Abstract
5-Aza-2′-deoxycytidine (5-azadC) is a DNA methyltransferase (DNMT) inhibitor increasingly used in treatments of hematological diseases and works by being incorporated into DNA and trapping DNMT. It is unclear what DNA lesions are caused by 5-azadC and if such are substrates for DNA repair. Here, we identify that 5-azadC induces DNA damage as measured by γ-H2AX and 53BP1 foci. Furthermore, 5-azadC induces radial chromosomes and chromatid breaks that depend on active replication, which altogether suggest that trapped DNMT collapses oncoming replication forks into double-strand breaks. We demonstrate that RAD51-mediated homologous recombination (HR) is activated to repair 5-azadC collapsed replication forks. Fanconi anemia (FA) is a rare autosomal recessive disorder, and deaths are often associated with leukemia. Here, we show that FANCG-deficient cells fail to trigger HR-mediated repair of 5-azadC-induced lesions, leading to accumulation of chromatid breaks and inter-chromosomal radial fusions as well as hypersensitivity to the cytotoxic effects of 5-azadC. These data demonstrate that the FA pathway is important to protect from 5-azadC-induced toxicity. Altogether, our data demonstrate that cytotoxicity of the epigenetic drug 5-azadC can, at least in part, be explained by collapsed replication forks requiring FA-mediated HR for repair.
INTRODUCTION
DNA methylation is controlled by a family of DNA methyltransferases (DNMT), enzymes that catalyze the transfer of a methyl moiety from S-adenosyl-l-methionine to the 5-position of cytosines in the CpG dinucleotide to silence gene expression (1). When entering the cell, 5-aza-2′-deoxycytidine (5-azadC) is phosphorylated to 5-azadCTP and incorporated into DNA during replication. As a result of the chemistry of the methyltrasferase reaction, the DNMT becomes covalently linked to DNA, in effect creating a protein–DNA cross-link (2–5). This results in depletion of soluble DNMT protein levels, which induces replication-dependent global demethylation and gene reactivation (6). Although there is a considerable amount of literature regarding the possible anti-cancer mode of action of 5-azadC, the molecular mechanism of cell death still remains unclear. There are two non-exclusive models: one involves the reactivation of silenced genes involved in cell growth, which is accompanied by cell cycle arrest and/or apoptosis (7). Another model emphasizes the importance of the formation of covalent DNMT–DNA adducts, which leads to DNA damage and cytotoxicity (3,8).
Epigenetic targets such as DNMT has become a promising new strategy for cancer treatments, especially hematologic malignancies such as myelodysplastic syndromes and acute myeloid leukemia (AML) (9,10). Many anti-cancer therapies are cytotoxicity based on their ability to cause DNA damage, e.g. ionizing radiation or chemotherapy drugs. The 5-azadC is a nucleoside analogue that when is incorporated into newly synthesized DNA, results in covalent trapping of DNMTs causing genome-wide protein–DNA cross links (2–5).
Fanconi anemia (FA) is a rare autosomal recessive disorder characterized by progressive pancytopenia, requiring bone marrow transplant, and associated congenital abnormalities, such as microcephaly, short stature, skeletal, cardiac and renal malformations. Mortality is associated with bone marrow failure and leukemia in the first 2 decades of life, with the majority of deaths occurring within 5 years after the onset of anemia (11,12). The disease is caused by mutations within a variety of genes associated with the FA pathway. Many genes have been linked in the pathogenesis of FA, and at least 15 genes, including a nine-gene FA nuclear core complex with associated downstream proteins, all function to promote genetic stability via repair of DNA following damage (13). One of these genes is the well-known breast cancer susceptibility gene BRCA2, while mutations in others such as PALB2, RAD51C or BRIP1 also pre-dispose to cancer. Germline mutations of these genes are associated with dysregulation of cell cycle and apoptosis (14,15), and chromosomal instability (14). It has been postulated that disruption of the FA complex and associated pathways are related with chromosomal and telomere instability characterized by impaired DNA double-strand break (DSB) repair leading to the inactivation of tumor suppressor genes and activation of oncogenes (16). The FA pathway in normal cells is not constitutively active but is inactivated by genetic or epigenetic mechanisms in a variety of cancers, including breast, ovarian, head and neck, cervical, pancreatic and lung carcinomas, rendering the affected tumors potentially hypersensitive to DNA cross-linking agents (17).
Here, we report that 5-azadC induces DNA damage that depends on active replication, which suggests that trapped DNMT collapses with oncoming replication forks into DSBs. We demonstrate that RAD51-mediated homologous recombination (HR) is involved in the repair of such DSBs, a process that is defective in FA cells. This agrees with a reduced cell survival and a potentiation of chromosomal abnormalities. To strengthen this, we also show that proteasome inhibition sensitizes against 5-azadC in wild-type, but not in FA cells. Furthermore, we report a preferential protection in FA cells when non-homologous end joining (NHEJ) inhibitors were used.
MATERIALS AND METHODS
Chemicals
The 5-azadC, aphidicolin (APH), NU7026 and MG132 were purchased from Sigma and diluted in dimethyl sulfoxide except 5-azadC that was diluted in phosphate buffered saline (PBS) (10 mM sodium phosphate (pH 7.4), 140 mM NaCl and 3 mM KCl). All of them were aliquoted and stored at −80°C before use.
Cell culture
The parental Chinese hamster ovary cell line AA8 was purchased from the American Type Culture Collection (ATCC), USA. The isogenic FANCG-deficient Chinese hamster ovary mutant KO40, derived from AA8, was kindly provided by Dr. Fabrizio Palitti (Department of Agrobiology and Agrochemistry, University of Tucsia, Viterbo, Italy) and has been previously described (18). Cells were routinely maintained as monolayers in McCoy’s 5A media (LONZA) supplemented with 10% fetal bovine serum, 2 mM L-glutamine and the antibiotics penicillin (50 U/ml) and streptomycin (50 µg/ml). Cells were cultured at 37°C in an atmosphere containing 5% CO2.
Colony formation
Cell survival following 5-azadC treatment was measured by clonogenic assay. AA8 and KO40 cells were plated at low density onto 10 cm Petri dishes. After 4 h, cultures were incubated for 24 h (two rounds of DNA replication approximately) in the presence of different concentrations of 5-azadC (1, 3.25, 7.5 and 15 µM). For combined experiments (5-azadC + inhibitor), cells were treated for 24 h with the NHEJ inhibitor NU7026 (10 µM) or the proteasome inhibitor MG132 (0.1 µM), respectively. These doses were chosen on the basis of its absence of cellular cytotoxicity. Then 5-azadC was discarded, and the cultures were further incubated for 24 h with each one of the inhibitors. After that, fresh media was added, and cells were allowed to grow from 7 to 10 days. Colonies were stained with methylene blue prepared in methanol (4 g/l). Surviving colonies made up of >50 cells per colony were counted. The data have been corrected according to cloning efficiencies of control cells.
