Abstract
The rpoS mRNA, which encodes the master regulator σS of general stress response, requires Hfq-facilitated base pairing with DsrA small RNA for efficient translation at low temperatures. It has recently been proposed that one mechanism underlying Hfq action is to bridge a transient ternary complex by simultaneously binding to rpoS and DsrA. However, no structural evidence of Hfq simultaneously bound to different RNAs has been reported. We detected simultaneous binding of Hfq to rpoS and DsrA fragments. Crystal structures of AU6A•Hfq•A7 and Hfq•A7 complexes were resolved using 1.8- and 1.9-Å resolution, respectively. Ternary complex has been further verified in solution by NMR. In vivo, activation of rpoS translation requires intact Hfq, which is capable of bridging rpoS and DsrA simultaneously into ternary complex. This ternary complex possibly corresponds to a meta-stable transition state in Hfq-facilitated small RNA–mRNA annealing process.
INTRODUCTION
To survive changes in the environment, bacteria have developed complicated mechanisms to respond to various stress conditions such as oxidative stress, UV irradiation, heat shock, hyperosmolarity, phosphosugar toxicity and change in iron concentration. Many of the stress response processes are mediated by small non-coding RNAs (sRNA). One major mechanism of sRNA regulation is base pairing to the target mRNA (1). RNA chaperon protein Hfq (host factor required for phage Qβ replication) is often required to facilitate base pairing between sRNA and target mRNA (2,3). Regulation of rpoS mRNA translation by DsrA sRNA at low temperature is one particular interesting example of sRNA-mediated stress response. The rpoS mRNA encodes the RNA polymerase subunit σS factor, which is the master regulator of the general stress response (4). The 5′ untranslated region (5′ UTR, 5′ leader) of rpoS mRNA forms a stem with the ribosome-binding site, blocking access of ribosomes to the mRNA. At lower temperatures, with the assistance of the RNA chaperon protein Hfq, DsrA anneals to the 5′ UTR of rpoS and unmasks the ribosome-binding site of the mRNA for effective ribosome binding and translation activation (5–8).
Hfq is bacterial homolog of eukaryotic Sm/Lsm family RNA-binding proteins (2). Eukaryotic Sm/Lsm proteins are involved in mRNA splicing (9–11). In Escherichia coli (Ec), Hfq is a homo-hexameric protein constituted by six subunits, with 102 amino acids in each subunit. A well-structured Sm fold is constituted by 1–65 amino acids (Hfq65, one amino-terminal α helix followed by five β strands), which forms ring-shaped hexamer with a central pore (12). A flexible but functional important tail is formed by 66–102 amino acids (13,14). One side of the ring on which the amino-terminal α helices lie is named the ‘proximal side’, while the opposite side is named the ‘distal side’. Proximal side and distal side binds to U-rich and A-rich single-stranded RNA (ssRNA) with high affinity, respectively (15). Interestingly, the lateral side of Hfq, which is rich in positively charged residues, is also reported to be involved in binding the ‘body’ of sRNAs (16).
Despite the central role of Hfq in sRNA regulation, the mechanism of how Hfq facilitates base pairing of DsrA sRNA to rpoS mRNA is not well understood. Hfq binding to U-rich sequences of DsrA with its proximal side has already been demonstrated in various studies (8,15,17,18). Recent works indicate that the binding of Hfq to an (AAN)4 motif in 5′ UTR of rpoS is critical for the regulation of rpoS by sRNAs. Hfq’s binding to this A-rich sequence may induce restructuring of the rpoS to promote the base pairing with DsrA. In addition, Hfq cannot stably bridge DsrA and rpoS if the complementary regions on both RNAs are not involved (19). Another study using mass spectroscopy shows that a poly (A) stretch A18 and DsrADII (nucleotides 23–60) may form 1:1:1 transient ternary complex with Hfq, although this ternary complex is unstable in solution (20). All these results support the scenario that in the early encounter stage of Hfq-facilitated base pairing of rpoS and DsrA, the (AAN)4 site tethers the distal face of Hfq to rpoS, leaving the proximal face available to engage in transient interactions with DsrA. Thus, a meta-stable ternary complex bridged by Hfq is formed. After this, two RNAs anneal to each other, while Hfq remains associated with one of the RNAs (likely rpoS). This latter ternary complex is not bridged by Hfq, but it is stable (3,19).
Multiple crystal structures of Hfq in complex with U-rich [AU5G (21), U6 (22), AU6A (23)] and A-rich [A15 (24), A7 (25), (AG)3A (26)] ssRNAs have been reported. Possibly owing to the instable nature of Hfq-bridged interaction between DsrA and rpoS, no structural evidence is yet available for the transient ternary complex DsrA•Hfq•rpoS. Nevertheless, structural and biochemical information for this transient ternary complex could be very helpful in understanding the mechanism of Hfq action.
