Abstract
Infected cell protein 8 (ICP8) from herpes simplex virus 1 was first identified as a single-strand (ss) DNA-binding protein. It is essential for, and abundant during, viral replication. Studies in vitro have shown that ICP8 stimulates model replication reactions, catalyzes annealing of complementary ssDNAs and, in combination with UL12 exonuclease, will catalyze ssDNA annealing homologous recombination. DNA annealing and strand transfer occurs within large oligomeric filaments of ssDNA-bound ICP8. We present the first 3D reconstruction of a novel ICP8–ssDNA complex, which seems to be the basic unit of the DNA annealing machine. The reconstructed volume consists of two nonameric rings containing ssDNA stacked on top of each other, corresponding to a molecular weight of 2.3 MDa. Fitting of the ICP8 crystal structure suggests a mechanism for the annealing reaction catalyzed by ICP8, which is most likely a general mechanism for protein-driven DNA annealing.
INTRODUCTION
Two-component recombinases are a class of protein complexes found in many linear double-strand DNA (dsDNA) viruses that infect bacteria to human cells. These recombinases catalyze single-strand DNA (ssDNA) annealing (SSA) homologous recombination reactions and are composed of a 5′–3′ exonuclease and an annealase that binds ssDNA and catalyzes its pairing and annealing with a homologous ssDNA molecule. Annealases are grouped into several protein superfamilies (1), and their functional conservation throughout evolution suggests an indispensable function. The founding member of the two-component recombinases is the red recombinase of phage λ, composed of λ exonuclease (λExo) and β protein (Redβ) (2). Human herpes simplex virus 1 (HSV-1) also uses a two-component recombinase, which is composed of UL12 exonuclease and ICP8 (infected cell protein 8) annealase. The similarities between the phage λ and HSV-1 two-component recombinases have been reviewed in Reuven et al. (3).
The classic recombinases, such as RecA, UvsX and Rad51, can be thought of as catalyzing two linked reactions: first a search for homology between an ssDNA and a dsDNA and formation of a stable joint, and second, the transfer of strands between the two DNAs. The annealases have not been found to catalyze the first stage, but they will carry out extensive strand transfer between two homologous DNAs if a stable joint is provided (4–8).
ICP8 is a 128 kDa protein homologous to >40 other ssDNA-binding proteins in Herpesviridae, including Kaposi’s sarcoma-associated herpesvirus (KSHV) Orf6 protein and BALF2 of Epstein–Barr virus (PSI-BLAST data not shown). ICP8 was initially observed to be essential for viral DNA replication (9) and then for transient replication of plasmids containing an HSV-1 origin (10), also to stimulate in vitro replication reactions (11), leading to its being termed a viral ssDNA binding protein (SSB). It was then observed that ICP8 will form regular helical filaments alone (11–13) or when bound to a DNA template (4,14,15), and the resemblance of these structures to RecA, UvsX and Rad51 filaments suggested that it might share similarities with those recombinases. This was supported by the fact that the classic SSBs (Escherichia coli SSB, T4 g32 protein and eukaryotic RPA) do not form regular helical filaments. Moreover, in reactions using homologous DNA templates in which the linear dsDNA partner is resected by HSV-1 UL12 nuclease, ICP8 will drive the transfer of strands between the paired DNAs (3,4) over distances as great as 7 kb. ICP8, however, will not form stable DNA joints in an adenosine triphosphate (ATP)-dependent manner as do RecA, UvsX and Rad51, but it rather relies on UL12 nuclease to expose homologous sequences (3). The findings that ICP8 has potent ssDNA annealing activity (15,16) and that other annealases will drive strand transfer when paired with a nuclease argues that ICP8 should be considered as an annealase with the added ability to form helical filaments as do RecA and Rad51.
ICP8 has been observed in various quaternary structures. It forms magnesium-dependent helical protein filaments in the absence of DNA. Based on negative-staining electron microscopy (EM) and 3D helical reconstruction, these filaments are double protein helices (13). We recently reported that KSHV Orf6 makes similar filaments (17). ICP8 forms thinner helical nucleoprotein filaments when it assembles onto ssDNA, which are likely single helices (4,14,15) unlike the protein-only filaments. In addition, ICP8 was also shown to form oligomeric rings (14) when bound to a short ssDNA. These rings are visually similar to those observed with RecT, Redβ, ERF and Rad52 annealases (18–21). Finally, ICP8 forms a thicker helical nucleoprotein filament as an intermediate during the catalysis of an annealing reaction between two long complementary ssDNA strands (15). These filaments, which are distinctly different from the protein-only or ssDNA-bound ones, were observed when the course of an ssDNA annealing reaction catalyzed by ICP8 was followed by EM. First, thin nucleoprotein filaments are generated on binding to complementary ssDNAs, which then associate with each other in a double-helix arrangement to form thicker nucleoprotein filaments. On extended incubation, these resolve into long dsDNA molecules free of ICP8. One interesting feature of these helical nucleoprotein filaments is that they frequently seem to start with a ring. Indeed, the thick double-helix annealing intermediate filaments can also be described as appearing like a stack of rings with the first ring having rotated away from the stack flat onto the EM support. Thus, these rings would represent a single turn of the ssDNA annealing machine. This was previously proposed for Rad52 (22), and for Dmc1, where it was suggested that such filaments originated from the rings (23).
Because of the irregular nature of the double-helix filaments (15), it has not been possible to generate a 3D structure that would aid in elucidating the mechanics of strand annealing. Recently, however, we have generated single turns of these filaments and report here the first 3D structure of this ICP8–ssDNA binary complex. Our findings and observations provide insights into the basic strand annealing mechanism. In addition, our fitting results can be used for predicting both the residues involved in protein–protein interactions during the formation of ICP8 filaments and also the ones on the surface exposed to the solution that may be involved in binding to other proteins.
