Abstract
All orthobunyaviruses possess three genome segments of single-stranded negative sense RNA that are encapsidated with the virus-encoded nucleocapsid (N) protein to form a ribonucleoprotein (RNP) complex, which is uncharacterized at high resolution. We report the crystal structure of both the Bunyamwera virus (BUNV) N–RNA complex and the unbound Schmallenberg virus (SBV) N protein, at resolutions of 3.20 and 2.75 Å, respectively. Both N proteins crystallized as ring-like tetramers and exhibit a high degree of structural similarity despite classification into different orthobunyavirus serogroups. The structures represent a new RNA-binding protein fold. BUNV N possesses a positively charged groove into which RNA is deeply sequestered, with the bases facing away from the solvent. This location is highly inaccessible, implying that RNA polymerization and other critical base pairing events in the virus life cycle require RNP disassembly. Mutational analysis of N protein supports a correlation between structure and function. Comparison between these crystal structures and electron microscopy images of both soluble tetramers and authentic RNPs suggests the N protein does not bind RNA as a repeating monomer; thus, it represents a newly described architecture for bunyavirus RNP assembly, with implications for many other segmented negative-strand RNA viruses.
INTRODUCTION
The Bunyaviridae family of segmented negative-stranded RNA viruses contains >330 viruses that are divided into five genera, namely, Hantavirus, Nairovirus, Orthobunyavirus, Phlebovirus and Tospovirus (1). Several of these viruses are emerging pathogens responsible for highly lethal infections of humans, including Crimean–Congo haemorrhagic fever nairovirus, Rift Valley fever phlebovirus and Sin Nombre hantavirus. In addition to inflicting serious human disease, bunyaviruses are also responsible for devastating diseases of animals and plants, such as the teratogenic Akabane orthobunyavirus (2) and the globally distributed tomato spotted wilt tospovirus (3).
Of these five genera, the Orthobunyavirus genus is the largest, currently comprising 48 classified species that are sub-divided into 18 distinct subtypes on the basis of serological characteristics (1). Orthobunyaviruses are arthropod-borne and include human pathogens, such as LaCrosse virus (4), Tahyna virus (5), Cache Valley virus (6) and Ngari virus (7), as well as the extensively studied prototypic bunyavirus member, Bunyamwera virus (BUNV). A further important addition to the Orthobunyavirus genus emerged in Europe in 2011 as the causative agent of a serious disease of cattle, sheep and goats, characterized by congenital defects and abortion of offspring (8). This virus was named Schmallenberg virus (SBV), and phylogenetic analysis suggests its emergence was not because of recent re-assortment events, although its relationship with other members of its serogroup is complex (9).
The orthobunyavirus genome comprises three segments of negative sense RNA named small (S) medium (M) and large (L). The S segment encodes the nucleocapsid protein (N) and non-structural protein (NSs) from overlapping open reading frames, whereas the M and L segments encode the membrane glycoproteins and RNA-dependent RNA polymerase (RdRp), respectively.
These RNA segments are each encapsidated with multiple copies of N to form complexes called ribonucleoproteins (RNPs), and this association is critical for gene expression by the viral polymerase. RNP formation is also required for segment packaging during assembly of new virus particles, mediated through direct interaction between N and the viral glycoproteins (10–12). The orthobunyavirus RNP is uncharacterized at the molecular level and consequently many fundamental aspects of RNP function are poorly understood, including the mechanism of RNA binding, its assembly pathway and quaternary structure and the mechanism by which the RdRp copies the N protein-protected RNA genome.
Molecular details of how bunyavirus N proteins encapsidate their cognate RNA genomes are best understood for Rift Valley fever virus (RVFV), and much of this information has resulted from determination of crystal structures of apo monomers (13) and hexamers (14), as well as RNA-bound multimers (15). These structures indicate that linkage between adjacent N molecules in the N multimer is mediated through a highly flexible α-helical N-terminal arm, which accounts for nearly all contact between adjacent N subunits. RNA is sequestered in a deep groove lining the internal surface of the multimer, with the RNA bases facing away from the solvent. Critically, the flexibility of the arm and its limited intermolecular contact allows formation of N tetramers, pentamers and hexamers both in solution and in crystals. However, the building block for segment RNP assembly is the monomer rather than the higher order multimers, and this is in good agreement with the non-helical and apparently flexible appearance of RVFV RNPs by electron microscopy (EM) (15).
Whether this same RNP assembly mechanism is common to other bunyaviruses is unknown, although the apparent diversity of N protein size and sequence across the Bunyaviridae family raises the possibility that there may be important and fundamental cross-genera differences (16,17). N protein crystal structures are available for only one other bunyavirus member, namely, CCHFV (16,18,19), which exhibits no structural homology with N from RVFV. However, the CCHFV N structures did not directly reveal N–N or N–RNA-binding surfaces, making interpretation of the nairovirus RNP assembly strategy difficult.
In this study, we report the crystal structure of the N protein from BUNV and SBV, which are important representatives of two different serotypes within the Orthobunyavirus genus (Supplementary Figure S1). The BUNV N structure includes RNA (derived from the bacterial expression host), directly identifying the RNA-binding surface within a deep positively charged channel, an assignment supported by both in vitro and in vivo functional analysis of N protein mutants. The orientation of RNA within the channel would render it inaccessible to a transcribing polymerase, implying that template-dependent RNA synthesis would require at least partial dissociation of the N–RNA complex. Both BUNV and SBV structures comprise N tetramers and reveal N- and C-terminal arms that contact different adjacent monomers within the multimer assembly. Comparison of the crystal structure of the N tetramer with electron micrographs of both soluble tetramers and authentic RNPs suggests that the assembled RNP does not comprise strings of individual monomers binding the viral RNA, a model that differs to that recently reported for RVFV.
MATERIALS AND METHODS
Expression and purification of the SBV N protein
A codon-optimized cDNA encoding the SBV N protein was chemically synthesized (Dundee Cell Products) using the published sequence of the S segment from SBV isolate BH80/11-1 (Accession number: HE649914). The cDNA was inserted into expression plasmid pET28(a) (Novagen) downstream of the coding sequence for both SUMO (type 3) and a 6xHis epitope, and the resulting plasmid pET28(a)-SUMO-SBV-N was transformed into Escherichia coli (E. coli) BL21 CodonPlus (DE3)-RIL cells.
Both SBV N and BUNV N purify primarily as tetramers. Also present are a mixture of larger oligomers as determined by size-exclusion chromatography (Supplementary Figure S2). The larger oligomers contain RNA, as judged by high Abs260/280 ratios, and they were not used further because of their heterogeneous nature. To produce SBV N tetramers containing RNA carried over from the expression host, cells were grown in 1 l of Luria–Bertani (LB) broth (Fisher Scientific) supplemented with 50 μg/ml of kanamycin and 35 μg/ml of chloramphenicol at 37°C until the OD600 reached 0.7. Expression of the SUMO-SBV-N fusion protein was induced using 1 mM isopropyl 1-thio-β-d-galactopyranoside (ITPG) for 36 h at 12°C. Cells were pelleted by centrifugation at 8000g for 30 min and resuspended in 25 ml of lysis buffer (500 mM NaCl, 25 mM imidazole, 2 mM MgCl2 and 100 mM Tris, pH 8.0). This and all subsequent purification steps were performed at 4°C. Cells were lysed by sonication after adding 200 μg of DNase 1, 200 μg of RNase A and 100 μg of chicken egg white lysozyme. Cell debris were pelleted by centrifugation at 23 000 g for 30 min. Soluble N within the supernatant was purified by metal affinity chromatography using a 5-ml HisTrap HP column (GE Healthcare) followed by four 20-ml washes using wash buffer (500 mM NaCl and 100 mM Tris, pH 8.0), with increasing imidazole concentrations (25, 50, 65 and 85 mM, respectively). Bound protein was eluted using 20 ml of elution buffer (500 mM NaCl, 500 mM imidazole and 100 mM Tris, pH 8.0). The eluted N protein was diluted with dialysis buffer (150 mM NaCl and 20 mM Tris, pH 8.0) to 0.3 mg/ml to prevent precipitation during dialysis, and the N-terminal 6xHis-SUMO tag was removed concurrently using U1p SUMO protease (200 μg of protease/20 mg of protein). Diluted protein was dialysed against 5 l of dialysis buffer overnight inside a 3.5 kDa MWCO dialysis membrane (Spectra/Por). The cleaved N-terminal tags were removed using a 5-ml HisTrap HP column, which was washed with 40 ml of wash buffer (500 mM NaCl, 50 mM imidazole and 100 mM Tris, pH 8.0), followed by 20 ml of elution buffer. The flowthrough and wash samples, which contain the protein, were pooled, diluted to 0.5 mg/ml with dialysis buffer and dialysed overnight. Removal of the N-terminal tag resulted in the addition of a single non-native serine residue at the N terminus. The cleaved protein was concentrated to 5 ml using 100 KDa MWCO Vivaspin20 concentrators (Generon) and subjected to size-exclusion chromatography using a Superdex S200 16/60 gel-filtration preparative column (GE Healthcare) equilibrated with low-salt buffer (150 mM NaCl and 20 mM Tris, pH 8.0). Fractions eluting as a single peak corresponding to a size of ∼112 kDa (i.e. tetramers) were pooled, concentrated to 8 mg/ml and stored at −80°C.