Here we report observations of a ternary complex bridged by Hfq between an A-rich Hfq-binding fragment of rpoS [rpoS-AA, nucleotides 366–400, containing an (AAN)4 and an A6 element] and a DsrA fragment (DsrAII, nucleotides 26–61, containing the AU6A U-rich Hfq-binding site) by electro-mobility shift assays (EMSAs). A crystal structure of Hfq ternary complex bound simultaneously to A7 and AU6A (nucleotides 28–35 of DsrA) at 1.8-Å resolution and a complex crystal structure of Hfq bound to A7 at 1.9-Å resolution were also acquired. The AU6A•Hfq•A7 is by far the first structure in which Hfq is bound simultaneously to A-rich and U-rich RNAs. Simultaneous binding of Hfq to an (AAN)3 segment of rpoS-AA and AU6A was further confirmed in solution NMR. The ternary complexes observed in our research may mimic the transient ternary complex of DsrA•Hfq•rpoS. In addition, we demonstrate that intact distal and proximal RNA-binding sites are essential for ternary complex formation. Mutant Hfq that cannot bridge ternary complex in vitro exhibited little activity in translation activation of rpoS in vivo. These observations suggest that Hfq does have the capacity to bridge a transient ternary complex by binding to rpoS on distal side and to DsrA on proximal side simultaneously, and this unstable ternary complex may be necessary for translation activation of rpoS mRNA. The implications of the transient ternary complex were discussed.
MATERIALS AND METHODS
Bacteria strains and plasmids
The hfq- BL21 (DE3) strain was constructed as described by Datsenko et al. (27). Full-length wild-type and mutant hfq genes were inserted into pBAD18-kan plasmid (28) under the control of inducible araBAD promoter. The rpoS-5′ UTR and green fluorescent protein variant optimized for maximal fluorescence when excited by ultra violet light (GFPuv) sequences were obtained by PCR amplification from Ec strain BL21 (DE3) and pGFPuv vectors (Clontech), respectively. These two segments were connected by overlapping PCR with a GSSG linker and subsequently inserted into pET-22b (+) vector (Novagen) with a preceding T7 promoter and Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible lac operator. Plasmids bearing the rpoS-5′ UTR-GFPuv fusion and full-length wild-type or mutant Hfq–coding sequences were transformed into the hfq- strain by electroporation (29).
Hfq purification, crystallization and structure determination
Recombinant full-length Hfq (HfqFL) and Hfq65 were over-expressed and purified from Ec as previously described (23). Uniformly 15N-labeled Hfq65 R16A/R17A sample was prepared by growing bacteria in LR medium supplemented with 15NH4Cl and purified by same procedure as non-labeled Hfq65. Hfq65 hexamer (0.2 mM) was mixed with 0.1–0.15 mM A7 together with 0.15–0.2 mM AU6A and then mixed with an equal volume of crystallization buffer (100 mM NaCl, 100 mM cacodylate, 12% polyethylene glycol (PEG) 8000 at pH 6.2). Crystal was obtained by hanging-drop vapor diffusion. Hfq65 hexamer (0.2 mM) with 0.15 mM A7 was crystallized by mixing an equal volume of 200 mM NH4Ac, 100 mM Tris and 26% 2-propanol at pH 7.9. The AU6A•Hfq•A7 crystal took the I121 space group and diffracted to 1.8-Å resolution. The Hfq•A7 crystal took the C121 space group and diffracted to 1.9-Å resolution. X-ray intensity data were collected at the Shanghai Synchrotron Radiation Facility using beamline BL17U and were merged and scaled with MOSFLM and SCALA in the CCP4 suite (30,31). Statistics of the structures are presented in Supplementary Table S1. Both Hfq•A7 and AU6A•Hfq•A7 structures were solved by molecular replacement by Phaser (32) using apo Ec Hfq structure (PDB ID 1HK9) as the search model. The Rwork and Rfree of Hfq•A7 structure were refined to 19.0 and 23.1%, respectively. For AU6A•Hfq•A7 structure, Rwork and Rfree were refined to 18.8 and 22.6%, respectively.
Coordinates
Coordinates and structure factors for the AU6A•Hfq•A7 and Hfq•A7 complexes have been deposited with the Protein Data Bank under the accession codes of 4HT8 and 4HT9, respectively.
Nuclear magnetic resonance
The assignments of resonance peaks for Hfq65 have been acquired previously (23). Four hundred microliters of 0.1 mM uniformly 15N-labeled Hfq65 R16A/R17A in NMR buffer (40 mM sodium phosphate, 40 mM NaCl, 1 mM EDTA, pH 6.5) containing 10% D2O was titrated with 5 μl of ∼8 mM rpoS-AC for four times and then 3.8 μl of ∼11 mM AU6A ssRNA for four times. After each titration, a 1H-15N HSQC spectrum was recorded on a Varian 700 M spectrometer at 42°C. Experiment data were processed using NMRPipe (33) and Sparky. The full titration spectra are shown in Supplementary Figure S1.
Fluorescence polarization
Lyophilized 5′-FAM (Carboxyfluorescein)-labeled RNA oligomers were purchased from Takara Bio, Inc., and dissolved in diethyl pyrocarbonate (DEPC)-treated water to a final concentration of 100 μM. Stock (100 μM) was diluted to 1 μM in dilution buffer (DB) (20 mM Tris, 100 mM NaCl at pH 8.0). Equilibrium dissociation constants of different RNAs and different HfqFL constructs were determined by measuring fluorescence polarization (FP) as previously described (23). HfqFL was first diluted to 20 times the highest concentration used in the binding system, then diluted 2-folds in succession till the lowest desired concentration was reached. Before the assay, 190 μl of 42 nM fluorescence-labeled RNA was mixed with 10 μl of protein stocks from the diluted series. Samples were then excited at 490 nm, and the FP at 526 nm was read using a SpectraMax M5 (Molecular Devices) plate reader at 22°C. All FP data were well fitted to a 1:1 binding model.
