Abstract
The Ntg1 and Mhr1 proteins initiate rolling-circle mitochondrial (mt) DNA replication to achieve homoplasmy, and they also induce homologous recombination to maintain mitochondrial genome integrity. Although replication and recombination profoundly influence mitochondrial inheritance, the regulatory mechanisms that determine the choice between these pathways remain unknown. In Saccharomyces cerevisiae, double-strand breaks (DSBs) introduced by Ntg1 at the mitochondrial replication origin ori5 induce homologous DNA pairing by Mhr1, and reactive oxygen species (ROS) enhance production of DSBs. Here, we show that a mitochondrial nuclease encoded by the nuclear gene DIN7 (DNA damage inducible gene) has 5′-exodeoxyribonuclease activity. Using a small ρ− mtDNA bearing ori5 (hypersuppressive; HS) as a model mtDNA, we revealed that DIN7 is required for ROS-enhanced mtDNA replication and recombination that are both induced at ori5. Din7 overproduction enhanced Mhr1-dependent mtDNA replication and increased the number of residual DSBs at ori5 in HS-ρ− cells and increased deletion mutagenesis at the ori5 region in ρ+ cells. However, simultaneous overproduction of Mhr1 suppressed all of these phenotypes and enhanced homologous recombination. Our results suggest that after homologous pairing, the relative activity levels of Din7 and Mhr1 modulate the preference for replication versus homologous recombination to repair DSBs at ori5.
INTRODUCTION
Eukaryotic cells contain mitochondrial DNA (mtDNA), which encodes essential components for oxidative phosphorylation-dependent adenosine triphosphate (ATP) production. As a byproduct, oxidative phosphorylation generates reactive oxygen species (ROS), which cause oxidative damage to mtDNA. MtDNA damage, such as double-strand breaks (DSBs), resulting in point mutations and deletions, which in turn lead to mitochondrial dysfunction and play a pathogenic role in the aging of post-mitotic tissues [see (1–4) for review]. However, ROS is not only harmful to mitochondrial function but is also critically involved in controlling mtDNA copy number [(5,6) and references cited therein]. MtDNA copy number increases or decreases considerably in response to the extent of oxidative stress and ATP demand (6–10). As described in detail later in the text, ROS induces a site-specific DSB at the mtDNA replication origin ori5, which is followed by homologous recombination or rolling-circle mtDNA replication. Homologous DNA recombination repairs DSBs, thereby helping to preserve mitochondrial genome integrity. Rolling-circle mtDNA replication promotes the establishment of homoplasmy, which is a genetic state in which all copies of the mitochondrial genome within a cell have the same sequence (11). Therefore, to understand mitochondrial genetic inheritance, it is important to study the coordination of mtDNA recombination and replication and the factors that select between the two pathways.
Our previous studies on Saccharomyces cerevisiae revealed that mtDNA copy number is controlled by a recombination-related mechanism that responds to oxidative stress. MtDNA is specifically modified at ori5, where the base-excision repair enzyme Ntg1 introduces a site-specific DSB (5,6), and mtDNA copy number is regulated by the ori5-specific DSBs introduced by Ntg1 (5,6). By the homologous pairing activity of Mhr1 (mitochondrial homologous DNA recombinase), a putative 3′-single-stranded DNA tail derived from the ori5-specific DSB forms a heteroduplex joint with the complementary sequence within a template circular double-stranded (ds) DNA molecule, and this 3′-terminus serves as a primer to initiate rolling-circle mtDNA replication, a type of DSB-induced DNA replication, on the circular dsDNA templates (12,13). Rolling-circle mtDNA replication produces mtDNA concatemers, which are tandem arrays of many mtDNA genome units (12). These concatemers are selectively transmitted to daughter cells where they are processed into circular monomers, ultimately generating homoplasmic cells (13).
MHR1 was identified as a gene required for homologous mtDNA recombination (8). In vitro, the Mhr1 protein acts as a recombinase to catalyze homologous DNA pairing through an intermediate DNA structure identical to the intermediate of RecA/Rad51-recombinase catalyzed homologous DNA pairing (12,14). Unlike RecA/Rad51, however, Mhr1 does not require ATP to catalyze homologous DNA pairing (12,15).
DSBs can initiate recombinational repair and break-induced DNA replication (BIR), using undamaged chromosomes as templates. In both cases, the DSB is processed by nucleases to generate recombinogenic 3′-single-strand ends that are potential substrates for RecA/Rad51-recombinases. During recombinational repair of DSBs, invasion of the 3′-single-strand end into an intact duplex leads to the formation of a heteroduplex joint with a displacement loop (D-loop), which then serves as an intermediate for the formation of a double-Holliday junction or for strand annealing with another end of the break, after limited DNA synthesis. In BIR events, invasion of the 3′-single-strand end into an intact duplex leads to D-loop formation, but continuous DNA synthesis does not generate double-Holliday junctions or strand annealing [see (16) for review]. BIR of linear chromosomal DNA can lead to duplication of chromosome arms. However, when circular mitochondrial DNA is a template, the result can be rolling-circle replication.
To initiate homologous pairing, exonucleolytic activity is required to generate a 3′-single-stranded tail from DSBs, but it is not known which exonuclease serves this function in mitochondria. In S. cerevisiae mitochondria, the major nuclease is Nuc1, which has both endo- and exonuclease activity, including 5′ exonuclease activity on dsDNA (17). However, disrupting NUC1 does not affect mtDNA recombination (18), nor does it increase the number of residual DSBs at ori5 (5), indicating that Nuc1 plays little or no role in mtDNA recombinational repair. Another candidate for the initiating nuclease is the Din7 protein. Din7 was identified as a DNA damage-induced mitochondrial nuclease (19), and it has an amino acid sequence homologous to that of Schizosaccharomyces pombe exonuclease I (Exo I: 20), a 5′–3′ exonuclease (21,22). Several studies have shown that overexpressing Din7 significantly increases the frequency of petite formation and mtDNA recombination (19,23–25), and these studies have suggested but not proven that Din7 is the exonuclease that generates 3′-single-stranded tails in mtDNA recombination.
To determine whether Din7 is involved in mtDNA recombination and rolling-circle mtDNA replication, we used a hypersuppressive (HS) ρ− mtDNA (HSC-1) as a model system (5). The HS ρ− mtDNA consists of a short mtDNA fragment (∼1 kb) that contains ori5, the most active of the four mtDNA replication origins and has a vast replication advantage over ρ+ mtDNA [75–85 kb (26,27)]. Various ρ− mtDNAs containing an origin (HS ρ−) have been used to study yeast mtDNA replication to avoid problems that would make it difficult to interpret the results (28,29). This approach was effective in our previous studies that identified the mechanism that controls the mtDNA copy number, described earlier in the text (6,13).
In this study, we investigated whether Din7 has 5′–3′ exonuclease activity, and asked whether it is a critical factor in Mhr1-dependent rolling-circle replication and recombination. We determined that Din7 and Mhr1 are required for both DSB-induced mtDNA replication and DSB repair at ori5. In addition, we found that the choice of mtDNA replication or repair after homologous DNA pairing at ori5 is regulated by the relative activities of Din7 and Mhr1, and that an imbalance in their activities causes deleterious effects leading to petite generation.
MATERIALS AND METHODS
Yeast strains, media and general genetic techniques
The yeast strains used in this study are listed in Table 1.
Table 1.