To produce SBV N tetramers containing no RNA, the same protocol was followed with the addition of a 150-ml wash with RNA removal buffer (2 M NaBr and 100 mM Tris, pH 8.0) before elution during the first metal affinity chromatography step. After elution, RNA removal was confirmed by checking the Abs260/280 ratio. Size-exclusion chromatography was performed using the gel filtration column equilibrated with high-salt buffer (2 M NaCl, 1 M NaBr and 20 mM Tris, pH 8.0).
Selenomethionine-substituted SBV N protein (SM-SBV N) was produced by transforming the pET28(a)-SUMO-SBV-N plasmid into E. coli B834 cells and growing a starter culture in 50 ml of LB supplemented with 50 μg/ml of kanamycin overnight. Cells were pelleted by centrifugation at 8000g for 10 min, washed by re-suspension in 25 ml of minimal medium (SelenoMet Nutrient Mix, Molecular Dimensions) and re-centrifuged. The supernatant was discarded, and the cells re-suspended in 25 ml of fresh minimal medium and washed again. The cells were finally re-suspended in 25 ml of fresh minimal medium and then added to 1 l of selenomethionine medium (SelenoMet Medium Base plus SelenoMet Nutrient Mix, Molecular Dimensions) supplemented with 50 μg/ml of kanamycin. The culture was incubated at 37°C for 45 min after which 30 mg of selenomethionine was added. Cells were grown until the OD600 reached 0.7, and expression of the SM-SBV N was induced using 1 mM ITPG. The cells were grown at 37°C overnight, and protein was purified as for SBV N purified without RNA.
Expression and purification of the BUNV N protein
The cDNA encoding the BUNV N protein was inserted into plasmid pET28(b) (Novagen) downstream of a 6xHis epitope and a thrombin cleavage site, as described previously (20). The resulting plasmid pET28(b)-BUNV-N was transformed into E. coli BL21 CodonPlus (DE3)-RIL cells. Protein expression and purification protocols for BUNV N with and without RNA carried over from the expression host, as well as selenomethionine-substituted BUNV N (SM-BUNV N), are the same as for SBV N protein with the following exception: wash buffer during the first metal affinity chromatography step contained 25, 50, 75 and 100 mM imidazole, respectively. The 6xHis tag was removed after dialysis by adding thrombin protease (200 μg of protease/20 mg of protein) to the protein sample and letting it stand (no shaking) at 12°C over night, followed by 3 h incubation at room temperature. Levels of RNA contamination were checked by ultraviolet absorbance. Protein with an Abs260/280 ratio <0.7 was deemed RNA free.
Crystallization
Initial crystallization trials were performed at room temperature with commercial screens using the sitting-drop vapour-diffusion method. Crystallization drops were set-up with the aid of an Oryx 6 Douglas crystallization robot (Douglas Instruments) with 250 nl of protein solution plus 250 nl of reservoir solution in MRC two-well crystallization microplates (Swissci) equilibrated against 60 µl of reservoir solution. Optimization trials were set-up using the hanging-drop vapour-diffusion method in 24-well Greiner plates by using 1 μl of protein solution plus 1 μl of reservoir solution equilibrated against 500 μl of reservoir solution.
Previously established crystallization conditions for BUNV N containing RNA (20) were further optimized, with the best being 10% (w/v) PEG 6000, 1.8 M NaCl and 100 mM sodium citrate, pH 5.7. Heavy atom soaks and co-crystallization trials were set-up to produce crystals suitable for anomalous dispersion experiments, and the best diffraction data were obtained from crystals grown in 11% (w/v) PEG 6000, 1.6 M NaCl, 4 mM zinc acetate and 100 mM sodium citrate, pH 5.7. The crystals were cryoprotected by stepwise transfers lasting 3 min each into reservoir solution with increasing concentrations [2, 4, 6, 8 and 10% (v/v)] of glycerol before being vitrified by submersion in liquid nitrogen. No crystals were obtained for BUNV N without RNA or for SM-BUNV N.
SBV N without RNA produced diffraction quality crystals in a variety of conditions, with the best diffraction data being obtained from crystals grown in 17% (w/v) PEG 10 000, 100 mM ammonium acetate and 100 mM Bis–Tris, pH 5.5. The crystals were cryoprotected by transfer into reservoir solution plus 20% (v/v) glycerol before being vitrified by submersion in liquid nitrogen. No diffraction quality crystals were obtained for SBV N containing RNA.
SM-SBV N without RNA crystallized in similar conditions to SBV-N without RNA, with the highest quality diffraction coming from crystals grown in 10% (v/v) glycerol, 13% (w/v) PEG 20 000, 340 mM ammonium acetate and 100 mM Bis–Tris, pH 5.5. The crystals were cryoprotected by direct submersion into liquid nitrogen.
Data collection, structure determination and refinement
X-ray data for BUNV N were recorded at beam line P13 of PETRA-III (Hamburg, Germany) at a wavelength of λ = 1.1267 Å (Zn K edge) to a maximum resolution of 3.2 Å. X-ray data for SBV N were collected at beam line I04 of the Diamond Light Source (Rutherford Appleton Laboratory, Harwell, UK) at a wavelength of λ = 0.9464 Å to a maximum resolution of 2.75 Å. X-ray data from several SM-SBV N crystals were collected at beam lines I04, I04-1 and I24 of the Diamond Light Source at a wavelength of λ = 0.9796 Å (Se K edge) to maximum resolutions varying from 3.3 to 3.5 Å.
X-ray data for the BUNV N crystal were integrated in space group C2 using XDS (21), whereas X-ray data for the SM-SBV N and SBV N crystals were integrated in space group P3 using iMOSFLM (22). All SM-SBV N and SBV N data sets collected were isomorphous. Space group identity was checked with POINTLESS (23), which corroborated C2 for BUNV N and indicated that the space group of the SBV N and SM-SBV N data sets could either be P31 or its enantiomorph P32. POINTLESS also indicated that the SBV N data set was twinned with an estimated twin fraction of 0.215 from the L-test. Reduced data from all crystals were then scaled with AIMLESS (23), whereas phasing was carried out using AUTOSOL (24). No anomalous signal could be found for Zn in the BUNV N data. The anomalous signal of individual data sets from SM-SBV N crystals was too low for phasing, and after further analysis of the data sets, the best sections of data from each individual set were selected and combined into a single data set using POINTLESS. The resulting data set was successfully phased with AUTOSOL using the single-wavelength anomalous dispersion (SAD) method with the correct enantiomorph being P32. AUTOSOL produced a density-modified map using RESOLVE (25), which was used with BUCCANEER (26) to build an initial model that showed a single tetramer in the asymmetric unit (AU). After some preliminary cycles of manual building in COOT (27) and structure refinement in REFMAC5 (28), the SM-SBV N crystal structure was used to solve the higher resolution data set from the native SBV N via rigid body refinement using REFMAC5. The structure was then fully refined after cycles of further manual building in COOT and refinement cycles in REFMAC5 using the amplitude-based twin refinement option, coupled with automatically generated local non-crystallographic symmetry restraints and TLS refinement. The globular domain of a monomer from the SBV N crystal structure was then used to solve the BUNV N data set using PHASER (29), which found eight monomers in the AU that correspond to two tetramers. The structure was fully refined after cycles of manual building in COOT and refinement cycles in REFMAC5, coupled with automatically generated local non-crystallographic symmetry restraints.