In vitro transcription of DsrAII and rpoS-AA fragment
The DNA templates for transcription of rpoS-AA were prepared by annealing complementary ssDNA oligmers of rpoS-AA-T7-up (5′-GAAATTAATACGACTCACTATAGGGAACAACAAGAAGTTAAGGCGGGGCAAAAAATA-3′) and rpoS-AA-T7-dn (5′-TATTTTTTGCCCCGCCTTAACTTCTTGTTGTTCCCTATAGTGAGTCGTATTAATTTC-3′) at 95°C for 1 min, followed by incubation on ice for ∼5 min. The DNA template for DsrAII transcription was constructed similarly with oligmers DsrA2-T7-up (5′-GAAATTAATACGACTCACTATAGGAATTTTTTAAGTGCTTCTTGCTTAAGCAAGTTTCA-3′) and DsrA2-T7-dn (5′-TGAAACTTGCTTAAGCAAGAAGCACTTAAAAAATTCCTATAGTGAGTCGTATTAATTTC-3′). DsrAII and rpoS-AA ssRNA fragments were then transcribed in vitro by T7 RNA polymerase. The names and sequences of RNA used in this research are listed in Supplementary Table S2. Transcription products were precipitated with isopropanol, dissolved in DEPC-treated water and then purified from native polyacrylamide gel. Final RNA products were dialyzed into DEPC-treated water and quantified by absorbance at 260 nm.
Fluorescence labeling of RNA
A thiol group was modified to the 3′ end of the in vitro–transcribed rpoS-AA through oxidization with sodium periodate (34). The –SH activated RNA was then labeled using DyLight 680 Maleimide (Thermo) as recommended by manufacturer. Labeled RNA was isopropanol precipitated and further purified from polyacrylamide gel. The concentration and labeling efficiency of RNA was determined by measuring OD260 and OD680. Typical labeling efficiency was ∼44%. 5′-DyLight 680–labeled DsrAII was purchased from Takara Bio, Inc.
Electrophoresis mobility shift assays
All RNA-binding reactions were performed in binding buffer (BB) (6.67 mM sodium phosphate, 50 mM NaCl, 0.33 mM EDTA, pH 7.0). Before use, all RNAs were refolded by heating to ∼98°C for 30 s, followed by incubation on ice for 5 min.
For the 3′-DyLight 680–labeled rpoS-AA, 10 μl of binding reaction system contained 2.5 μl of 40 nM 3′-DyLight 680–labeled rpoS-AA, 5 μl of 78 nM HfqFL hexamer and 2.5 μl of DsrAII at various concentrations. Specifically, DsrAII was first diluted to 500 nM, and then followed by successive 2-fold dilutions to a final concentration of 31.25 nM. In the assay of Hfq mutants in bridging ternary complex, the final concentration of DsrAII was 62.5 nM. Reactions were incubated at room temperature for 30 min and resolved on 4% native polyacrylamide gels unless stated otherwise.
For the 5′-DyLight 680–labeled DsrAII, 10 μl of binding reaction system contained 2.5 μl of 20 nM 5′-DyLight 680–labeled DsrAII, 5 μl of 156 nM HfqFL hexamer and 2.5 μl of rpoS-AA at various concentrations. Specifically, rpoS-AA was first diluted to 2000 nM, and then followed by successive 2-fold dilutions to a final concentration of 31.25 nM. In the assay of Hfq mutants in bridging ternary complex, the final concentration of rpoS-AA was 125 nM. Reactions were incubated at room temperature for 30 min and resolved on 6% native polyacrylamide gels.
Gels were scanned in an Odyssey Infrared Imaging System using the 700-nm channel for detection. Each experiment performed on a same gel was repeated at least three times.
Western blotting
Overnight cultures of bacteria bearing GFPuv reporter (and HfqFL) plasmids were diluted 100× in ‘Luria–Bertani’ media and further grown at 30°C with appropriate antibiotics in the presence of l-arabinose (0.0225%) and IPTG (100 μM) for 8 h with agitation. Antibiotics concentrations used were 100 μg/ml for ampicillin and 10 μg/ml for kanamycin. For analysis, 3 ODs of each bacterial culture (1 OD is the total bacteria in 1 ml of culture, the OD600 of which is 1.0 for 1-cm light path) were collected and suspended in 100 μl of 2× SDS–PAGE loading buffer (0.1 M Tris–HCl, 20% glycerol, 4% SDS, 0.2% bromophenol blue, pH 6.8). The suspensions were heated to ∼98°C for 10 min and centrifuged at 16 400 g for 10 min. Three microliters of resulting supernatants were separated by SDS–PAGE and subjected to western blotting. The membrane was probed with mouse monoclonal anti-GFP antibody (Sigma Cat# G1546). Equal loading across lanes was verified by detecting GroEL with antibody purchased from Abcam (Cat# ab82592) (35,36). Antibody–antigen complex was detected using West Pico mouse IgG detection kit (Thermo) and visualized using ImageQuant LAS 4000 (GE). The experiments were carried out in triplicates, with similar results.