MATERIALS AND METHODS
Formation of ICP8–ssDNA rings and EM
ICP8 protein was overexpressed and purified as reported previously (15,24). The protein was >95% pure as determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Coomassie staining and did not contain detectable levels of nuclease activity. The nucleoprotein rings were formed by incubating 7.8 μM ICP8 monomers with a 38mer oligonucleotide (3.2 μM) (GGATCTCCATGGGGGATGGAGAGCAAGTGAAGACCTTG)(25) at 37°C in 20 mM Tris–HCl pH 7.5 for 1 h and then adding MgCl2 to 5 mM final concentration in a total volume of 50 μl, followed by incubation at 4°C overnight. The ring preparation was mounted onto glow-charged thin carbon films supported by 400 mesh copper grids, stained with 2% uranyl acetate and imaged at 50 000× under low-dose conditions using a Philips CM12 EM. Images were captured on Kodak Electron SO-163 film and scanned using an Imacon Flextight, Model 848 scanner at 2000 dpi.
3D reconstruction, fitting, docking and rendering
3D reconstruction was done by following the single particle workflow provided in the EMAN2 (26) software package, except for particle picking, which was done in EMAN (27). The majority of particles observed were rings consisting of nine subunits. Some minor instances of particles with 8–11 subunits, in addition to a distinct trimeric assembly, were also observed. Only particles appearing to have nine subunits and a regular structure were included in the analysis. This resulted in a data set of ∼7500 final particles of ∼20 000 particles originally automatically selected.
For fitting the available X-ray crystal structure of ICP8 (PDB code 1URJ) (28) into the reconstructed EM map, a PDB file containing the N-terminal domain of one monomer together with the C-terminal domain of the other interacting monomer was used. Chimera (29) and Situs 2.6 (colores, collage and pdbsymm modules) (30) were used to perform rigid body fitting. For fitting in Chimera, the map was first segmented to 18 sub-volumes (Supplementary Figure S4), and two copies of the generated PDB were fitted into one symmetrical unit. After this, symmetrical copies of the fitted PDBs were generated using C9 symmetry. Finally, the fit was optimized using the unsegmented whole map and a sequential symmetrical fitting algorithm. For Colores fitting, Sculptor (31) was used for examining the fits. After the fitting was completed, the fitted crystal structure was superimposed with a model containing the modeled ssDNA as in Mapelli et al. (28) to visualize the putative location of ssDNA substrate. The dimensions of the ring were measured using RasMol (32).
To assess whether our presented fit was ambiguous at this resolution, we computed a Z-score using two different methodologies. First, crystal structures of proteins with monomer molecular weights similar to ICP8 were randomly selected from the PDB database and fitted into one symmetrical unit of the reconstructed map using Chimera and sequential fitting. The population of cross-correlation scores resulting from these fits was compared with the cross-correlation score of the actual fit (Figure 3d and e) to obtain a Z-score. As a second method, e2fhstat.py distributed with the EMAN2 package was used with default parameters to assess the goodness of fit using the entire map.
Figure 3.
Fitting the X-ray crystal structure. Eighteen copies of a C-terminal truncated ICP8 monomer [(28) and ‘Materials and Methods’ section] were fit into the reconstructed double-ring map, nine on top and nine at the bottom. (a–c) They are the top, side and the bottom views, respectively; red and magenta are the N-terminal domains, and blue and green are the C-terminal domains. Cyan and orange monomers were colored such that to show individual monomers with their N- and C-termini. Yellow shows two monomers fitted into one symmetrical unit. (d and e) They show one symmetrical unit containing two monomers isolated from the map. d shows the side view facing the next symmetrical unit, and e shows the side that faces to the center of the ring. Red and cyan are the N- and C-terminal domains, respectively, fitted into one symmetrical unit composed of top and bottom densities.
For ligand docking, the PatchDock web server (33) was used with its default parameters. An ICP8 tetramer fitted to two symmetrical units of the reconstructed map (as described earlier in the text) was used as the receptor. As ligands, two dsDNA PDBs were generated using 3D-DART web server (34). One was a 20-bp dsDNA, referred to as ‘straight dsDNA’ in this article. The other was generated by creating a 144-bp circular dsDNA based on the size of the reconstructed ring, and then trimming it to a 20-bp segment using PyMol (35), and referred to as bent or curved dsDNA. The relevant results shown in this article were ranked sixth of the 20 622 for the straight dsDNA and first of the 21 098 for the curved dsDNA. The 3D figures were rendered using Chimera (29) and POVRay (36) or ePMV (37) and Blender (38).