Yeast strains used in this study
| Abbreviation | Strains | Nuclear genotype | Mitochondrial genotype | Source |
|---|---|---|---|---|
| mhr1-1 | FL67-1423 | α his1 trp1 can1 mhr1-1 | ρ+ ω+ ens2 Chl321R | Ling et al. (8) |
| mhr1-1/pYES2/CT | FL67-1423/pYES2/CT | α his1 trp1 can1 mhr1-1 pYES2/CT(URA3) | ρ+ ω+ ens2 Chl321R | Ling et al. (8) |
| mhr1-1/pYES2/CT-DIN7 | FL67-1423/pYES2/CT-DIN7 | α his1 trp1 can1 mhr1-1 | ρ+ ω+ ens2 Chl321R | Ling et al. (8) |
| pYES2/CTDIN7(DIN7, URA3) | ||||
| WT | YKN1423 | α leu2 ura3 met3 | ρ+ ω+ Δens2 Oli2R | Ling and Shibata (12) |
| WT/pYES2/CT | YKN1423/pYES2/CT | α leu2 ura3 met3 MHR1 pYES2/CT(URA3) | ρ+ ω+ Δens2 Oli2R | Ling and Shibata (13) |
| WT/pYES2/CT-DIN7 | YKN1423/pYESCT-DIN7 | α leu2 ura3 met3 MHR1 | ρ+ ω+ Δens2 Oli2R | This study |
| pYES2/CT-DIN7(DIN7, URA3) | ||||
| WT/pYES2/CT-din7Δ | YKN1423/pYESCT-din7Δ | α leu2 ura3 met3 MHR1 | ρ+ ω+ Δens2 Oli2R | This study |
| pYES2/CT-din7Δ (din7Δ, URA3) | ||||
| WT/pYESMHR1 | YKN1423/pYESMHR1 | α leu2 ura3 met3 MHR1 | ρ+ ω+ Δens2 Oli2R | Ling and Shibata (12) |
| pYESMHR1 (MHR1, URA3) | ||||
| WT/pESC-DIN7 + MHR1 | YKN1423/pESC-DIN7 + MHR1 | α leu2 ura3 met3 MHR1 | ρ+ ω+ Δens2 Oli2R | This study |
| pESC-DIN7 + MHR1 (DIN7, MHR1, URA3) | ||||
| WT/(ENS2) | IL1666b-5010 | a leu2 ura3 trp1 can1 MHR1 | ρ+ ω+ ENS2 EryR AntR | Ling et al. (8) |
| WT/(ens2) | IL1666b-5011 | a leu2 ura3 trp1 can1 MHR1 | ρ+ ω+ ens2 EryR AntR | Ling et al. (8) |
| WT HS ρ− | YKN1423C-1 | α leu2 ura3 met3 | HS [ori5] ρ− mtDNA | Ling et al. (5) |
| Δmhr1 HS ρ− | YKN1423C-1Δmhr1 | α leu2 ura3 met3 mhr1::LEU2 | HS [ori5] ρ− mtDNA | This study |
| Δdin7 HS ρ−/pYESMHR1 | YKN1423C-1Δdin7 | α leu2 ura3 met3 din7::URA3 | HS [ori5] ρ− mtDNA | This study |
| Δdin7 Δmhr1 HS ρ− | YKN1423C-1Δdin7 Δmhr1 | α leu2 ura3 met3 mhr1::LEU2 din7::URA3 | HS [ori5] ρ− mtDNA | This study |
| WT HS ρ−/ pYES2/CT | YKN1423C-1/ pYES2/CT | α leu2 ura3 met3 pYES2/CT(URA3) | HS [ori5] ρ− mtDNA | This study |
| WT HS ρ−/pYES2/CT-DIN7 | YKN1423C-1/pYES2/CT-DIN7 | α leu2 ura3 met3 | HS [ori5] ρ− mtDNA | This study |
| pYES2/CT-DIN7(DIN7, URA3) | ||||
| WT HS ρ−/pYESMHR1 | YKN1423C-1/pYESMHR1 | α leu2 ura3 met3 | HS [ori5] ρ− mtDNA | This study |
| pYESMHR1(MHR1, URA3) | ||||
| WT HS ρ−/ | YKN1423C-1/ | α leu2 ura3 met3 pES-DIN7 + MHR1 | HS [ori5] ρ− mtDNA | This study |
| pESC-DIN7 + MHR1 | pESC-DIN7 + MHR1 | (DIN7, MHR1, URA3) | ||
| Δmhr HS ρ−/pYES2/CT | YKN1423C-1Δmhr1/ pYES2/CT | α leu2 ura3 met3 mhr1::LEU2 | HS [ori5] ρ− mtDNA | This study |
| pYES2/CT (URA3) | ||||
| Δmhr1 HS ρ−/ | YKN1423C-1Δmhr1/ | α leu2 ura3 met3 ntg1::LEU2 | HS [ori5] ρ− mtDNA | This study |
| pYES2/CT-DIN7 | pYES2/CT-DIN7 | pYES2/CT-DIN7(DIN7, URA3) | ||
| WTρ0 | YKN1423C-1ρ0 | α leu2 ura3 met3 | ρ0 | This study |
Media preparations were largely identical to those previously described (8,12,13). SGlycerol medium consisted of 2% glycerol and 0.7% yeast nitrogen base without amino acids, which contains all essential vitamins and inorganic salts necessary for the cultivation of yeasts except histidine, methionine, tryptophan and a source of carbohydrate (Difco™; Becton Dickinson, Sparks, MD, USA) in 50 mM KH2PO4 (pH 6.25). RGal medium consisted of 2% raffinose, 2% galactose and 0.7% yeast nitrogen base without amino acids. YPGly medium consisted of 3% glycerol, 2% bacto-peptone and 1% yeast extract in 50 mM KH2PO4 buffer (pH 6.25). Synthetic Dextrose (SD) minimal medium consisted of 2% glucose and 0.7% yeast nitrogen base without amino acids.
The general genetic techniques used in this study are described in (30). Yeast transformation was performed using the lithium-acetate method (31).
Radioactive materials
[γ-32P]ATP (specific activity, 3000 Ci/mmol), [α-32P]dCTP (specific activity, 3000 Ci/mmol) and [α-32P]dGTP (specific activity, 3000 Ci/mmol) were obtained from PerkinElmer Japan Co., Ltd. (Yokohama, Japan).
Nucleotides
The 2′-deoxynucleoside 5′-triphosphates were purchased from Amersham Pharmacia Biotech Inc. (Piscataway, NJ, USA).
Enzymes
T4 DNA polymerase and mung bean nuclease were purchased from Takara Bio, Inc. (Shiga, Japan), and proteinase K was purchased from Promega Corporation (Madison, WI, USA).
Antibodies and immunoblot analysis
An anti-porin monoclonal antibody was purchased from Invitrogen (Life Technologies, CA, USA), and anti-6× His-tag antibody was purchased from MBL (Medical and Biological Laboratories Co., Ltd, Nagoya, Japan). A 16mer peptide (CGGESNSFNKKVEQPL) of Din7 was designed and synthesized by Scrum Inc. (Tokyo, Japan). The peptide was used for rabbit immunization to prepare an anti-Din7 rabbit serum for this study by RRC (Research Resources Center, RIKEN Brain Science Institute, Japan). Immunoblot analyses were performed with a sodium dodecyl sulfate–polyacrylamide gel electrophoresis system as described previously (12).
Construction of plasmids harboring DIN7, DIN7Δ, MHR1 and both DIN7 and MHR1
pYESMHR1, which encodes Mhr1 without a tag, was constructed as described previously (13). To construct pYES2/CT-DIN7, which encodes Din7 with a C-terminal 6× His tag, the 1.29-kb open reading frame (ORF) of DIN7 without a stop codon was polymerase chain reaction (PCR) amplified using whole cellular DNA from the wild-type strain (YKN1423) as a template and inserted under control of the GAL1 promoter between the KpnI and XbaI sites of pYES2/CT (Invitrogen). To generate pESC-DIN7 + MHR1, two fragments were PCR amplified using total DNA from the wild-type strain (YKN1423) as a template: the 1.3-kb ORF of DIN7, without a stop codon, encoding Din7 with a C-terminal Flag tag, and the 0.68-kb ORF of MHR1, without a stop codon, encoding Mhr1 with a C-terminal Myc tag. These two fragments were subcloned into the pESC-URA vector, using the NotI and SpeI sites to place Flag-tagged DIN7 under control of the GAL10 promoter and the BamHI and SalI sites to place Myc-tagged MHR1 under control of the GAL1 promoter (Stratagene). To construct pYES2/CT-din7Δ, which encodes a deleted Din7 lacking 118 amino acids at the C-terminus, the 0.98-kb ORF of din7Δ lacking a stop codon was PCR amplified using pYES2/CT-DIN7 as a template and placed under control of the GAL1 promoter between the KpnI and XbaI sites of pYES2/CT (Invitrogen).
Petite production
Wild-type and mhr1-1 ρ+ cells harboring pYES2/CT, pYES2/CT-DIN7 or pESC-DIN7 + MHR1 were grown from an initial density of 1.0 × 105 cells/ml to 1.0 × 107 cells/ml in glycerol medium (SGlycerol) supplemented with amino acids, and then used as pre-cultures. An aliquot from each pre-culture (2.0 × 105 cells) was transferred to 2 ml of RGal medium supplemented with required amino acids and cultivated for 18−20 generations (2 days) at 30°C. At a concentration of 1 × 107 cells/ml, a portion of the cells was treated with 10 µM hydrogen peroxide. The treated and untreated cells were diluted, plated on SD plates and incubated at 30°C for 3 days. The colonies were replica-plated onto YPGly plates to test their respiration proficiency. Petite cells formed colonies only on SD plates, but not on YPGly plates.
Preparation of mitochondrial extracts
Pre-cultures of wild-type ρ+ cells transformed with the empty pYES2/CT vector, pYES2/CT-DIN7 or pYES2/CT-din7Δ were grown at 30°C for 3 days in 1 l of raffinose medium supplemented with amino acids. The cells were transferred to 1 l of RGal medium and cultivated at 30°C for 11 h to induce Din7 expression under control of the GAL1 promoter. Mitochondrial extracts were obtained after sonicating mitochondria isolated from the resulting cells, as described previously (12), and then adjusted to the same protein concentration by determining the porin levels through immunoblot analysis using an anti-porin antibody.
DNA substrates to examine 5′–3′ or 3′–5′ exonuclease activity
The 5′-32P-labeled pUC119/HincII dsDNA fragments were prepared by labeling HincII-linearized, dephosphorylated pUC119 DNA fragments with [γ-32P]ATP to a specific activity of 1.3–2.0 × 106 cpm/pmol, using T4 polynucleotide kinase from the MEGALABEL™ kit (Takara Bio, Inc.).
The 3′-32P-labeled pUC119/BamHI dsDNA fragments were prepared by labeling BamHI-linearized pUC119 DNA fragments with [α-32P]dCTP, [α-32P]dGTP, dATP and dTTP to a specific activity of 1.7–3.0 × 106 cpm/pmol, using T4 DNA polymerase.
Assay for exonuclease activities in mitochondria
The 5′-labeled pUC119/HincII dsDNA fragments (75 nM) and the 3′-labeled pUC119/BamHI dsDNA fragments (87 nM) were incubated at 37°C for 15 min with various amounts (0, 4, 20, 32 and 50 µg/ml in protein) of mitochondrial extracts in a 10-µl reaction solution containing 1× λ exonuclease buffer (New England BioLabs Inc., MA, USA): 67 mM glycine–KOH (pH 9.4), 2.5 mM MgCl2 and 50 µg/ml of bovine serum albumin. After removing proteins with proteinase K, the DNA was separated on a 1% agarose gel. Signals corresponding to the 5′-32P-labeled pUC119/HincII or 3′-32P-labeled pUC119/BamHI dsDNA fragments were quantitated using a Fuji BAS 2500 image analyzer, as previously described (5,6).
Detection of single-stranded DNA generation
The pUC119/HincII dsDNA fragments (104 µM) were incubated at 37°C for 15 min with various amounts (0, 4, 20, 32 and 50 µg/ml in protein) of mitochondrial extracts. Mung bean nuclease was used to digest the resulting single-stranded DNA molecules. After removing proteins with proteinase K, the DNA was separated on a 1% agarose gel. The signals for the pUC119/HincII fragments and the single-stranded DNAs were visualized by staining with ethidium bromide, which fluoresces under ultraviolet light when intercalated into DNA.
Genetic crosses to detect the functions of Din7 and Mhr1 in mitochondrial recombination
The effects of Din7 and Mhr1 were detected as described previously (8,32).