Analysis of RNA binding by orthobunyavirus N proteins
The RNA-binding ability of wild-type N proteins from BUNV and SBV, as well as BUNV N protein mutants, was examined in vitro using fluorescence polarization (FP). Orthobunyavirus N proteins without RNA were expressed and purified as described earlier in the text, but with the following changes: after metal affinity chromatography, the protein samples were dialysed and concentrated to 1 mg/ml. The 6xHis tags were not removed, and the protein samples were not subjected to size-exclusion chromatography (protein purified by this quick method has an Abs260/280 ratio of ∼1.0, indicating that the majority of RNA had been removed). FP assays were performed as described previously (30). Briefly, a 3′-fluorescein-labelled (3′-Fl) RNA representing the 5′-terminal 48 nt of the BUNV S segment anti-genome (BUNV48AG) was synthesized (5′-AGUAGUGUACUCCACACUACAAACUUGCUAUUGUUGAAAAUCGCUGUG-3′-Fl; Dharmacon Research). Reactions were carried out in 20 mM Tris, pH 7.5, 150 mM NaCl and 0.01% (v/v) Triton X-100, with 20 nM 3′-Fl-labelled RNA and increasing concentrations of protein (from 1.4 nM to 13 μM). Experiments were performed in triplicate, and readings were taken using an EnVision Multilabel Plate Reader (Perkin-Elmer). Data were expressed as the fraction of RNA bound, plotted against protein concentration and fitted by non-linear least squares regression using OriginPro 8.6 (OriginLab) to the following quadratic equation:
where Θ is the fraction of RNA bound, Kd is the dissociation constant, Ro is the total RNA concentration and Po is the total protein concentration.
Purification and visualization of BUNV RNPs
BUNV was purified using iodixanol density gradient centrifugation, based on a previously described method (31). BHK-21 cells were infected with BUNV at a multiplicity of infection (MOI) of 0.01 and incubated for 4 days at 32°C. Virus-containing medium was collected and clarified at 3200g for 10 min at 4°C. The supernatant was further clarified through a 0.2-µm filter to remove cell debris, and then diluted 10-fold with 50% (w/v) PEG 6000 dissolved in cold TEN buffer (0.01 M Tris, pH 7.4, 0.1 M NaCl and 1 mM ethylenediaminetetraacetic acid). Virus was precipitated for 1.5 h at 4°C with moderate stirring, following by centrifugation at 3200g for 10 min at 4°C. The pelleted virus was then resuspended in 0.5 ml of cold TEN buffer and loaded on top of a 5–25% continuous iodixanol gradient. The gradient was prepared by sequentially layering 0.9 ml of 25, 20, 15, 10 and 5% (v/v) Optiprep iodixanol density gradient medium (Sigma-Aldrich) diluted in TEN buffer with protease inhibitor cocktail (Roche) and overnight incubation to allow interface diffusion. Centrifugation of virus preparation was performed at 250 000 g in an AH-650 swing rotor (Thermo Scientific) for 1.5 h at 4°C. Fractions of 1 ml were collected from the bottom of the gradient by penetrating the centrifuge tube. Purified BUNV was located by analysing each fraction by sodium dodecyl sulphate–polyacrylamide gel electrophoresis with silver staining or western blotting using an anti-BUNV N antibody. The enrichment and infectivity of purified virus was also confirmed by plaque assay. RNPs were spilled from infectious virus by treatment with saponin (Sigma-Aldrich) as previously described (32). Treated RNPs were added to glow-discharged carbon-coated EM grids, stained with 4% uranyl acetate, and then imaged on a FEI Tecnai-F20 microscope using low-dose protocols and a dose of ∼40 e-/Å2. Micrographs were recorded with a Gatan US4000 CCD camera at a calibrated magnification of ×84 800, giving a final object sampling of 1.77 Å/pixel.
Visualization of purified SBV tetramers and image processing
The purified SBV N protein was incubated with an unlabelled version of the 48mer RNA BUNV48AG, described earlier in the text. N protein and RNA were mixed at a molar ratio of 12:1 in 20 mM Tris, pH 7.5 and 150 mM NaCl, using a quantity of protein that would lead to a final tetramer concentration of 2.5 µM. The same microscope and procedure were used as described earlier in the text. A data set of 460 particles was interactively selected, and the images were excised into 256 × 256 pixel boxes, band-pass filtered (between 20 and 250 Å) and the pixel values normalized to a constant mean and standard deviation using SPIDER (33). This data set was then centred by mass and subjected to multivariate statistical classification in IMAGIC (34). All data were then re-aligned to a single, representative class average and then reclassified. All classes show complexes with apparent 4-fold symmetry (note that this 4-fold symmetry was not applied during averaging).
Analysis of mutant N protein activity using the BUNV replicon system
The ability of mutant BUNV N proteins to support RNA synthesis was examined using the BUNV replicon system, which has been previously described (35,36). Briefly, functional BUNV RNPs were assembled in confluent BHK-21 monolayers by infection with vaccinia virus recombinant vTF7-3, followed by transfection with cDNAs expressing either wild-type or mutant N proteins, L protein or the BUN-S(ren) replicon template. The source of BUNV N protein mutants was temperature-sensitive (ts) viruses generated by 5-fluorouracil treatment (37), and corresponding N protein cDNAs were inserted into expression plasmid pTM1. Two mutants were analyzed: the ts19 N protein possessed a C-terminal extension (SKQGSRGLIN) because of a single nucleotide deletion within the authentic stop codon, whereas the ts63 N protein contained a methionine to threonine change at residue position 150 (M150T). Transfected cells were treated with actinomycin-D (Sigma-Aldrich) and 3H uridine (Perkin-Elmer) to metabolically label BUNV-specific RNAs, which were harvested 20 h post-transfection. All cell growth, infection, transfection and labelling were performed at 33°C. RNAs were separated using agarose–urea electrophoresis and visualized by fluorography and autoradiography.
Phylogenetic analysis of protein sequences
ClustalW (38,39) was used to align the amino acid sequences of the N proteins of BUNV and SBV with 16 other N protein sequences from six serogroups within the Orthobunyavirus genus. The alignment was performed using default settings for multiple sequence alignment analyses. The accession numbers of the N proteins used in the analysis are listed along with their corresponding serogroup: Anopheles A serogroup: Anopheles A virus (ACN43212). Bunyamwera serogroup: Bunyamwera virus (NP047213), Batai virus (CAA51843), Cache Valley virus (CAA51845), Maguari virus (AAA57147) and Germiston virus (AAA87603). Bwamba serogroup: Bwamba virus (ACE07176). California serogroup: La Crosse virus (AAA42782), Jamestown Canyon virus (ABS29152) and Tahyna virus (AAC55340). Simbu serogroup: Akabane virus (BAA24201), Oropouche virus (AAP69950), Sathuperi virus (CCG93489), Shamonda virus (YP006590077), Simbu virus (CCG93497) and Schmallenberg virus (CCF55031). Wyeomyia serogroup: Sororoca virus (AEZ35275) and Wyeomyia virus (AEZ35275). CLUSTAL_X (38,39) was used to analyze the resultant alignment and predict bootstrap confidence values of the alignment using the neighbour-joining algorithm. A total of 1000 bootstrap trials were conducted on each submitted alignment file, and 111 were used as a random number generator seed. Crimean–Congo haemorrhagic fever virus N protein sequence (Accession number: AAB48503.1) was assigned as an outlier and used to root the resulting phylogenetic tree. The graphical representation of the phylogeny was created using TreeView (40). Bootstrap confidences are indicated at each node.
RESULTS
Structure of the apo Schmallenberg N protein
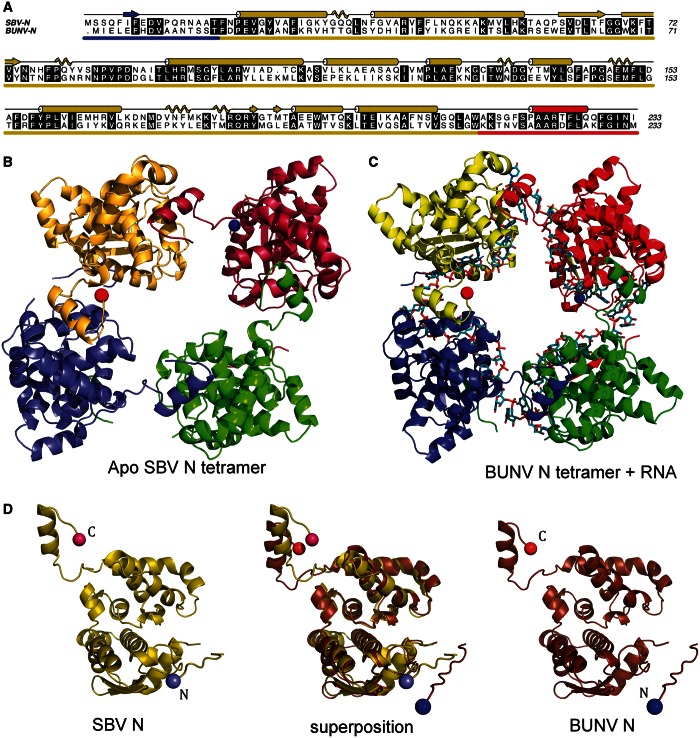
To investigate the mechanisms of RNA binding and RNP assembly for members of the Orthobunyavirus genus, we solved the crystal structures of the N protein (without RNA) from SBV and BUNV N–RNA complex. The proteins exhibit only 37% amino acid identity (Figure 1A) and are classified in different serogroups of this diverse genus (Supplementary Figure S1). We first solved the structure of the SBV N protein by using SAD to phase data to 3.28 Å from crystals of protein labelled with selenomethionine (see Table 1). The resulting structure was then used to solve data from a higher resolution (2.75 Å) data set obtained from an isomorphous SBV N crystal grown from native protein.