Chromosome analysis
Exponential growing AA8 or KO40 cells were cultured for 24 h in the presence of different concentrations of 5-azadC (3.25, 7.5 and 15 µM). After that, cells were incubated with fresh media for 12 h before mitotic arrest. For combined experiments (5-azadC + APH), cells were treated for 12 h with 5-azadC. After that, cells were thoroughly washed and 0.5 µM of APH was added for a further 12 h. Then, APH was discarded, and cultures were allowed to recover in fresh media for 6 h until the addition of colcemid to arrest cells in metaphase. To prepare metaphase spreads, cells were treated with 2 × 10−7 M of colcemid for 2 h and 30 min. Cells were collected and incubated in hypotonic solution (0.075 M KCl) for 2 min, fixed in methanol: acetic acid (3:1) and dropped onto microscope slides. Slides were stained with 3% Giemsa/Sörensen’s buffer for 5 min and mounted in D.P.X. (Sigma). Two hundred complete metaphases were evaluated in each experimental point. Images were taken with a Nikon eclipse 50i microscope equipped with a Nikon DS-Fi1 camera and using the 100-fold magnification objective (numerical aperture 1.25) and the NIS Elements 3.0 adquisition software (Nikon).
To visualize sister chromatid exchanges (SCE), cells were incubated with 10 µM of bromodeoxyuridine (BrdU, Sigma) along with increasing doses of 5-azadC (0.25, 0.5 and 1 µM) for 24 h. Then, cells were washed and allowed to repair for 12 h before addition of colcemid. Differential staining of BrdU-substituted sister chromatids was obtained by the fluorescence-plus-Giemsa technique as reported elsewhere (19).
Immunofluorescence labeling and microscopy (Foci detection)
Cells were seeded on coverslips the day before being treated with 5-azadC (0.25, 0.5 and 1 μM) for 24 h. After that, cells were washed with PBS and incubated for 30 s with cold 0.1% Triton X-100 in PBS to pre-extract soluble protein unlocated in foci. Afterwards, cells were fixed with 4% formaldehyde in PBS for 10 min at room temperature. The primary antibodies used were a rabbit polyclonal antibody α-RAD51 (H-92, Santa Cruz) or α-53BP1 (H-300, Santa Cruz) and a mouse monoclonal α-γH2AX (Upstate). Secondary fluorescent antibodies were an Alexa fluor 488-conjugated goat anti-mouse and an Alexa fluor 555-conjugated goat anti-rabbit (Invitrogen). DNA was stained with 100 nM of 4,6 diamidino-2-phenylindole for 15 min. Slides were mounted in anti-fade media (Vectashield). Immunofluorescence was observed using a Nikon eclipse 50i microscope with a 40-fold magnification objective. Cells with ≥10 foci were scored as positive. At least 200 nuclei were analyzed for each treatment. Micrographs were taken using a Leica AF6000 microscope equipped with a Leica DFC 350 FX camera and using the Application Suite Advanced Fluorescence adquisition software (Leica) and a 63-fold magnification objective (numerical aperture 1.4). Images were bright-contrast processed using Adobe Photoshop 5.0 software (Adobe).
Statistical analysis
For the determination of significance of the difference between the means, Student’s t-test was used. Statistical treatment and plotting of the results were performed using the Sigma Plot and MS Excel for Windows XP software. The results come from at least two independent experiments and are presented as mean ± standard deviation (SD) of the mean. Differences were considered significant when *P < 0.05 or **P < 0.01.
RESULTS
5-azadC causes replication-dependent strand breaks resulting in chromatid breaks and radial fusion chromosomes
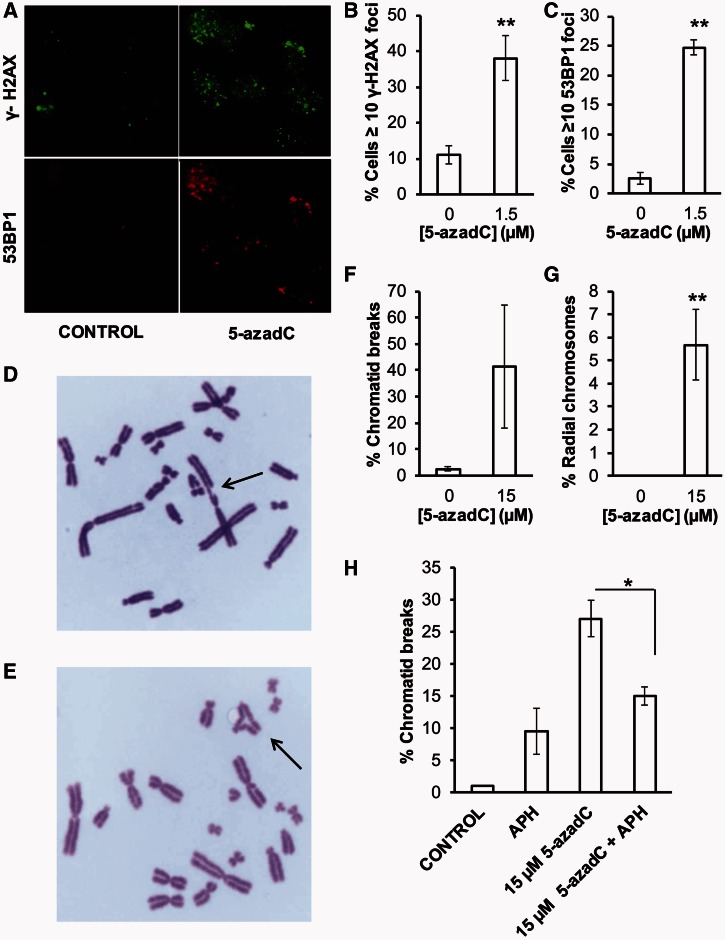
It has been previously shown that cytoxicity of 5-azadC to mammalian cells can be mediated through covalent DNMT-DNA adducts, which in turn cause DNA damage that activates ATR signaling (3,20). Here, we find that 5-azadC treatment produces γ-H2AX foci (Figure 1A and B), which has also been reported earlier (3). It is established that γ-H2AX foci can form also in the absence of DSBs (21), whereas 53BP1 foci formation are more strictly associated with DSBs. Here, we find that 5-azadC also induces 53BP1 foci (Figure 1A and C), suggesting that DSBs may be formed after 5-azadC treatments.
