Abstract
Guanine-rich RNA sequences can fold into non-canonical, four stranded helical structures called G-quadruplexes that have been shown to be widely distributed within the mammalian transcriptome, as well as being key regulatory elements in various biological mechanisms. That said, their role within the 3′-untranslated region (UTR) of mRNA remains to be elucidated and appreciated. A bioinformatic analysis of the 3′-UTRs of mRNAs revealed enrichment in G-quadruplexes. To shed light on the role(s) of these structures, those found in the LRP5 and FXR1 genes were characterized both in vitro and in cellulo. The 3′-UTR G-quadruplexes were found to increase the efficiencies of alternative polyadenylation sites, leading to the expression of shorter transcripts and to possess the ability to interfere with the miRNA regulatory network of a specific mRNA. Clearly, G-quadruplexes located in the 3′-UTRs of mRNAs are cis-regulatory elements that have a significant impact on gene expression.
INTRODUCTION
With the recent discovery that >90% of the human genome is actively transcribed, the view of the transcriptome has completely changed (1). Cells rely significantly on post-transcriptional regulation mechanisms to express a certain set of genes at a precise time, localization and magnitude. Therefore, exhaustive characterization of post-transcriptional regulatory elements is required for a better understanding of gene expression.
Guanine-rich nucleic acids have the ability to fold into a non-canonical four-stranded helical structure called a ‘G-quadruplex’ (G4). In this structure, four co-planar guanines interact with one another through Hoogsteen base pairs and are stabilized by the presence of monovalent metal cations, usually potassium, that are stacked one over the other and form the core of the structure (2–4). Genome-wide bioinformatic studies looking at the distribution of potential intramolecular G4, consisting of four consecutive runs of three of more guanines intercalated with connecting loop sequences, have been reported (5,6). Enrichment in G4 motifs has been associated with telomeres, gene promoters, ribosomal DNA, recombination hotspots and both the 5′- and 3′-untranslated regions (UTRs) of mRNAs, suggesting a potential regulatory role for these structures in many processes (7–12). G4 structures found in DNA have been the subject of considerable study; however, considering that the RNA version of this structure is generally more stable than its DNA counterpart, RNA should be more prone to fold into a G4 structure. G-quadruplexes found within the cellular transcriptome attracted a lot of attention recently [for a recent review see (13)]. The most studied RNA G4 structures are those located in the 5′-UTR of mRNA, which have been shown to be translational repressors (7,14–16). A recent study also revealed that a G4 structure located in the 3′-UTR of two dendritic mRNAs can dictate their localization in neurites (17). Another one reported a G4 structure found in the 3′-UTR of the PIM1 mRNA acting as translational repressor (18). Moreover, RNA G4 structures have been reported to modulate the alternative splicing of the TP53 gene (encoding the p53 protein) and the hTERT gene (encoding the telomerase reverse transcriptase) (19,20). In the case of the TP53 gene, an RNA G4 structure present downstream of the gene was reported to be crucial in maintaining an accurate 3′-end processing and function under conditions of stressing DNA damage (21). To date, this is the only reported indication that an RNA G4 structure located downstream of a gene may impact the polyadenylation (PA) process.
Polyadenylation is a fundamental processing step of mRNA maturation, and it is essential for its export, stability and translation. The pre-mRNA is cleaved 10–30 nt downstream of the PA signal (AAUAAA and its polymorphic variants) and then an untemplated poly-A stretch is added (22–24). Most 3′-UTR also contain alternative polyadenylation (APA) signals (25), the use of which creates the deletion of large portions of the 3′-UTRs, as well as cis- and trans-acting regulatory elements. This 3′-UTR shortening may affect mRNA stability, translational efficiency, nuclear export and cytoplasmic localization (26). For example, it has been reported that both the increase in stability and the translational efficiency of shorter mRNA isoforms is derived in part from the loss of microRNA-mediated repression (27). A higher incidence of APA and 3′-UTR shortening was observed in cancer cells, suggesting a pervasive role for APA in oncogene activation without genetic alteration (28). Clearly, a better understanding of the factors modulating APA is imperative.
Here, a robust approach, including in silico, in vitro and in cellulo experiments, that permitted the exploration of G4s located in human mRNA 3′-UTRs is presented. Specifically, two 3′-UTR G4s were studied in their different natural contexts, revealing several roles for these structures. Particular attention was focused on the modulation of APA by the G4 structure and on its impact on 3′-UTR mRNA shortening and gene expression.
MATERIALS AND METHODS
The sequences of the oligonucleotides used in this study are shown in Supplementary Table S1.
Bioinformatics
The human 3′-UTR databases were derived from sequences taken from UTRdb (UTRfull release 1 and UTRef release 9) (29). Potential G-quadruplex (PG4) sequences were identified using the algorithm mentioned in the text and the program RNAMotif (30). The results were exposed to various homemade Perl scripts (i.e. to keep only PG4 distanced by a minimum of 10 nt and to gather the proper information and values) and manually curated to obtain the PG4 databases in an Excel file format. When a 3′-UTR PG4 was located in a gene that generates more than one transcript with the same 3′-UTR, each transcript was considered individually and was counted as one more PG4 (Supplementary Data sets S1 and S2). Gene ontology and disease association were performed using the complementary 3′-UTR PG4 results from UTRef and the Database for Annotation, Visualization and Integrated Discovery (DAVID) web-accessible program version 6.7 (Supplementary Data sets S3–S5) (31). The database of putative APA units containing PG4 elements was constructed using homemade Perl scripts (Supplementary Data set S5).
RNA synthesis and labeling
RNAs for the in vitro experiments were synthesized by transcription using T7 RNA polymerase as described both previously (30). Briefly, two overlapping oligonucleotides (2 µM each) were annealed, and double-stranded DNA was obtained by filling in the gaps using purified Pfu DNA polymerase in the presence of 5% dimethyl sulfoxide. The duplex DNA containing the T7 RNA promoter sequence followed by the PG4 sequence was then ethanol precipitated. After dissolution of the polymerase chain reaction (PCR) product in ultrapure water, run-off transcriptions were performed in a final volume of 100 µl using purified T7 RNA polymerase (10 µg) in the presence of RNase OUT (20 U, Invitrogen), pyrophosphatase (0.01 U, Roche Diagnostics) and 5 mM nucleotide triphosphates (NTP) in a buffer containing 80 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES)–KOH, pH 7.5, 24 mM MgCl2, 40 mM dithiotreitol (DTT) and 2 mM spermidine. The reactions were incubated for 2 h at 37°C followed by a DNase RQ1 (Promega) treatment at 37°C for 20 min. The RNA was then purified by phenol:chloroform extraction followed by ethanol precipitation. RNA products were fractionated by denaturing (8 M urea) 10% polyacrylamide gel electrophoresis (PAGE; 19:1 ratio of acrylamide to bisacrylamide) using 45 mM Tris–borate, pH 7.5, and 1 mM ethylenediaminetetraacetic acid (EDTA) solution as running buffer. The RNAs were detected by ultraviolet shadowing, and those corresponding to the proper sizes of the PG4s were excised from the gel and the transcripts eluted overnight at room temperature in a buffer containing 1 mM EDTA, 0.1% sodium dodecyl sulfate and 0.5 M ammonium acetate. The PG4s were then ethanol precipitated, dried, dissolved in water and analyzed by spectrometry at 260 nm to determine their concentration.