RESULTS
Hfq simultaneously binds to rpoS mRNA and DsrA sRNA
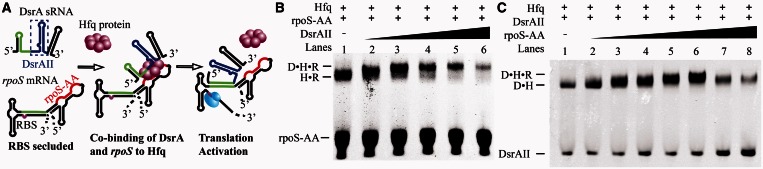
In the process of Hfq-facilitated base pairing between DsrA and rpoS, an intermediate ternary complex in which Hfq simultaneously binds to DsrA and rpoS on proximal and distal sides, respectively, has been suggested crucial for the activity of Hfq (Figure 1A). Because Hfq cannot stably bridge DsrA and rpoS if the two RNAs are not base paired (3,19), to capture the transient ternary complex bridged by Hfq between DsrA and rpoS, we selected two non–base-paired RNA fragments, DsrAII and rpoS-AA, to represent DsrA and rpoS for further investigation. DsrAII, a 37-nt portion of DsrA, contains neither the A-rich sequence preceding AU6A nor the region for base pairing with rpoS (besides the few nucleotides required for Hfq binding, it also contains one additional G residue from the T7 promoter at the 5′ end). In contrast, rpoS-AA represents nucleotides 366–400 of rpoS, which contains the A-rich Hfq-binding tract but not the region recognized by DsrA (Figure 1A).
Figure 1.
Co-binding of rpoS and DsrA to Hfq. (A) Co-binding of Hfq to DsrA sRNA and rpoS mRNA is a possible mechanism of Hfq in mediating DsrA-dependent rpoS translation activation. The A-rich Hfq-binding sequence on rpoS, rpoS-AA [nucleotides 366–400, containing an (AAN)4 and an A6 element], is colored red. The fragment containing U-rich Hfq-binding site and stem loop II of DsrA, DsrAII (nucleotides 26–61, containing the AU6A U-rich Hfq-binding site), is shown in blue. Regions on both RNAs for base paring to each other is colored in green. In EMSA experiment using HfqFL and fluorescence-labeled RNAs, we have observed (B) a supershift to Hfq•rpoS-AA (rpoS-AA was labeled with fluorescent probe) complex on addition of DsrAII and (C) a supershift to DsrAII•Hfq (DsrAII was labeled with fluorescent probe) complex on addition of rpoS-AA, suggesting that a DsrAII•Hfq•rpoS-AA ternary complex may form. Unbound rpoS-AA RNA migrates as two bands (Supplementary Figure S4). Brightness, contrast and gamma adjustments were applied to the whole image. Full images of Figure 1B and C showed in Supplementary Figure S5.
EMSAs, fluorescence-labeled rpoS-AA may form 1:1 complex with HfqFL, at lower protein concentration (Figure 1B lane 1, Supplementary Figure S2). Incubation of DsrAII together with HfqFL and rpoS-AA induced a supershift to the Hfq•rpoS-AA band (Figure 1B lanes 2–5). Fluorescence-labeled DsrAII can also form 1:1 complex with Hfq (Figure 1C lane 1, Supplementary Figure S3A). Binding of rpoS-AA induced a supershift to DsrAII•Hfq complex band (Figure 1C lanes 2–6). These observations suggest the formation of a ternary complex containing Hfq, DsrAII and rpoS-AA. Because DsrAII and rpoS-AA do not contain base-pairing regions, we did not observe the interaction between these two RNA fragments in the absence of Hfq (Figure 6D and E lane 2). Consequently, the ternary complex we observed is essentially formed by simultaneous binding of Hfq to both RNAs. At high DsrAII (or rpoS-AA) concentrations (>10-fold molar excess to rpoS-AA in Figure 1B lane 6, or DsrAII in Figure 1C lanes 7 and 8), the bands corresponding to this ternary complex start to fade, indicating that the RNA binding on two distinct sides of Hfq may affect each other.
Figure 6.