RESULTS
Visualization of a ring-like structure formed by ICP8 binding to a single-stranded oligonucleotide
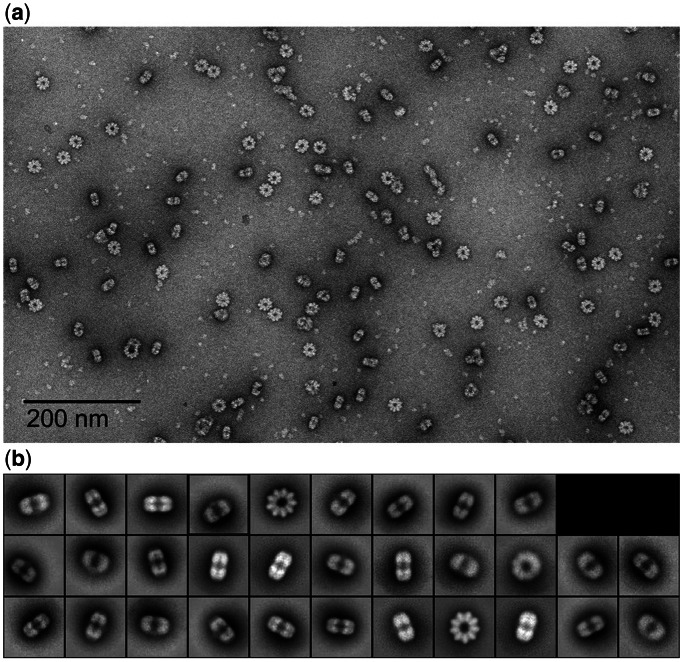
ICP8–DNA filaments that form during an annealing reaction often start with a ring, with the rest of the filament appearing as a stack of rings (15). To generate a helical filament, a closed ring must assume a split lock-washer conformation as shown for Redβ annealase of phage λ (39). Calculation of the amount of ssDNA in one ring formed by ICP8 suggested that each might contain ssDNA in the range of 140–160 nt, and using a 158 nt ssDNA, we found that ICP8 will assemble into rings containing 9–10 monomers (14). Further optimization using smaller ssDNAs showed that incubating ICP8 with 38-nt oligomers of several different sequences produced a homogeneous population of particles. Fields stained with uranyl acetate (Figure 1a) revealed particles consistent with different views of a ring-like structure possessing a ∼250 Å diameter and a central hole. These structures would have a molecular weight of at least a megadalton making them an ideal candidate for single-particle reconstruction.
Figure 1.
Raw data and 2D class averages. (a) A field of ring-like particles formed by incubating ICP8 with a 38-nt ssDNA oligomer and staining with uranyl acetate (see ‘Materials and Methods’ section). (b) The reference-free 2D class averages. The major classes are views from the top showing a ring of 9-fold symmetry and rectangular side views with a stack of two densities.
Single-particle reconstruction was performed using standard methods in EMAN2 (see ‘Materials and Methods’ section). The results clearly revealed 9-fold symmetry of the particles in the top view. Although the particles appeared to be two stacked rings from the side views, owing to visually detectable asymmetry between these rings, only a single 9-fold rotational symmetry was imposed (Figure 1b). It was also apparent from the images that, although some tilted views were visible, there was a strong preference for the top and side views, as is common for particles with a toroidal shape. After generating an initial model from these 2D class averages (Supplementary Figure S1), convergence in the 3D refinements was reached in eight iterations (Supplementary Figure S2, top). Reprojections, 2D class averages and raw particles were consistent with each other (Supplementary Figure S3, right). The Fourier shell correlation (FSC) curve used for resolution determination was generated using e2refine_evenodd.py, which, unlike traditional FSC tests, performs two completely independent refinements with different initial models on independent halves of the data, a so-called ‘gold-standard’ resolution test (40). The resolution of the reconstructed map was determined as 22 Å based on this curve at 0.143 cut-off threshold (Supplementary Figure S2, bottom).
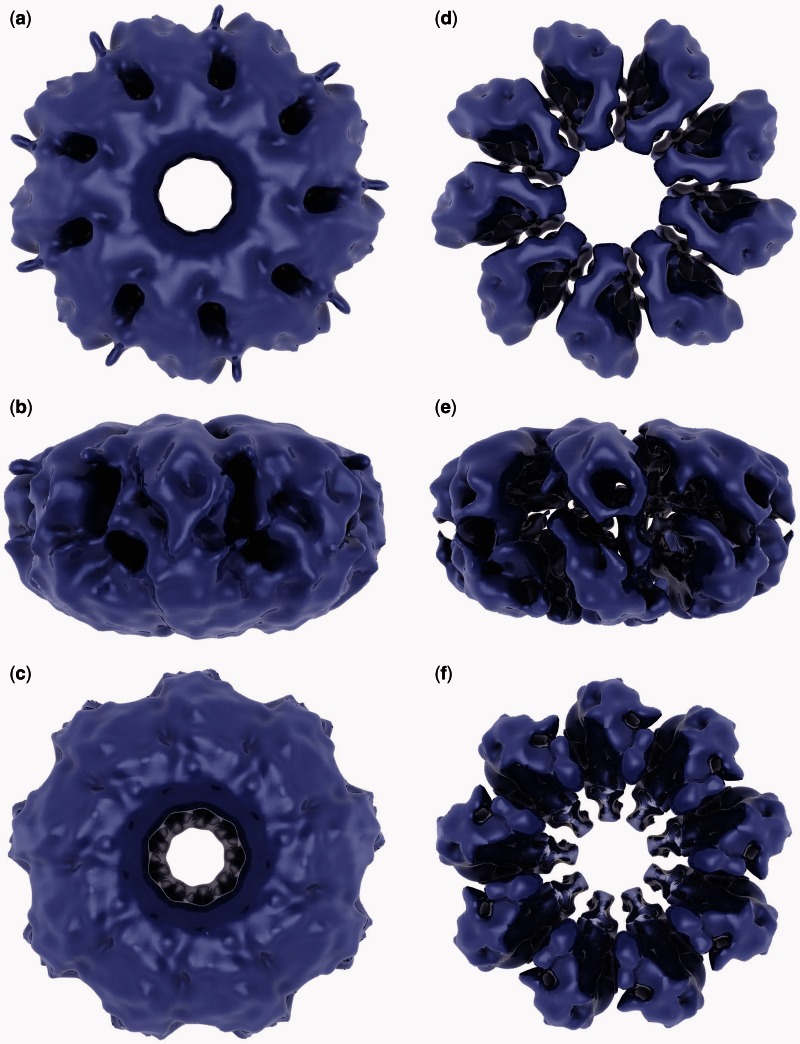
The 3D oligomeric structure of ICP8 in complex with ssDNA is ∼250 Å in diameter and 135 Å in height, and it consists of two nonameric rings stacked on top of each other (like a sliced bagel) (Figure 2). In Figure 2, a–c were rendered at a threshold covering the volume of the fitted crystal structure, and they show the overall shape of the reconstructed map. In d–f, densities were rendered at a higher threshold to clearly show both the two stacks of densities at top and bottom and the individual densities within those. Therefore, this oligomeric ring, which we initially suspected to have 9 subunits, is indeed composed of a stack of two rings with 9-fold rotational symmetry (Figure 2e) with a combined molecular weight of 2.3 MDa. These two stacked rings will be referred to as top (Figure 2a) and bottom (Figure 2c) rings. Although the two rings have similar features, their overall shapes are visibly different. The holes in the middle of the top and bottom rings are slightly different in size. When the map is visualized at a low threshold, small protrusions extending toward the outside of the ring are seen on the top ring (Figure 2a), whereas they are missing on the bottom (Figure 2c). Although it is possible that this may be noise, considering their size and location, we speculate that these may represent the