To produce 5′-end-labeled RNA molecules, 50 pmol of purified transcripts were dephosphorylated at 37°C during 1 h in the presence of 1 U of Antarctic phosphatase (New England BioLabs) in a final volume of 10 µl containing 50 mM Bis–propane (pH 6.0), 1 mM MgCl2, 0.1 mM ZnCl2 and RNase OUT (20 U, Invitrogen). The enzyme was inactivated by 5-min incubation at 65°C. Dephosphorylated RNAs (5 pmol) were 5′-end radiolabeled using 3 U of T4 polynucleotide kinase (Promega) for 1 h at 37°C in the presence of 3.2 pmol of [γ-32P]adenosine triphosphate (6000 Ci/mmol; New England Nuclear). The reactions were stopped by the addition of formamide dye buffer (95% formamide, 10 mM EDTA, 0.025% bromophenol blue and 0.025 xylene cyanol), and the RNA molecules were then purified by 10% polyacrylamide–8 M urea gel electrophoresis. The bands containing the 5′-end-labeled RNAs were detected by autoradiography, and those corresponding to the correct sizes were excised and recovered as described earlier in the text.
Circular dichroism spectroscopy and thermal denaturation
Detailed procedures are as described previously (7). All circular dichroism (CD) experiments were performed using 4 µM of the appropriate RNA transcript dissolved in 50 mM Tris–HCl (pH 7.5) either in the absence of monovalent salt or in the presence of 100 mM of LiCl, NaCl or KCl. Before all CD measurements, all samples were heated in a water bath at 70°C for 5 min and then slow-cooled to room temperature for >1 h. CD spectroscopy experiments were performed using a Jasco J-810 spectropolarimeter equipped with a Jasco Peltier temperature controller in a 1-ml quartz cell with a path length of 1 mm. The CD scans were recorded ranging from 220 to 320 nm at 25°C at a rate of 50 nm min−1 and with a 2 s response time, 0.1-nm pitch and 1-nm bandwidth. The means of at least three wavelength scans were collected. Substraction of the buffer was not required, as control experiments in the absence of RNA showed negligible curves. CD melting curves were recorded by heating the samples from 25°C to 90°C at a controlled rate of 1°C min−1 and monitoring a 264-nm CD peak every 0.2 min. Melting temperature (Tm) values were calculated using ‘fraction folded’ (θ) versus temperature plots.
In-line probing
In-line probings were performed as described previously (7). Trace amounts of 5′-end-labeled RNA (<1 nM) were heated at 70°C for 5 min and then slow-cooled to room temperature for >1 h in buffer containing 50 mM Tris–HCl (pH 7.5) and with either no monovalent salt or 100 mM LiCl, NaCl or KCl in a final volume of 10 µl. After the slow cooling, the volume of each sample was adjusted to 20 µl such that the final concentrations were 50 mM Tris–HCl (pH 7.5), 20 mM MgCl2 and either no salt or 100 mM of either LiCl, NaCl or KCl. The reactions were incubated for 40 h at room temperature, and then 20 µl of formamide loading buffer (95% formamide and 10 mM EDTA) was added to each sample. For alkaline hydrolysis, 5′-end-labeled RNA was dissolved in 5 µl of water, 1 µl of 1 N NaOH was added and the reactions were incubated for 1 min at room temperature before being quenched by the addition of 3 µl of 1 M Tris–HCl (pH 7.5). The RNA in each sample was then ethanol precipitated and dissolved in formamide loading buffer. The RNase T1 ladder was prepared using 5′-end-labeled RNA dissolved in 10 µl of buffer containing 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2 and 100 mM LiCl. The reactions were incubated for 2 min at 37°C in the presence of 0.6 U of RNase T1 (Roche Diagnostic), and they were then quenched by the addition of 20 µl of formamide loading buffer. All of resulting samples were fractionated on denaturing (8 M urea) 10% polyacrylamide gels. The gels were subsequently dried, visualized by exposure to phosphorscreens (GE Healthcare) and the radioactivity quantified using the SAFA software as described previously (7,32).
Cell culture
HEK293T cells (human embryonic kidney) were cultured in T-75 flask (Sarstedt) in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate and an antibiotic–antimycotic drug mixture (all purchased from Wisent) at 37°C in a 5% CO2 controlled atmosphere in a humidified incubator.
Plasmids construction
The Fluc–LRP5 and Fluc–FXR1 constructions were built based on 3′-UTR sequences from the NCBI database [i.e. NM_002335 and NM_005087, respectively; UTRdb Locus 3HSAA093364 (UTRfull) and 3HSAR019368 (UTRef), respectively]. The full-length 3′-UTR of LRP5 was reconstituted in vitro by the filling in of multiple overlapping oligonucleotides and various PCR steps. For the FXR1 constructions, a plasmid containing the FXR1 3′-UTR was purchased from plasmID DF/HCC DNA Resource Core (HsCD00334849) and was used as template for PCR amplification with the proper forward and reverse oligonucleotides (Supplementary Table S1). The 3′-UTRs harboring either the wild-type (wt) or the G/A-mutant G4 versions were synthesized for each candidate. Site-directed mutagenesis was used to build constructions with alternative polyadenylation signal (PAS) mutations (LRP5 AAUAAA to ACUAAC and FXR1 AUUAAA to ACUAAC) and FXR1 miRseed mutation (UGUGCAAU to CCUGUUAG). The list of oligonucleotides used for each candidate is shown in Supplementary Table S1. The reconstituted 3′-UTRs were double digested with XbaI and BamHI for the LRP5 constructions and XbaI and SalI for the FXR1 constructions. Digestion products were inserted into the pGL3 control vector plasmid (Promega) previously digested with the same enzymes. DNA sequencing of each construction confirmed the insertion of the correct sequence.
DNA transfection
Typically, HEK293T cells (6 × 105) were seeded in six-well plates. The cells were co-transfected 24 h later with both the specific pGL3-control plasmid (firefly luciferase, Fluc) and the pRL-TK plasmid (renilla luciferase, Rluc) (Promega) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After an additional 24 h, 10% of the cells were used to measure the Rluc and Fluc activities using the Dual-luciferase Reporter Assay kit (Promega). Total cellular RNA was extracted from the remaining cells (90%) using TriPure Isolation Reagent (Roche Applied Science) according to the manufacturer’s protocol. Harvested total RNA was used for RNaseH/northern blot hybridization and RNA protection assay experiments.
To test the impact of the G4-specific ligand (PhenDC3), HEK293T cells (6 × 104) were seeded in 48-well plates and co-transfected 24 h later as described previously. Various concentrations of PhenDC3 were then added to the cells 4 h after the transfection. All of the cells were collected 24 h latter and subjected to dual-luciferase assays.
To investigate the impacts of an inhibitor specific for miR-92b and of an irrelevant inhibitor control, HEK293T cells (6 × 104) were seeded in 48-well plates. Twenty-four hours later, the cells were initially transfected with 100 nM (final concentration) of either the specific miR-92b inhibitor or the irrelevant inhibitor control using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Two hours post-transfection, the cells were then co-transfected with the specific Fluc and Rluc constructions using Lipofectamine 2000 as described earlier in the text. All of the cells were collected 24 h post-transfection and subjected to dual-luciferase assays.
Dual-luciferase assays
Twenty-four hours after the transfection of HEK293T cells (see Supplementary Methods), 10% fraction of the transfected cells was lysed in 150 µl of passive lysis buffer and used to measure both the firefly (Fluc) and renilla luciferase (Rluc) activities using the Dual-luciferase Reporter Assay kit according to the manufacturer’s protocol in a test tube using a GloMax 20/20 luminometer (Promega). For each lysate, the Fluc value was divided by the Rluc value. The ratios obtained for the wt version were compared with those obtained with the G/A-mutant version of each candidate and/or constructions harboring specific mutations (e.g. AltPAS-mut and miRseed-mut). Both the mean values and the standard deviations were calculated from at least three independent experiments.