Figure 1.
DNA damage induced by 5-azadC. (A) DNA damage response induced by 5-azadC. AA8 cells were grown on coverslips, treated with 5-azadC for 24 h (1.5 µM) and fixed for analysis of nuclear γ-H2AX or 53BP1 foci by inmunofluorescence. Original magnification 630X. Quantification of γ-H2AX (B) or 53BP1 (C) foci was evaluated in 200 nuclei for each treatment. Cells with ≥10 foci were scored as positive. (D and E) Chromosomal abnormalities induced by 5-azadC. Exponential growing AA8 cells were cultured for 24 h in the presence of 5-azadC (15 µM), washed and allowed to recover for 12 h before mitotic arrest. Two hundred metaphases were analyzed for chromosomal abnormalities in each experimental point. Representative micrographs of AA8 metaphases treated with 5-azadC (7.5 µM). Arrows point to a chromatid break (D) and a radial fusion chromosome (E). Original magnification 1000X. Their respective quantifications are plotted on (F and G). (H) Influence of APH on the induction of chromatid breaks by 5-azadC. AA8 cells were treated for 12 h with 5-azadC (15 µM), washed and allowed to repair in free media or in media containing APH (0.5 µM) for 12 h as described in ‘Materials and Methods’ section. Each bar represents the mean and the SD from three independent experiments. Differences were statistically significant (*P < 0.05, **P < 0.01 according Student’s t-test).
DSBs are highly toxic and may cause chromatid breaks if they persist into mitosis or radial-fusion chromosomes if chromatid breaks are aberrantly repaired. Figure 1D–G shows that control metaphases showed only low percentages of abnormalities, whereas 5-azadC treatment induced an increase in chromosomal aberrations, including chromatid breaks and radial chromosomes, which may explain toxic effects caused by 5-azadC.
Furthermore, we wanted to determine how 5-azadC may cause DSBs and chromatid breaks. A lack of induction of isochromatid breaks (data not shown) seems to indicate that 5-azadC behave as a typical S-phase dependent agent. In hypothesis, trapped DNMT may obstruct replication forks causing replication collapse and DSBs. It is well documented that replication-blocking DNA lesions can cause replication fork collapse and thereby lead to the formation of DNA DSBs (22). To test this, we determined chromosome damage by 5-azadC while transiently inhibiting DNA synthesis using APH, using a dose previously shown to completely inhibit replication progression in the same cells (23), to test whether the chromatid breaks were a result of collapsed replication forks. We found a decreased amount of chromatid breaks in agreement with this hypothesis (Figure 1H). Overall, our findings are in line with a hypothesis that 5-azadC-induced DSBs arising as a consequence of trapped DNMT collapses oncoming replication forks into DSBs.
Repair of 5-azaC induced DSBs requires HR
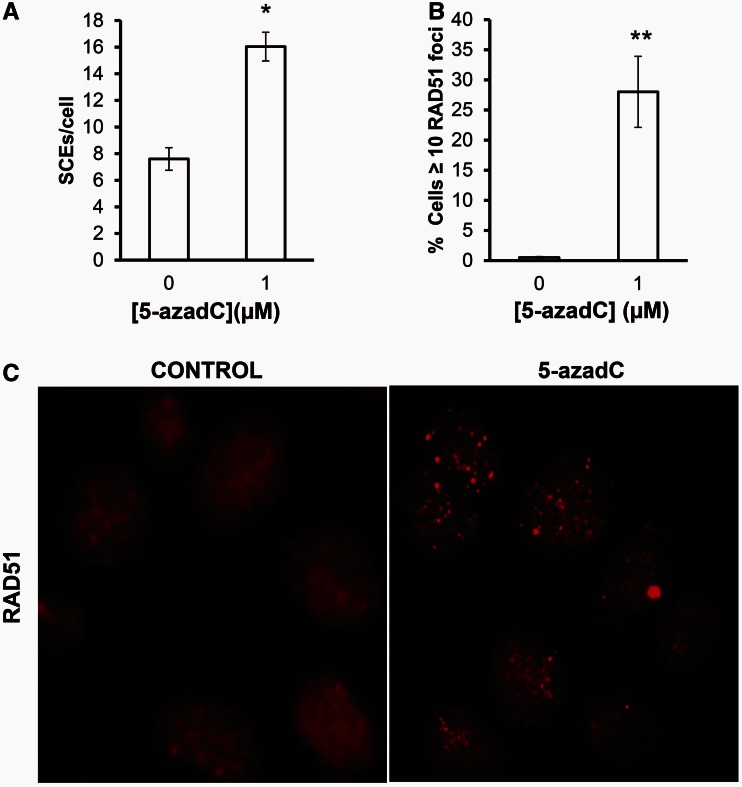
Our data suggest that DSBs arising as a consequence of 5-azadC incorporation are generated during the second round of DNA replication. It is well accepted that replication-induced DSBs are potent substrates for HR giving rise to RAD51-mediated SCEs formation (24–26). To test this directly, we treated cells with increasing doses of 5-azadC and scored SCEs or RAD51 foci formation. We found that the frequency of SCEs is increased after treatment with 5-azadC (Figure 2A). Furthermore, we also find an increase in RAD51 foci formation (Figure 2B and C), which altogether support a model where replication forks collapsed by DNMT–DNA adducts trigger HR for repair. Furthermore, cells deficient in BRCA2, a protein that interacts with RAD51 and promotes its loading to single-stranded DNA, are hypersensitive to the toxic lesions induced by 5-azadC (Supplementary Figure S1).
Figure 2.
The 5-azadC induces lesions that are repaired by HR. (A) SCEs in AA8 cells after a 24 h treatment with 5-azadC (1 µM). Cells were incubated with 10 µM of BrdU in the 5-azadC for two cell cycles, washed and allowed to repair for 12 h before metaphase arrest. Differential staining of BrdU-substituted sister chromatids was obtained by fluorescence-plus-Giemsa technique. A number of 50 complete metaphases with well-preserved chromosome morphology were scored from two independent experiments. (B) The 5-azadC induced RAD51 foci in AA8 cells. Cells growing on coverslips were treated with 5-azadC (1 µM) for 24 h and fixed for analysis of nuclear RAD51 foci by inmunofluorescence. Foci were evaluated in 200 nuclei per treatment. Cells with ≥10 foci were scored as positive. Each bar represents the mean and the SD from three independent experiments. Differences were statistically significant (*P < 0.05, **P < 0.01 according Student’s t-test). (C) Representative micrographs of either control and 5-azadC-treated AA8 cells showing RAD51 foci.