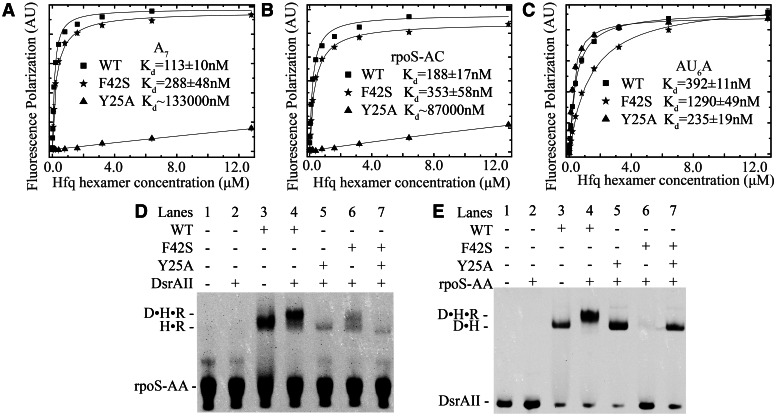
Both the distal and proximal sides are required for ternary complex formation. (A) A7 binding affinity to HfqFL is significantly decreased by Y25A mutation, while F42S has no obvious effect. (B) rpoS-AC binding affinity to HfqFL is significantly decreased by Y25A mutation, while F42S has no obvious effect. (C) AU6A binding affinity to HfqFL is significantly decreased by F42S mutation, while Y25A has no obvious effect. Data points of wild-type, F42S and Y25A HfqFL are shown as filled rectangular, star and triangle, respectively. Curve fitting results using 1:1 binding model are shown as black lines. EMSAs with fluorescence-labeled rpoS-AA (D) and DsrAII (E) show that wild-type HfqFL may bridge DsrA and rpoS, while mutation on either distal or proximal side abolishes ternary complex formation. A 1:1 mixture of distal- and proximal-side mutants also fails to bridge ternary complex (R: rpoS-AA, D: DsrAII, H: HfqFL). Brightness, contrast and gamma adjustments are applied to the whole image.
Crystal structure of AU6A•Hfq•A7 and Hfq•A7 complex
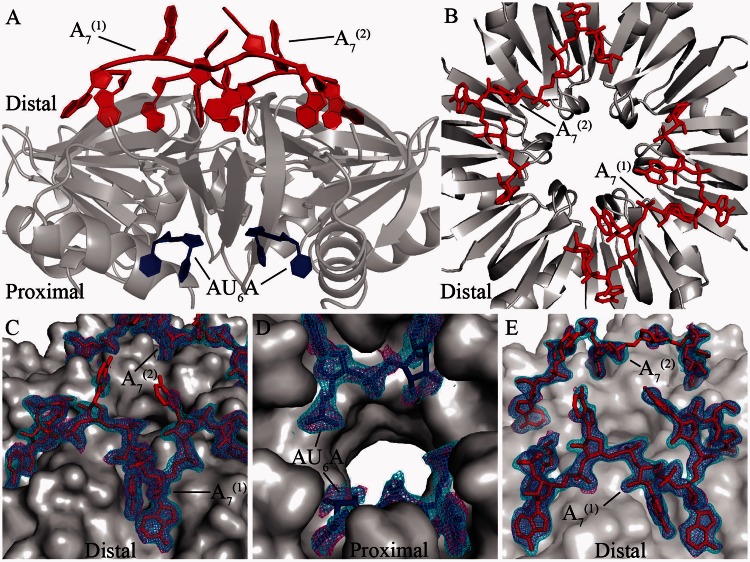
Hfq can bind to A-rich or U-rich ssRNA fragment using its distinct sides, indicating that Hfq is capable of simultaneously binding two types of short RNA strands. Ternary complex in which Hfq binds A-rich fragments on distal side and U-rich fragments on proximal side has been widely assumed (2,3,15,18,19,24,37–39). However no such kind of ternary complex structure has yet been reported. In the present research, we used Hfq65 to co-crystallize with a poly (A) fragment A7, or A7 together with AU6A ssRNA, and two high-resolution complex structures were obtained. The final structure model of the AU6A•Hfq•A7 ternary complex was refined to Rwork and Rfree values of 18.8 and 22.6%, respectively, at 1.8-Å resolution. The Hfq•A7 structure was refined to Rwork and Rfree values of 19.0 and 23.1%, respectively, at 1.9-Å resolution. The statistics of these two structures are shown in Supplementary Table S1. In AU6A•Hfq•A7 complex structure, each asymmetric unit contains three Hfq subunits, one A7 strand and 2 uridine nucleotides. The biological relevant assembly was generated according to crystallographic symmetry (Figure 2A). Two A7 strands are also observed bound on each Hfq hexamer in the Hfq•A7 structure (Figure 2B). Clear electron densities were observed for RNA fragments in both AU6A•Hfq•A7 and Hfq•A7 structures (Figure 2C–E).
Figure 2.
Global structures of AU6A•Hfq•A7 and Hfq•A7 complexes. In the AU6A•Hfq•A7 crystal, each asymmetric unit contained half of the Hfq hexamer. Biologically relevant assembly was generated according to crystallographic symmetry. (A) In the AU6A•Hfq•A7 structure, two A7 (red) molecules and one AU6A (blue) molecule are bound to each Hfq hexamer (gray) on distal and proximal sides, respectively. (B) Two A7 (red) molecules are bound to distal side of Hfq hexamer (gray) in the Hfq•A7 structure. (C) Clear density maps are observed for the two A7 molecules (red) in the AU6A•Hfq•A7 structure. (D) Part of AU6A in the AU6A•Hfq•A7 structure. (E) Electron densities for the two A7 molecules in the Hfq•A7 structure are also clearly observed. Difference maps Fo–Fc before inclusion of RNAs are shown as purple mesh (contoured at 2.0 σ), and 2Fo–Fc densities are shown as cyan mesh (contoured at 1.0 σ). The statistics of these two structures are shown in Supplementary Table S1.