RNase H/northern blot hybridizations and ribonuclease protection assays
Total cellular RNA was extracted from the remaining 90% of the transfected HEK293T cells using the TriPure Isolation Reagent (Roche Applied Science) according to the manufacturer’s protocol. The extracted RNA (20 µg) was snap cooled in water in the presence of 300 ng of both a Fluc-specific DNA oligonucleotide (see Supplementary Table S1 for sequences) and oligo-(dT)12–18 (Invitrogen), or of only the Fluc-specific oligonucleotide in a volume of 10 µl. After the snap cooling, RNase H 10× reaction buffer, 1 U of RNase H enzyme (Ambion) and water were added so as to obtain final concentrations of 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT and 25 µg/ml of bovine serum albumin in a total volume of 15 µl. The samples were then incubated at 37°C for 1 h, and the reactions were stopped by the addition of 15 µl of iced-cold formamide loading dye. 32P-radiolabeled ladder was synthesized by in vitro transcription from the plasmid pPD1 as described previously (33). Both the RNA samples (30 µl) and the ladder were fractionated on 6% denaturing (8 M urea) PAGE gels. Northern blots were hybridized using 32P-5′-end-labeled either Fluc or 7SL RNA-specific DNA probes (see Supplementary Table S1 for the sequences) for 18 h at 42°C. The membranes were washed, exposed to a phosphorscreen (GE Healthcare) and analyzed using a Typhoon apparatus (GE Healthcare) for detection and quantification. Precise PA sites were determined by 3′-rapid amplification of cDNA ends (RACE) experiments.
Ribonucleic protection assays (RPA) were performed using 10 µg of total RNA extract and the RPA III™ Kit (Ambion) as recommended by the manufacturer. Fluc- and Rluc-specific probes with 15-nt 5′- and 3′-overhangs were transcribed from PCR products (see Supplementary Table S1 for the primers′ sequences) for both the pGL3 and pRL-TK plasmids (Promega), which contain the Fluc and Rluc genes, respectively.
Statistical analysis
Analysis of a single data set was done with a one-sample t-test to examine whether the means differed from the hypothetical value of 1. Comparison analysis was performed using an unpaired two-tailed t-test, assuming that the two populations had the same variances. All calculations were performed using GraphPad Prism 5.0, and P < 0.05 was considered as being significant.
RESULTS
Potential G-quadruplex sequences within human 3′-UTRs
A database of potential G-quadruplex (PG4) sequences located in the 3′-UTRs of known human mRNAs was constructed using the procedure described previously (7,8). PG4 sequences were identified on both strands using an algorithm that searches for the sequence Gx-N1–7-Gx-N1–7-Gx-N1–7-Gx, where x ≥ 3 and N is any nucleotide (A, C, G or U). The PG4s located on the template strands correspond to tracks of cytosines in the sequences database, whereas those located on the complementary strands, which can be found in mRNAs, are tracks of guanosines. The analysis was performed on the 33 694 3′-UTRs obtained from the UTRef collection (Table 1 and Supplementary Data set S1; data obtained for the UTRFull collection can be found in Supplementary Data set S2). A total of 8903 PG4 sequences were retrieved in 5046 (15.0%) 3′-UTRs. Each 3′-UTR contains at least one PG4, but it may possess more. An unequal distribution of the PG4s between the two strands was observed (55.2% on the template DNA strand versus 44.8% on the complementary mRNA strand). A similar bias was observed in studies looking at the distribution of 5′-UTR PG4 sequences, and it suggests potential biological repercussions (7,8). The number of PG4 per 3′-UTR (ratio PG4/3′UTR) differs between the strands, with values of 1.55 for the template and 1.42 for the complementary strands (Table 1), respectively, suggesting that the cell is better able to deal with consecutive G4 structures within a given 3′-UTR on the DNA template strand than in the mRNA. Finally, the PG4 density in 3′-UTRs was estimated to be 0.130/kbase and 0.105/kbase for the template and complementary strands, respectively, which corresponds to a 2-fold enrichment as compared with the entire human genome (0.057/kbase using the same algorithm) (8).
Table 1.
Incidence of potential G-quadruplexes in a human 3′-UTR database
| Template strand | Complementary strand | Total | |
|---|---|---|---|
| Number of 3′UTR | 33 694 | ||
| Number of 3′UTR with PG4 (%) | 3163 (9.4) | 2794 (8.3) | 5046 (15.0) |
| 3′UTR with 1 PG4 (%) | 2079 (65.7) | 1973 (70.6) | 2986 (59.2) |
| 3′UTR with 1 PG4 (%) | 1084 (34.3) | 821 (29.4) | 2060 (40.8) |
| Number of PG4 | 4917 | 3986 | 8903 |
| % of PG4 | 55.20 | 44.80 | |
| Ratio PG4/3′-UTR | 1.55 | 1.42 | 1.76 |
| PG4 density | 0.130/kbase | 0.105/kbase | 0.235/kbase |
A second bioinformatic analysis was performed to estimate the biological impact of these 3′-UTR PG4s. Gene ontology analysis revealed a significant enrichment in the number of PG4s in several categories of genes, including those involved in certain biological processes (e.g. neuron differentiation) and pathways (e.g. MAPK signaling pathway), to name two examples (Table 2 and Supplementary Data set S3). Moreover, analysis of the 3′-UTR PG4s in the OMIM database revealed that 314 of these mRNA can be related to 573 different diseases, including cancer (Supplementary Data set S4). Thus, the 3′-UTR PG4 sequences are widely distributed within the transcriptome and are potentially involved in various cellular mechanisms and diseases.
Table 2.
Gene ontology analysis
| Category | Term | P-value |
|---|---|---|
| Biological process | Neuron differentiation | 9.41 × 10−09 |
| Regulation of transcription | 1.46 × 10−07 | |
| Cell projection organization | 3.11 × 10−06 | |
| Neuron development | 5.67 × 10−05 | |
| Neuromuscular process | 6.21 × 10−05 | |
| Molecular function | Transcription regulator activity | 8.67 × 10−09 |
| Sequence-specific DNA binding | 1.27 × 10−08 | |
| Transcription factor activity | 3.26 × 10−08 | |
| Protein kinase activity | 6.68 × 10−06 | |
| Voltage-gated channel activity | 1.76 × 10−04 | |
| Ion binding | 4.75 × 10−04 | |
| Cellular component | Plasma membrane part | 1.16 × 10−07 |
| Plasma membrane | 1.91 × 10−07 | |
| Synapse | 3.45 × 10−06 | |
| Cell junction | 4.36 × 10−06 | |
| Pathway | MAPK signaling pathway | 8.94 × 10−10 |
| Neurotrophin signaling pathway | 9.98 × 10−05 | |
| ErbB signaling pathway | 2.31 × 10−04 | |
| Glioma | 3.92 × 10−04 |
In vitro testing of G-quadruplex formation in LRP5
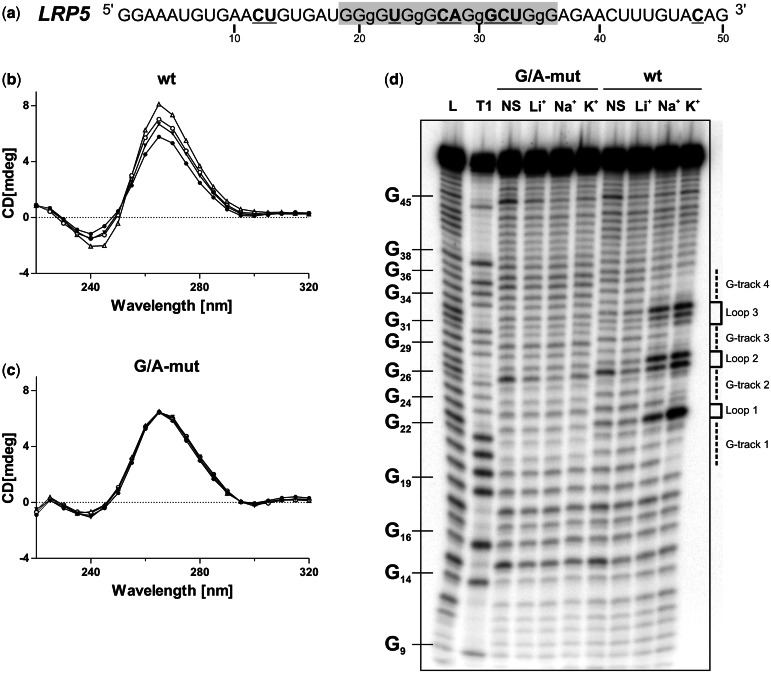
Initially the PG4 located in the 3′-UTR of the low-density lipoprotein receptor-related protein 5 (LRP5) mRNA was studied as a model candidate. This PG4 sequence possesses small loops, a high number of guanosines and a low number of cystosines in flanking sequences; consequently, it possesses a strong predisposition to fold into a G4 structure (Figure 1a). Moreover, the full-length LRP5 3′-UTR is relatively short (203 nt), which significantly simplifies both the manipulations and the analysis of the data.