Southern hybridization analyses
Whole cellular DNA was digested with restriction endonucleases and then analyzed by standard gel electrophoresis on a 1.5 or 2.0% agarose gel (FMC, USA) at 2.0 V/cm in Tris-Acetate EDTA (TAE) buffer (40 mM Tris–acetate and 10 mM ethylenediaminetetraacetic acid, pH 8.0) at room temperature. Unless stated otherwise, we loaded 6−15 µg of whole cellular DNA that had been treated with or without restriction enzymes into each well of the gel. The λ/HindIII DNA (New England Biolabs) and 100-bp ladder (Invitrogen) were used as size markers. The DNA in the gel was transferred to a Hybond-N+ membrane (Amersham Biosciences) in 10× SSC (Sodium chloride and Sodium Citrate solution; 1.5 M NaCl and 0.15 M sodium citrate) by capillary blotting. The HS ρ− mtDNA signals were detected by Southern hybridization using 32P-labeled BglII-digested 1.1-kb HSC-1 ρ− mtDNA (5) as a probe: HS ρ− mtDNA fragments were excised from the pUC119HSC-1 vector and labeled with [a-32P]dCTP (PerkinElmer Japan) using the Ready-To-Go DNA-Labeling Kit (Pharmacia). The relative mtDNA copy number was determined by comparing the signal for HS ρ− mtDNA signal with the signal for the nuclear NUC1 gene, detected using a 32P-labeled 0.99-kb fragment containing NUC1 as the probe, as described previously (5,6).
Estimation of mtDNA copy number and DSB at ori5
When the effects of hydrogen peroxide were analyzed, wild-type, Δmhr1, Δdin7 and Δdin7Δmhr1 double-mutant cells were grown in Yeast Peptone Dextrose (YPD) medium at 30°C from an initial concentration of 1.0 × 105 cells/ml to 1.0 × 107 cells/ml (standard conditions). Cultures of the indicated mutants were divided into two samples and then washed with 1× PBS (phosphate-buffered saline). One sample was resuspended in PBS, whereas the other was treated with 10 μM hydrogen peroxide at 30°C for 1 h. The untreated and hydrogen peroxide-treated cells were subjected to respiratory function tests as previously described (8).
To analyze the effects of Din7 overproduction, WT HS ρ−/pYES2/CT, WT HS ρ−/pYES2/CT-DIN7, WT HS ρ−/pYESMHR1, WT HS ρ−/pESC-DIN7 + MHR1, Δmhr1 HS ρ−/pYES2/CT and Δmhr1 HS ρ−/pYES2/CT-DIN7 strains were grown at 30°C in raffinose medium supplemented with required amino acids from an initial concentration of 1.0 × 105 cells/ml to 1.0 × 107 cells/ml. Then, the cells were transferred to RGal medium supplemented with required amino acids and cultivated for 12 h at 30°C to induce DIN7 expression under control of the GAL1 promoter. Whole cellular DNA was isolated as previously described (8). Southern hybridization analyses were performed as described earlier in the text. To measure DSBs at ori5, we calculated the ratio of the signal for the 0.8-kb DNA fragment derived from the unique DSB at ori5 to the signal for the unit-size 1.1-kb HS ρ− mtDNA.
Two-dimensional gel electrophoresis analyses
Two-dimensional (2D) gel electrophoresis and detection of the 32P signals in the Southern hybridization analysis were performed as previously described (5). Signals for supercoiled closed-circular and nicked-circular multimers were determined by comparing the 2D gel profiles of DNA samples treated or untreated with 1.0 U/µl (∼10 µg) of calf thymus topoisomerase I (Takara Bio, Inc.) at 37°C for 30 min. After topoisomerase treatment, in the first-dimension run the closed-circular DNAs (monomers and multimers) migrated at the same velocity as the corresponding nicked-circular DNAs, whereas in the second-dimension run, the closed-circular DNAs migrated much faster.
RESULTS
Din7 overexpression increases mitochondrial 5′–3′ exonuclease activity
In a previous study, a C-terminal 6× His-tagged Din7 was partially purified from yeast cells overexpressing DIN7 and shown to have nuclease activity, based on the trichloroacetic acid-soluble nucleotide levels after Din7 was incubated with uniformly 3H-labeled λ DNA (19). However, this analysis did not indicate whether Din7 is an exonuclease or endonuclease. We, therefore, sought to determine whether Din7 has exonuclease activity on dsDNA and generates 3′-single-stranded tails (Figure 1A). Because we could not identify a purification procedure that prevented the degradation of Din7 overexpressed in Escherichia coli, we used cell-free extracts from mitochondria isolated from cells overexpressing Din7 encoded on pYES2/CT-DIN7.
Figure 1.
Overproduction of Din7 enhances 5′−3′ dsDNA exonuclease activity in mitochondria. (A) Principle of the 5′−3′ and 3′−5′ exonuclease activity assay. The 5′- and 3′-32P-labels were preferentially removed by 5′−3′ and 3′−5′ exonuclease activity, and they are resistant to 3′−5′ and 5′−3′ exonucleases, respectively. (B) Schematic diagram of the construction of the Din7 deletion mutant (Din7Δ). Immunoblot analysis of overproduced C-terminally 6× His-tagged Din7 or Din7Δ in mitochondrial extracts (bottom). Mitochondria were isolated from cells bearing WT/pYES2/CT, WT/pYES2/CT-DIN7 and WT/pYES2/CT-din7Δ (overproducing Din7Δ with a C-terminal 6× His tag). The signals of overproduced Din7 and Din7Δ were detected using a monoclonal anti-6× His-tag antibody. As a control, the levels of porin (a constitutively expressed protein, used here as a control) were determined by immunoblot analysis using a monoclonal anti-porin antibody to adjust the protein concentrations of mitochondrial extracts from all three strains to the same level. (C and E) Analyses of 5′−3′ or 3′−5′ exonuclease activity in mitochondrial extracts. The 5′- or 3′-32P-labeled pUC119/HincII dsDNA fragments were incubated at 37°C for 15 min with increasing concentrations of mitochondrial extracts. After removing the proteins by treating the extracts with proteinase K, the DNA molecules were separated on a 1% agarose gel. (D and F) Quantitative representation of the 5′−3′ and 3′−5′ exonuclease activities in mitochondrial extracts, detected in (C) and (E). Signals relative to untreated DNA are plotted. ds, double-stranded; ss, single-stranded. Open circles, incubated with mitochondrial extracts from wild-type cells (cells without Din7 overproduction); closed circles, incubated with mitochondrial extracts from Din7-overproducing cells; closed triangles, incubated with mitochondrial extracts from Din7Δ-overproducing cells. Each bar represents the results of at least two independent experiments. (G) The generation of single-stranded DNA by Din7. HincII-linearized pUC119 DNA (104 µM) was treated at 37°C for 5 min with increasing amounts of mitochondrial extracts derived from cells of WT/pYES2/CT, WT/pYES2/CT-DIN7 or WT/pYES2/CT-din7Δ in the same buffer used for detection of λ exonuclease activity. Then, each reaction solution (10 µl) was separated into duplicate aliquots. Proteinase K was added to one aliquot to stop the reaction. The second aliquot from each set was treated at 37°C for 10 min with 2.25 U mung bean nuclease followed by addition of proteinase K to stop the reaction. Mung bean nuclease digests only the single-stranded DNA, leaving the double-stranded DNA intact. Samples were then electrophoresed on a 1% agarose gel and stained with ethidium bromide. Lane 1, 1-kb plus ladder used as a size marker; lane 2, HincII-linearized pUC119 DNA; lane 3, HincII-linearized pUC119 DNA treated in a reaction mixture without mitochondrial extracts; lanes 4–7, HincII-linearized pUC119 DNA treated with increasing amounts of mitochondrial extracts; lane 8, HincII-linearized pUC119 DNA treated only with mung bean nuclease; lanes 9–12, samples treated as in lanes 4–7, treated additionally with mung bean nuclease. The samples treated with mitochondrial extracts overexpressing DIN7 contain both single-stranded DNA fragments with discrete sizes and double-stranded DNA because discrete DNA signals smaller than those of the double-stranded DNA in the middle were removed by mung bean nuclease treatments.
We first investigated whether Din7-dependent exonuclease activity is present in yeast mitochondria. Cells were transfected with either pYES2/CT-DIN7, encoding tagged Din7, or pYES2/CT-din7Δ, encoding a tagged and truncated Din7 protein (Din7Δ) lacking 118 amino acids at the C-terminus; the transformants overexpressed either of the proteins fused to a C-terminal 6× His tag under control of the GAL1 promoter on galactose induction (Figure 1B). The overproduction of tagged Din7 in pYES2/CT-DIN7 or pYES2/CT-din7Δ-bearing cells under GAL1 induction (Din7 or Din7Δ-overproducing cells) was confirmed by immunoblot analysis with an anti-6× His-tag antibody (Figure 1B). We equalized the protein levels in the mitochondrial extracts based on the amounts of porin present (Figure 1B).
To determine whether Din7 has 5′ or 3′ exonuclease activity, we used 5′-32P-labeled HincII-linearized or 3′-32P-labeled BamHI-linearized pUC119 double-stranded DNA as a DNA substrate. The DNA was incubated with increasing amounts of mitochondrial extracts from cells not overproducing Din7, Din7-overproducing cells or Din7Δ-overproducing cells (Figure 1C and D). In this assay, signals from 5′-32P-labeled double-stranded DNA decreased significantly, but they did not change in size or sharpness in the presence of increasing amounts of mitochondrial extracts from Din7-overproducing cells, whereas this decrease was much less apparent in the presence of extracts derived from cells not overproducing Din7 or cells overproducing Din7Δ (Figure 1C and D). The absence of changes in the size or sharpness of the labeled DNA signals excludes the presence of endonucleolytic activities on double-stranded DNA. By contrast, signals from the 3′-32P-labeled double-stranded DNA did not significantly decrease, when treated with mitochondrial extracts from Din7Δ-overexpressing or control cells, but the DNA signals were tailed only when the DNA was treated with mitochondrial extracts from Din7-overexpressing cells (Figure 1E and F). These results suggest that the extracts prepared from Din7-overexpressing cells contain 5′ exonuclease activity on double-stranded DNA, but those from control cells do not, and that the digestion of DNA by the 5′ exonuclease activity in the extracts from Din7-overexpressing cells was limited, probably because of the presence of various proteins that bind to double-stranded DNA in the extracts.