DNA entering or exiting the ring (see ‘Discussion’ section). To eliminate the possibility that these features might have been generated because of the imposed symmetry, the refinement process was repeated without imposing any symmetry (C1). In the resulting map, both longer and shorter protrusions were observed, all of which come out from locations close to the space between symmetrical units, i.e. between subunits (data not shown). Of the nine spaces between symmetrical units, four had a protrusion at the position as seen in Figure 2a, suggesting that these protrusions likely are not simply an artifact produced by the imposed symmetry.
Figure 2.
Reconstructed map of the ICP8–ssDNA complex. (a–c) The density was rendered at a cut-off threshold used for fitting the crystal structure. C9 symmetry is clearly visible, which was imposed during the reconstruction based on the 9-fold symmetry apparent in the top and/or bottom views of raw particles that became better defined in the reference-free 2D class averages (Figure 1). (d–f) The volume was rendered at a higher threshold to show each symmetrical unit visible in the top and bottom views in d and f, respectively. In addition, at this threshold, two densities stacked on top of each other are visible in the side view shown in e.
Fitting the crystal structure of ICP8 into the reconstructed map
The crystal structure of ICP8 with a 60 amino acid C-terminal truncation (of 1196 total amino acids) has been solved by Mapelli et al. (28). When this structure was fitted into the reconstructed map, 18 copies of the monomer were fitted into two stacks of densities with C9 symmetries forming the top and bottom rings (Figure 3a–c). The result obtained from Chimera is used throughout the figures. The fitting result obtained using Situs (30) (not shown) was similar to the one from Chimera, placing the two dimers into one symmetrical unit in the same orientations as seen in Figure 3d and e, placing the putative ssDNA-binding sites of the top and bottom monomers facing each other in the middle of the ring. Full-length ICP8 was used in this study for preparation of the binary complex rings and reconstructing the map, but the crystal structure used for fitting was obtained from a 60 residue C-terminal truncated version. Therefore, it is expected that there should be some extra density in the map to accommodate the amino acids missing from the crystal structure. Indeed, such extra density exists in the map around the area corresponding to the truncated C-terminal domain of the fitted crystal structure (Supplementary Figure S5).
The cross-correlation score for the fit from Chimera was 0.9, and the Z-score calculated by fitting the crystal structures of proteins randomly selected from the PDB database with sizes similar to ICP8 (see ‘Materials and Methods’ section for details) was 5.78. The scores from e2fhstat.py were 2.98 SD above the mean (percentile rank of 99) for real space correlation and 2.62 SD above the mean (percentile rank 99) for atom inclusion. As the PDB file that was prepared from the crystal structure and used for fitting contained the N-terminus from one monomer and the C-terminus from the interacting monomer, it was necessary to check the distance between the N- and C-termini for a single monomer to make sure they were close enough to each other. The measured distances from the two halves of the ring were 13.9 and 14.1 Å compared with the distances from the published crystal structure, which were 19.2 and 16.1 Å. Therefore, the distance in the fitted structure is suitable for accommodating the length of the flexible loop between the N- and C-termini, which is invisible in the crystal structure.
To further validate our fit, we compared the conformation of the fitted monomers with the locations of the monomers in the crystal packing relative to each other. We saw that a pair in the crystal packing matches the conformation of two ICP8 monomers fitted to one symmetrical unit of the map well, supporting our fit (Supplementary Figure S6a and b). The slight conformational difference between the two may be attributed to either the relatively low resolution of the reconstructed map, or a conformational change in the dimer on ssDNA binding.