FANCG-mediated HR is required for 5-azadC survival
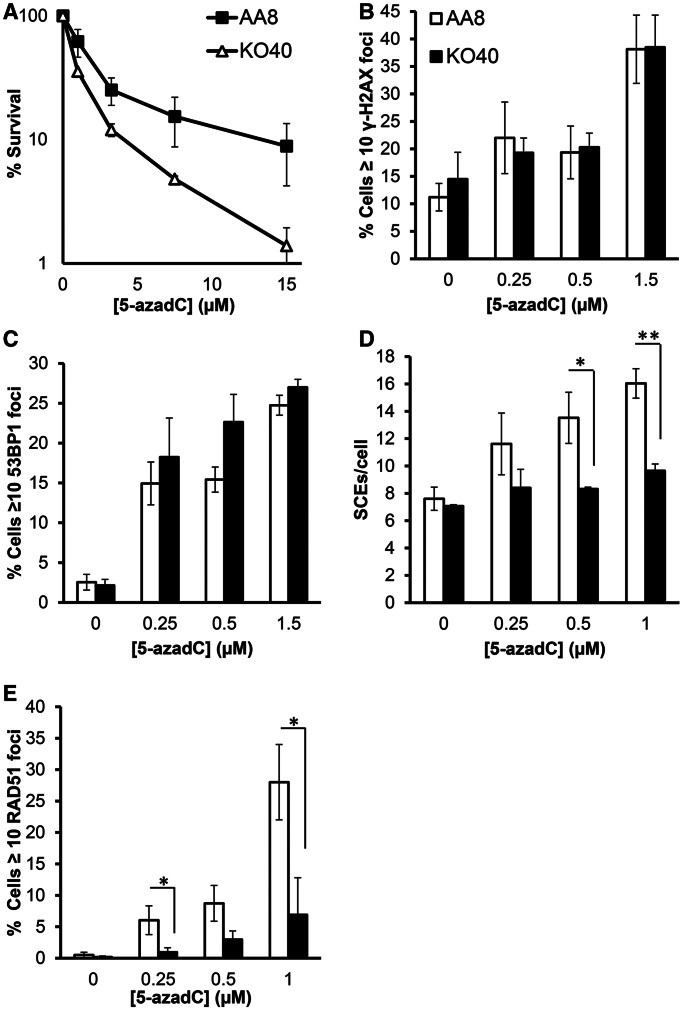
The FA proteins are known to play a critical role in repair at obstructed replication forks, especially of crosslinks (27). Here, we tested 5-azadC sensitivity in the absence the FA core complex using the FANCG mutant KO40 cell line (18). Results show that KO40 cells were more sensitive to 5-azadC treatment, with a significant decrease in cell survival to all doses tested compared with its isogenic and parental cell line AA8. The sensitization ranged from 2 to 10 times for the doses of 3.25 to 15 µM, respectively (Figure 3A). These results demonstrate that FANCG-deficient cells are sensitive to 5-azadC-mediated cytotoxicity. Furthermore, using human fibroblasts deficient in FANCD2, a central key protein of the FA pathway, we found that they were also sensitive to 5-azadC as compared with corrected cells (Supplementary Figure S2).
Figure 3.
Cells deficient in the FA pathway are hypersensitive to 5-azadC. (A) Cell survival after a 24 h treatment with increasing concentrations of 5-azadC. AA8 and KO40 cells were seeded on Petri dishes and allowed to attach for 4 h; subsequently, they were treated with increasing concentrations of 5-azadC. Then media was changed and cells were allowed to form colonies. (B and C) Quantification of γ-H2AX and 53BP1 foci in AA8 and KO40 cells after 24 h treatment with increasing concentrations of 5-azadC. (D) Differential induction of SCEs by 5-azadC in AA8 and KO40 cells. Cells were treated with 10 µM BrdU together with increasing concentrations of 5-azadC (0.25–1 µM) as described in ‘Materials and Methods’ section. (E) RAD51 foci formation in AA8 and KO40 cells after 24 h of 5-azadC treatment. Data show the mean and the SD from 2–3 independent experiments. Differences were statistically significant (*P < 0.05, **P < 0.01 according Student’s t-test).
To test the molecular reason for the increased toxicity of FANCG-deficient cells, we determined the amount of DNA lesions induced by 5-azadC in KO40 and AA8 cells. Both AA8 and KO40 cells showed the same amount of increase of γ-H2AX and 53BP1 foci on 5-azadC treatment (Figure 3B and C), suggesting that the lesions are produced irrespectively of FA status.
Next, we wanted to asses whether the differences observed in survival could be the result of the HR repair defects in FANCG-deficient cells. Here, we found clear differences between AA8 and KO40 in the yield of SCEs induced by 5-azadC, where only the WT AA8 cells are able to induce SCE and the KO40 cells are defective in 5-azadC-induced HR (Figure 3D). The same results were obtained when we evaluate the residual foci formation of RAD51 as a marker of HR after 5-azadC treatment (Figure 3E). Overall, these data revealed that FA pathway promotes the repair of DNA lesions induced by 5-azadC by a process mediated by HR.