Figure 1.
Crystal structures of the apo SBV N protein and BUNV N protein in complex with RNA. (A) Sequence alignment of SBV N and BUNV N with secondary structure assignments for SBV shown above in cartoon form (β-strands as arrows; α-helices as cylinders; 310 helices as coils). Conserved residues are highlighted. The N-terminal arm is underscored in blue, globular domain in yellow and C-terminal domain in red. (B) SBV N tetramer in ribbons representation. (C) BUNV N tetramer (ribbons) bound to RNA (cyan, sticks). In both B and C, the N- and C- termini of the yellow monomer are marked with a blue and red sphere, respectively (note that a small part of the N-terminal arm in both SBV and BUNV N is not visible in electron density; therefore, it is not built as part of the models). (D) Protein monomers of SBV N (left), BUNV N (right) and the superposition of both (centre). Figure 1A has been made using A-line (41). Figure 1 B–D and all other figures were made using PyMol (Version 1.5.0.4 Schrödinger).
Table 1.
Data collection and refinement statistics
| Data set | SM-SBV Na | SBV N | BUNV N |
|---|---|---|---|
| Wavelength (Å) | 0.9796 | 0.9464 | 1.1267 |
| Space group | P32 | P32 | C2 |
| Cell parameters (Å,°) | a = 80.17 | a = 81.03 | a = 328.24 |
| b = 80.17 | b = 81.03 | b = 88.72 | |
| c = 128.47 | c = 128.55 | c = 88.72 | |
| α = 90.00 | α = 90.00 | α = 90.00 | |
| β = 90.00 | β = 90.00 | β = 94.30 | |
| γ = 120.00 | γ = 120.00 | γ = 90.00 | |
| Total reflections | 660 751 (47587) | 77 156 (11 307) | 157 101 (15 674) |
| Unique reflections | 14 225 (2925) | 24 459 (3580) | 47 586 (4503) |
| Resolution (Å) | |||
| Overall | 190.58–3.28 | 61.59–2.75 | 128.47–3.20 |
| (High-resolution shell) | (3.54–3.28) | (2.90–2.75) | (3.30–3.20) |
| Rmerge (%)b | 56.9 (132.4)a | 7.7 (56.2) | 5.1 (59.2) |
| Rpim (%)d | 11.6 (43.6) | 7.7 (55.7) | 4.8 (55.4) |
| Rcryst (%)d | 23.08 | 16.41 | 24.68 |
| Rfree (%)c | 25.72 | 20.13 | 29.68 |
| Completeness (%) | 99.8 (99.2) | 99.8 (100) | 96.9 (99.0) |
| Redundancy | 46.4 (16.3) | 3.2 (3.2) | 3.3 (3.5) |
| I/σ(I) | 13.9 (2.0) | 8.7 (2.0) | 13.2 (1.9) |
| VM (Å3/Da) | 2.31 | 2.31 | 2.41 |
| Mol. per AU | 4 | 4 | 8 |
| Reflections working set | 13 481 | 23 228 | 44 762 |
| Free R-value set (number of reflections) | 5.0% (713) | 5.0%(1219) | 5.1% (2383) |
| RMSD bond lengths (Å) | 0.005 | 0.010 | 0.006 |
| RMSD bond angles (°) | 0.996 | 1.291 | 1.327 |
| Number of atoms used in refinement | |||
| Non-hydrogen atoms | 7048 | 7182 | 16 571 |
| Protein atoms | 7064 | 7158 | 14 811 |
| Nucleic acid atoms | 0 | 0 | 1760 |
| Water molecules | 16 | 24 | 0 |
| Mean B value (Å2) | |||
| Total | 69.1 | 78.9 | 139.1 |
| Protein atoms | 69.1 | 79.0 | 139.1 |
| RNA atom | N/A | N/A | 158.2 |
| Water molecules | 44.9 | 59.7 | N/A |
| Ramachandran plot statistics (%) | |||
| Preferred region | 90.60 | 95.15 | 91.61 |
| Allowed region | 6.68 | 3.83 | 6.67 |
| Outliers | 2.72 | 1.01 | 1.72 |
aThis data set was only partially refined until the structure was good enough to serve as a molecular replacement model for the other two data sets. SM = selenomethionine.
bRmerge is defined as 100 × Σ∣I − <I>∣/Σ I, where I is the intensity of the reflection.
cRcryst = Σ∣∣Fo∣ − ∣Fc∣ ∣/Σ∣Fo∣, where Fo and Fc are observed and calculated structure factors, respectively.
dThe high Rmerge values of this data set reflect its very high redundancy; therefore, the Rpim values have also been included, as they are a better measure of the intrinsic quality of highly redundant data.
The crystals contain a single tetramer per AU, and the structure reveals that the SBV N protein is composed of a large globular domain of ∼200 residues with short N- and C-terminal arms extending away from the globular domain (Figure 1B and D). The globular domain comprises 10 α-helices, 6 short 310 helices and 2 short β strands. A short section of five amino acids (residues 12–16) that links the N-terminal arm to the globular domain could not be modelled because of lack of electron density. The SBV N tetramer does not display true 4-fold symmetry, with the tetramer ring slightly ‘squashed’ into a rhombus with 2-fold symmetry (Figure 1).
The N-terminal arm is in contact with an adjacent protomer within the tetramer, whereas the C-terminal arm contacts a different protomer and buries a significant surface area of an α-helix within the interface. This C-terminal helix seems to be the major driving force of oligomerization. PISA (42) analysis determines that formation of the SBV tetramer buries 8360 Å2 of surface area with a ΔG of complex formation of −74.6 kcal/mol. A single C-terminal helix interaction buries 2030 Å2 of surface area with ΔG of −18.6 kcal/mol. The C-terminal helix is buried in a hydrophobic pocket of the adjacent protomer, with three phenylalanine side chains stacked against hydrophobic side chains in the pocket (Supplementary Figure S3). However, if the C-terminal helix is removed from each monomer, then PISA suggests that there are no stable oligomeric species in the crystal assembly. This emphasizes the importance of the C-terminal arm interaction and suggests the interaction mediated by the N-terminal arm is less significant.
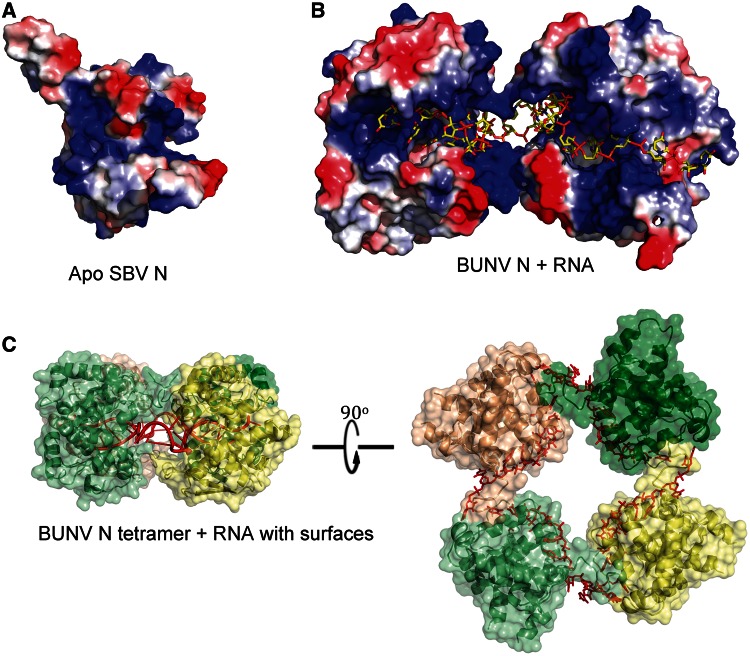
Inspection of an electrostatic surface of the SBV N monomer (Figure 2A) reveals a large electropositive groove, which is suggestive of an RNA-binding surface. Within the tetramer, this groove lines the inside of the central cavity of the tetramer, suggesting that RNA could be wrapped within this central cavity when RNPs are assembled.
Figure 2.
RNA binding by orthobunyavirus N proteins. (A) Electrostatic surface of a SBV N protomer reveals the electropositive RNA-binding groove. (B) Two molecules of BUNV N bound to RNA (as seen from inside the tetramer; RNA is yellow sticks representation). (C) BUNV N bound to RNA (red) with solvent accessible surface shown (yellow/greens, transparent).