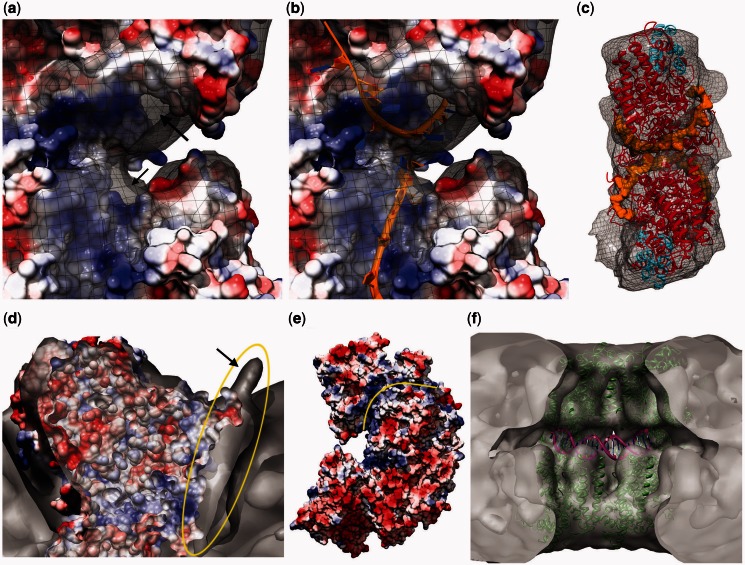
When a surface-charge map is generated from the fitted PDB, it becomes clear that the positively charged patches (represented as blue) found in the N-terminus of the top and bottom monomers come together, meeting in the middle of the ring. Supplementary Figure S6c shows this arrangement using a single symmetrical unit fitted with two monomers to the top and bottom. On superimposing the reconstructed map with this surface-charge model, extra densities became visible around both the top and bottom monomers (Figure 4a). These extra densities could be accounted for by including the modeled ssDNA as in Mapelli et al. (28) (Figure 4b). It became apparent that the ssDNAs that would be associated with the top and bottom halves of the ring would be facing each other in the middle of the map (Figure 4c and Supplementary Figure S6e). Considering the function of ICP8 as an annealase, this arrangement of the modeled DNA suggests a mechanism for the annealing, discussed later in the text (‘Discussion’ section).
Figure 4.
Proposed locations of DNA in the oligomeric ring structure. (a) It shows a side view of the putative ssDNA-binding sites of a top and bottom monomer pair fitted into one symmetrical unit (i.e. close-up side view of the ssDNA-binding surfaces shown in Supplementary Figure S6c) superimposed with the reconstructed map showing the extra density left unfilled after fitting. This is visible around the positively charged surface shown in blue (arrows point to parts of these densities visible from this angle). (b) It shows the ssDNAs (rendered as orange ribbons) modeled as published previously (28), fitting into the extra density seen in a. (c) It shows an isolated symmetrical unit with the fitted PDB (red and cyan for N- and C-termini, respectively) and the modeled ssDNAs bound to the top and bottom monomers (rendered orange, space filling). The view point corresponds to the side that faces to the center of the ring. (d) It is a clipped top view rendering showing the extra density (circled yellow) left unfilled around the positively charged (blue) surface after fitting the crystal structure. This unfilled density extends to the outside of the ring, creating a small protrusion (arrow). (e) It shows the side view surface-charge representation of two monomers fitted into one symmetrical unit. The positively charged path (blue) on the top monomer (marked by a yellow line) coincides with the extra density shown in d. This positively charged path starts from the proposed ssDNA-binding site (facing left here, shown in a as a close-up from the other side) and extends all the way to the outside of the monomer, coinciding with the small protrusion shown in d. Hence, this extra density is suggested to represent the DNA entering into or exiting from the oligomeric ring through a port formed between two monomers placed next to each other. (f) It shows the PatchDock result obtained by docking a bent 20-bp dsDNA molecule (see ‘Materials and Methods’ section) to a fitted ICP8 tetramer composed of two symmetrical units.
Docking dsDNA onto the fitted structure
In the reconstructed model, additional extra density was observed around the positively charged patch extending toward the outside of the ring, meeting with the small protrusions observed in the reconstruction (Figure 4d and e and Supplementary Figure S6d). Based on this, we hypothesized that these densities may belong to DNA that extends toward the outside of the ring and/or may have an entrance/exit port between the symmetrical units. To test this hypothesis, a straight random-sequence 20-bp dsDNA was docked to a fitted tetramer composed of two symmetrical units, using PatchDock (33). One of the highest scoring solutions showed a position for this docked dsDNA molecule coinciding with the density protruding from the surface of the ring (Supplementary Figure S6f). Once the two ssDNAs are annealed forming a dsDNA, there should be enough space between the top and bottom rings to accommodate it. To test this, a 20-bp curved dsDNA (see ‘Materials and Methods’ section) was docked similarly using PatchDock to determine the predicted location of the dsDNA forming on annealing. In the highest scored complex (Figure 4f), the dsDNA was docked into the available space between the top and bottom rings, fitting into the cleft formed by the top and bottom monomers coming together. This would be the predicted location a dsDNA should occupy, once it has formed on annealing of the two ssDNA molecules predicted to be right above and below this location.
DISCUSSION
We have reconstructed a 3D structure of full-length ICP8 in complex with ssDNA in the form of two nonameric rings, one on top of the other, which we propose represents one turn of the large nucleoprotein filaments formed during an ssDNA annealing reaction (15). The reconstructed volume is composed of two back-to-back 9-fold symmetric rings, corresponding to a molecular weight of ∼2.3 MDa. This structure is consistent with previously published structures of ICP8 and other annealases, as well as with classic recombinases catalyzing strand invasion (SI). Fitting the crystal structure of ICP8 into the reconstructed ring suggests a mechanism for the annealing phase of homologous recombination that occurs in all life forms from viruses to humans.
Euler angle distribution (Supplementary Figure S3, left) is consistent with our initial observation that the particles were preferentially oriented either flat or on their sides, with tilted views being less common because of the way in which a ring would attach onto a flat surface (see ‘Results’ section). It may be possible to avoid this issue using cryoEM, which may also have the added value of reconstructing a higher resolution structure. Efforts in this regard will be pursued in the future.