Then, we treated HincII-linearized pUC119 DNA (without labeling) with increasing amounts of mitochondrial extracts from cells containing the empty pYES2/CT vector (i.e. not overexpressing Din7), Din7-overproducing cells or Din7Δ-overproducing cells. We observed that the treatment of the double-stranded DNA molecules with increasing amounts of mitochondrial extracts from Din7-overproducing cells produced discrete DNA fragments, which disappeared after additional treatment with mung bean nuclease (Figure 1G, middle). Mung bean nuclease endonucleolytically degrades single-stranded DNA but does not affect double-stranded DNA (33). No such activity was obvious in mitochondrial extracts derived from cells not overproducing Din7 or cells overproducing Din7Δ (Figure 1G, top and bottom). These results suggest that, although the activity is limited as described earlier in the text, mitochondria have a Din7-dependent exonuclease activity that generates single-stranded fragments from double-stranded DNA.
Together, these results support the observation that the 5′–3′ exonuclease activity we detected is derived from Din7.
Epistasis analysis of Δdin7 and Δmhr1 in DSB repair and the requirement for Din7 and Mhr1 in oxidative stress-induced mtDNA replication
Although overexpressing Din7 increased petite formation and DIN7 disruption did not alter mtDNA stability (19), we observed a slight deficiency in Mhr1-dependent polar recombination: i.e. we observed a decrease in the transmission ratio of the ω+-linked chloramphenicol-resistant marker, from 98% in the presence of DIN7 to 81% in its absence [F.L., unpublished manuscript and see (8)]. To investigate the role of Din7 in repairing DSBs at ori5 and regulating mtDNA copy number, we used wild-type, Δdin7, Δmhr1 and Δdin7Δmhr1 cells that contained HS ρ− mtDNA (HSC-1; Table 1). The HSC-1 ρ− mtDNA is a 1.1-kb mtDNA segment containing a replication origin, ori5 (DDB/EMBL/GenBank accession number AB182994). For these experiments, whole cellular DNA was prepared, treated with restriction endonuclease BglII and then subjected to gel electrophoresis. Next, mtDNA and nuclear DNA were detected by Southern hybridization using the 1.1-kb HSC-1 ρ− mtDNA and a fragment of the NUC1 0.99-kb ORF as probes, as previously described (5,6). The amount of DSBs in each sample was calculated based on the ratio between the signal of a 0.8-kb DNA fragment that is obtained in BglII-digested DNA only when a DSB is present at ori5 and the signal of a larger fragment that is always present in BglII-digested DNA (unit size 1.1-kb HSC-1 ρ− mtDNA, Figure 2A). The amount of mtDNA (i.e. mtDNA copy number) in each sample was expressed as the ratio of the signal of the 1.1-kb ρ− mtDNA to the signal from a BglII digest-derived 3.2-kb DNA fragment containing the NUC1 gene.
Figure 2.
Analysis of DSBs at ori5 and mtDNA copy number in Δdin7 and Δmhr1 mutant cells. (A) Physical map of the ori5 and DSB sites at ori5 in HSC-1, the HS ρ− mtDNA used in this study. The ori5 sequence is framed with an open box. The DSB site is indicated by an arrow. (B) Example of a gel profile, showing an analysis of the DSB at ori5 and mtDNA copy number. The wild-type and indicated mutant cells containing the HS ρ− mtDNA were grown to log phase in YPD medium and then divided into two samples. One sample was resuspended in PBS, whereas the other was treated with 10 µM hydrogen peroxide for 1 h at 30°C. Whole cellular DNA (∼10 µg) isolated from wild-type and mutant yeast cells was digested with BglII, separated by electrophoresis on a 2% agarose gel and transferred to Hybond-N+ membranes (GE Healthcare). The HS ρ− mtDNA on the membranes was detected by Southern hybridization using 32P-labeled HS ρ− mtDNA and the NUC1 gene as probes, as described previously (5). WT, wild-type; NUC1, 3.2-kb DNA fragment containing 0.99-kb NUC1 gene; 1.1-kb, unit size of BglII-digested HS ρ− mtDNA (1.1 kb); 0.8-kb (DSB) and 0.8-kb DNA fragment derived from the unique DSB at ori5 by the BglII-digest; asterisks indicates mtDNA containing single-stranded regions resistant to BglII digestion. (C) Quantitative representation of the DSBs at ori5 and mtDNA copy numbers. The amount of DSBs at ori5 in HS ρ− mtDNA in the indicated cells was calculated based on the signals from the 0.8-kb DNA fragment, generated only when a DSB is present, relative to the signals from the unit-sized 1.1-kb DNA of HS ρ− mtDNA. MtDNA copy numbers were expressed based on the signals from unit-sized 1.1-kb HS ρ− mtDNA relative to those from the 3.2-kb DNA fragment containing the NUC1 gene. Both signals were normalized against those of HS ρ− mtDNA isolated from the wild-type cells without hydrogen peroxide treatment and plotted. Each bar represents the results of at least two independent experiments. (D) Din7 expression level in hydrogen peroxide-treated cells. Wild-type cells containing HS ρ– mtDNA were grown to log phase in 2 l of YPD medium and then divided into two samples. One sample was resuspended in PBS, and the other was treated with 10 µM hydrogen peroxide for 1 h at 30°C. Then, mitochondria were isolated, and mitochondrial extracts were prepared for immunoblot analysis using an anti-Din7 rabbit serum prepared for this study. Note: to detect optimal signals for the mitochondrial outer-membrane protein porin, the prepared mitochondrial extracts were diluted 100-fold before being used in immunoblot analysis using a monoclonal anti-porin antibody.
First, we analyzed the amount of DSBs at ori5 and the mtDNA copy number in each strain. As shown in Figure 2B and C, there were more DSBs in Δdin7, Δmhr1 and Δdin7Δmhr1 cells than in wild-type cells, whereas the mtDNA copy number was lower in the single and double mutants than in the wild-type. Because a limited degree of oxidative stress increases mtDNA copy number, dependent on the function of Ntg1 and Mhr1 (5,6), we exposed the cells to a low concentration of hydrogen peroxide. The hydrogen peroxide treatment resulted in a 2.2-fold increase in the mtDNA copy number and a 1.3-fold increase in the amount of DSB in wild-type cells, as previously reported [Figure 2B and C (6)]. Although Din7 is a damage-inducible enzyme, immunoblot analysis using peptide-based rabbit serum against Din7 revealed that hydrogen peroxide treatment did not increase Din7 levels under these experimental conditions (Figure 2D).
The same hydrogen peroxide treatment did not induce an increase in the mtDNA copy number in Δdin7, Δmhr1 or Δdin7Δmhr1 cells, but it did increase the amount of DSBs at ori5 in these mutant cells (Figure 2B and C). The increase in DSBs at ori5 in Δdin7 cells was larger than that in Δmhr1 cells, but the same as that in Δdin7Δmhr1 double-mutant cells with or without hydrogen peroxide treatment (Figure 2B and C). We confirmed that there was a similar increase in ROS levels after hydrogen peroxide treatment in wild-type, Δdin7, Δmhr1 and Δdin7Δmhr1 cells (Supplementary Figure S1). Thus, Δdin7 is epistatic to Δmhr1 with respect to repair of DSBs at ori5, i.e. both Din7 and Mhr1 act in the same pathway. Furthermore, these results show that Din7 is strictly required for the ROS-triggered increase in mtDNA copy number through Mhr1-dependent mtDNA replication, as well as for DSB repair. These results and those described in the preceding section support a model in which the 5′–3′ exonuclease activity of Din7 helps generate 3′-single-stranded DNA tails that are required for Mhr1-mediated homologous pairing with intact double-stranded DNA, which acts as a template in both DSB repair at ori5 and rolling-circle mtDNA replication.
Effects of overexpressing Din7 on residual DSB levels at ori5 and on mtDNA copy number
As described earlier in the text, Din7 is required for DSB repair and mtDNA replication. Although MHR1 is constitutively expressed, Din7 is induced by DNA damage; however, DIN7 overexpression is harmful to mitochondria, as shown by elevated petite formation (19). We speculated that changes in the relative amounts of Din7 to Mhr1 would cause genetic effects that would reveal new functions of these proteins. Therefore, we transformed wild-type and Δmhr1 HS ρ− cells with the pYES2/CT (negative control) and pYES2/CT-DIN7 (Din7-overproduction) vectors. The transformed cells were grown in raffinose medium to avoid the effects of glucose, which can decrease mtDNA copy number (34,35). Then, the cells were transferred into RGal medium and cultivated for 11 h to induce Din7 expression. Din7 overproduction clearly induced a higher amount of DSBs at ori5 in both wild-type and Δmhr1 cells after the 11-h incubation (Figure 3A).
Figure 3.