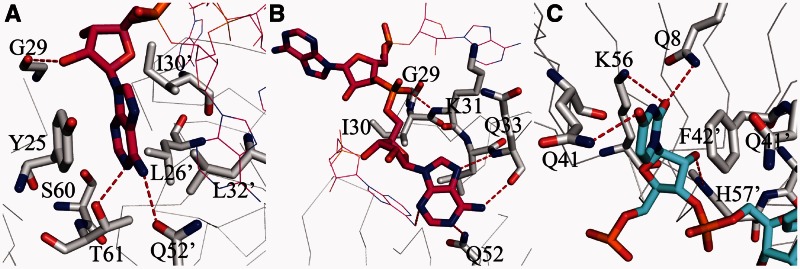
In the AU6A•Hfq•A7 structure, the overall binding of A7 is similar to the reported A15 binding to Hfq, exhibiting an A-R-N recognition pattern (24). The first, fourth and seventh adenosines of A7 insert into the ‘R’ sites, stacking against side chains of Y25 of one Hfq subunit and L26, I30 and L32 of an adjacent subunit (Figure 3A). The second and fifth adenosines bind to the ‘A’ sites, forming hydrogen bonds with Nε of Q52 as well as backbone amide hydrogen and carbonyl oxygen of Q33. The rest two adenosines bind on the ‘N’ sites where the adenine bases do not directly interact with Hfq (Figure 3B). Thus, the two A7 strands on the distal side occupy all ‘R’ sites. On the proximal side, four uridines bind to the canonical uridine-binding pockets, stacking against side chains of F42 and Q41 from two adjacent Hfq subunits. Hydrogen bonds with Q8, Q41, K56 and H57 are also observed (Figure 3C). However, other nucleotides of AU6A, which do not bind in the canonical uridine-binding pockets, are not resolved.
Figure 3.
Details of RNA binding on the distal and proximal sides of Hfq. The binding of A7 to the distal side of Hfq exhibits an A-R-N recognition pattern in both AU6A•Hfq•A7 and Hfq•A7 structures. (A) Adenosine inserts into ‘R’-binding site, stacking against side chains of Y25, L26’, I30’ and L32’ (where ’ denotes residues from an adjacent Hfq subunit). Hydrogen bonds with T61 and Q52’ are also observed. (B) In the ‘A’ site, adenine base forms hydrogen bonds to backbone atoms of Q33 and K31. ‘N’-site adenine base does not directly interact with Hfq. (C) The observed uridines bind to the proximal-side canonical uridine-binding pocket, stacking against side chains of Q41 and F42’. Hydrogen bonds with Q8, Q41, K56 and H57’ are also observed.
Structural comparison reveals possible changes in RNA binding on proximal side
In both Hfq•A7 and AU6A•Hfq•A7 structures, all six ‘R’ sites (24) on the distal side of Hfq are fully occupied. The conformations of A7 in these two structures are very similar, indicating that the binding of AU6A causes no significant effect on the binding of A7 (Figure 4A). However, the fully occupation of ‘R’ sites by two A7 strands on the distal side prevented AU6A from binding simultaneously to two different Hfq hexamers as in AU6A•Hfq•ADP structure (23), resulting in evident changes in the binding of AU6A on proximal side between AU6A•Hfq•A7 and AU6A•Hfq•ADP structures (Figure 4B). In the AU6A•Hfq•ADP structure, four canonical pockets for uridine binding (PUs) near F42 and Q41 are occupied, leaving two PUs empty. 5′-adenosine nucleotide of AU6A is bound to ‘R’ site on distal side of another Hfq hexamer. However, in AU6A•Hfq•A7 structure, no electron density is observed for nucleotides that are not bound in PU. The four PUs in AU6A•Hfq•A7 structure are occupied by four uridines, while the same PUs are occupied by three uridines and one adenosine in AU6A•Hfq•ADP structure. Clearly the inaccessibility of distal side to AU6A in AU6A•Hfq•A7 structure prevents the inter-hexamer–binding mode observed in AU6A•Hfq•ADP complex. The differences of observed nucleotides of AU6A between these two structures possibly indicate a prominent change in AU6A binding on the proximal side. However, because the remaining nucleotides of AU6A were invisible, to better understand how AU6A binds differently to Hfq when the inter-hexamer–binding mode is prohibited, further structural information will be required. In contrast, the binding of RNAs to Hfq does not cause significant structural changes to the protein. The root-mean-square deviation between backbone atoms of Hfq proteins in AU6A•Hfq•A7 and in Hfq•A7 is ∼0.42 Å, while that between AU6A•Hfq•A7 and AU6A•Hfq•ADP is ∼0.47 Å. The root-mean-square deviation between backbone atoms of Hfq in AU6A•Hfq•ADP and apo Hfq is ∼0.55 Å (23).
Figure 4.
Comparison of RNA binding in Hfq•A7, AU6A•Hfq•A7 and AU6A•Hfq•ADP structures. Hfq is shown as semi-transparent gray surface. (A) A7 binding is not significantly altered by AU6A binding. A7 strands in AU6A•Hfq•A7 and Hfq•A7 complex structures are shown as red and green sticks, respectively. The structures of A7 in these two complex structures are very similar. (B) AU6A binding on proximal side is different between AU6A•Hfq•A7 and AU6A•Hfq•ADP structures, in which AU6A are colored in blue and yellow, respectively. Nucleotides of AU6A that are not bound in canonical uridine-binding pockets are not observable in AU6A•Hfq•A7 structure.