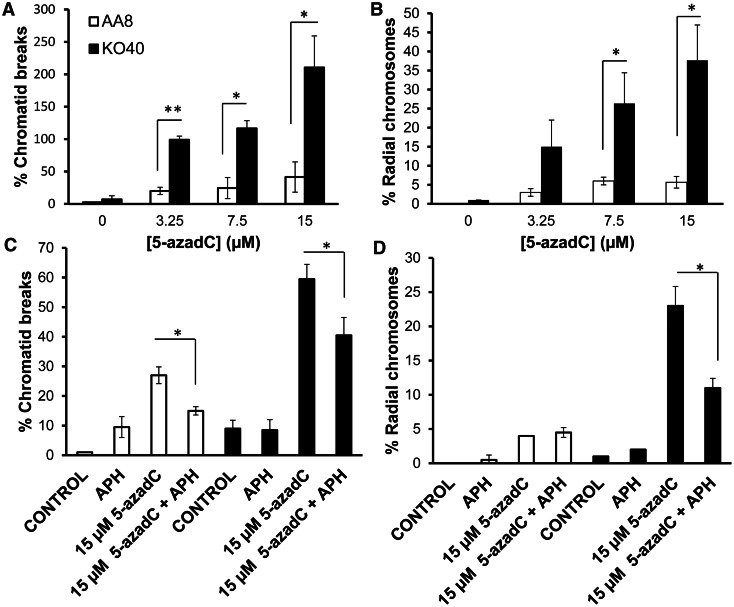
Next, we tested chromosome analysis on AA8 and KO40 cells cultured in the presence of 5-azadC. Control metaphases showed only low percentages of abnormalities (Figure 4A and B). Concerning 5-azadC treatment, there was a dose-dependent increase in chromosomal aberrations, including chromatid breaks and radial chromosomes for both AA8 and KO40 cells. However, only FANCG-deficient cells showed a dramatic increase in chromosome aberrations as compared with the parental cells AA8. Metaphases of KO40 treated with the highest dose of 5-azadC showed severe aberrations (Figure 4A and B). We also wanted to determine whether the increase in chromatid breaks and radial chromosomes were of the same origin, as the one occurring in wild-type cells, that is, derived following replication. To test this, 5-azadC-treated cells were washed and treated with APH, which prevents the subsequent collision of replication forks with DNMT–DNA adducts. We found that the 5-azadC-induced chromatid breaks and radial fusion chromosomes were reversed by preventing replication forks collide into the DNMT-trapped complex on DNA (Figure 4C and D). Altogether, these data suggest that the FA pathway is involved in HR repair of replication-associated DSBs formed after collapse of replication forks that are obstructed by DNMT–DNA adducts. Failed repair leads to chromatid breaks and formation of radial chromosomes.
Figure 4.
Deffects in the FA pathway potentiate the level of chromosome abnormalities induced by 5-azadC. (A and B) Quantification of chromatid breaks and radial fusion chromosomes in AA8 and KO40 cells. Cells were treated for 24 h with increasing concentrations of 5-azadC, washed and allowed to repair for 12 h before mitotic arrest. To test whether the induction of chromosome aberrations has the same origin in both cell lines, we analyzed the effect of replication inhibition by APH (0.5 µM) on the levels of chromatid breaks (C) and radial fusion chromosomes (D). Exponential AA8 and KO40 cells were treated with 5-azadC for 12 h, washed and allowed to recover for 12 h in the presence of 0.5 µM of APH. Replication inhibition contributes to a reduction in chromosomal aberrations in both cell lines. Data show the mean and the SD from three independent experiments. Differences were statistically significant (*P < 0.05, **P < 0.01 according Student’s t-test).
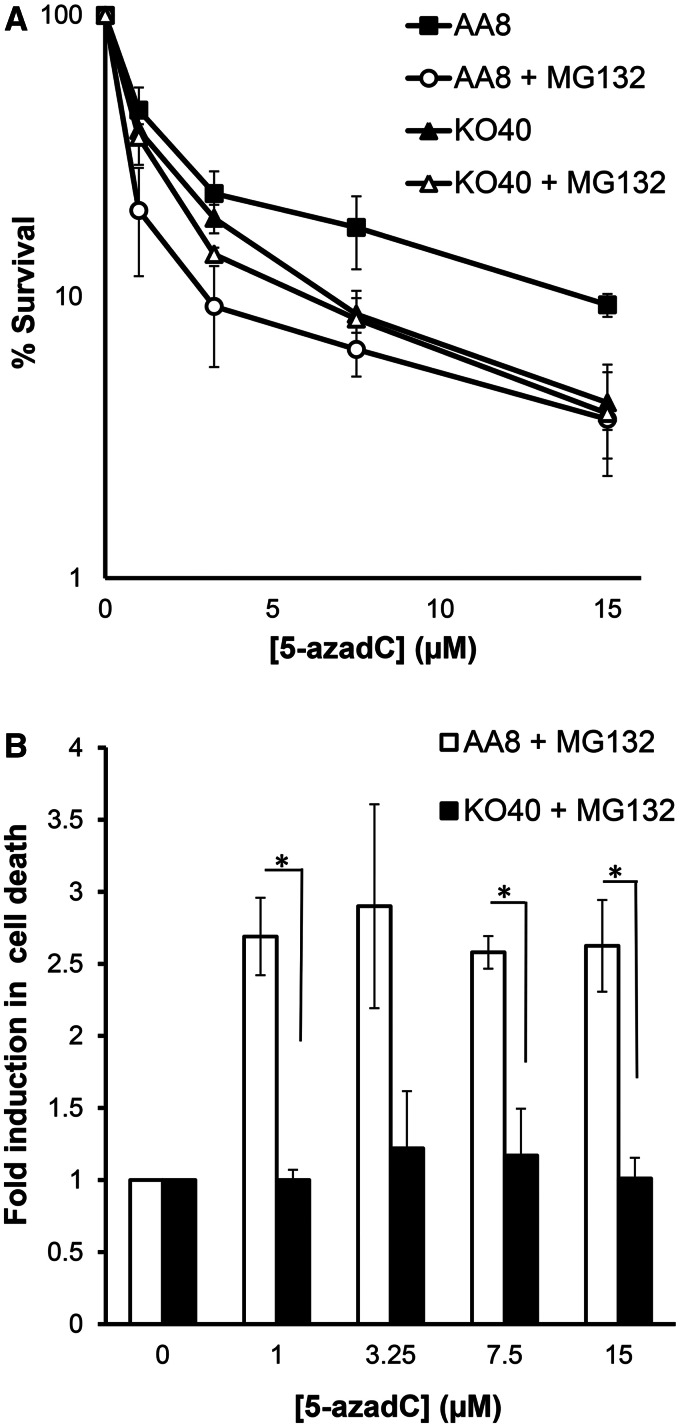
Proteasome inhibitor MG132 sensitizes wild-type, but not FA defective cells to 5-azadC
Previous reports have shown that proteasome function is required for activation of the FA pathway (28), as well as to catalyze HR (29). To test whether proteasome inhibition affects 5-azadC induced cell death, combined experiments were performed where AA8 and KO40 cells were co-treated with the proteasome inhibitor MG132 at non-toxic dose (Supplementary Figure S3). MG132 sensitized AA8 cells treated with 5-azadC to a similar level as observed in FA cells (Figure 5A). The sensitization effect was 2.5-fold across all doses used (Figure 5B). Interestingly, no evidence of sensitization was observed for KO40 cells, and hence an epistatic effect is observed between FANCG and the proteasome inhibitor MG132. This finding demonstrates that proteasome is required to promote cell survival after 5-azadC treatment. Also, the data point to that, directly or undirectly, proteasome and FA pathway work in the same pathway to promote survival. Overall, these data also strengthen the overall finding that FA-mediated HR is required for survival after 5-azadC treatment.