Effects of overproducing DIN7 on DSBs at ori5 and mtDNA copy number. (A) Example of a gel analysis of DSBs at ori5 and mtDNA copy number in the indicated cells, with or without Din7 overproduction. DIN7 expression under control of the GAL1 promoter was induced in wild-type and Δmhr1 HS ρ− cells, both of which harbored the DIN7 expression vector, pYES2/CT-DIN7, by growing the cells for 12 h at 30°C in galactose-containing RGal medium. Control cells contain the empty pYES2/CT vector. Cellular DNA (∼10 µg) was isolated from cells, digested with BglII and analyzed by 2% agarose gel electrophoresis and Southern hybridization, using 32P-labeled HS ρ− mtDNA and the NUC1 gene as probes. (B) Changes in DSBs at ori5 and mtDNA copy number in wild-type and Δmhr1 HS ρ− cells during growth under Din7 overproduction. The HS ρ− wild-type and Δmhr1 cells harboring either the DIN7 expression vector or the empty vector were grown for the indicated times after the induction of DIN7 expression. Whole cellular DNA was isolated and subjected to Southern hybridization analysis as in (A). The amount of DSBs at ori5 in HS ρ− mtDNA and the mtDNA copy number were measured and calculated as in Figure 2B. Top, DSBs at ori5; bottom, copy number of HS ρ− mtDNA. ○, HS ρ− wild-type cells harboring pYES2/CT; •, HS ρ− wild-type cells harboring pYES2/CT-DIN7; ▵, HS ρ− Δmhr1 cells harboring pYES2/CT; ▴, HS ρ− Δmhr1 cells harboring pYES2/CT-DIN7. Each symbol represents the results of at least two independent experiments. (C) Immunoblot analysis of Din7 and Mhr1 expression in DIN7-induced cells. Cell-free extracts were prepared from HS ρ− wild-type and Δmhr1 cells both of which harbor either the empty pYES2/CT vector or the DIN7 expression vector pYES2/CT-DIN7 and grown for the indicated times under the conditions that induced DIN7 expression. Din7 with a C-terminal 6× His tag, Mhr1 and porin (a constitutively expressed control protein) was detected using an anti-His-tag antibody, an anti-Mhr1 serum (12) and a monoclonal anti-porin antibody, respectively.
Quantification of the results revealed that the relative signals of DSBs at the ori5 site increased 7-fold and the mtDNA copy number increased 2.8-fold, in parallel with the galactose induction in wild-type HS ρ− cells harboring pYES2/CT-DIN7 after an 11-h incubation. However, this increase was not observed in wild-type HS ρ− cells that did not overproduce Din7 (Figure 3B). During this incubation period, Din7 increased to the same level independent of the presence of MHR1, whereas the level of Mhr1 remained constant in MHR1 cells (Figure 3C). In the absence of Mhr1 (Δmhr1 cells), DSBs at ori5 increased as in the MHR1 cells, but there were no obvious changes in mtDNA copy number (Figure 3B). Thus, higher Din7 expression increased DSBs at ori5 in either the presence or absence of Mhr1, whereas the increase in mtDNA copy number depended on Mhr1.
When considering the effects of DSBs, it is important to note the following effects on the amount of DSBs, which are the residual DSBs after (i) DSBs at ori5 are generated by Ntg1 in response to the extent of oxidative stress and also by an unidentified mechanism; (ii) DSBs are suppressed because of recombinational repair; and (iii) the broken DNA is possibly diluted by replication of intact mtDNA replication. Thus, the observed increase in DSBs at ori5 in Din7-overproducing cells is likely a consequence of generation by Din7 of excess long single-stranded tails from broken ends, exceeding the capacity of DSB repair by Mhr1-dependent recombination, because Mhr1 is not also overexpressed and is, therefore, present at an insufficient level.
Enhanced circular multimer formation and DSB repair at ori5 when MHR1 and DIN7 are co-overexpressed
If the aforementioned suggestion is valid, Mhr1 overproduction in Din7-overexpressing cells would suppress the residual DSBs by enhancing recombinational repair, which decreases mtDNA replication by reducing the number of broken ends available for initiation. To test this hypothesis, we constructed pESC-DIN7 + MHR1, in which DIN7 and MHR1 were placed under control of the GAL1 and GAL10 promoters, respectively. The cells transformed with this plasmid were grown in raffinose medium, transferred into RGal medium and cultivated further. We analyzed whole cellular DNA isolated from these cells by 2D electrophoresis followed by Southern hybridization analysis. The signals of HS ρ− mtDNA containing ori5 represent the following molecular forms: concatemers appear as a continuous major arc, single-stranded DNA appears as a line and circular double-stranded multimers and monomers appear as discrete dots (Figure 4A-a). We calculated the ratios of the signals from each molecular species relative to the total signals of monomers or concatemers as shown in Figure 4B. The relative ratio of concatemers to monomers increased by 2.7- and 4.2-fold in cells overproducing Din7 and Mhr1, respectively, compared with cells harboring the empty pYES2/CT vector (Figure 4A-b, A-c, A-d and B, top). As expected from the 5′–3′ exonuclease activity of Din7, there was a 4.5-fold increase in the ratio of single-stranded DNA to monomers in Din7-overproducing cells, but this increase was completely suppressed by co-overexpression of Mhr1 (Figure 4A-e and B, middle).
Figure 4.
Effects of overproducing Din7 and Mhr1 on the levels of molecular species of HS ρ− mtDNA. Wild-type HS ρ− cells harboring the control plasmid pYES2/CT, the MHR1 expression plasmid pYESMHR1, the DIN7 expression plasmid pYES2/CT-DIN7 or the pESC-DIN7+MHR1 plasmid expressing both DIN7 and MHR1 were cultivated for 11 h at 30°C in RGal medium to induce gene expression. Whole cellular DNA was isolated from the cells, and ∼20 µg of cellular DNA was separated by 2D gel electrophoresis and transferred to a Hybond-N+ membrane. HS ρ− mtDNA on the membrane was detected by Southern hybridization analysis using 32P-labeled HS ρ− mtDNA as a probe. (A-a) Schematic diagram of the molecular species of HS ρ− mtDNA separated by 2D gel electrophoresis. The first dimension separates DNA species according to mass, whereas the second dimension, which is run in the presence of ethidium bromide, fractionates molecules according to both mass and shape. The signals of the DNA species are indicated: continuous arc, concatemers (linear tandem multimers of varying continuous lengths); ssDNA, single-stranded DNA. Signals for supercoiled circular and nicked-circular double-stranded monomers and multimers were assigned by comparing A-e and A-f as described in the ‘Results’ section. 1C, 2C, 3C, 4C and 5C represent supercoiled closed-circular monomers, dimers, trimers, tetramers and pentamers, respectively. 1N, 2N, 3N, 4N and 5N represent nicked-circular monomers, dimers, trimers, tetramers and pentamers, respectively. Circular multimers are products of crossing-over–type homologous recombination (5,36). Signals corresponding to concatemers, supercoiled and nicked-circular monomers were measured and corrected by subtracting the background of an area of equivalent size that lacked radioactivity. (A-b, A-c, A-d and A-e) 2D gel profiles of HS ρ− mtDNA that were isolated from wild-type cells harboring pYES2/CT, pYESMHR1, pYES2/CT-DIN7 and pESC-DIN7+MHR1, respectively. (B) The top shows the amounts of concatemers relative to supercoiled closed circular monomers. The middle shows the amounts of single-stranded DNA molecules relative to supercoiled closed-circular monomers. The bottom shows the amounts of circular multimers relative to concatemers. The numbers obtained from these calculation were directly plotted. (C) The bottom is an example of a 1D gel profile of BglII-digested DNA with probes for 32P-labeled HS ρ− mtDNA and the NUC1 gene, as described for Figure 2B. The DSBs at ori5 (top) and the HS ρ− mtDNA copy number (middle) were calculated, normalized against those obtained from wild-type cells harboring HS ρ− mtDNA and plotted as described in Figure 2C. (B and C) Each bar represents the average of three independent experiments. (D) Levels of overproduced Din7 and Mhr1 as measured by immunoblot analysis. Mitochondrial extracts were prepared from wild-type HS ρ− cells harboring the control plasmid pYES2/CT, the MHR1 expression plasmid pYESMHR1, the DIN7 expression plasmid pYES2/CT-DIN7 or the pESC-DIN7 + MHR1 plasmid, which were cultivated for 11 h at 30°C in 2 l of RGal medium to induce gene expression. Din7 and Mhr1 expression levels were detected by immunoblot analysis using anti-Din7 rabbit serum and anti-Mhr1 rabbit serum, respectively. As a control, porin was detected using a monoclonal anti-porin antibody.
As expected from the enhanced recombination repair in cells that overproduced both Din7 and Mhr1, the ratio of circular multimers to concatemers increased by 2.3-fold compared with control cells (Figure 4B, bottom), and the elevated DSB level at ori5 with Din7-overproduction was remarkably suppressed (Figure 4C, top and bottom; Figure 4D). In addition, the content of the circular multimers changed. In 2D gel profiles of mtDNA isolated from wild-type cells, faint, discrete dot-like signals were detected alongside a series of dot-like signals representing circular multimers (Figure 4A-b). These signals increased when Din7, Mhr1 or especially both were overproduced, and two series of discrete dot-like signals could be clearly detected (Figure 4A-c, A-d, and A-e). To identify which series represented closed-circular supercoiled or closed-circular double-stranded DNA, we treated DNA that was isolated from cells that overproduced both Din7 and Mhr1 with eukaryotic topoisomerase I and then performed 2D gel electrophoresis. The 2D gel profile showed a series of signals that migrated faster in the first-dimension run (1C, 2C, 3C, 4C, 5C and so forth) before topoisomerase treatment and were shifted to the left relative to the concatemer signals. Each member of the shifted series was located just beneath each member of the series of signals for 1N, 2N, 3N, 4N and 5N (compare Figure 4A-f with A-e). Thus, this analysis clearly showed that the signals that were shifted by topoisomerase I treatment represented supercoiled, closed-circular, double-stranded DNA (1C, 2C, 3C, 4C and 5C), whereas those that did not shift represented nicked-circular double-stranded DNA (1N, 2N, 3N, 4N and 5N). The supercoiled closed-circular forms are the results of either intermolecular recombination between circular forms or intramolecular recombination between tandemly repeated sequences in concatemers of HS ρ− mtDNA (5,37); thus, they represent the final product of crossing-over–type homologous recombination. By contrast, the nicked-circular DNA is an intermediate form in this process. Thus, simultaneous overexpression of Din7 and Mhr1 enhances homologous recombination.
