expression. The functional role of HIPK2 is restricted to proliferating myoblasts and to the initial steps of myogenesis, as the increased caspase activity that is characteristic for the ongoing differentiation process (51) leads to the C-terminal truncation of this kinase. In line with these findings, a previous study showed that differentiation of C2C12 cells resulted in decreased HIPK2 levels (48), but the rabbit polyclonal antibody used in this study did not allow to detect the truncated version of the kinase. The functional role of HIPK2 may not be only restricted for its ability to influence posttranslational modifications of MEF2C. As HIPK2 participates in bone morphogenic protein and transforming growth factor beta signaling (59) and both pathways regulate muscle differentiation (60,61), it will be interesting to study whether the impact of HIPK2 on muscle differentiation also relies on these processes. It may well be possible that muscle differentiation is not only regulated by HIPK2, but also by the highly related kinases HIPK1 and HIPK3, which often exert overlapping roles. As the double knockout of the genes encoding Hipk1 and Hipk2 is embryonically lethal (2), it will be interesting to investigate the phenotype of a muscle cell–specific double knockout of both kinases in the future.
HIPK2 as a novel component of the MEF2–HDAC complex
The molecular mechanisms used by HIPK2 to corepress MEF2C-dependent transcription are schematically displayed in Figure 7C and involve HIPK2–HDAC3-mediated deacetylation of MEF2C, which will impair its transcriptional activity (39,62). The functional consequences of HIPK2-triggered MEF2C phosphorylation are currently unknown. Published work documents activating MEF2C phosphorylations, as exemplified by p38, BMK1 or MyLC kinase (63). On the other hand, there are also examples for inhibitory phosphorylations affecting MEF2 activity: Cdk5-mediated phosphorylation of MEF2C in its transactivation domain inhibits its transcriptional activity (42), and MEF2C phosphorylation at Ser98 and Ser110 allows docking of the prolyl-isomerase Pin1, which in turn decreases MEF2C stability and activity (64). The kinase mediating these phosphorylations is not known, but as Ser98 and Ser110 are directly flanked by proline, they could be good substrates for HIPK2, a kinase that can also interact with Pin1 (3).
Here we show that expression of wild-type HIPK2 leads to the complete recruitment of HDAC4 to subnuclear speckles. The underlying mechanism is not known and could either rely on direct protein–protein interactions or alternatively on HIPK2-dependent phosphorylation of HDAC4. Accordingly, a previous study showed that ERK1/2-mediated phosphorylation of HDAC4 leads to its nuclear translocation (54), but other examples show that phosphorylation can also keep HDAC4 in the cytoplasm by creating docking sites for 14-3-3 proteins (65). Another possibility for HIPK2-mediated control of HDAC4 localization comes from the fact that both proteins can be SUMOylated (66,67) and that HIPK2 contains a functional SUMO-interacting motif (SIM) (68,69). SIMs allow non-covalent binding to SUMO proteins and thus can act as a docking motif to allow the assembly of nuclear bodies hosting macromolecular multi-protein complexes (70). As also HDAC3 can be recruited to HIPK2 via SUMO–SIM binding, this mode of protein–protein interactions may be a more widely used principle to attract HDACs. While the general role of HDAC4 for muscle differentiation is well documented (30,31), its biochemical role in this process is not well understood. Biochemical experiments showed that HDAC4 does not function as a deacetylating enzyme and rather interferes with gene expression by acting as a scaffold. HDAC4 recruits a corepressor complex consisting of HDAC3, SMRT and N-CoR (71). As HDAC4 has the ability to bind the SUMO E3 conjugating enzyme Ubc9, it can enhance the inhibitory SUMOylation of MEF2C and MEF2D (40). A further example for a scaffolding function of HDAC4 comes from its association with kinase activities, which were identified as Cdk5 (42) and HIPK2 (this study). It is currently unclear how HIPK2 instructs HDAC3 to deacetylate MEF2C, possibly by phosphorylation of MEF2C or a phosphorylation-induced increase in the enzymatic activity of HDAC3. Alternatively, the kinase may help to bring HDAC3 and MEF2C into close proximity, which is consistent with the notion that HIPK2 1-916 is enzymatically fully active (8), but fails to efficiently mediate MEF2C deacetylation.