Hfq65 may form ternary complex with A-rich and U-rich ssRNA in solution
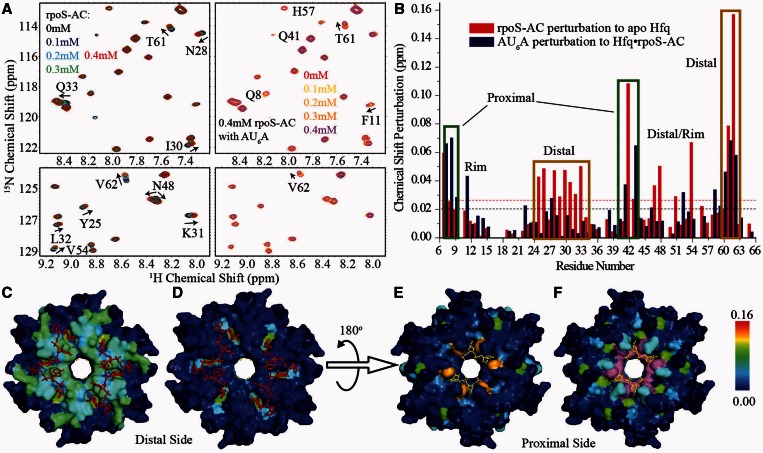
In AU6A•Hfq•A7 ternary crystal structure, we observed simultaneous binding of Hfq to A7 and AU6A. It has been reported that two elements on rpoS, A6 and (AAN)4 are possible Hfq-binding sites (19,40). Because A6 is only one adenosine shorter than A7, it is very likely that A6 could also form ternary complex with AU6A and Hfq in a similar way as A7 does. In contrast, the (AAN)4 element is not a poly (A) sequence very similar to A7. We, therefore, examined whether Hfq may also bridge (AAN)4 element and AU6A into a ternary complex using solution NMR (Figure 5). A fragment containing the first nine nucleotides of the (AAN)4 element, 5′-AACAACAAG-3′ (rpoS-AC, nucleotides 369–377), was selected in this study. The equilibrium dissociation constant of rpoS-AC with HfqFL is ∼200 nM as determined in FP experiments (Figure 6B). We used a uniformly 15N-labeled Hfq65 mutant, Hfq65 R16A/R17A, in NMR titration to avoid protein aggregation on RNA binding (23). rpoS-AC was first titrated into 0.1 mM Hfq hexamer to a 4:1 rpoS-AC:Hfq final molar ratio. Subsequently, AU6A was titrated into the same sample containing 0.1 mM Hfq hexamer and 0.4 mM rpoS-AC. 1H-15N HSQC spectra were recorded for each titration point. Selected regions on the HSQC spectrum and full spectrum are shown in Figure 5A and Supplementary Figure S1, respectively. The chemical shift changes of resonance peaks between the first and the last spectra are summarized in Figure 5B as column bars.
Figure 5.
Hfq could form ternary complex with rpoS-AC and AU6A in solution. rpoS-AC and AU6A were sequentially titrated into 0.1 mM Hfq hexamer. (A) Selected regions on 1H-15N HSQC spectrum of Hfq on rpoS-AC (left) and subsequent AU6A (right) titration. (B) Chemical shift differences between first and last titration points are presented in the column bars. Red and blue bars correspond to rpoS-AC and subsequent AU6A titrations, respectively. rpoS-AC binding to Hfq causes chemical shift perturbations on distal side of Hfq (C), while the following AU6A titration results in minor changes on this side (D). Proximal side is slightly perturbed by rpoS-AC binding (E) but evidently perturbed by subsequent AU6A titration (F). Hfq is colored according to chemical shift changes in blue-to-red gradient. F42 and H57 disappeared on AU6A titration and are colored in purple. Unassigned residues are colored in dark blue. A7 and observed nucleotides on AU6A are shown as red and yellow sticks, respectively.
As expected, the binding of rpoS-AC mainly perturbed the distal side of Hfq (Figure 5C). Residues involved in A7 binding on distal side, for instance Y25, N28, I30, Q33, K47, S60 and T61, exhibited prominent chemical shift changes. One residue on proximal side, F42, was also evidently perturbed, consistent with our previous observation that A7 titration to Hfq also caused large chemical shift change on this residue (23). Subsequent titration of AU6A caused marginal extra changes on distal-side residues (Figure 5D), indicating that the binding of rpoS-AC to Hfq is not disrupted by the addition of AU6A. On the contrary, resonance peaks of residues on the proximal side are significantly affected by AU6A titration. Residues Q8, Q41, Q52 and V43 are prominently perturbed, while F42 and H57 basically disappeared. These residues are either located near the proximal binding site or directly involved in U-rich sequence binding. Interestingly, several residues in the groove of ‘R’ site, S60, T61 and V62, also exhibited chemical shift changes on AU6A binding. Based on the fact that AU6A titration shifted resonance peaks of these residues further away from, instead back to, apo-state Hfq, it is highly possible that these chemical shift changes correspond to a ternary complex state, which differs from Hfq•rpoS-AC binary complex. Residues on the outer rim of Hfq, L7 and F11, also showed large chemical shift changes, similar to our previous observations (23). These results demonstrate that AU6A titration into Hfq•rpoS-AC complex does not cause dissociation of rpoS-AC from distal side of Hfq. Hfq could simultaneously bind to rpoS-AC on distal side and AU6A on proximal side to form an AU6A•Hfq• rpoS-AC ternary complex in solution.