Figure 5.
Proteasome and FA pathway work in the same route to promote cell survival in 5-azadC-treated cells. AA8 and KO40 cells were cotreated with 5-azadC and the proteasome inhibitor MG132 (0.1 µM) according to ‘Materials and Methods’ section. Then cultures were allowed to grow (7–10 days) for analysis of colony-forming efficiency (A). Data show that proteasome catalytic activity is necessary for promoting cell survival of those cells treated with 5-azadC; however, no evidence of sensitization was observed for KO40 cells. Data were plotted as fold increase in cell death (B). Each bar represents the mean and the SD from two independent experiments. Differences were statistically significant (*P < 0.05, according Student’s t-test).
DNA-protein kinase inhibition protects from the deleterious effect induced by 5-azadC
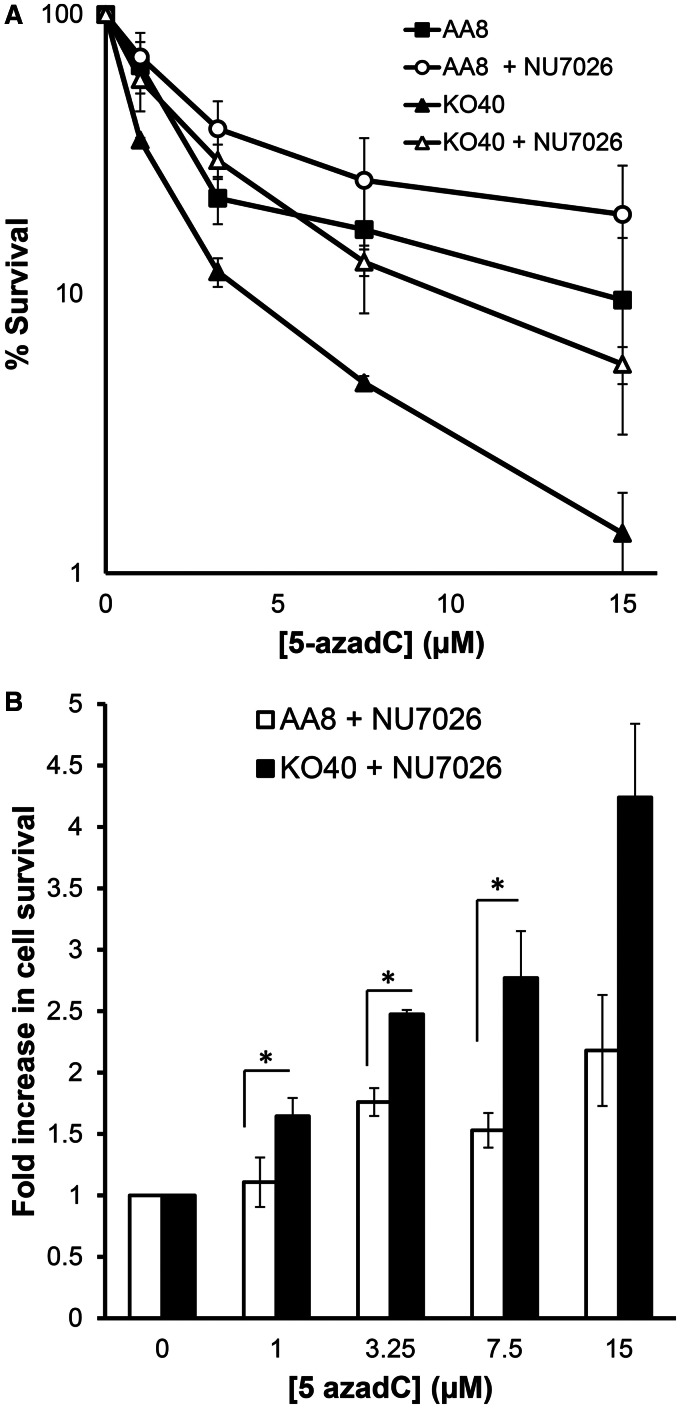
It is well accepted that HR repairs a broader spectrum of lesions that occur at stalled replication forks, whereas NHEJ is believed to have a more important role in the repair of two-ended DSBs (30,31). However, the relationship between HR and NHEJ is complex, and it has previously been demonstrated that the repair defect in FA cells reverts by inhibition of DNA-protein kinase and that the cells then are no longer sensitive to crosslinkers (32). The underlying mechanism is not entirely clear, but it is possible that inhibition of NHEJ prevent error-prone repair, which can be toxic.
Here, we used the DNA-protein kinase inhibitor NU7026 to test whether inhibition of NHEJ affects the response to 5-azadC. Interestingly, we show an increase in cell survival in both AA8 and KO40 cells after treatment with 5-azadC (Figure 6A) in the presence of a non-toxic dose of NU7026 (Supplementary Figure S3), compared with those cells that received only the treatment with 5-azadC. Also, we show that the increase in cell survival (represented as fold increase) was clearly higher for FANCG-deficient cells than for AA8 cells (Figure 6B). Furthermore, the sensitization effect was increased with increasing doses (Figure 6B). Our findings are consistent with the interpretation given by others (32) that FA protects from the promiscuous action of NHEJ during the repair of lesions induced by crosslink agents. In this respect, the severe cytotoxicity that 5-azaC produced in KO40 cells can be significantly suppressed by inhibiting NHEJ.
Figure 6.
Inhibition of NHEJ protects from the deleterious effect induced by 5-azadC. AA8 and KO40 cells in exponential growth were plated at low density onto 10 cm Petri dishes and treated with 5-azadC in combination with NU7026 (10 µM) according to ‘Materials and Methods’ section. Then cultures were allowed to grow (7–10 days) for analysis of colony-forming efficiency (A). Data, plotted as fold increase in cell survival, show that the increase in cell survival was clearly higher for FANCG-deficient cells than for AA8 cells (B). Each bar represents the mean and the SD from two independent experiments. Differences were statistically significant (*P < 0.05, according Student’s t-test).