In conclusion, we identify HIPK2 as a new component of the multi-protein complex that is stably assembled with MEF2 transcription factors and restricts its activity in non-differentiating myoblasts. The repressive function of HIPK2 is alleviated by its caspase-mediated cleavage, a fate that is shared with other components of this complex such as HDAC4 (28).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figure 1.
ACKNOWLEDGEMENTS
We thank all colleagues who provided expression constructs that made this work possible. We are also grateful to Dr Silvia Soddu (Rome, Italy) for HIPK2 Adenoviruses, Dr Gergana Dobreva (Bad Nauheim, Germany) for C2C12 cells and both colleagues as well as Dr Xiang-Jiao Yang (Montreal, Canada) for helpful discussions.
FUNDING
German Research Foundation projects [SCHM 1417/7-1, SCHM1417/9-1, DFG SCHM 1417/8-1, SFB/TRR81]; Excellence Cluster Cardio-Pulmonary System (ECCPS); [EXC 147/2]. Funding for open access charge: University and Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Isono K, Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, Nakagawara A, Koseki H. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol. Cell. Biol. 2006;26:2758–2771. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saul VV, de la Vega L, Milanovic M, Krüger M, Braun T, Fritz-Wolf K, Becker K, Schmitz ML. HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J. Mol. Cell Biol. 2013;5:27–38. doi: 10.1093/jmcb/mjs053. [DOI] [PubMed] [Google Scholar]
- 4.Calzado MA, de la Vega L, Moller A, Bowtell DD, Schmitz ML. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat. Cell Biol. 2009;11:85–91. doi: 10.1038/ncb1816. [DOI] [PubMed] [Google Scholar]
- 5.Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J. Biol. Chem. 2008;283:4682–4689. doi: 10.1074/jbc.M708873200. [DOI] [PubMed] [Google Scholar]
- 6.Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell. 2007;25:739–750. doi: 10.1016/j.molcel.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Shima Y, Shima T, Chiba T, Irimura T, Pandolfi PP, Kitabayashi I. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol. Cell Biol. 2008;28:7126–7138. doi: 10.1128/MCB.00897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gresko E, Roscic A, Ritterhoff S, Vichalkovski A, del Sal G, Schmitz ML. Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J. 2006;25:1883–1894. doi: 10.1038/sj.emboj.7601077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi CY, Kim YH, Kim YO, Park SJ, Kim EA, Riemenschneider W, Gajewski K, Schulz RA, Kim Y. Phosphorylation by the DHIPK2 protein kinase modulates the corepressor activity of Groucho. J. Biol. Chem. 2005;280:21427–21436. doi: 10.1074/jbc.M500496200. [DOI] [PubMed] [Google Scholar]
- 10.Lee W, Andrews BC, Faust M, Walldorf U, Verheyen EM. Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev. Biol. 2009;325:263–272. doi: 10.1016/j.ydbio.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 11.Inoue T, Kagawa T, Inoue-Mochita M, Isono K, Ohtsu N, Nobuhisa I, Fukushima M, Tanihara H, Taga T. Involvement of the Hipk family in regulation of eyeball size, lens formation and retinal morphogenesis. FEBS Lett. 2010;584:3233–3238. doi: 10.1016/j.febslet.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Chalazonitis A, Tang AA, Shang Y, Pham TD, Hsieh I, Setlik W, Gershon MD, Huang EJ. Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J. Neurosci. 2011;31:13746–13757. doi: 10.1523/JNEUROSCI.1078-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattangadi SM, Burke KA, Lodish HF. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood. 2010;115:4853–4861. doi: 10.1182/blood-2009-07-235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtsu N, Nobuhisa I, Mochita M, Taga T. Inhibitory effects of homeodomain-interacting protein kinase 2 on the aorta-gonad-mesonephros hematopoiesis. Exp. Cell Res. 2007;313:88–97. doi: 10.1016/j.yexcr.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 15.D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 17.Hikasa H, Sokol SY. Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J. Biol. Chem. 2011;286:12093–12100. doi: 10.1074/jbc.M110.185280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hailemariam K, Iwasaki K, Huang BW, Sakamoto K, Tsuji Y. Transcriptional regulation of ferritin and antioxidant genes by HIPK2 under genotoxic stress. J. Cell Sci. 2010;123:3863–3871. doi: 10.1242/jcs.073627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracaglia G, Conca B, Bergo A, Rusconi L, Zhou Z, Greenberg ME, Landsberger N, Soddu S, Kilstrup-Nielsen C. Methyl-CpG-binding protein 2 is phosphorylated by homeodomain-interacting protein kinase 2 and contributes to apoptosis. EMBO Rep. 2009;10:1327–1333. doi: 10.1038/embor.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada D, Perez-Torrado R, Filion G, Caly M, Jammart B, Devignot V, Sasai N, Ravassard P, Mallet J, Sastre-Garau X, et al. The human protein kinase HIPK2 phosphorylates and downregulates the methyl-binding transcription factor ZBTB4. Oncogene. 2009;28:2535–2544. doi: 10.1038/onc.2009.109. [DOI] [PubMed] [Google Scholar]
- 21.Roscic A, Moller A, Calzado MA, Renner F, Wimmer VC, Gresko E, Ludi KS, Schmitz ML. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell. 2006;24:77–89. doi: 10.1016/j.molcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Choi CY, Kim YH, Kwon HJ, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 1999;274:33194–33197. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- 23.de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell. 2012;46:472–483. doi: 10.1016/j.molcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 24.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 28.Paroni G, Mizzau M, Henderson C, Del Sal G, Schneider C, Brancolini C. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell. 2004;15:2804–2818. doi: 10.1091/mbc.E03-08-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 30.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 31.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett AM, Tonks NK. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997;278:1288–1291. doi: 10.1126/science.278.5341.1288. [DOI] [PubMed] [Google Scholar]
- 33.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 34.Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 35.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mal A, Harter ML. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl Acad. Sci. USA. 2003;100:1735–1739. doi: 10.1073/pnas.0437843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol. Cell. Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nebbioso A, Manzo F, Miceli M, Conte M, Manente L, Baldi A, De Luca A, Rotili D, Valente S, Mai A, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10:776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell. Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, Yang XJ. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 43.Ehrhardt C, Schmolke M, Matzke A, Knoblauch A, Will C, Wixler V, Ludwig S. Polyethylenimine, a cost-effective transfection reagent. Signal Transduct. 2006;6:179–184. [Google Scholar]
- 44.Renner F, Saul VV, Pagenstecher A, Wittwer T, Schmitz ML. Inducible SUMO modification of TANK alleviates its repression of TLR7 signalling. EMBO Rep. 2011;12:129–135. doi: 10.1038/embor.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno R, Sobotzik JM, Schultz C, Schmitz ML. Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon. Nucleic Acids Res. 2010;38:6029–6044. doi: 10.1093/nar/gkq439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierantoni GM, Bulfone A, Pentimalli F, Fedele M, Iuliano R, Santoro M, Chiariotti L, Ballabio A, Fusco A. The homeodomain-interacting protein kinase 2 gene is expressed late in embryogenesis and preferentially in retina, muscle, and neural tissues. Biochem. Biophys. Res. Commun. 2002;290:942–947. doi: 10.1006/bbrc.2001.6310. [DOI] [PubMed] [Google Scholar]
- 47.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 48.Iacovelli S, Ciuffini L, Lazzari C, Bracaglia G, Rinaldo C, Prodosmo A, Bartolazzi A, Sacchi A, Soddu S. HIPK2 is involved in cell proliferation and its suppression promotes growth arrest independently of DNA damage. Cell Prolif. 2009;42:373–384. doi: 10.1111/j.1365-2184.2009.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doxakis E, Huang EJ, Davies AM. Homeodomain-interacting protein kinase-2 regulates apoptosis in developing sensory and sympathetic neurons. Curr. Biol. 2004;14:1761–1765. doi: 10.1016/j.cub.2004.09.050. [DOI] [PubMed] [Google Scholar]