It is surprising that co-overproduction of Mhr1 with Din7 suppressed the increase in the mtDNA copy number in Din7-overproducing cells (Figure 4C, middle and bottom). This observation suggests that after homologous pairing, an increase in Mhr1 activity relative to Din7 activity switches the pathway from mtDNA replication to mtDNA recombination and vice versa.
Effects of overproducing Din7 and Mhr1 on Endo.SceI-induced mitochondrial homologous recombination
As described previously, Din7 overproduction increased DSBs at ori5, as well as mtDNA copy number. However, these results significantly differ from those observed after hydrogen peroxide treatment. Din7 overproduction increased DSBs to a much greater extent than it increased mtDNA copy number (Figures 3B and 4C), whereas hydrogen peroxide treatment increased mtDNA copy number much more than it increased DSBs (Figure 2C). On the other hand, overproduction of Din7 or disruption of the gene encoding Dun1 (DNA-damage uninducible), a suppressor of Din7, extensively increases petite formation (19,23) and enhances recombination within the mitochondrial cyt b gene in yeast (23). These observations indicate that Din7 overproduction enhances recombination between imperfectly homologous sequences (23). Considering these effects of Din7 overproduction, it would be interesting to determine whether Din7 overproduction in cells with normal mtDNA (i.e. in ρ+ cells) induces abnormalities in mtDNA that explain petite formation. To address this question, we analyzed Din7-enhanced recombination.
We previously showed that Mhr1 is required for mitochondrial homologous recombination induced by mitochondrial sequence-specific Endo.SceI (8). To examine the effects of Din7 overproduction on Endo.SceI-induced mitochondrial homologous recombination, as shown in Figure 5A, we crossed MATa OliS (an oligomycin-sensitive allele of the Oli2 locus) bearing a mitochondrially encoded subunit (Ens2) of Endo.SceI [a heterodimeric endonuclease different from I-SceI (38)] with MATα OliR Δens2 cells harboring the Din7-overproduction vector. In this cross, Endo.SceI in zygotic cells (Endo.SceI+) introduced a specific DSB within the OliR allele in mtDNA to initiate polarized homologous mtDNA recombination (unidirectional gene conversion-type homologous mtDNA recombination) to replace the OliR sequence with that of OliS, thereby yielding an OliS recombinant. Unlike diploid nuclear genomic DNA in zygotes produced by mating, the mitochondrial genomic DNA of zygotes contains mtDNA pools of unequal sizes derived from each parent, and the transmission ratios of the a-parent and a-parent genetic markers to the progeny are, therefore, unequal. The transmission ratios of genetic markers from each parent are equal and independent of genetic loci or mutation types. Only markers involved in polarized homologous mtDNA recombination, in this case OliS from the a parent, have larger (or smaller) transmission ratios than other markers (reference markers), in this case, AntR and EryR from the a parent (recombination-positive cross 1 versus recombination-negative cross 2 is shown in Figure 5B; the observed numbers of colonies derived by cross experiments are shown in Table 2). The difference in the transmission ratio of the observed marker compared with the reference markers indicates the extent of polarized homologous recombination that occurs at the mtDNA locus being tested.
Figure 5.
Effects of overproducing Din7 and Mhr1 on mitochondrial homologous DNA recombination. (A) Schematic diagrams of Endo.SceI-induced homologous recombination at the Oli2-ENS2 locus. The donor mitochondrial genome contains the Oli2S (oligomycin-sensitive) gene lacking the Endo.SceI cutting site; ENS2, which encodes the smaller subunit of Endo.SceI (indicated by the filled boxes); and control markers, EryR (erythromycin-resistant) and AntR (antimycin-resistant) in loci distant from the Oli2 locus. The recipient mitochondrial genome carries the Oli2R (oligomycin-resistant base-substitution mutant) gene containing an Endo.SceI cutting site and a deletion mutation of ENS2 (Δens2), and it is EryS and AntS. After the mating of parent cells, each of which has the donor or recipient mitochondrial genome, Endo.SceI induces polar homologous recombination between the donor mitochondrial genome and the recipient mitochondrial genome to generate Oli2S, ENS2, EryS and AntS recombinant mitochondrial genomes, in which the region containing Oli2R and Δens2 in the recipient genome is replaced by the donor sequence. On the other hand, no Endo.SceI-induced recombination product is generated when the donor mitochondrial genomes carry ens2, a mutant of ENS2 mutant that is defective because of the insertion GC clusters in the gene (indicated with a filled box containing a hatched box inside). Because the recipient genome has the Endo.SceI site but the donor mitochondrial genome does not, gene conversion-type homologous recombination occurs in a strictly unidirectional fashion, resulting in polar homologous recombination. Endo.SceI-induced polar recombination increases the marker frequency in zygotic progeny (indicated as the transmission ratio) of the donor-derived Oli2S but not the frequencies of the control markers, EryR and AntR (8,32). (B) Effects of overexpressing DIN7 and MHR1 on Endo.SceI-induced polar homologous recombination. The transmission ratios of Oli2S and the reference markers (EryR and AntR) of the donor mitochondrial genome were calculated from the results shown in Table 2. The difference between the transmission ratio of Oli2S and those of EryR and AntR in cross 1 indicates the extent of polar homologous recombination induced by Endo.SceI in a cross involving wild-type cells that do not overexpress Din7 and Mhr1.
Table 2.
Effects of overproducing Din7, Mhr1 and both Din7 and Mhr1 on Endo.SceI-induced gene conversion at the Oli2 locus
| Crosses mating type α [mitochondrial genotype] × mating type a [mitochondrial genotype] | Numbers of colonies with Ery/Ant/Oli markers |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RRS | RSR | SRR | RRR | SSS | RSS | SRS | SSR | Total | ||
| 1 | WT/pYES2/CT [Δens2 Oli2R] × WT/(ENS2)[ENS2 EryR AntR] | 48 | 8 | 5 | 1 | 91 | 23 | 16 | 170 | 362 |
| 2 | WT/pYES2/CT [Δens2 Oli2R] × WT/(ens2) [ens2 EryR AntR] | 52 | 16 | 8 | 7 | 6 | 17 | 14 | 275 | 395 |
| 3 | WT/pYES2/CTDIN7 [Δens2 Oli2R] × WT/(ENS2) [ENS2 EryR AntR] | 34 | 8 | 0 | 0 | 208 | 17 | 37 | 67 | 371 |
| 4 | WT/pYES2/CTDIN7 [Δens2 Oli2R] × WT/(ens2) [ens2 EryR AntR] | 59 | 16 | 5 | 5 | 10 | 20 | 6 | 319 | 330 |
| 5 | WT/pYESMHR1 [Δens2 Oli2R] × WT/(ENS2) [ENS2 EryR AntR] | 44 | 7 | 0 | 0 | 188 | 38 | 39 | 64 | 380 |
| 6 | WT/pYESMHR1 [Δens2 Oli2R] × WT/(ens2) [ens2 EryR AntR] | 30 | 20 | 12 | 2 | 12 | 3 | 17 | 114 | 210 |
| 7 | WT/pESDIN7 + MHR1 [Δens2 Oli2R] × WT/(ENS2) [ENS2 EryR AntR] | 27 | 5 | 0 | 0 | 161 | 25 | 66 | 56 | 340 |
| 8 | WT/pESDIN7 + MHR1 [Δens2 Oli2R] × WT/(ens2) [ens2 EryR AntR] | 16 | 17 | 8 | 8 | 3 | 9 | 42 | 146 | 249 |
‘Endo.SceI’ is an abbreviated designation of Endonuclease SceI, a mitochondrial sequence-specific endonuclease. ENS2 is a mitochondrial gene encoding a subunit of Endo.SceI. ens2 encodes a non-functional subunit of Endo.SceI because of the insertion of GC clusters in the ENS2 coding region. Δens2 encodes a partially deleted subunit of Endo.SceI, and it is non-functional. Ery, erythromycin; Ant, antimycin; Oli, oligomycin. R, resistant; S, sensitive. EryR, resistant to erythromycin; AntR, resistant to antimycin; Oli2R, resistant to oligomycin at the mitochondrial Oli2 locus.
The transmission ratios of OliS in cells overproducing Din7 increased from 49% (without overproduction of any protein) to ∼80% in the presence of Ens2 (Figure 5B, cross 1 versus cross 3), whereas the transmission ratios of OliS in the absence of Ens2 (ens2 × Δens2) were 20−30%, similar to the transmission ratios of the reference markers, AntR and EryR, for a-cells (Figure 5B, cross 2 and cross 4). Thus, increased Din7 levels enhance Endo.SceI-induced polarized mitochondrial homologous recombination. The additional overproduction of Mhr1 did not further increase recombination at the Oli2 locus, as indicated by the transmission ratio of OliS (Figure 5B, cross 7), and Mhr1 overproduction without Din7 overproduction resulted in the same level of recombination. It is likely that 80% transmission is the saturation level of recombination observed in this system (Figure 5B, cross 5).