Structure of the BUNV N protein with RNA
We then used the 2.75 Å SBV N structure as a model for molecular replacement to phase the 3.2 Å data set from BUNV N. Unlike the SBV tetramers, which do not obey 4-fold symmetry, the BUNV crystals contain two tetramers per AU each of which does display 4-fold symmetry (Figure 1C).
Despite extensive treatment with RNaseA during the purification protocol, each tetramer contains RNA bound within the electropositive groove described for SBV earlier in the text. The RNA, which originates from the expression host (43), is wrapped inside the tetramer with the RNA-binding groove of each monomer facing the centre of the tetrameric ring (Figure 2B), presumably protecting the RNA from RNaseA digestion. The SBV N protein used to generate apo crystals was treated with high salt to remove RNA during purification, but the high-salt wash was not performed on the BUNV N protein before the generation of this crystal form.
Each BUNV N protein monomer buries 9–10 bases within the RNA-binding groove (Figure 2B and C), and there is electron density visible for 44 nt within each tetramer, in close agreement with our previous estimate (43). The precise identities of individual bases are not discernable because the RNA electron density is 4-fold averaged due to the 4-fold symmetry of the tetramer. In any case, the bound RNA within the crystal is likely of random sequence; previous studies have shown that RNA bound to purified soluble recombinant tetramers contains no specific or consensus sequences (43). These two factors, along with the 3.2 Å resolution, limit how much precise detail can be described of the N–RNA interaction. It is, however, apparent that charge and hydrogen bond interactions will play a key role in the mechanism of RNA recognition by BUNV N protein, as there is little evidence in our structure of the base stacking interactions that can characterize RNA binding for some other proteins (15,30). The only side chain in the N protein that seems to stack against a base in this complex is Y176. There are two deep cavities within the groove lined by hydrophobic side chains, but these do not seem to be explored by RNA. The groove is, however, lined with residues that could hydrogen bond to bases, and there are many positively charged side chains (Figure 2) close to RNA backbone phosphates, e.g. K55, H93, R94, K127 and K179, which would provide affinity for RNA via electrostatic interactions. The final refined 2Fo − Fc density and omit map densities for RNA are presented in Supplementary Figure S4.
Structural comparison of the BUNV and SBV N proteins
Despite the fact that the SBV and BUNV N proteins are from distinct serogroups of the Orthobunyavirus genus (Supplementary Figure S1), the structure of the monomeric N protein is remarkably similar (Figure 1D) with an RMSD of 1.3 Å for 208 α-carbons. As for the SBV N monomers, each BUNV N monomer also contains a small flexible region that could not be modelled between the N-terminal arm and the globular domain of each monomer, which comprises residues 11–15. There are minor differences in loop positions between the two N proteins, but the biggest differences are in the positions of the N- and C-terminal arms; the α-carbon in C-terminal residue 233 is transposed by 6.25 Å when the models are superposed (Figure 1D), whereas the α-carbons in the last residue of the globular domain (L211) are only 0.69 Å apart. The α-carbons in residue 3 are 8.74 Å apart. Despite these differences, and the shifted arrangement of the four monomers, C-terminal arms in each structure establish functionally equivalent N–N interactions, and they likely represent the biggest driving force for oligomerization of the orthobunyavirus N proteins.
Multimer interactions
Tetramerization of both apo SBV N protein and BUNV N–RNA complex is mediated via burial of the C-terminal helix into an adjacent protomer in the tetramer (Supplementary Figure S3). The loop connecting the globular domain to the C-terminal helix adjusts its position; therefore, it seems to be flexible, to allow different conformations and potentially different tetrameric arrangements of four N protomers. This is reminiscent of the N-terminal arm of the RVFV N protein where the arm contacts the neighbouring N protomer to mediate oligomerization (15). However, the RVFV N protein can adopt tetrameric, pentameric and hexameric arrays in solution and in crystals because of the remarkable flexibility of this N-terminal arm (15). For SBV and BUNV, we only observe tetramers both in solution via gel filtration and in crystals, and perhaps this is a reflection of reduced flexibility of the N- and C-terminal arms.
Again, PISA analysis of subunit interactions suggests that the BUNV N tetramer bound to RNA is the only stable assembly. Unlike the SBV N crystals where a single tetramer is the content of each AU, in the BUNV N crystals, each AU contains two tetramers each bound to RNA. The tetramer:tetramer interface is mediated through an unusual β-sheet with contributions from four molecules (Supplementary Figure S5). Despite the reasonable hydrogen bonding across the strands of this β-sheet that sits between tetramers in the BUNV crystals, PISA suggests that this interface is not stable. Somewhat similar N-terminal interactions are seen in the SBV N protein structure; thus, there is the possibility that this interaction is used during one of the N protein functions. However, with the C-terminal helix attached to the globular domain via a flexible linker (Figure 1B), similar to the arm in RVFV that mediates a variety of oligomeric interactions, there remains the possibility that the assembly in our crystals, either apo or bound to short pieces of E. coli RNA, is not the same as in native RNPs. In RVFV, the crystal oligomers bound to short RNA do not represent the established RNP model (15).
Structural similarity between other members of the Bunyaviridae family
To provide insight into bunyavirus phylogenetics and to identify common structural features responsible for N protein functions, we compared the N protein structures of BUNV and SBV with all structures in the PDB (which includes several N protein structures from related segmented negative-stranded RNA viruses) using the DALI server (44).
Using SBV N as the search model, the top score from DALI has a Z-score of only 4.9, suggesting that there is not a close structural homologue in the current PDB. In this context, we classify the new SBV and BUNV N protein structures as representing a new fold. The top solution suggests a limited structural homology with a Maz-G-type protein (PDB 1VMG). However, with an RMSD of 3.5 Å for only 66 residues, the similarity is low.
The DALI search does reveal limited but clear structural homology with the N protein of RVFV (PDB 4H5O), with a Z-score of 4.0 and RMSD of 7.3 Å for 142 residues (Supplementary Figure S6). A similar homology was also detected for the closely related phlebovirus, Toscana virus (PDB 4H5L). The most extensive region of structural homology comprises five helices and some connecting loops buried within the globular core. Despite this rather isolated structural homology, the overall multimer architecture of the RVFV and orthobunyavirus N proteins was similar, with the vast majority of inter-subunit interactions arising from an α-helix within an extended terminal arm, rather than through interactions involving the larger globular domains. Notably, a critical difference is that the RVFV N multimer relies on one arm (two helices in the N-terminal extension), whereas in the SBV and BUNV N crystals, the multimers involve contacts mediated by both N- and C-terminal arms. As described earlier in the text, both the N- and C-terminal arms in both SBV and BUNV N seem to be rather flexible and could allow for a variety of oligomeric assemblies, which is likely to have important consequences for assembly of the RNP (Figures 1B and 2 and Supplementary Figure S2).
A remote match is detected for both the coiled-coil domain of the N protein from Sin Nombre Hantavirus (PDB 2IC6; Z-score 2.0) and the CCHFV N protein (PDB 4AQF; Z-score 2.2), but on inspection, these appear to align little more than a single helix.
Analysis of N protein residues involved in RNA binding
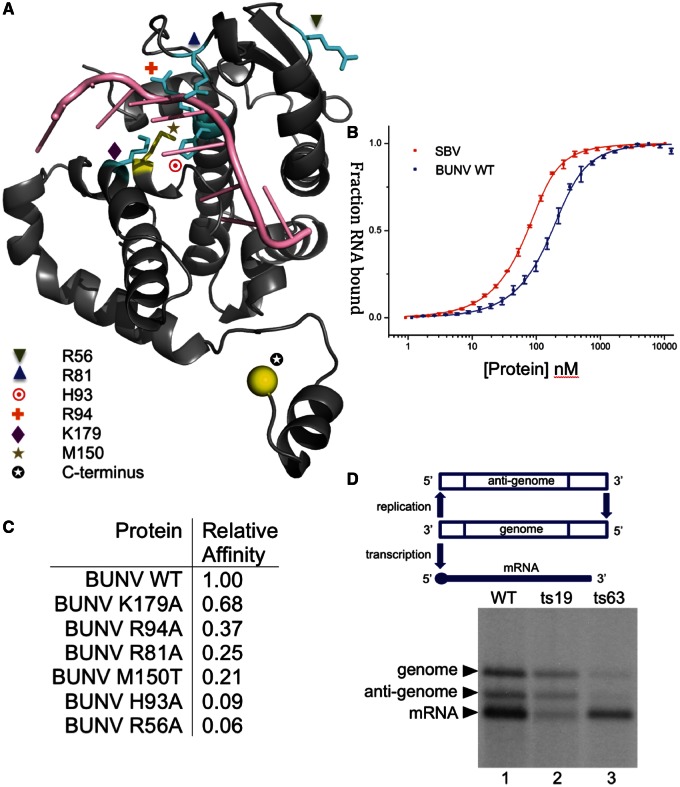
As described earlier in the text, the structure of the BUNV N–RNA complex identifies nine nucleotides in contact with each monomer within a deep positively charged channel (Figure 2). We assessed the RNA-binding contribution made by selected conserved residues within this groove using fluorescence polarization.