DISCUSSION
The 5-azadC (decitabine) and 5-azaC (azacitidine) are extensively used in myelodysplastic syndrome treatment and increasingly used in experimental treatment of AML (9,10). Both are inhibitors of DNMT, but the cytotoxic mechanism remains somewhat unclear and can involve both activation of apoptosis and DNA damage. Here, we confirm earlier findings that 5-azadC causes γ-H2AX foci (3,20), which is a general marker for DNA damage and not specific for DSBs. Adding to this, we observe that 53BP1 foci are induced by 5-azadC treatment, which is a more stringent marker for DSBs. Also, we observe an increase in chromatid breaks and radial chromosomes, which if left unrepaired are toxic to cells. Altogether, our data strongly suggest that DSB formation is formed after 5-azadC treatment. The next question is how DSB may form after 5-azadC treatment? It is well established that DNMT is covalently linked to DNA on 5-azadC treatment (2–5), which may become an obstacle during DNA replication, which could lead to collapse of replication forks and DSBs. It is well established that the topoisomerase I poison camptothecin traps topoisomerase I onto DNA, which is converted to a DSB at replication forks (26,33), which in turn trigger HR for repair (26). In analogy, we use APH to stop ongoing replication in cells with incorporated 5-azadC that traps DNMT, to test whether this would reduce the amount of chromatid breaks. Indeed, we observe that the number of chromatid breaks is reduced, demonstrating that the toxic DSBs formed after 5-azadC treatment requires ongoing replication. These data suggest that 5-azadC causes replication collapse to form DSBs. Normally, such breaks are repaired by RAD51-mediated HR to repair and restart replication (34), which leaves an SCE (26). Our data strongly suggest that HR is activated to repair 5-azadC lesions as both RAD51 foci, and SCEs are induced by this treatment. This is further strengthen by the observation that KO40 cells, defective in FANCG, which is required for HR repair at collapsed replication forks (18), are hypersensitive to 5-azadC treatment and cannot trigger RAD51-mediated repair and fail to induce SCEs. We observe an increase in unrepaired chromatid breaks in FANCG defective cells, which is the logical consequence by failure to activate HR repair. We also observe an increase in radial chromosomes in FANCG defective cells, clearly demonstrating the link between unrepaired chromatid breaks and the formation of radial chromosomes. In absence of HR, it is highly likely that NHEJ will eventually fuse DSBs. If breaks occur at replication forks, only single DNA ends would be present and fusion with another end would result in formation of chromosome aberrations, such as radial chromosomes.
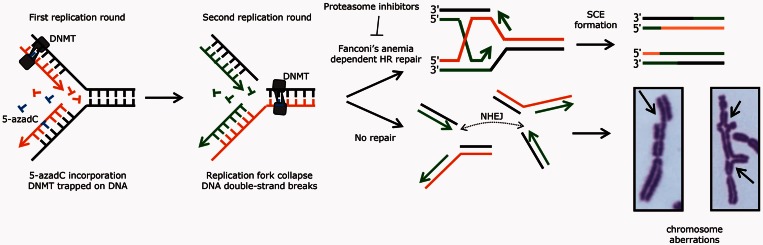
Altogether, our data point to a model to explain the effects of 5-azadC, where incorporated 5-azadC traps DNMT onto DNA, which becomes an obstacle to the second round of replication and results in a collapsed replication fork with a DSB (Figure 7). Such replication-associated DSB is normally repaired by RAD51-mediated HR, which results in an SCE. However, in absence of repair, chromatid breaks accumulate, and NHEJ fuses DNA ends that results in radial chromosomes, which will break during mitosis (Figure 7).
Figure 7.
Proposed model explaining the repair of 5-azadC induced DSBs. The 5-azadC is incorporated into DNA during the first round of replication. Once incorporated, DNMT becomes covalently trapped in the attempt to methylate this analogue creating protein–DNA adducts, which, if are not repaired will provoke the stall of the replication forks in the second round of replication, creating DSBs. These replication-associated DSBs need Fanconi’s Anemia dependent HR to be properly resealed, a process that ends in a SCE formation. In the absence of the Fanconi pathway, or in the presence of proteasome inhibitors, these DSBs left unrepaired (which will end in chromatid type break, image on the left) or misjoined, this latter being a consequence of the error-prone side of NHEJ, giving rise to the formation of radial fusion chromosomes (right).
It is well established that proteasome inhibitors impair both the activation of FA pathway (28), as well as to catalyze HR (29). The likely mechanism is that the FA pathway requires a mono- ubiquitination of the FANCD2 and FANCI subunits to promote HR repair (35). Similarly, the HR pathway requires RNF8 mediated ubiquitination to promote HR repair (36). As proteasome inhibitors prevent degradation of ubiquitinated proteins, the level of free ubiquitin is rapidly depleted in cells, which will impair both FA and HR pathways. Here, we found that the proteasome inhibitor MG132 potentiate the toxic effects of 5-azadC treatment only in wild-type AA8 cells, but not in the FANCG-defective cells. This can be explained by HR is impaired by MG132 in wild-type cells, which increase the sensitivity of 5-azadC to the same level as the FANCG-defective cells. As the proteasome inhibitor does not potentiate FANCG-defective cells, it demonstrates that the sensitization is mediated through inactivation of the FA–HR pathway.
These results may have important clinical implications. Resistance is a major difficulty with 5-azadC treatments, and analogy can be made to melphalan resistance in multiple myeloma. Melphalan is a crosslinker and binds covalently to DNA and cause obstruction to oncoming replication forks, replication collapse and DSBs, much in the same matter as we demonstrate for 5-azadC. Furthermore, FANCG-mutated cells are sensitive to melphalan (37). Resistance to melphalan is demonstrated to be mediated by increased HR by overexpression of Fanconi’s anemia genes (38). Bortezomib is a proteasomal inhibitor used in first line in combination with melphalan to treat multiple myeloma (39). The mechanism of creating a clinical benefit with the combination of bortezomib and melphalan is likely mediated by preventing resistance through activation of the FA pathway to trigger HR repair (40). Here, we suggest that bortezomib may be useful in combination with 5-azadC, to prevent emerging resistance. Indeed, a recent phase I clinical study bortezomib was used in combination with 5-azadC in AML patients, and a complete remission was observed in 5 of 10 previously untreated patients (41), demonstrating that this may be an interesting future option that may be tested.
Fanconi’s anemia patients often develop AML, and treatment options are often limited owing to the intrinsic sensitivity to DNA damaging agents. As 5-azadC is a more modern and targeted treatment option, it may be suggested as an alternative treatment to these patients. In one reported case, a Fanconi’s anemia patient with an AML derived from healthy donor cells received complete remission after a 5-azaC treatment (42). Although we do not discourage, we would recommend cautious use of 5-azadC in treatment of AML in Fanconi’s anemia patients, as we show that cells defective in the Fanconi’s anemia pathway are hypersensitive to 5-azadC.