Hydrogen peroxide treatment promoted petite formation induced by Din7 overproduction
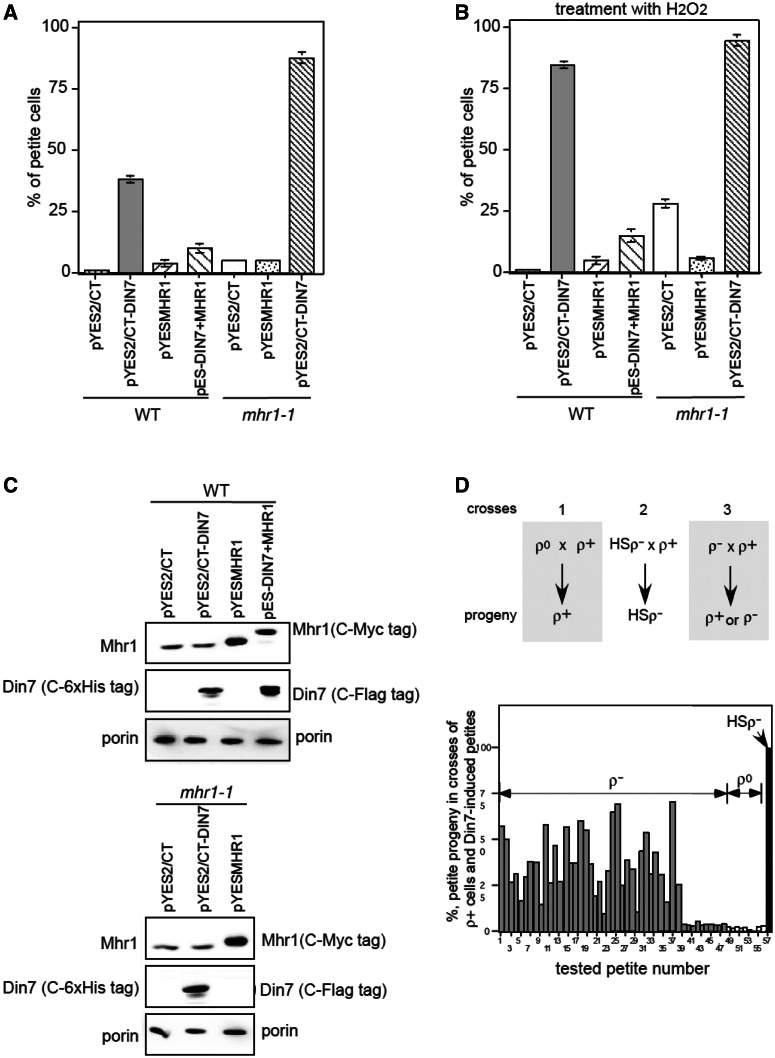
Next, we further analyzed the events at the mtDNA level induced by Din7 overproduction under various genetic backgrounds in ρ+ cells. Overproducing Din7 by introducing the pYES2/CT-DIN7 vector increased petite formation to 38% and the mhr1-1 mutation further increased petite formation to 88% (Figure 6A), indicating that enhanced petite formation is independent of Mhr1, which is required for major mtDNA recombination (8). Hydrogen peroxide treatment (10 µM) and Din7 overproduction induced an additive effect on petite formation (compare Figure 6B with A). Co-expression of Mhr1 in ρ+ cells that were overproducing Din7 and were either left untreated or treated with hydrogen peroxide restored mtDNA stability, i.e. the level of petite formation was suppressed to 10−14% (Figure 6A and B). Immunoblot analysis revealed increases in Din7 and Mhr1 expression levels (Figure 6C). All of these results suggest that petite formation induced by Din7 overproduction is independent of Mhr1-dependent homologous mtDNA recombination.
Figure 6.
Petite formation and loss of the ori5 region from respiration-proficient (ρ+) cells induced by Din7 overproduction and suppression by co-overexpression of Mhr1. (A and B) Effects of overproducing Din7 and Mhr1 on petite formation. Wild-type ρ+ cells harboring pYES2/CT, pYES2/CT-DIN7, pYESMHR1 or pESC-DIN7 + MHR1 and mhr1-1 ρ+ cells harboring pYES2/CT, pYESMHR1 or pYES2/CT-DIN7 were grown in SGlycerol medium, transferred to RGal medium and then cultivated for 18−20 generations (2 days) at 30°C to induce overproduction. A portion of the cells was treated with 10 μM hydrogen peroxide. Then, the treated cells (B) and untreated cells (A) were plated on SD plates and allowed to form colonies at 30°C for 3 days. The colonies were replica-plated onto YPGly plates to examine the respiration proficiency. The percentage of petite colonies among the total colonies on SD plates was calculated and plotted. Each bar represents the average of three independent experiments. (C) Din7 and Mhr1 expression levels were analyzed by immunoblot analysis of mitochondrial extracts derived from a portion of the cells cultivated under the conditions described in A. As a control, the mitochondrial outer-membrane protein porin was detected. (D) Analysis of the mtDNA types in the progeny of crosses between ρ+ and petite cells induced by Din7 overproduction. When ρ+ cells were crossed with ρ+ cells, almost all of the resulting progeny contained ρ+ mtDNA (cross 1). When HS-ρ– cells were crossed with ρ+ cells, all resulting progeny had HS ρ– mtDNA (cross 2). When ρ– cells were crossed with ρ+ cells, the progeny contained either ρ– or ρ+ mtDNA (cross 3). After crossing ρ– cells obtained by Din7 overproduction with ρ+ cells, the fraction of petite cells in each clone of the zygotic progeny was measured and is plotted in the bottom.
Finally, we examined the types of petite progeny produced by Din7 overproduction by crossing them with ρ+ cells. In the cross between ρ+ and ρ0 (mtDNA-free) cells, almost 100% of the progeny was respiration-proficient (Figure 6D, cross 1). By contrast, if HS ρ− cells were crossed with ρ+ cells, almost all of the progeny inherited HS ρ− mtDNA; therefore, they had no respiration capacity (Figure 6D, cross 2). If moderate ρ− cells were crossed with ρ+ cells, the progeny included a valuable and moderate fraction of respiration-deficient cells (Figure 6D, cross 3). When each of the petite clones derived from Din7-overproducing ρ+ cells (total number, 54; numbers 55, 56 and 57 are WT ρ0, WT and WT HS ρ-, respectively) was crossed with a ρ+ cell, 11% of the petite progeny had no mtDNA (ρ0), 89% contained moderate ρ− and none harbored HS ρ− mtDNA (Figure 6E). As previously described, Din7 overproduction increased DSBs at the ori5 site (Figures 2 and 3) and in the single-stranded DNA region (Figure 4B, middle). Because single-stranded DNA is rather sensitive to degradation in vivo, we assumed that petite formation is associated with the deletion of the mtDNA region that includes ori5. The petite formation with preferential loss of active replication origins, such as ori5, is likely a consequence of Mhr1-independent mtDNA rearrangements caused by DSBs at ori5 that are induced by Din7 overproduction.
DISCUSSION
In this study, we showed that Din7 has 5′–3′ exonucleolytic activity on double-stranded DNA (Figure 1). We also demonstrated the effects of the presence, absence or overproduction of Din7, together with those of Mhr1 and hydrogen peroxide treatment, on two pathways initiated by a DSB at ori5: homologous DNA recombinational repair of the DSB and DSB-induced replication leading to rolling-circle DNA replication. These effects are summarized in Table 3.
Table 3.
Summary of the effects of presence, absence or overproduction of Din7 and/or Mhr1, with or without H2O2 treatment
 |
Dark gray boxes: basal level in DIN7 MHR1 cells. Conditions: B, the basal level; 0, null or untreated; OP, overproduced; +, H2O2-treated.
Results: Gray boxes, DSB in mtDNA; light gray boxes, mtDNA copy number; empty boxes, not tested.
+, the basal level; +/−, slightly reduced; ++/−, slightly enhanced; ++, +++, ++++, +++++++, enhanced to the extent indicated.
All tests were carried out in HS ρ– cells.
Din7 produces 3′-single-stranded tails before homologous pairing
Homologous recombination for DSB repair and rolling-circle DNA replication both require homologous pairing of a 3′-single-stranded tail and template dsDNA [Figure 7 (39,40)]. In this study, we showed that Din7 has 5′−3′ exonuclease activity, as suggested by its homology with S. pombe ExoI (Figure 1). In addition, Din7 was required for the repair of ROS-induced or spontaneous DSBs at ori5 and for the ROS-induced increase in mtDNA copy number (Figure 2). We previously showed that ROS-induced Ntg1-dependent DSBs at ori5 trigger an increase in mtDNA copy number (5,6). Considering these previous observations, the results presented here support a model in which the 5′−3′ exonuclease activity of Din7 is responsible for generating 3′-single-stranded tails involved in both Mhr1-dependent homologous mtDNA recombination and rolling-circle mtDNA replication induced by Ntg1-dependent DSBs at ori5 (Figure 7). In eukaryotic nuclei and in eubacteria, the 3′-single-stranded tails are generated from DSB sites by complex machinery, such as the Mre11-Rad50-Xrs2 complex in eukaryotes [see (41) for a review], the multimeric helicase/nuclease RecB-RecC-RecD complex in E. coli or the concerted actions of RecJ nuclease and RecQ DNA helicase in the RecF pathway of eubacteria [see (42) for a review]. In yeast nuclei, the 5′−3′ exonuclease ExoI functions in mitotic recombination in parallel with the Mre11-Rad50-Xrs2 complex (43,44). On the other hand, in the recombination system of E. coli phage λ, a 5′−3′ exonuclease (λ exonuclease, Redα) is the major enzyme that generates the 3′ tail for homologous recombination. We previously showed that Mhr1-dependent rolling-circle mtDNA replication generates concatemers that are specifically transmitted to daughter cells and then processed into circular monomeric mtDNA in daughter-cell mitochondria; this process is similar to the later phase of DNA replication and DNA packaging into phage particles in the phage λ lytic cycle [(12) and references therein]. The results presented here show that Din7 is a counterpart to λ exonuclease in homologous recombination and replication of the bacteriophage DNA.
Figure 7.
A model for the cooperative mechanism involving Din7 and Mhr1 in repair of DSB at ori5 and the DSB-induced rolling-circle replication via homologous DNA recombination. The DSB at ori5, a mtDNA replication origin, is induced by Ntg1, which detects ROS-mediated–specific modifications at ori5. Din7 has 5′−3′ double-stranded DNA exonuclease activity and processes the DSB to 3′-single-stranded DNA tails. Mhr1 pairs one or both of the 3′-single-stranded DNA tails with a homologous sequence of intact circular mtDNA to form heteroduplex DNA joints. When the amount of Din7 relative to that of Mhr1 is at a proper level, Mhr1 binding to the tail limits single-stranded tailing. The bound Mhr1 promotes homologous pairing, which is followed by a rolling-circle replication pathway that increases the mtDNA copy number, as well as by homologous recombination to repair the DSB and minimize the number of residual DSBs. When the amount of Din7 is higher relative to Mhr1, enhanced single-stranded tailing prevents homologous recombinational repair of the DSB but increases the initiation of rolling-circle replication to increase the mtDNA copy number because any single-stranded tail can serve as primer to initiate rolling-circle mtDNA replication. Preventing Mhr1-dependent recombinational repair of the DSB increases the number of residual DSBs and Mhr1-independent error-prone DSB repair, which would cause petite formation. In the absence of Din7 (Δdin7 cells), Mhr1-dependent homologous pairing induced by a DSB at ori5 is limited, resulting in a decrease in the mtDNA copy number and an increase in the number of residual DSBs.