RNA binding by N protein was detected using a 3′-fluorescein-labelled RNA representing the 5′-terminal 48 nt of the BUNV S segment anti-genome (BUNV48AG). Binding isotherms are shown for apo wild-type SBV N and wild-type BUNV N (Figure 3B). The RNA-binding affinity of SBV N protein (21.3 ± 1 nM) seems to be higher than that for BUNV N (62.9 ± 5 nM). Both N proteins displayed high affinity for the BUNV48AG RNA, whereas mutants were weaker binders. Point mutants within or close to the RNA-binding groove were generated, purified without removing the His-tag and tested for RNA binding. The affinity relative to the wild-type BUNV N (purified the same way) is shown in Figure 3C. Alanine mutants within the RNA-binding groove reduced RNA-binding affinity, as predicted. This highlights that, unsurprisingly, residues in the RNA-binding groove are critical for N protein function.
Figure 3.
Structure–function analysis. (A) Structure of the BUNV N protein monomer bound to RNA shown with residues highlighted in cyan that have been mutated and tested for RNA-binding affinity, and mutant residues that generate temperature-sensitive viruses in yellow tested in the replicon assay. (B) Fluorescence anisotropy RNA-binding assay. Wild-type (WT) BUNV N, SBV N and mutant BUNV N proteins were tested for binding to a fluorescein-labelled 48mer. (C) Table of affinities of BUNV N mutants and SBV N wild-type relative to the BUNV N WT. (D) The replicon assay tests relative activity of the N protein in the replication and transcription of a model BUNV segment; ts19 shows considerably diminished transcription activity, whereas ts63 shows dramatically reduced replication. See text for details of mutations.
In addition to these RNA-binding assays described earlier in the text, two previous studies have analysed mutations within the BUNV N protein (45,46). We have mapped the mutations that have an effect on either virus viability or RNA synthesis onto our BUNV N structure (Supplementary Figure S7), and find that the mutants can be grouped into three categories: large buried hydrophobics are likely to disrupt the overall N protein fold; residues with side chains close to RNA within the binding groove are likely to affect RNA binding; mutation in the C-terminal helix is likely to affect oligomerization. Thus, our BUNV N structure allows the interpretation of our new mutagenesis data, as well as extant observations.
Identification of N protein residues in transcription and replication
To explore the relationship between the structure of the N protein and its critical function in forming the RNP template for RNA synthesis, we wanted to map the position of residues within mutant N proteins with interesting phenotypes onto the N–RNA crystal structure (Figure 3A). Mutant N proteins were derived from temperature-sensitive (ts) BUNV obtained by previous chemical mutagenesis of live virus in cell culture (37). Sequence analysis identified two ts viruses with altered N proteins, and the effect of these changes on promoting mRNA transcription and RNA replication from assembled RNPs was tested in the BUNV replicon system (Figure 3D).
BUNV ts19 possessed a C-terminal extension of 10 residues (SKQGSRGLIN), and it exhibited replication ability equivalent to wild-type, but it showed considerably diminished transcription activity. This severe perturbation of RNP function is consistent with the critical role of the C-terminal helical arm in promoting N–N interaction as identified in the tetramer structures. Although the helical arm that mediates this interaction is preserved in this mutant, the additional residues provided by the extension are likely to negatively impact the N–N association driven by the burial of this helix. The observation that this extension differentially affects transcription rather than replication identifies the C-terminal arm domain as an important element in maintaining RNP function and, in particular, transcription.
In contrast, N protein mutant BUNV ts63, which possessed a methionine to threonine change at residue position 150 (M150T), exhibited equivalent transcription activity to wild-type but resulted in dramatically reduced replication. Residue 150 is located with the side chain facing the RNA in the binding groove, where incorporation of the smaller and less hydrophobic side chain of threonine in place of methionine would potentially impact the overall architecture of the cavity and thus interfere with its ability to sequester RNA. When expressed and purified, recombinant M150T mutant BUNV N displays an ∼5-fold reduction in RNA-binding affinity in the polarization assay (Figure 3). The phenotype of the M150T mutation is consistent with the important role of the RNA-binding groove in RNA synthesis, which, when taken together with mutant ts19 described earlier in the text, shows that distinct regions within the N protein differentially influence transcription versus replication activities. Unfortunately, purification of BUNV N with modifications at the C-terminus (to test effects on RNA binding and oligomerization) proved unsuccessful, possibly because disruption of the tetramer could cause protein instability in vitro.
Biological relevance of the tetramer
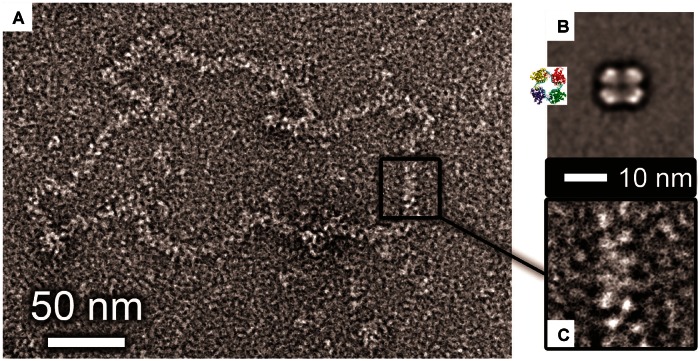
The SBV N protein exclusively formed tetramers within crystals, and the tetramer is also the predominant multimeric form of both RNA-bound and RNA-free SBV N in solution, as determined by gel filtration. Direct observation of soluble SBV N protein tetramers bound to synthetic 48mer RNA using EM followed by single-particle reconstruction (Figure 4B) reveals its diameter is 10 nm. In the crystal lattice, the SBV N tetramer exhibits 2-fold symmetry, with dimensions of 9.5 and 10.5 nm. This close correlation suggests that the tetramer conformation is indeed favoured in solution over alternative multimeric states, and that RNA binding may induce a conformational change converting the 2-fold symmetric apo tetramer into a 4-fold symmetric complex. An alternative possibility is that the crystal lattice has trapped one of many conformations of a flexible apo tetramer.
Figure 4.
EM of purified tetramers and native RNPs. (A) Micrograph of negative-stained RNP purified from BUNV virus particles in cell culture. (B) Single particle average of purified SBV tetramers bound to synthetic 48mer RNA (inset shows the crystal structure of the BUNV N tetramer bound to RNA). (C) Zoomed in view of a section of native RNP at the same scale as B (the 10-nm scale bar applies to both B and C).
Gel filtration and EM analysis also show the BUNV N protein in complex with RNA is a tetramer in solution, as it is in crystals, again providing strong evidence that the tetramer is a stable and preferred multimer.
To understand the biological relevance of the orthobunyavirus tetramer and to understand the quaternary arrangement of N in the authentic BUNV RNP, we compared the dimensions of both soluble and crystalline SBV N tetramers with that of BUNV RNPs harvested from purified virus. These BUNV RNPs are commonly found in a closed loop conformation, mediated by previously described interactions of the RNA termini (47–51). The BUNV RNPs seem somewhat flexible, being imperfect circles, and the RNP has a constant width along its length, which is ∼10 nm (Figure 4A). The 10-nm width of the BUNV RNP is inconsistent with it comprising single-BUNV N protein monomers individually bound to the RNA, which measure only 4 nm within the crystal lattice. Instead, the BUNV RNP width matches that of both the EM reconstructed and the crystalline tetramer. Therefore, we propose that the orthobunyavirus RNPs possess an RNP structure that is based on a repeating N protein multimer, and not a repeating monomer as has been recently reported for the related bunyavirus RVFV, a phlebovirus (15).
DISCUSSION
The mechanism by which the RNP of orthobunyaviruses is able to simultaneously organize and protect the viral RNA genome, and yet present the RNA bases for polymerization is poorly understood. To provide much needed insight into these seemingly conflicting but critical attributes, we solved the crystal structures of the N protein from two distantly related orthobunyaviruses. We have established that these proteins represent a new RNA-binding fold. The SBV structure is at higher resolution and thus provides the greater detail of specific residues involved in the protein–protein interactions, whereas the BUNV structure is at lower resolution but allows us to visualize the location of bound RNA.