We previously reported that the diameter of the annealing intermediate filaments was ∼280 Å (15). Using the images from that study, we calculated the height of each turn (helical pitch) as being ∼140 Å. This agrees well with our 3D reconstructed ring with a 250 Å diameter and 135 Å height, further supporting the idea that the annealing intermediate filaments are stacks of such rings. Being a ring, the reconstructed structure is not only similar to previously observed structures of other ATP-independent annealases catalyzing single-strand annealing reactions (such as Rad52, Redβ, ERF and RecT) (18–21) but also to some of the structures published for ATP-dependent recombinases catalyzing double-SI (such as RadA, PfRad51, Sak and hDmc1) (41–45). Here, however, the reconstructed ICP8–ssDNA ring is a stack of two rings, whereas only a single ring was formed by Rad52, Redβ and ERF when they were not bound to their DNA substrates.
Based on the fitting results, the interaction of the C-terminal domain of one ICP8 molecule with the N-terminal domain of another one observed in the crystal structure is responsible for the arrangements of the monomers forming the top and bottom rings. Such an interaction was observed previously in oligomers formed by hDmc1 and Rad52 (22,43). Moreover, it was shown that mutating the residues at the monomer–monomer interaction interface disrupts the function of ICP8 (46). To form a stable complex, the top and bottom rings must be held together by either protein–protein interactions between two N-terminal domains of ICP8 as observed in the crystal packing, or by the hydrogen bonds between the ssDNAs attached to these two rings.
Of the limited number of available crystal structures of annealases, none of them crystallized with a top–bottom dimer arrangement as in our ICP8 fitting result. However, when we examined the crystal packing of classical recombinases RecA (3CMT) (47), PfRad51 (1PZN) (48) and RadA (3NTU), we observed dimers with a conformation similar to our top–bottom ICP8 dimers fitted into one symmetrical unit of the density map. The most striking similarity among these and ICP8 is that the DNA-binding sites face each other on the two monomers forming such a dimer (Supplementary Figure S7). This would allow searching for productive base pairing between two ssDNA molecules for annealing and forming a dsDNA molecule. This is similar to the trans-searching model recently proposed for Rad52 (49). In the same report, the authors concluded that the most efficient annealing took place when each of the two ssDNA strands was wrapped onto Rad52, and that homology search and annealing occur between two Rad52–ssDNA binary complexes, as in case of ICP8 reported here.
The presence of two DNA-binding sites seems common among recombinases. As an example, Rad52 was shown to have two DNA-binding sites in its DNA-binding groove, with the deeper site binding to ssDNA and the outer site to dsDNA (50). The authors also proposed a mechanism explaining how Rad52 may catalyze both SSA and SI using both of these DNA-binding sites. Likewise, human Dmc1 was shown to have two basic patches predicted to be DNA-binding sites, and mutational analysis has shown that both the inner and outer basic patches are involved in ssDNA binding, whereas the inner basic patch is only involved in dsDNA binding (45). The authors suggested that the outer patch may guide ssDNA to the inner patch. The DNA-binding site of ICP8 was initially identified as residues 803–849 (51). Later, the crystal structure of ICP8 showed a cleft containing, or flanked by, well-conserved residues (Tyr543, Asn551, Arg772, Lys774, Arg776, Tyr988, Phe998 and Asn1002) that were proposed to interact with ssDNA. When the surface-charge model of the crystal structure of ICP8 is examined, a positive patch corresponding to residues 800–880, which is outside and above the cleft proposed to be the ssDNA-binding site, can be observed. This agrees with the consensus DNA-binding site (residues 803–849). In light of the discovery of a secondary DNA-binding site on other recombinases, combined with earlier literature supporting the presence of two separate binding sites on ICP8 for ssDNA and dsDNA (52,53), we predict this region as the secondary DNA-binding site of ICP8 (Supplementary Figure S8). Both of these DNA-binding sites may be involved in catalysis of DNA annealing, as suggested for Rad52. This mechanism would not require ATP; therefore, it should be fundamentally different from the mechanism of SI proteins, such as RecA-like classic recombinases.