- 50.Kuranaga E, Miura M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007;17:135–144. doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl Acad. Sci. USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang XH, Zhang L, Mitch WE, LeDoux JM, Hu J, Du J. Caspase-3 cleaves specific 19 S proteasome subunits in skeletal muscle stimulating proteasome activity. J. Biol. Chem. 2010;285:21249–21257. doi: 10.1074/jbc.M109.041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon JW, Pagiatakis C, Salma J, Du M, Andreucci JJ, Zhao J, Hou G, Perry RL, Dan Q, Courtman D, et al. Protein kinase A-regulated assembly of a MEF2{middle dot}HDAC4 repressor complex controls c-Jun expression in vascular smooth muscle cells. J. Biol. Chem. 2009;284:19027–19042. doi: 10.1074/jbc.M109.000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, Richon VM, Wang AH, Yang XJ, Rifkind RA, Marks PA. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc. Natl Acad. Sci. USA. 2000;97:14329–14333. doi: 10.1073/pnas.250494697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nat. Genet. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- 56.Soddu S, Blandino G, Scardigli R, Coen S, Marchetti A, Rizzo MG, Bossi G, Cimino L, Crescenzi M, Sacchi A. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J. Cell Biol. 1996;134:193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rinaldo C, Prodosmo A, Siepi F, Soddu S. HIPK2: a multitalented partner for transcription factors in DNA damage response and development. Biochem. Cell Biol. 2007;85:411–418. doi: 10.1139/O07-071. [DOI] [PubMed] [Google Scholar]
- 58.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 59.Harada J, Kokura K, Kanei-Ishii C, Nomura T, Khan MM, Kim Y, Ishii S. Requirement of the co-repressor homeodomain-interacting protein kinase 2 for ski-mediated inhibition of bone morphogenetic protein-induced transcriptional activation. J. Biol. Chem. 2003;278:38998–39005. doi: 10.1074/jbc.M307112200. [DOI] [PubMed] [Google Scholar]
- 60.Katagiri T, Akiyama S, Namiki M, Komaki M, Yamaguchi A, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 inhibits terminal differentiation of myogenic cells by suppressing the transcriptional activity of MyoD and myogenin. Exp. Cell Res. 1997;230:342–351. doi: 10.1006/excr.1996.3432. [DOI] [PubMed] [Google Scholar]
- 61.Massague J, Cheifetz S, Endo T, Nadal-Ginard B. Type beta transforming growth factor is an inhibitor of myogenic differentiation. Proc. Natl Acad. Sci. USA. 1986;83:8206–8210. doi: 10.1073/pnas.83.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol. Cell. Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al Madhoun AS, Mehta V, Li G, Figeys D, Wiper-Bergeron N, Skerjanc IS. Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C. EMBO J. 2011;30:2477–2489. doi: 10.1038/emboj.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magli A, Angelelli C, Ganassi M, Baruffaldi F, Matafora V, Battini R, Bachi A, Messina G, Rustighi A, Del Sal G, et al. Proline isomerase Pin1 represses terminal differentiation and myocyte enhancer factor 2C function in skeletal muscle cells. J. Biol. Chem. 2010;285:34518–34527. doi: 10.1074/jbc.M110.104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gresko E, Moller A, Roscic A, Schmitz ML. Covalent modification of human homeodomain interacting protein kinase 2 by SUMO-1 at lysine 25 affects its stability. Biochem. Biophys. Res. Commun. 2005;329:1293–1299. doi: 10.1016/j.bbrc.2005.02.113. [DOI] [PubMed] [Google Scholar]
- 67.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de la Vega L, Frobius K, Moreno R, Calzado MA, Geng H, Schmitz ML. Control of nuclear HIPK2 localization and function by a SUMO interaction motif. Biochim. Biophys. Acta. 2011;1813:283–297. doi: 10.1016/j.bbamcr.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 69.Sung KS, Lee YA, Kim ET, Lee SR, Ahn JH, Choi CY. Role of the SUMO-interacting motif in HIPK2 targeting to the PML nuclear bodies and regulation of p53. Exp. Cell Res. 2011;317:1060–1070. doi: 10.1016/j.yexcr.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 70.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.