Consequences of buried RNA
The crystal structure of the BUNV N–RNA complex shows the RNA to be buried within a narrow cleft, with the bases facing away from the solvent. This deep sequestration of RNA within the binding channel would prevent the RNA bases from interacting with other nucleotides via Watson–Crick base pairing interactions. Perhaps the most significant consequence of this finding is that the RNP must surely disassemble into its N and RNA components to expose the RNA for copying during both replication and transcription by the viral RNA-dependent RNA polymerase (RdRp). It is highly likely that this disassembly process is transient to maintain the high level of RNA protection that the RNP architecture affords. Presumably the high-energetic cost of stripping and replacing the N protein along the entire length of the RNP during copying is off set by the gains in RNA protection and in preventing the annealing of template and nascent strands.
This deep shielding of the RNA also raises interesting questions concerning other aspects of bunyavirus molecular biology, and one of the most fundamental of these is the mechanism that drives the closed circular morphology observed for many bunyaviral RNPs (47–49,51,52). The current model posits that circularization is driven by base paring between nucleotides at opposite ends of the template (48,50,53), which is clearly not compatible with the inaccessibility of the RNA bases in the RNP. One possible explanation to account for circularization is that the termini of the RNPs are partially unencapsidated, thus allowing the exposed bases to interact. This strategy would seem risky, given the functional sensitivity of terminal promoter sequences and the cellular response to dsRNA. However, BUNV is known to be able to repair partial deletions at its termini that could arise through nuclease attack (54), thus restoring wild-type sequence and functionality.
The inaccessibility of the RNA bases also presents problems for how a newly synthesized BUNV RdRp is able to recognize a suitable template strand in readiness for further RNA synthesis, a recognition process thought to be at least partially sequence specific. In the case of influenza virus, the viral polymerase is involved in a specific interaction with both RNP termini (55–57), which is driven by both sequence and structural elements (58–61). This interaction is compatible with the relative availability of the RNA within the influenza virus NP–RNA complex, as demonstrated by its sensitivity to chemical or enzymatic attack (61,62). This differs with what we have revealed for BUNV, in which the deep RNA shielding must surely prevent RdRp access to any promoter sequences in its encapsidated state. The mechanism by which a newly made RdRp identifies a BUNV RNA template must be either sequence independent or, as described earlier in the text, the RNA sequence required for RdRp binding must be exposed by disassembly. A ‘parked’ RdRp bound to the termini could also afford the same protection to viral RNA as encapsidation by N protein.
An additional problem for the bunyavirus RdRp in the context of the BUNV N–RNA structure we present here is in the ‘prime and realign’ mechanism of transcription initiation (63,64). This process, which seems common to all bunyaviruses, is thought to involve complementary base pairing between the 3′-end of the viral template and a donor cellular mRNA (prime) before any polymerization by the RdRp. Primer binding is followed by polymerization and an adjustment of primer and template position (align), as evidenced by the presence of template-specific reiterated sequences in the nascent mRNA. It is difficult to see how the initial primer-binding event can occur unless the RNA template is first released from the groove in N. Exposed nucleotides at the termini would provide convenient explanations for RNP circularization, template recognition and prime and realign transcription.
The architecture of the orthobunyavirus RNP
Our results show that the assembled BUNV RNP released from infectious virus is too wide to simply represent individual N protein monomers joined end-to-end. Instead the RNP has a width that corresponds to that of the assembled tetramer, or at least a similar size oligomer (see later in the text). This contrasts with the model that has recently been proposed for RVFV, in which the RNP seems to comprise individual monomers linked by a single flexible arm that results in a similarly flexible RNP chain (15). This conclusion is also supported by the gross physical characteristics of their corresponding RNPs, with those from RVFV appearing extremely narrow, flexible and kinked, whereas those from BUNV appearing wider and more rigid. RVFV N–RNA complexes crystallize as tetramers (width 9 nm), pentamers (9.8 nm) and hexamers (10.5 nm); thus, different oligomeric forms of RVFV N in crystals do not reflect the authentic RNP, and this may be true for BUNV N. Our results define that the quaternary structure of the BUNV RNP is different to that from RVFV, and they indicate that bunyaviruses as a group have adopted multiple approaches to achieve a functional RNP architecture.
The close correlation between the widths of the RNP and the tetramer presents the possibility that the RNP comprises repeating stacked tetramers. However, this raises several interesting questions regarding how this may be achieved. First, BUNV N tetramers possess an internal RNA-binding channel that follows a planar path and in stacked tetramers a short sequence of RNA would be required to leave this channel then link to the channel of an adjacent tetramer. This could result in gross differences in RNA nucleotide encapsidation states throughout the viral genome. An alternative possibility that better accommodates the repeating tetramer is that each could form a split ring forming a proto-helix, in which stacked tetramers would possess a similarly helical RNA channel and remove the need for potentially unprotected RNA linkage between adjacent tetramer units. However, this scenario would require a relatively large torsion to generate sufficient rise with only 4 N molecules per repeating turn.
Recently, the structure of the native influenza virus RNP has been elucidated using cryo-EM (65,66), which revealed the RNP comprises anti-parallel chains of repeating N proteins to form a rod-shaped double helix. A further possibility to explain the apparent width of the BUNV RNP is that it forms a similar double stranded assembly. The individual strands of such a double helix could comprise a chain of repeating monomers linked by the C-terminal arm, with the RNA still buried within the channel, but following a more exposed path around the external surface. The dimensions of this type of double-stranded assembly would correspond to the observed RNP width of two monomers, as we have described. However, such anti-parallel RNP chains would be expected to form rod-shaped structures rather than the circular RNPs we observe; therefore, this possibility is unlikely.
Although there are several models that could fit our data, we can conclude that the RNPs we observe by EM clearly do not correspond to the monomers on a string RVFV model. We cannot exclude the possibility that the BUNV RNP is able to form more than one conformation. Perhaps the wide chain architecture that we have observed represents a condensed RNP, which is also able to adopt a more relaxed conformation reminiscent of the slender RVFV RNPs. These different RNP conformations could perform distinct functions within the virus life cycle, such as RNA synthesis and packaging.
BUNV N protein mutants
The BUNV N–RNA structure allowed us to investigate the structural basis for the interesting phenotypes exhibited by N protein mutations ts19 and ts63, identified from viable but growth restricted ts viruses. Both these mutations impacted RNA synthesis but in different ways; the extended C-terminal arm of ts19 resulted in near wild-type levels of replication, but considerably reduced transcription, whereas the M150T change of mutant ts63 displayed high levels of transcription, but severely diminished replication. Mapping of these mutations onto the N–RNA structure reveals they lie within functionally significant regions, although the structural basis for these interesting phenotypes is not immediately clear. However, these results firmly establish that the N protein is able to significantly influence other aspects of RNP function outside of RNA encapsidation.
In addition, the BUNV N–RNA structure we present here allows interpretation of our previous work in which we analysed the role of conserved positively charged amino acids in RNA binding and BUNV-specific RNA synthesis (46). We previously showed that mutant R94A was unable to bind RNA, and it was poorly active in both RNA replication and mRNA transcription. This phenotype is consistent with the position of R94 at the rim of the RNA-binding channel, where it likely participates in electrostatic interactions with the RNA phosphate backbone, an assertion that is further supported by the reduced affinity of the R94A mutant for RNA, as described here. Additionally mutant K179A also displayed reduced levels of RNA synthesis, and at a qualitative level, it exhibited moderately reduced RNA binding compared with wild-type N. Residue K179 is located within the wall of the binding groove, and this position is consistent with this both its phenotype, and our quantitative analysis of RNA binding reported here. Significantly, mutations at both these positions were previously shown to be lethal to recovery of infectious virus (45), showing the importance of these residues to RNP function. Several other N protein mutants were reported to be lethal for virus rescue, and a selection of these are also known to be defective in replicon-based RNA synthesis. Many of these mutations map to sites within and surrounding the RNA-binding groove, and one to the C-terminal helix mediating oligomerization, strongly implicating these residues as critical for RNA-binding activities. The provision of these high-resolution structures paves the way for a comprehensive structure: function analysis, to decipher the role of all functionally important N protein residues.
In conclusion, the N protein structures we present here offer important insight into the mechanism of orthobunyavirus RNA recognition, N–N multimerization and assembly of the RNP complex across the entire genus, with potential implications for all segmented negative-stranded RNA viruses. They will provide a critical resource for further analysis of many diverse aspects of the orthobunyavirus life cycle, as well as providing a possible target for structure-based drug design to interfere with critical RNP functions.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the protein data bank (www.pdb.org) under PDB ID codes 3ZLA (BUNV N protein) and 3ZL9 (SBV N protein).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7 and Supplementary References [35–38].