Figure 1.
LRP5 3′-UTR PG4 folds into a G4 structure in vitro. (a) Sequence and numbering of the wt LRP5 PG4 used in the in vitro experiments. The lowercase guanosines (g) correspond to those mutated to adenosines in the G/A-mutant version. Nucleotides that were hydrolyzed significantly more in the presence of KCl during the in-line probing are both in bold and underlined. (b, c) CD spectra for the LRP5 PG4 sequence using 4 µM of either the wt (b) or the G/A-mutant (c) versions performed either in the absence of salt (closed circle) or in the presence of 100 mM of either LiCl (inverted closed triangle), NaCl (open circle) or KCl (open triangle). (d) Autoradiogram of a 10% denaturing polyacrylamide gel of the in-line probing of the 5′-end-labeled LRP5 wt and G/A-mutant PG4 versions performed either in the absence of salt (NS), or in the presence of 100 mM of either LiCl, NaCl or KCl. Lanes L and T1 correspond to alkaline hydrolysis and RNase T1 mapping of the wt version, respectively. The positions of the guanosines are indicated on the left of the gel, whereas the domains of the G4 structure are indicated on the right.
Initially, a sequence that exceeded the LRP5 PG4 by ∼15 nt at both ends was examined to evaluate its ability to fold into a G4 structure in vitro (Figure 1a). A G/A-mutant version, created by the substitution of several guanosines for adenosines (i.e. to prevent the formation of a G4 structure), was also synthesized for use as a negative control. First, G4 formation was monitored by CD, a conventional method for which the four-stranded helical structures possess a typical spectrum. Because of the presence of the ribose residue, an RNA G4 structure is forced to adopt a parallel topology that is characterized by the appearances of both a negative peak at 240 nm and a positive one at 264 nm (34). The CD spectra for both the wt (Figure 1b) and G/A-mutated (Figure 1c) versions were initially recorded either in the absence of salt or in the presence of 100 mM LiCl (two conditions that do not support the formation of G4 structures), and then in the presence of 100 mM of either NaCl or KCl (two conditions that favor the formation of such structures). A significant transition through a characteristic parallel G4 structure was observed only for the wt version, especially in the presence of KCl. This supports the folding into a G4 structure within the LRP5 3′-UTR. Second, thermal denaturation analyses were performed. The formation of a G4 should lead to a significant increase in stability that is accompanied by a higher Tm value for the RNA species in question (35). When the experiment was performed, significant increases in the Tm were only observed for the wt version in the presence of both NaCl and KCl (Table 3). The presence of LiCl only induced a small increase in the Tm value because of stabilization of the RNA structure caused by a counter ion effect of the cations that attenuated the repulsion of the negatively charged phosphate backbone. Third, in-line probing analyses, which require only trace amounts of RNA (<1 nM) favoring the formation of the G4 unimolecular topology that is most likely representative of that found within mRNAs (7), were performed. This method differs from both the CD and thermal denaturation methods, both of which require relatively large amounts of RNA (i.e. in the low µM range). During the incubation, the more flexible and single-stranded nucleotides have a higher tendency to undergo a non-enzymatic cleavage of their phosphodiester bonds through the in-line nucleophilic attack of the 2′-oxygen on the adjacent phosphorus center (36). On the formation of the G4 structure, the nucleotides located in the loops should bulge out and, therefore, be more susceptible to in-line spontaneous cleavage. The LRP5 PG4-derived sequences demonstrated this phenomenon. More specifically, the bands corresponding to the nucleotides located in the predicted loops, that is to say between the guanosine tracks (e.g. U23, C27, A28 and U33), became drastically more intense only for the wt version in the presence of either NaCl or KCl (Figure 1d). Quantifications’ of the intensity of each band in presence of either LiCl or KCl indicated that the nucleotides that became more susceptible to hydrolysis in the presence of potassium were all proposed to be located in single-stranded regions within the G4 structure (Figure 1a, bold and underlined nucleotides). All of the results obtained from the three distinct methods demonstrated that the LRP5 PG4 sequence folds, in vitro, into a stable unimolecular G-quadruplex at physiological KCl concentrations (i.e. 100 mM).
Table 3.
Thermal denaturation analyses
| 3′ UTR | No salt | Li+ | Na+ | K+ | |
|---|---|---|---|---|---|
| LRP5 | wt | 39.1 ± 2.1 | 51.3 ± 0.7 | 69.0 ± 0.6 | >90 |
| mut | 37.2 ± 1.9 | 49.0 ± 0.1 | 51.1 ± 1.8 | 47.3 ± 1.3 | |
| FXR1 | wt | 35.3 ± 0.6 | 57.1 ± 0.1 | 80.3 ± 0.1 | >90 |
| mut | 40.3 ± 0.1 | 52.1 ± 1.0 | 55.1 ± 0.9 | 48.8 ± 0.3 | |
Values shown are the means ± SD of two independent experiments.