The amounts and relative levels of Din7 and Mhr1 determine the preference for mtDNA replication or recombination after homologous pairing
Because the amount of the Ntg1-dependent and -independent DSBs initially present at ori5 is likely to be independent of Din7, which is an exonuclease but lacks endonucleolytic activity (Figure 1), the observed Mhr1-dependent increase in DSB level, single-stranded DNA regions and mtDNA copy number resulting from Din7 overproduction (Table 3) indicates that overproduced Din7 generates longer single-stranded DNA regions through its exonucleolytic activity, but that these longer single-stranded regions prevent repair of DSBs and enhance mtDNA replication. The suppression of the aforementioned effects of Din7 overproduction by co-overproduction of Mhr1 (Table 3) suggests that the overproduced Mhr1 binds to the single-stranded DNA region to prevent further exonucleolytic attack by Din7, and that under these conditions, the bound Mhr1 preferentially stimulates recombinational repair of the DSB rather than mtDNA replication. However, a slight increase in mtDNA copy number on Mhr1 overproduction (Table 3) indicates that after the production of a DSB at ori5, Mhr1 activity preferentially promotes mtDNA replication when Din7 is limiting. Thus, a balance between three factors determines the preference for mtDNA replication (to increase mtDNA copy number) or recombination (to repair DSBs): (i) Din7-mediated exonucleolytic degradation, which generates single-stranded DNA tails from the DSB termini; (ii) termination of the Din7 exonucleolytic activity by bound Mhr1; and (iii) homologous pairing by bound Mhr1.
The balance between Din7 and Mhr1 helps determine mtDNA integrity and stability. Because co-overproducing Din7 and Mhr1 greatly reduced the increase in mtDNA copy number compared with hydrogen peroxide treatment (Figure 2C versus 4C), it is likely that controlling the mtDNA copy number at ori5 is primarily mediated by Ntg1-catalyzed DSB at the origin.
Petite formation is caused by excessive exonucleolytic degradation by Din7, which is suppressed by Mhr1-mediated homologous DNA recombination
Din7 overproduction induces petite formation, which has been attributed to enhanced recombination (19,23). Din7 enhances mtDNA recombination in ρ+ cells, but also induces mtDNA rearrangements and instability, which is detected as petite formation (Figures 5 and 6). Such petite formation is not caused by enhanced Mhr1-dependent homologous recombination, but instead by Mhr1-independent recombination, because the petite production induced by Din7 overproduction was not decreased or even enhanced by the absence of Mhr1 (Figure 6A and B). Because ROS enhances such recombination, this recombination is likely to be induced by unrepaired DSBs at ori5 (Figure 6).
In this study, we found that the balance between the exonuclease activities of Din7 and Mhr1-mediated homologous pairing influence the relative preference for mtDNA recombination versus rolling-circle mtDNA replication after homologous pairing. Furthermore, we determined that these processes affect both mtDNA copy number and mtDNA stability. It is likely that other factors influence the preference for or selection of specific pathways after homologous pairing, and that mitochondria contain a system that regulates this choice. These hypothetical systems will be examined in future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Life Science Foundation of Japan (to F.L.); Grants-in-Aid for Scientific Research (C) [18570168, 20570171, 23510237] from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to F.L.); RIKEN Strategic Research Program (to F.L.); RIKEN (to F.L.); JST-CREST (to F.L.). Funding for open access charge: Grants-in-Aid for Scientific Research (C) [23510237] from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to F.L.).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Calabrese V, Scapagnini G, Giuffrida Stella AM, Bates TE, Clark JB. Mitochondrial involvement in brain function and dysfunction: relevance to aging, neurodegenerative disorders and longevity. Neurochem. Res. 2001;26:739–764. doi: 10.1023/a:1010955807739. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 5.Ling F, Hori A, Shibata T. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho−] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 2007;27:1133–1145. doi: 10.1128/MCB.00770-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009;37:749–761. doi: 10.1093/nar/gkn993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulery TL, Jang SH, Jaehning JA. Glucose repression of yeast mitochondrial transcription: kinetics of derepression and role of nuclear genes. Mol. Cell. Biol. 1994;14:1160–1170. doi: 10.1128/mcb.14.2.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling F, Makishima F, Morishima N, Shibata T. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 1995;14:4090–4101. doi: 10.1002/j.1460-2075.1995.tb00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 10.Wei YH, Lee CF, Lee HC, Ma YS, Wang CW, Lu CY, Pang CY. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4977 BP-deleted mitochondrial DNA. Ann. N.Y. Acad. Sci. 2001;928:97–112. doi: 10.1111/j.1749-6632.2001.tb05640.x. [DOI] [PubMed] [Google Scholar]
- 11.Dujon B. Mitochondrial genetics and functions. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. New York, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 505–635. [Google Scholar]
- 12.Ling F, Shibata T. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 2002;21:4730–4740. doi: 10.1093/emboj/cdf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling F, Shibata T. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell. 2004;15:310–322. doi: 10.1091/mbc.E03-07-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda T, Ito Y, Terada T, Shibata T, Mikawa T. A non-canonical DNA structure enables homologous recombination in various genetic systems. J. Biol. Chem. 2009;284:30230–30239. doi: 10.1074/jbc.M109.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling F, Yoshida M, Shibata T. Heteroduplex joint formation free of net topological change by Mhr1, a mitochondrial recombinase. J. Biol. Chem. 2009;284:9341–9353. doi: 10.1074/jbc.M900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haber JE. DNA recombination: the replication connection. Trends Biochem. Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 17.Zassenhaus HP, Hofmann TJ, Uthayashanker R, Vincent RD, Zona M. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 1988;16:3283–3296. doi: 10.1093/nar/16.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morishima N, Nakagawa K, Shibata T. A sequence-specific endonuclease, Endo.SceI, can efficiently induce gene conversion in yeast mitochondria lacking a major exonuclease. Curr. Genet. 1993;23:537–541. doi: 10.1007/BF00312648. [DOI] [PubMed] [Google Scholar]
- 19.Fikus MU, Mieczkowski PA, Koprowski P, Rytka J, Sledziewska-Gojska E, Ciesla Z. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mieczkowski PA, Fikus MU, Ciesla Z. Characterization of a novel DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, which is a structural homolog of the RAD2 and RAD27 DNA repair genes. Mol. Gen. Genet. 1997;253:655–665. doi: 10.1007/s004380050369. [DOI] [PubMed] [Google Scholar]
- 21.Szankasi P, Smith GR. A single-stranded DNA exonuclease from Schizosaccharomyces pombe. Biochemistry. 1992;31:6769–6773. doi: 10.1021/bi00144a017. [DOI] [PubMed] [Google Scholar]
- 22.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 23.Koprowski P, Fikus MU, Dzierzbicki P, Mieczkowski P, Lazowska J, Ciesla Z. Enhanced expression of the DNA damage-inducible gene DIN7 results in increased mutagenesis of mitochondrial DNA in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2003;269:632–639. doi: 10.1007/s00438-003-0873-8. [DOI] [PubMed] [Google Scholar]
- 24.Phadnis N, Sia RA, Sia EA. Analysis of repeat-mediated deletions in the mitochondrial genome of Saccharomyces cerevisiae. Genetics. 2005;171:1549–1559. doi: 10.1534/genetics.105.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mookerjee SA, Sia EA. Overlapping contributions of Msh1p and putative recombination proteins Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting events in the mitochondrial genome of Saccharomyces cerevisiae. Mutat. Res. 2006;595:91–106. doi: 10.1016/j.mrfmmm.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Blanc H, Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc. Natl. Acad. Sci. USA. 1980;77:3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- 28.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Anal. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 29.Shadel GS, Seidel-Rogol BL. Diagnostic assays for defects in mtDNA replication and transcription in yeast and humans. Methods Cell Biol. 2007;80:465–479. doi: 10.1016/S0091-679X(06)80023-3. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, New York, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 31.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa K, Morishima N, Shibata T. An endonuclease with multiple cutting sites, Endo.SceI, initiates genetic recombination at its cutting site in yeast mitochondria. EMBO J. 1992;11:2707–2715. doi: 10.1002/j.1460-2075.1992.tb05336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghangas GS, Wu R. Specific hydrolysis of the cohesive ends of bacteriophage lambda DNA by three single strand-specific nucleases. J. Biol. Chem. 1975;250:4601–4606. [PubMed] [Google Scholar]
- 34.Li Z, Ling F, Shibata T. Glucose repression on RIM1, a gene encoding a mitochondrial single-stranded DNA-binding protein, in Saccharomyces cerevisiae: a possible regulation at pre-mRNA splicing. Curr. Genet. 1998;34:351–359. doi: 10.1007/s002940050406. [DOI] [PubMed] [Google Scholar]
- 35.Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorimer HE, Brewer BJ, Fangman WL. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol. Cell. Biol. 1995;15:4803–4809. doi: 10.1128/mcb.15.9.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacAlpine DM, Perlman PS, Butow RA. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 2000;19:767–775. doi: 10.1093/emboj/19.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watabe H, Iino T, Kaneko T, Shibata T, Ando T. A new class of site-specific endo-deoxyribonucleases: Endo.SceI isolated from a eukaryote, Saccharomyces cerevisiae. J. Biol. Chem. 1983;258:4663–4665. [PubMed] [Google Scholar]
- 39.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 40.Shibata T, Ling F. DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion. 2007;7:17–23. doi: 10.1016/j.mito.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 42.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 43.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11: 2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.