ACKNOWLEDGEMENTS
The authors thank the beam line scientists at the Diamond light source, as well as Matthew Groves at the PETRA synchrotron, Hamburg, Germany. Additional thanks are given to Dr Arwen Pearson and Dr Chi Trinh for support with data collection and comments on this manuscript. They also thank Dr Falko Steinbach, Dr Ashley Banyard and Dr Nicholas Johnson (AHVLA, UK) for providing the SBV N-gene construct.
FUNDING
Wellcome Trust through a jointly held project grant [WT091783MA to J.N.B. and T.A.E.], a project grant [WT084332MA to J.N.B.] and a studentship [086774/Z/08/Z to K.C.D.]; BBSRC and The Health Protection Agency through a studentship (to S.J.T.); EPSRC through a White Rose studentship (to D.S.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist A, Schmaljohn CS, Tesh RB. Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: classification and nomenclature of viruses: Ninth report of the International Committee on Taxonomy of Viruses. 2012. pp. 725–741. [Google Scholar]
- 2.Parsonson IM, Della-Porta AJ, Snowdon WA. Congenital abnormalities in newborn lambs after infection of pregnant sheep with Akabane virus. Infect. Immun. 1977;15:254–262. doi: 10.1128/iai.15.1.254-262.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappu HR, Jones RA, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009;141:219–236. doi: 10.1016/j.virusres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Thompson WH, Kalfayan B, Anslow RO. Isolation of California Encephalitis Group Virus from a Fatal Human Illness. Am. J. Epidemiol. 1965;81:245–253. doi: 10.1093/oxfordjournals.aje.a120512. [DOI] [PubMed] [Google Scholar]
- 5.Bardos V, Medek M, Kania V, Hubalek Z. Isolation of Tahyna virus from the blood of sick children. Acta Virol. 1975;19:447. [PubMed] [Google Scholar]
- 6.Sexton DJ, Rollin PE, Breitschwerdt EB, Corey GR, Myers SA, Dumais MR, Bowen MD, Goldsmith CS, Zaki SR, Nichol ST, et al. Life-threatening Cache Valley virus infection. N. Engl. J. Med. 1997;336:547–549. doi: 10.1056/NEJM199702203360804. [DOI] [PubMed] [Google Scholar]
- 7.Gerrard SR, Li L, Barrett AD, Nichol ST. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 2004;78:8922–8926. doi: 10.1128/JVI.78.16.8922-8926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann B, Scheuch M, Hoper D, Jungblut R, Holsteg M, Schirrmeier H, Eschbaumer M, Goller KV, Wernike K, Fischer M, et al. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg. Infect. Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goller KV, Hoper D, Schirrmeier H, Mettenleiter TC, Beer M. Schmallenberg virus as possible ancestor of Shamonda virus. Emerg. Infect. Dis. 2012;18:1644–1646. doi: 10.3201/eid1810.120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overby AK, Pettersson RF, Neve EP. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J. Virol. 2007;81:3198–3205. doi: 10.1128/JVI.02655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribeiro D, Borst JW, Goldbach R, Kormelink R. Tomato spotted wilt virus nucleocapsid protein interacts with both viral glycoproteins Gn and Gc in planta. Virology. 2009;383:121–130. doi: 10.1016/j.virol.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Kohl A, Li P, Elliott RM. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J. Virol. 2007;81:10151–10160. doi: 10.1128/JVI.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl Acad. Sci. USA. 2010;107:11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferron F, Li Z, Danek EI, Luo D, Wong Y, Coutard B, Lantez V, Charrel R, Canard B, Walz T, et al. The hexamer structure of the rift valley Fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7:e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc. Natl Acad. Sci. USA. 2012;109:19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter SD, Surtees R, Walter CT, Ariza A, Bergeron E, Nichol ST, Hiscox JA, Edwards TA, Barr JN. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J. Virol. 2012;86:10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 2011;92:2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, et al. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc. Natl Acad. Sci. USA. 2012;109:5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Dutta S, Karlberg H, Devignot S, Weber F, Hao Q, Tan YJ, Mirazimi A, Kotaka M. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. J. Virol. 2012;86:12294–12303. doi: 10.1128/JVI.01627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers JW, Zhou Q, Green TJ, Barr JN, Luo M. Purification, crystallization and preliminary X-ray crystallographic analysis of the nucleocapsid protein of Bunyamwera virus. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 2006;62:361–364. doi: 10.1107/S1744309106006397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins HT, Malkova B, Edwards TA. Kinked beta-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc. Natl Acad. Sci. USA. 2011;108:18266–18271. doi: 10.1073/pnas.1105211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novoa RR, Calderita G, Cabezas P, Elliott RM, Risco C. Key Golgi factors for structural and functional maturation of bunyamwera virus. J. Virol. 2005;79:10852–10863. doi: 10.1128/JVI.79.17.10852-10863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arstila P. Characteristics of vesicular stomatitis virus envelopes released with saponin. J. Gen. Virol. 1974;24:319–326. doi: 10.1099/0022-1317-24-2-319. [DOI] [PubMed] [Google Scholar]
- 33.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 34.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 35.Barr JN, Elliott RM, Dunn EF, Wertz GW. Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology. 2003;311:326–338. doi: 10.1016/s0042-6822(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 36.Dunn EF, Pritlove DC, Jin H, Elliott RM. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology. 1995;211:133–143. doi: 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- 37.Iroegbu CU, Pringle CR. Genetic interactions among viruses of the Bunyamwera complex. J. Virol. 1981;37:383–394. doi: 10.1128/jvi.37.1.383-394.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Bond CS, Schuttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 42.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Mohl BP, Barr JN. Investigating the specificity and stoichiometry of RNA binding by the nucleocapsid protein of Bunyamwera virus. RNA. 2009;15:391–399. doi: 10.1261/rna.1367209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eifan SA, Elliott RM. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J. Virol. 2009;83:11307–11317. doi: 10.1128/JVI.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter CT, Bento DF, Alonso AG, Barr JN. Amino acid changes within the Bunyamwera virus nucleocapsid protein differentially affect the mRNA transcription and RNA replication activities of assembled ribonucleoprotein templates. J. Gen. Virol. 2011;92:80–84. doi: 10.1099/vir.0.024240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hewlett MJ, Pettersson RF, Baltimore D. Circular forms of Uukuniemi virion RNA: an electron microscopic study. J. Virol. 1977;21:1085–1093. doi: 10.1128/jvi.21.3.1085-1093.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardigon N, Vialat P, Girard M, Bouloy M. Panhandles and hairpin structures at the termini of germiston virus RNAs (Bunyavirus) Virology. 1982;122:191–197. doi: 10.1016/0042-6822(82)90388-9. [DOI] [PubMed] [Google Scholar]
- 49.Pettersson RF, von Bonsdorff CH. Ribonucleoproteins of Uukuniemi virus are circular. J. Virol. 1975;15:386–392. doi: 10.1128/jvi.15.2.386-392.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J. Virol. 1989;63:122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samso A, Bouloy M, Hannoun C. Circular ribonucleoproteins in the virus Lumbo (Bunyavirus) C. R. Acad. Sci. 1975;280:779–782. [PubMed] [Google Scholar]
- 52.Obijeski JF, Bishop DH, Palmer EL, Murphy FA. Segmented genome and nucleocapsid of La Crosse virus. J. Virol. 1976;20:664–675. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barr JN, Wertz GW. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3'- and 5'-terminal sequences. J. Virol. 2004;78:1129–1138. doi: 10.1128/JVI.78.3.1129-1138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter CT, Barr JN. Bunyamwera virus can repair both insertions and deletions during RNA replication. RNA. 2010;16:1138–1145. doi: 10.1261/rna.1962010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fodor E, Seong BL, Brownlee GG. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. Gen. Virol. 1993;74(Pt. 7):1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 56.Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin-Benito J, Area E, Ortega J, Llorca O, Valpuesta JM, Carrascosa JL, Ortin J. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheong HK, Cheong C, Choi BS. Secondary structure of the panhandle RNA of influenza virus A studied by NMR spectroscopy. Nucleic Acids Res. 1996;24:4197–4201. doi: 10.1093/nar/24.21.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew' conformation. J. Gen. Virol. 1999;80(Pt. 10):2565–2572. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- 60.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad. Sci. USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baudin F, Bach C, Cusack S, Ruigrok RW. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. The 5' ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin H, Elliott RM. Non-viral sequences at the 5' ends of Dugbe nairovirus S mRNAs. J. Gen. Virol. 1993;74:2293–2297. doi: 10.1099/0022-1317-74-10-2293. [DOI] [PubMed] [Google Scholar]
- 65.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. The structure of native influenza virion ribonucleoproteins. Science. 2012;338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 66.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.