The LRP5 3′-UTR G-quadruplex influences gene expression in cellulo
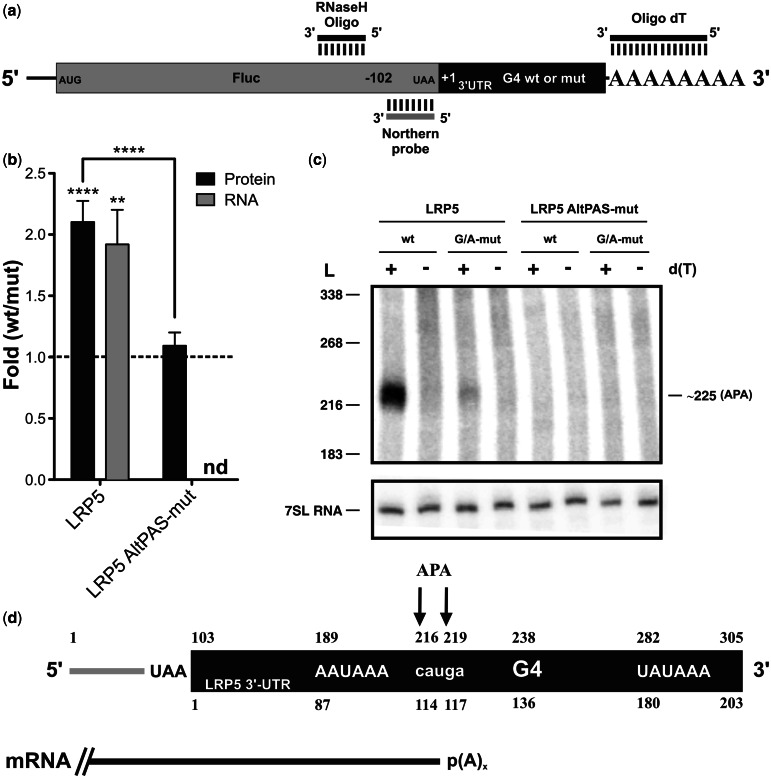
The full-length LRP5 3′-UTR was cloned downstream of the firefly luciferase reporter gene (Fluc) to verify its ability to affect gene expression (Figure 2a). During the cloning, the SV40 late PA signal in the pGL3 vector was removed from the construction. Only the natural PAS of the LRP5 3′-UTR, which is located 24 nt away from the PA site and corresponds to the polymorphic sequence UAUAAA, was kept. HEK293T cells were then co-transfected by both Fluc–LRP5 constructions (i.e. either with the wt or the G/A-mutant versions of the G4 structure) and a plasmid containing the renilla luciferase gene (Rluc) for normalization of the transfection efficiency. The cells were harvested 24 h post-transfection, lysed and luciferase activity assays performed to estimate gene expression. The ratio of the luciferase activities (value of the wt 3′-UTR divided by that of the G/A-mutant version) showed a 2-fold increase (Figure 2b), indicating that the formation of the G4 structure significantly enhanced the luciferase expression level.
Figure 2.
The LRP5 3′-UTR G4 structure in cellulo. (a) Schematic representation of the Fluc–LRP5 construction. The Fluc-coding sequence is shown in gray, whereas the LRP5 3′-UTR is shown in black. The binding regions of the oligonucleotides used for the RNase H hydrolysis, as well as the luciferase-specific probe, are illustrated. (b) Gene expression levels of the different LRP5 constructs either at the protein level (black) or the mRNA level (gray). The x-axis identifies the constructions used and the y-axis the fold difference (i.e. wt result divided by G/A-mutated result) (for LRP5 protein n = 3, whereas for mRNA n = 5, for LRP5 AltPAS-mut protein n = 4, nd indicates not detectable). Error bars, mean ± SD, **P < 0.01 and ****P < 0.0001. (c) Northern blot hybridization of RNA samples subjected to an RNase H hydrolysis in the presence of a Fluc-specific DNA oligonucleotide and either in the absence (−) or the presence (+) of oligo-dT. The numbers on the left refer to the sizes of a molecular RNA ladder, whereas that on the right is the estimated size of the detected transcript. 7SL RNA was probed as internal control. (d) Schematic view of the RNA product resulting from the RNase H hydrolysis. The upper numbers correspond to the numbering from the 5′-end of the digestion product, whereas lower ones refer to the start of the LRP5 3′-UTR. The arrows map the different PA sites as determined by 3′-RACE, and the mRNA produced is depicted in black.
RNA samples were also extracted from the cells, and RNase H treatment coupled to northern blot hybridization was performed to verify whether a correlation existed between the amounts of cellular proteins and mRNAs. Briefly, DNA oligonucleotides that specifically bind to a region 102-nt upstream of the Fluc gene’s stop codon were annealed to the mRNA (Figure 2a), and the resulting RNA/DNA heteroduplex was then hydrolyzed by RNase H treatment. This removed the 5′-end of the Fluc–LRP5 mRNAs, thereby permitting fractionation of the remaining 3′-ends by denaturing PAGE electrophoresis followed by northern blot hybridization using a probe specific for the remaining part of the Fluc-coding sequence regardless of the sequence of the 3′-UTR. The RNase H hydrolysis was performed in either the absence or the presence of oligo-d(T), which caused the heterogeneous polyadenylated products to collapse into discrete products. A single well-defined band was observed only in the presence of oligo-d(T) for both the wt and G/A-mutant versions, indicating that they correspond to polyadenylated RNAs (Figure 2c). The wt version produced more mRNA as compared with the G/A-mutant, although the abundance of 7SL RNA (used as a loading control) remained invariable. The differences in the mRNA levels were in good agreement with what was observed at the protein level (Figure 2b). A representation of the RNase H cleavage product for the Fluc–LRP5 3′-UTR is shown in Figure 2d illustrating the 102 nt from the RNase H cleavage site to the Fluc stop codon, the restriction site and the full-length LRP5 3′-UTR, which starts at position 103. The distance from the RNase H cleavage site to the LRP5 PA site was estimated, by comparison with an RNA ladder, to be 220 nt, which was unexpected (see later in the text). To confirm this evaluation, a 3′-RACE experiment was performed, permitting resolution at the nucleotide level. Two close, but distinct, PA sites were detected, generating fragments of 216 and 219 nt in size (i.e. corresponding to positions 114 and 117 of the LRP5 3′-UTR; Figure 2d), thus validating the previous observation. These bands were not produced from the promoter-distal PA site located at position 305 (according to NCBI), but instead from an APA unit situated around positions 216 and 219 and under the control of an AAUAAA PAS located at position 189. This observation suggested that the G4 acts as a downstream PA regulatory element that enhances the efficiency of the APA unit, although it is excluded from the produced mature isoform. To test this hypothesis, new constructions possessing a mutated AAUAAA PAS (AltPAS-mut), which inactivates this APA unit, were synthesized for both the wt and the G/A-mutant G4 versions. No difference was observed in the luciferase activity levels, and no PA was detected in the LRP5 3′-UTR or in its vicinity (Figure 2b and c). The very low amount of luciferase protein produced, which was unaffected by either the presence or the absence of the G4 structure, potentially came from a PAS present in the pGL3 vector (located ∼3000-nt downstream the LRP5 3′-UTR) that was impossible to detect by the RNase H/northern blot experiment under the used conditions. The absence of PA at the promoter-distal site could be due to the absence of its own downstream regulatory elements, as they are located outside the LRP5 3′-UTR; therefore, they are not included in the LRP5 constructions. These elements might be required to observe a PA driven by the uncommon and less efficient UAUAAA PAS. Together, these results demonstrated that the G4 structure located within the LRP5 3′-UTR acts as a downstream regulatory element, and that it positively modulates the use of an internal PA unit.
The 3′-UTR G-quadruplexes seem to be frequently associated with alternative polyadenylation units
The human 3′-UTR mRNA database was revisited to identify PG4 sequences potentially involved in the regulation of an APA unit. Each PG4 sequence was examined for the presence, within the first 100 nt upstream (an arbitrarily chosen distance), of either a typical human PAS (i.e. AAUAAA) or the most common single polymorphism (i.e. AUUAAA). This analysis revealed the presence of 75 and 39 3′-UTR PG4s possessing near upstream AAUAAA and AUUAAA PAS, respectively, that formed putative APA units (Supplementary Data set S5). This yielded 108 individual mRNAs that include such putative APA site susceptible to G4 stimulation. Moreover, they could potentially be linked to 22 different diseases (Supplementary Data set S5). This suggests that the case of LRP5 is not isolated, and that 3′-UTR G4 structures may be noteworthy cis-acting elements for the regulation of APA.
A G-quadruplex structure promotes FXR1 3′-UTR shortening
To further evaluate the role of G-quadruplexes as positive regulatory elements for APA units, a second candidate was studied. The fragile X-related mental retardation autosomal homolog 1 (FXR1) gene produces an mRNA with a 3′-UTR 870 nt in length that possesses both a PG4 sequence and a putative internal APA unit located around position 250 (Figure 3a; note that the numbering from the positions of the FXR1 3′-UTR differs because the 102 upstream nucleotides of the Fluc-coding sequence and the restriction site are also considered). Initially, the ability of the FXR1 3′-UTR PG4 sequence to fold into a G-quadruplex in vitro was assessed. The same three methods described earlier in the text were used, and all agreed that it adopts a G4 structure in the presence of a physiological concentration of KCl (Supplementary Figure S1).