Here, we demonstrate that 5-azadC causes replication-associated DSBs, likely owing to DNMT–DNA adducts obstructing replication fork progression, which in turn trigger RAD51-mediated HR involving the Fanconi’s anemia pathway. This likely represent an important cytotoxic mechanism for 5-azadC. However, our data are not in disagreement with that 5-azadC may deplete DNMT and reduce cancer burden through activation of genes independently of DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3, Supplementary Methods and Supplementary References [43,44].
ACKNOWLEDGEMENTS
The authors thank Dr. Fabrizio Palitti for his generous gift of KO40 cells and Dr. Jordi Surralles for providing human FANCD2 deficient cell lines. They also thank Felipe Cortés-Ledesma for helpful discussion and critical reading of the manuscript.
FUNDING
Spanish Ministry of Education and Science [BFU2007-61301]; Junta de Andalucía [BIO-120, Spain]; the Swedish Cancer Society; the Swedish Children’s Cancer Foundation; the Swedish Research Council; the Swedish Pain Relief Foundation; and the Söderberg Foundation (in part). M.L.O. was supported by a mobility fellowship from Plan Propio of University of Seville, Spain. Funding for open access charge: Swedish Research Council supports publication charges.
Conflict of interest statement. None declared.
REFERENCES
- 1.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 2.Christman JK. 5-azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 3.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell. Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl Acad. Sci. USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods. 2005;2:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 6.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Cancer Cell. 2004;2:62–72. [PubMed] [Google Scholar]
- 7.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 10.Klimek VM, Dolezal EK, Tees MT, Devlin SM, Stein K, Romero A, Nimer SD. Efficacy of hypomethylating agents in therapy-related myelodysplastic syndromes. Leuk Res. 2012;36:1093–1097. doi: 10.1016/j.leukres.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy AW, Hart WR. Multiple squamous-cell carcinomas in fanconi's anemia. Cancer. 1982;50:811–814. doi: 10.1002/1097-0142(19820815)50:4<811::aid-cncr2820500432>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 12.Alter BP. Cancer in fanconi anemia, 1927-2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 13.Collins N, Kupfer GM. Molecular pathogenesis of fanconi anemia. Int. J. Hematol. 2005;82:176–183. doi: 10.1532/IJH97.05108. [DOI] [PubMed] [Google Scholar]
- 14.Bagby GC. Genetic basis of fanconi anemia. Curr. Opin. Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 16.D'Andrea AD. The fanconi road to cancer. Genes Dev. 2003;17:1933–1936. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 17.Lyakhovich A, Surralles J. Disruption of the fanconi anemia/BRCA pathway in sporadic cancer. Cancer Lett. 2006;232:99–106. doi: 10.1016/j.canlet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 18.Tebbs RS, Hinz JM, Yamada NA, Wilson JB, Salazar EP, Thomas CB, Jones IM, Jones NJ, Thompson LH. New insights into the fanconi anemia pathway from an isogenic FancG hamster CHO mutant. DNA Repair. 2005;4:11–22. doi: 10.1016/j.dnarep.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Cortés F, Morgan WF, Wolff S. Effect of exogenous thymidine on sister-chromatid exchange frequency in Chinese hamster ovary cells with bromodeoxyuridine- and chlorodeoxyuridine-substituted chromosomes. Mutat. Res. 1987;192:277–282. doi: 10.1016/0165-7992(87)90069-8. [DOI] [PubMed] [Google Scholar]
- 20.Kiziltepe T, Hideshima T, Catley L, Raje N, Yasui H, Shiraishi N, Okawa Y, Ikeda H, Vallet S, Pozzi S, et al. 5-azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol. Cancer Ther. 2007;6:1718–1727. doi: 10.1158/1535-7163.MCT-07-0010. [DOI] [PubMed] [Google Scholar]
- 21.Elvers I, Johansson F, Groth P, Erixon K, Helleday T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 2011;39:7049–7057. doi: 10.1093/nar/gkr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groth P, Orta ML, Elvers I, Majumder MM, Lagerqvist A, Helleday T. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 2012;40:6585–6594. doi: 10.1093/nar/gks315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orta ML, Mateos S, Cantero G, Wolff LJ, Cortes F. Protection of halogenated DNA from strand breakage and sister-chromatid exchange induced by the topoisomerase I inhibitor camptothecin. Mutat. Res. 2008;637:40–48. doi: 10.1016/j.mrfmmm.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 27.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 2007;67:7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 29.Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 30.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundin C, Schultz N, Arnaudeau C, Mohindra A, Hansen LT, Helleday T. RAD51 is involved in repair of damage associated with DNA replication in mammalian cells. J. Mol. Biol. 2003;328:521–535. doi: 10.1016/s0022-2836(03)00313-9. [DOI] [PubMed] [Google Scholar]
- 32.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of fanconi anemia. Mol. Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 33.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 35.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 37.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, DeMarzo AM, Offerhaus GJ, Isacoff WH, Hruban RH, et al. In vivo therapeutic responses contingent on fanconi anemia/BRCA2 status of the tumor. Clin. Cancer Res. 2005;11:7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 38.Chen Q, Van der Sluis PC, Boulware D, Hazlehurst LA, Dalton WS. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood. 2005;106:698–705. doi: 10.1182/blood-2004-11-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, Kropff M, Spicka I, Petrucci MT, Palumbo A, Samoilova OS, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N. Engl. J. Med. 2008;359:906–917. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- 40.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- 41.Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld AK, Whitman S, Walker A, Klisovic R, Byrd JC, Santhanam R, et al. Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood. 2012;119:6025–6031. doi: 10.1182/blood-2012-03-413898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gustafsson B, Moell J, Leblanc K, Barbany G, Söderhäll S, Winiarski J. Donor cell-derived acute myeloid leukemia after second allogenic cord blood transplantation in a patient with Fanconi anemia. Pediatr. Transplant. 2012;16:241–245. doi: 10.1111/j.1399-3046.2011.01584.x. [DOI] [PubMed] [Google Scholar]
- 43.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castillo P, Bogliolo M, Surralles J. Coordinated action of the Fanconi anemia and ataxia telangiectasia pathways in response to oxidative damage. DNA Repair. 2011;10:518–525. doi: 10.1016/j.dnarep.2011.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.