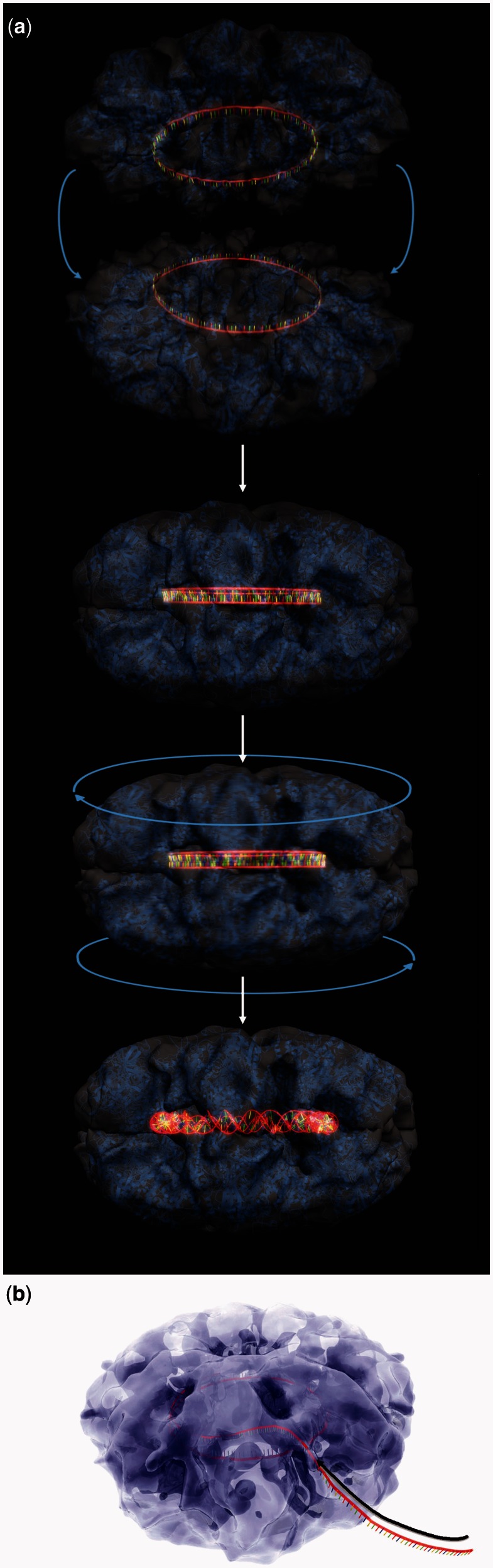
Several structural models for the mechanism of DNA annealing and strand transfer by ICP8 can be envisioned. The model most frequently considered in the literature begins with the annealase or recombinase assembling onto ssDNA to generate a long helical nucleoprotein filament, as the thin filaments we observed previously (15). If these ssDNA–protein interactions are strong, the ssDNA would be assumed to be immobile within the filament. Hence, two filaments, each containing an ssDNA, must slide over each other for sequence searching and annealing to take place (Figure 5a and Supplementary Video). At some point in this process, the two extended ICP8–ssDNA filaments must undergo a structural transition that leads to a chain of rings, and a further conformational change that converts the rings into a lock-washer conformation. This would allow the rings to fuse to each other, transforming the chain of rings into a continuous double-helical nucleoprotein filament, as the thick we previously observed (15), with the two halves sliding over each other to catalyze search for homology and annealing. If the new protein–protein contacts made during the sliding can energetically compensate for the bonds broken, then annealing may not require an external energy input. Energetic requirements in a similar, although simpler, case were discussed in detail (54–56), and may provide insights into the much more complex situation of ICP8–DNA filaments searching for homology.
Figure 5.
Artistic renderings depicting the models for two proposed mechanisms for annealing. (a) It shows frames from the Supplementary Video: ssDNA molecules are first bound by ICP8, and then annealing follows catalyzed by the docking and sliding of these two rings. (b) It shows the ssDNA entering a pre-formed oligomeric ICP8 ring.
In an alternative model, segments of the helical protein filaments may form as we observed previously (13), which then would provide a scaffold into which ssDNA can enter (Figure 5b). A similar model was proposed for human Dmc1 (45). Implicit in this model is the possibility that the ssDNA is able to slide within the filament to facilitate the search for homology and base pairing. Such sliding also may not require energy. Here, too, a reorganization to generate the active conformation for annealing must occur, which may be facilitated by the ssDNA being able to slide within the protein structure.
In a cell undergoing lytic HSV-1 replication, a portion of the ICP8 is thought to be complexed with the HSV-1 alkaline nuclease UL12 (57) (also our unpublished results and S. Weller, personal communication). The UL12 + ICP8 system is homologous to λExo + Redβ two-component recombinase, where it has been shown that λExo generates the ssDNA substrate for Redβ, and being in complex with λExo, Redβ is kept close to the site of generation of ssDNA that will be the substrate for the following annealing reaction. As a result, λExo effectively loads Redβ onto the ssDNA generated by λExo (manuscript in preparation) (58). In context of SSA catalyzed by two-component recombinases, a chain-of-rings structure should form when annealase-bound ssDNA tails of two homologous dsDNA breaks being processed by the exonuclease come in contact.
Based on our findings and previous work of others, we would summarize the following common themes used by recombinases: (i) formation of rings and filaments, with the possibility of filaments generated from rings; (ii) presence of two DNA-binding sites, allowing the catalysis of both SSA and SI; (iii) formation of a symmetrical unit by forming a dimer with a pseudo 2-fold symmetry (two monomers facing each other), bringing the ssDNA-binding sites, and therefore the two ssDNAs (substrates for the annealing reaction), together for catalyzing strand annealing. Therefore, the mechanisms we propose have the potential to be general mechanisms used by all annealases found in bacteriophages to humans.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8 and Supplementary Video.
ACKNOWLEDGEMENTS
The authors thank Ludovic Autin (ePMV), Stefan Birmanns (Sculptor), Manuel Esguerra (Blender), Jeff Roach (UNC Computation Clusters), Smaranda Willcox (ICP8 purification) and Willy Wriggers (Situs) for their help. They also thank Sandra Weller and Paul A. Tucker for critically reading the manuscript and their input. They greatly appreciate the early help in this study from Paul Tucker and Eleni Mumtsidu at the European Molecular Biology Laboratory Hamburg Unit.