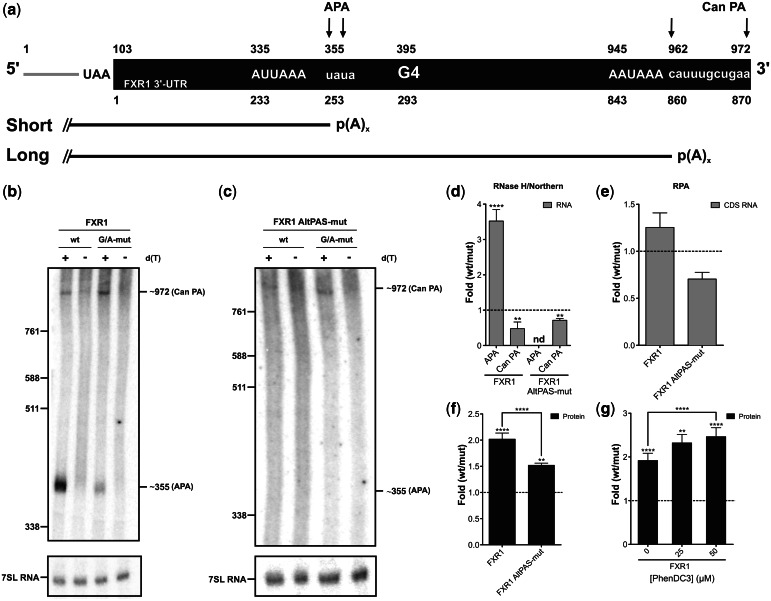
Figure 3.
The FXR1 3′-UTR G4 structure in cellulo. (a) Schematic representation of the Fluc–FXR1 transcripts resulting from the RNase H hydrolysis. The upper numbers correspond to the numbering from the 5′-end of the hydrolyzed product, whereas lower ones refer to the start of the FXR1 3′-UTR (black part). The arrows map the different PA sites as determined by the 3′-RACE experiments [alternative (APA) and canonical (Can PA) sites]. The short and long mRNA isoforms produced are shown. (b, c) Northern blot hybridizations of the RNA samples previously subjected to RNase H hydrolysis in either the absence (−) or the presence (+) of oligo-dT. The numbers on the left refer to the sizes of a molecular RNA ladder, whereas those on the right are the estimated sizes of the two isoforms. 7SL RNA was probed as an internal control. (d–f) Gene expression levels of constructs either at the mRNA level as determined by northern blot hybridization (for FXR1 n = 5, whereas for FXR1 AltPAS-mut n = 3; nd indicates not detectable) (d), by RNase protection assay (FXR1 and FXR1 AltPAS-mut n = 3) (e) (gray) or at the protein level as determined by luciferase assay (FXR1 n = 7, FXR1 AltPAS-mut n = 3) (f) (black). The x-axis identifies the constructions used and the y-axis the fold difference (wt result divided by G/A-mutated result). (g) Luciferase assays in the presence of various concentrations of PhenDC3 (0–50 µM; n = 3). Error bars, mean ± SD, **P < 0.01 and ****P < 0.0001.
Subsequently, the full-length FXR1 3′-UTR was cloned downstream of the Fluc gene to verify its impact on gene expression. According to the primary sequence of the 3′-UTR, two mRNA species could be synthesized: one long isoform produced from the canonical PA site (AAUAAA PAS located 28-nt upstream of the predicted cleavage site) and a shorter isoform produced from an APA site [AUUAAA PAS located 60-nt upstream of the FXR1 3′-UTR G4 (Figure 3a)]. The in cellulo experiments were performed as described earlier in the text. RNase H/northern blot hybridization analysis confirmed the detection of both isoforms for both the wt and G/A-mutant versions only in the presence of oligo-d(T) (Figure 3b). The shorter polyadenylated RNA species was estimated to be ∼355 nt and the longer one ∼970 nt. These lengths correlated, respectively, with the positions of the alternative and canonical PA units. In agreement, 3′-RACE results indicated that the cleavage sites of both the alternative and the canonical PA units were located at positions 355–357 and 962–970, respectively (Figure 3a). Quantification of the intensities of the bands for both isoforms revealed an ∼3-fold increase in the presence of the G4 structure for the shorter isoform, whereas for the longer isoform, a decrease of the same magnitude (∼3-fold) was observed, under the same condition (Figure 3d). The FXR1 G4 structure seems to significantly affect the short/long ratio of the produced mRNA isoforms. To investigate whether only the ratio between both isoforms was affected or whether the levels of total mRNA also varied, a second mRNA quantification was performed. An RPA using probes that covered regions within the coding sequences of both the Fluc and Rluc genes (for normalization purposes) were performed. This approach permitted the quantification of the level of global mRNA synthesis without discriminating between the different isoforms. Almost no difference was observed, indicating that the FXR1 G4 structure did not affect the global quantity of mRNA, but instead affected only the short/long isoform ratio (Figure 3e). Taking into account these values and their standard deviation, the northern blot and RPA results are concordant, suggesting that no read through occurred. Briefly, the magnitude of the increase of the PA occurring at the APA site is compensated by the decrease of PA occurring at the Can PA site, resulting in no significant difference in the total amount of mRNA produced. These results suggest that the AAUAAA PAS at the distal PA site is sufficient to drive complete PA in the absence of its downstream elements, and that this is so even for the G/A-mutant FXR1 G4 version. Interestingly, at the protein level, luciferase activity was increased by 2-fold in the presence of the FXR1 G4 structure (Figure 3f). These experiments demonstrated that the FXR1 G4 structure influences gene expression at the protein level primarily by affecting the ratio between the short and the long mRNA isoforms without affecting the global mRNA level.
Afterward, new constructions in which the AUUAAA PAS was mutated (AltPAS-mut) in both G4 contexts (i.e. for both the wt and G/A-mutant versions) were synthesized to verify whether the FXR1 G4 structure positively modulates the efficiency of the APA unit. The insertion of this mutation completely abolished the activity of the APA unit and the synthesis of the shorter isoform (Figure 3c). Interestingly, a significant decrease in the quantity of the long isoform was still observed in the presence of the G4 structure (Figure 3c and d). Quantification of the mRNA produced at the canonical PA site, based on the RNase H/northern blot hybridization analysis, correlates with the total mRNA detected by the RNase protection assays for the alternative PAS mutants (Figure 3d and e). At the same time, the luciferase activity assays showed a smaller increase (1.5-fold) with the inactive APA unit constructions as compared with the active ones (2-fold) (Figure 3f). This observation suggests that approximately half of the increase at the protein level in presence of the G4 structure is due to the stimulation of the APA unit. Moreover, the effect of the G4 structure on the amount of mRNA synthesized at the downstream canonical PA site (30% decrease) seems likely to be independent of the use of the alternative site that is located upstream (Figure 3d and e). Importantly, the experiment suggests that a smaller amount of mRNA harboring the FXR1 G4 structure produced a larger amount of protein than did a larger amount of mRNA lacking the G4 structure. This represents an original characterization of this phenomenon.
To obtain support for the conclusion that the effects observed on gene expression were due to the presence of G4 structure, the impact of a G-quadruplex-specific ligand on gene expression was tested. Specifically, PhenDC3 is a bisquinolinium-derived compound with both a strong G4 stabilizing ability and selectivity (37,38). The luciferase activity was observed to increase with increasing PhenDC3 concentrations, thus providing additional evidence that the FXR1 G4 structure directly contributes to the differences observed in gene expression (Figure 3g).
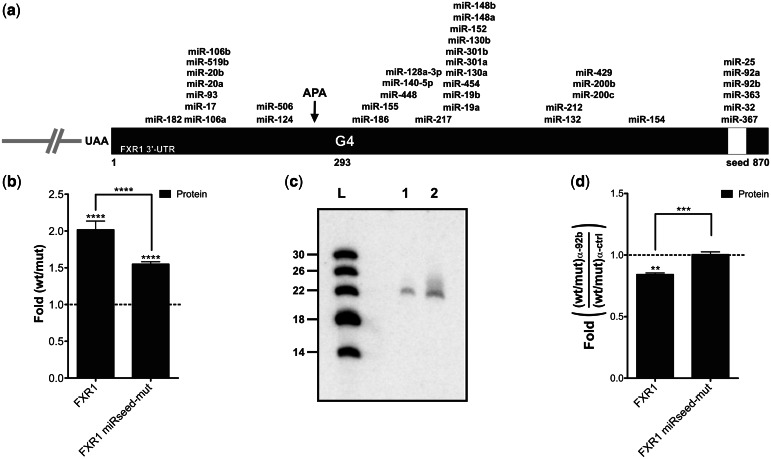
Finally, the impact of the FXR1 3′-UTR shortening was then investigated in terms of its microRNA regulatory network, which is also known as trans-factor elements regulating both mRNA stability and translation efficiency (39). First, the mirSVR software was used to map the predicted microRNA-binding sites present in the FXR1 3′-UTR (Figure 4a) (40). Only sites with a mirSVR score <−0.5 were considered. The loss of all of the predicted microRNA-binding sites located downstream of the APA unit during the 3′-UTR shortening process should likely lead to a modification of the microRNA-mediated regulation of the mRNA. The FXR1 3′-UTR has already been shown to be the target of various microRNAs, especially a seed region that is shared between six different microRNAs located at position 813 (Figure 4a; yellow box) (41). To test whether the increase in gene expression caused by the variation of the short/long isoform ratio driven by the FXR1 G4 structure came from the loss of this negative regulatory element, constructions in which the conserved and shared seed region was mutated (FXR1 miRseed-mut) were synthesized. The mutation of the seed region led to a reduction of the effect (50%) of the FXR1 G4 as measured by luciferase activity (Figure 4b). The same decrease was observed for FXR1 AltPAS-mut, suggesting an important role for this region in gene expression because of the modulation of the APA site (Figure 3f). Moreover, northern blot experiment was used to detect the expression of three microRNAs proposed to bind to this seed region (i.e. miR-92b, miR-363 and miR 367). Only miR-92b was detected from HEK293T RNA extracts (Figure 4c). To enhance the role of miR-92b in this phenomenon, experiments using either a miR-92b inhibitor, or an irrelevant inhibitor control, were performed with the constructions. The effects of the FXR1 G4 (wt/mut) observed onto protein synthesis in the presence of the irrelevant inhibitor (i.e. 2.2- and 1.5-fold increase for FXR1 and FXR1 miRseed-mut constructions, respectively) were used to set the 1-fold ratio in Figure 4d. A decrease of >15% was observed for the natural FXR1 context in the presence of the miR-92b-specific inhibitor, whereas constructions harboring the miRseed-mutation remained unaffected (Figure 4d). These results support the hypothesis that most of the impact on gene expression caused by the FXR1 3′-UTR mRNA shortening promoted by the G4 structure come from the modification of its microRNA regulatory network.
Figure 4.
FXR1 3′-UTR shortening and the microRNAs regulatory network. (a) Schematic representation of the FXR1 3′-UTR. The numbering refers to the position from the start site of the FXR1 3′-UTR. All predicted microRNA target sites with a mirSVR score <−0.5 according to the miRanda algorithm are shown (40). The white region corresponds to the predicted shared microRNA seed region that was mutated in the FXR1 miRseed-mut constructions. (b) Gene expression levels of different FXR1 constructs at the protein level as determined by luciferase assays. The x-axis identifies the constructions used and the y-axis the fold difference (wt result divided by G/A-mutated result) (for both FXR1 and miRseed-mut n = 4). (c) Northern blot hybridization for the detection of miR-92b performed using either 5 µg (lane 1) of small RNAs (<200 nt) or 50 µg of total RNA (lane 2) extracted from untransfected HEK293T cells. The numbers on the left refer to the sizes of a molecular RNA ladder of 5′-end labeled in vitro transcripts (lane L). (d) Gene expression levels of different FXR1 constructs at the protein level as determined by luciferase assays in the presence of either 100 nM miR-92b inhibitor or of irrelevant control inhibitors. The x-axis identifies the constructions used and the y-axis the fold difference (ratio wt on G/A-mutated version obtained in the presence of the miR-92b inhibitor divided by that obtained in presence of the control inhibitor) (both FXR1 and miRseed-mut n = 3). **P < 0.01, ***P < 0.001 and ****P < 0.0001.
DISCUSSION
In contrast to DNA G4 structures, the importance of both the presence and the impacts of RNA G4 structures in biology remains to be elucidated and appreciated. The bioinformatic analysis reported here is in agreement with a previous one showing that PG4 sequences are found in thousands of human 3′-UTRs (8), including in numerous mRNAs of proteins related to both human diseases and to various cellular processes (Tables 1 and 2 and Supplementary Data sets S1–S4). Although the previous bioinformatics study mostly focused on both 5′- and 3′-UTR PG4s’ occurrences, distribution and positioning, the one presented here strictly concentrate on 3-UTR PG4s with an emphasis on their potential biological roles. For example, it was recently reported that two dendritic mRNAs, PSD-95 and CaMKIIa, possessed 3′-UTR G4s with the ability to act as specific localization signals, targeting these RNAs to cortical neuritis (17). Moreover, the FMRP protein, already known to bind G4 structures, has been suggested to act as one of the trans-acting factors in this phenomenon (42). In the present study, the PG4 sequences found in the 3′-UTRs of the LRP5 and FXR1 mRNAs were demonstrated to fold into G4 structures in vitro in the presence of a physiological concentration of KCl. Once in their 3′-UTR’s natural context, and cloned downstream of a luciferase reporter gene, both of these G4 structures were shown to increase gene expression by 2-fold (Figures 2b and 3f). These increases were associated with a more efficient PA at sites located few nucleotides upstream of the G4 structures (Figures 2 and 3).
In metazoans, a PA unit is composed of various RNA elements located near its cleavage site (22). Among the most common downstream elements are the U/GU-rich and the G-rich auxiliary elements. In light of the results presented here, the 3′-UTR G4 structures most likely act as downstream auxiliary elements that enhance the productivity of APA sites. To emphasize the involvement of a G4 structure in this phenomenon, another experiment was performed (unpublished data). Constructions harboring the LRP5 3′-UTR wherein the LRP5 G4 was substituted with a new G4 structure retrieved from the 3′-UTR of the TTYH1 mRNA (NM_020659 in position 208 of its 3′-UTR, see Supplementary Data set S1) were tested in luciferase assay experiments. A 3-fold increase in gene expression was observed between a TTYH1 wt and G/A-mutant G4 version, suggesting that the phenotype observed is not attributable to the LRP5 G4 primary sequence, but it can be restored using a substitute G4 structure. Although, the results obtained from the experiment using the PhenDC3-specific G4 stabilizing ligand also strongly support the implication of the G4 structure in the phenomenon observed, we cannot completely rule out the possibility that the changes at the level of the primary sequence between the wt and G/A-mutant might partially play a role in the observed effects. Two prevalent models have been proposed for the functionality of such auxiliary elements (43). First, these elements could promote processing efficiency by maintaining the core PAS in an unstructured form, thus enabling a better assembly of the general PA factors. In this regard, the extreme stability of RNA G4 structures may be a favorable characteristic. Additionally, in-line probing results typically showed that the regions flanking the G4 structure become both more flexible and single-stranded on its formation (Figure 1a and d and Supplementary Figure S1a and d) (7). Second, the auxiliary elements could interact with specific proteins, which would in turn stimulate the assembly of the general PA factors on the pre-mRNA. For example, it has been reported that a G4 structure, located in 3′ of the p53 gene, was essential in maintaining the efficient 3′-end processing of the pre-mRNA under stress-induced DNA damage throughout the interaction with heterogeneous nuclear RNP H/F (21). Undoubtedly, many characteristics of the G4 structure make it a suitable candidate to act as a PA auxiliary element.