FUNDING
National Institutes of Health (NIH) [ES13773, GM31819 and CA019014 to J.D.G. and GM080139 to S.J.L.]. Some of the molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco [NIH P41 RR001081]. Funding for open access charge: NIH [GM31819 to J.D.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vellani TS, Myers RS. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J. Bacteriol. 2003;185:2465–2474. doi: 10.1128/JB.185.8.2465-2474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reuven N, Staire A, Myers R, Weller S. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 2003;77:7425–7458. doi: 10.1128/JVI.77.13.7425-7433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bortner C, Hernandez TR, Lehman IR, Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J. Mol. Biol. 1993;231:241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 5.Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc. Natl Acad. Sci. USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 7.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda promotes strand exchange. J. Mol. Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 9.Conley AJ, Knipe DM, Jones PC, Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J. Virol. 1981;37:191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challberg MD. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl Acad. Sci. USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Donnell ME, Elias P, Funnell BE, Lehman IR. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J. Biol. Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 12.Makhov AM, Taylor DW, Griffith JD. Two-dimensional crystallization of herpes simplex virus type 1 single-stranded DNA-binding protein, ICP8, on a lipid monolayer. Biochim. Biophys. Acta. 2004;1701:101–108. doi: 10.1016/j.bbapap.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Makhov AM, Sen A, Yu X, Simon MN, Griffith JD, Egelman EH. The bipolar filaments formed by herpes simplex virus type 1 SSB/recombination protein (ICP8) suggest a mechanism for DNA annealing. J. Mol. Biol. 2009;386:273–279. doi: 10.1016/j.jmb.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol. 1996;258:789–799. doi: 10.1006/jmbi.1996.0287. [DOI] [PubMed] [Google Scholar]
- 15.Makhov AM, Griffith JD. Visualization of the annealing of complementary single-stranded DNA catalyzed by the herpes simplex virus type 1 ICP8 SSB/recombinase. J. Mol. Biol. 2006;355:911–922. doi: 10.1016/j.jmb.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Dutch RE, Lehman IR. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J. Virol. 1993;67:6945–6949. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozgur S, Damania B, Griffith J. The Kaposi’s sarcoma-associated herpesvirus ORF6 DNA binding protein forms long DNA-free helical protein filaments. J. Struct. Biol. 2011;174:37–43. doi: 10.1016/j.jsb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thresher RJ, Makhov AM, Hall SD, Kolodner R, Griffith JD. Electron microscopic visualization of RecT protein and its complexes with DNA. J. Mol. Biol. 1995;254:364–371. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- 19.Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc. Natl Acad. Sci. USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poteete AR, Sauer RT, Hendrix RW. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J. Mol. Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- 21.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 22.Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- 23.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez TR, Lehman IR. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J. Biol. Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 25.Mumtsidu E, Makhov AM, Konarev PV, Svergun DI, Griffith JD, Tucker PA. Structural features of the single-stranded DNA-binding protein of Epstein-Barr virus. J. Struct. Biol. 2008;161:172–187. doi: 10.1016/j.jsb.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 28.Mapelli M, Panjikar S, Tucker PA. The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding. J. Biol. Chem. 2005;280:2990–2997. doi: 10.1074/jbc.M406780200. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 31.Birmanns S, Rusu M, Wriggers W. Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J. Struct. Biol. 2011;173:428–435. doi: 10.1016/j.jsb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 33.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dijk M, Bonvin AM. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37:W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schrodinger,LLC. (2010). http://www.pymol.org/citing.
- 36. Persistence of Vision Pty Ltd. (2004). 3.6 ed. Persistence of Vision Pty. Ltd. Williamstown, Victoria, Australia. http://www.povray.org/documentation/view/3.6.0/203/
- 37.Johnson GT, Autin L, Goodsell DS, Sanner MF, Olson AJ. ePMV embeds molecular modeling into professional animation software environments. Structure. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blender foundation. (2011). Blender v2.59, Stichting Blender Foundation, Amsterdam, the Netherlands.
- 39.Erler A, Wegmann S, Elie-Caille C, Bradshaw CR, Maresca M, Seidel R, Habermann B, Muller DJ, Stewart AF. Conformational adaptability of Redbeta during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol. 2009;391:586–598. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S, Yu X, Seitz E, Kowalczykowski S, Egelman E. Archaeal RadA protein binds DNA as both helical filaments and octameric rings. J. Mol. Biol. 2001;314:1077–1162. doi: 10.1006/jmbi.2000.5213. [DOI] [PubMed] [Google Scholar]
- 42.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okorokov AL, Chaban YL, Bugreev DV, Hodgkinson J, Mazin AV, Orlova EV. Structure of the hDmc1-ssDNA filament reveals the principles of its architecture. PLoS One. 2010;5:e8586. doi: 10.1371/journal.pone.0008586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson JY. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 2008;18:1142–1146. doi: 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, Shibata T, Kurumizaka H, Yokoyama S. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol. Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 46.Bryant KF, Yan Z, Dreyfus DH, Knipe DM. Identification of a divalent metal cation binding site in herpes simplex virus 1 (HSV-1) ICP8 required for HSV replication. J. Virol. 2012;86:6825–6834. doi: 10.1128/JVI.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 48.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010;38:2917–2930. doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kagawa W, Kagawa A, Saito K, Ikawa S, Shibata T, Kurumizaka H, Yokoyama S. Identification of a second DNA binding site in the human Rad52 protein. J. Biol. Chem. 2008;283:24264–24273. doi: 10.1074/jbc.M802204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang YS, Hall JD. Characterization of a major DNA-binding domain in the herpes simplex virus type 1 DNA-binding protein (ICP8) J. Virol. 1990;64:2082–2089. doi: 10.1128/jvi.64.5.2082-2089.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee CK, Knipe DM. An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J. Virol. 1985;54:731–738. doi: 10.1128/jvi.54.3.731-738.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruyechan WT, Weir AC. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J. Virol. 1984;52:727–733. doi: 10.1128/jvi.52.3.727-733.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 55.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor—operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 56.Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor—operator interaction: equilibrium measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 57.Thomas MS, Gao M, Knipe DM, Powell KL. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 1992;66:1152–1161. doi: 10.1128/jvi.66.2.1152-1161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolun G. More than the sum of its parts: physical and mechanistic coupling in the phage lambda Red recombinase. Dissertations from ProQuest. 2007 paper 2590. Department of Biochemistry and Molecular Biology, Miller School of Medicine, University of Miami, Miami, FL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.