Over 100 mRNAs were shown to harbor putative APA units composed of either an AAUAAA or an AUUAAA PA signal and a 3′-UTR PG4 (Supplementary Data set S5). This is most likely an underestimation, as there are many other known variant PA signals in mammalian cells (44,45), and the distance used here (100 nt) is minimal considering that G-rich regulatory elements located as far as 440-nt downstream of the core PA site of a mRNA have already been shown to be critical for efficient 3′-end processing (46). In addition to these facts, an enrichment of PG4 sequences located in the first 10% of the 3′-UTRs (i.e. near downstream stop codons) was observed, suggesting that the deletion of larger sequences is favored (Supplementary Figure S2). Most likely only the ‘tip of the iceberg’, in terms of G4 structures that may act as auxiliary APA elements, has been revealed.
The study of two different candidates permitted evaluation of the impact of the 3′-UTR G4 in two distinct contexts. In the case of the LRP5 3′-UTR, the PA unit containing the G4 structure was the only efficient one. The modulation of its efficiency by the G4 directly determined the level of mRNA produced and, consequently, the level of protein synthesized (Figure 2b). The impact of the G4 promoting APA was significantly different in the FXR1 3′-UTR environment, and it provides a quick overview of how complex this mechanism can be. The FXR1 3′-UTR contains both an alternative and a canonical PA units resulting in the production of a short and a long isoform, respectively (Figure 3). A tight coordination between both PA units was observed. The mRNA with a wt G4 structure favored the short isoform, whereas an mRNA with the G/A-mutated version accumulated more of the long isoform. The overall impact of the modification of this short/long isoform ratio was an increase in the level of protein produced in the presence of the G4 structure. This observation is in accordance with the notion that an mRNA with a shorter 3′-UTR is usually both more stable and more actively translated than is one with a longer 3′-UTR (28). Moreover, living cells use shortened 3′-UTRs to increase the expression of various genes during specific processes, such as proliferation and oncogene activation, without genetic alteration (27,28). The better translational efficiency is a consequence of the loss of 3′-UTR repressive elements, mainly microRNA-binding sites. In agreement with this, the loss of a shared microRNA seed region located in position 813 of the FXR1 3′-UTR seemed to be responsible for the better translational properties of the shorter isoform, this in a process in which miR-92b has a significant role (Figure 4). With 3′-UTR mRNA shortening attracting a lot of attention recently (28,27), the G4 structures located in 3′-UTR may gain in popularity as an RNA motif to study for a better understanding of this phenomenon.
The characterization of the FXR1 3′-UTR also demonstrated that the amount of mRNA synthesized at the level of the downstream canonical PA site seems to be independent of the use of the alternative upstream site. Indeed, a decrease of mRNA produced and polyadenylated at the level of the canonical site, in the presence of the G4, was still observed in the FXR1 AltPAS-mut construction, where the activity of the APA site was shut down (Figure 3a and c–e). On the basis of this observation, it is tempting to speculate that the 3′-UTR G4 sequence may act also as a transcriptional termination element; however, additional physical support is required to confirm this hypothesis. That said, it is supported by studies reporting that G4s that form in the nascent RNA transcript stimulate mitochondrial transcription termination (47), and that G-rich regions were shown to form an R-loop, which can act as a transcriptional pause site important in transcriptional termination in mammalian cells (47,48).
In summary, this study demonstrates that G4 structures are abundant within 3′-UTRs, and that these RNA motifs seem to have diverse contributions to mRNA processing events, such as APA. In fact, looking at the G4 structures of two independent 3′-UTRs revealed that their impacts are considerably more complex than initially believed. This is nicely illustrated by the demonstration that the 3′-UTR G4 structure of the FXR1 mRNA stimulates APA and, consequently, leads to 3′-UTR shortening which in turn impairs its microRNA regulation and, ultimately, gene expression. In brief, G4 structures emerge as important cis-acting elements present in 3′-UTRs with important impacts on both APA and gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–2 and Supplementary Data sets 1–5.
ACKNOWLEDGEMENTS
The authors are grateful to the laboratory of Marie-Paule Teulade-Fichou at the Institut Curie (France) for providing the PhenDC3 ligand, Dominique Levesque for technical assistance and François Bachand for critical discussions. J.D.B. conceptualized the study, designed and performed the experiments. J.D.B and J-P.P analyzed the data and wrote the manuscript together.
FUNDING
Canadian Institutes of Health Research (CIHR) [CIHR: MOP-44022 to J-P.P.]; CIHR Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Awards (to J.D.B.). J-P.P. held the Canada Research Chair in Genomics and Catalytic RNA and now holds the Chaire de l’Université de Sherbrooke en structure et génomique de l’ARN. J-P.P. is member of the Centre de Recherche Clinique Étienne-Le Bel. Funding for open access charge: CIHR.
REFERENCES
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 3.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC Publishing; 2006. p. 301. [Google Scholar]
- 5.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaudoin JD, Perreault JP. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;38:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani P, Yadav VK, Das SK, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Du Z, Zhao Y, Li N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009;37:6784–6798. doi: 10.1093/nar/gkp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 13.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 14.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji X, Sun H, Zhou H, Xiang J, Tang Y, Zhao C. Research progress of RNA quadruplex. Nucleic Acid Ther. 2011;21:185–200. doi: 10.1089/nat.2010.0272. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora A, Suess B. An RNA G-quadruplex in the 3′ UTR of the proto-oncogene PIM1 represses translation. RNA Biol. 2011;8:802–805. doi: 10.4161/rna.8.5.16038. [DOI] [PubMed] [Google Scholar]
- 19.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcel V, Tran PLT, Sagne C, Martel-Planche G, Vaslin L, Teulade-Fichou M-P, Hall J, Mergny J-L, Hainaut P, Van Dyck E. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 21.Decorsière A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 24.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 25.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 27.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 2008;3:1395–1401. doi: 10.1038/nprot.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaudry D, Busière F, Lareau F, Lessard C, Perreault JP. The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 1995;23:745–752. doi: 10.1093/nar/23.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 37.Halder K, Largy E, Benzler M, Teulade-Fichou MP, Hartig JS. Efficient suppression of gene expression by targeting 5′-UTR-based RNA quadruplexes with bisquinolinium compounds. Chembiochem. 2011;12:1663–1668. doi: 10.1002/cbic.201100228. [DOI] [PubMed] [Google Scholar]
- 38.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheever A, Blackwell E, Ceman S. Fragile X protein family member FXR1P is regulated by microRNAs. RNA. 2010;16:1530–1539. doi: 10.1261/rna.2022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalziel M, Nunes NM, Furger A. Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol. Cell. Biol. 2007;27:1568–1580. doi: 10.1128/MCB.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc. Natl Acad. Sci. USA. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.