Intact distal and proximal RNA-binding sites are essential in bridging ternary complex
The AU6A•Hfq•A7 crystal structure and NMR titration experiments show that Hfq binds A7 or rpoS-AC on the distal side and AU6A on the proximal side to form a ternary complex, indicating that intact distal and proximal RNA-binding sites may be functionally necessary for Hfq. To verify whether this is indeed the case, we generated mutations on distal side (Y25A) and proximal side (F42S) to disrupt A-rich and U-rich RNA binding, respectively. Y25 stacks with adenosine bases bound in ‘R’ sites on distal side (Figure 3A), while F42 stacks with uracil bases on proximal side (Figure 3C). In FP experiments, the binding affinities of Hfq to A7 and rpoS-AC are significantly lowered by Y25A but not F42S mutation (Figure 6A and B). On the contrary, AU6A, which binds mostly on proximal side, is prominently affected by F42S but not Y25A mutation (Figure 6C). Accordingly, binding affinity of DsrAII to Hfq, which was determined by EMSA, is reduced by F42S mutation but not by Y25A (Supplementary Figure S3). These Hfq mutants were further tested for their ability in bridging rpoS-AA and DsrAII into ternary complex (Figure 6D and E). In absence of Hfq, rpoS-AA and DsrAII do not form duplex (lane 2). Wild-type Hfq can form binary complex with labeled RNAs (lane 3), and bridge rpoS-AA and DsrAII into ternary complex (lane 4). However, neither Y25A nor F42S mutant bridges ternary complex (lanes 5 and 6). A 1:1 mixture of Y25A and F42S mutants still cannot bridge ternary complex (lane 7). These results indicate that intact distal and proximal RNA-binding sites on a same Hfq hexamer are required for DsrAII•Hfq•rpoS-AA ternary complex formation, further suggesting that the ternary complex we observed is indeed formed by simultaneous binding of Hfq to the two RNAs.
Activating rpoS translation in vivo requires intact Hfq, which can bridge rpoS and DsrA ternary complex
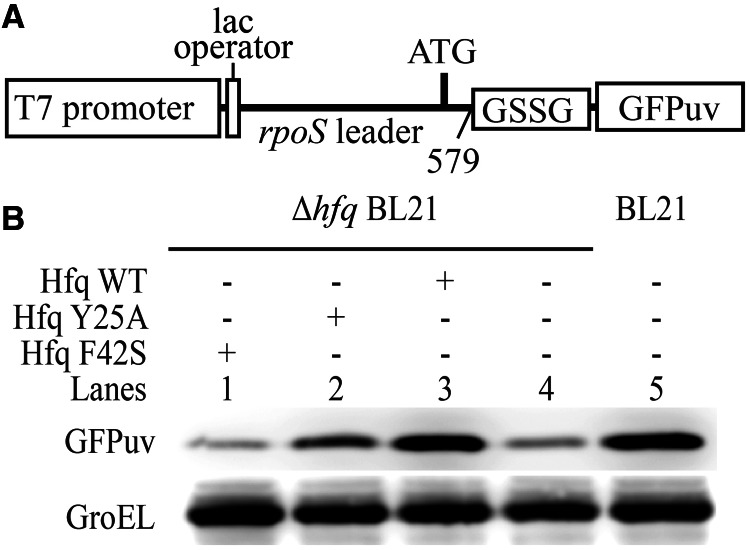
Hfq bound simultaneously to rpoS and DsrA is likely an important intermediate for translation activation of rpoS by DsrA (3,19). To evaluate the importance of this ternary complex in translation activation of rpoS, we tested whether the mutations that cannot bridge ternary complex in vitro (Figure 6D and E) would affect rpoS translation. A reporter system was therefore constructed in which the coding sequence of GPFuv was fused to 5′ UTR (nucleotides 1–579) of rpoS mRNA with a GSSG spacer (Figure 7A). This fusion, together with plasmids bearing full-length wild-type or mutant Hfq constructs, was transformed into hfq− Ec strain. Bacteria were then cultured at low temperature to test the efficiency of Hfq mutants in facilitating translation activation of rpoS mRNA by DsrA sRNA. Translation of the reporter GFPuv protein was detected in western blot with anti-GFP antibodies. Anti-GroEL antibody was used to detect GroEL as loading control (Figure 7B). Our results demonstrate that the expression level of GFPuv in wild-type BL21 is very similar to that in hfq− strain transformed with wild-type Hfq (lanes 3 and 5). In hfq− strain carrying empty pBAD vector, expression of GFPuv can barely be detected (Lane 4). This suggests that the exogenous wild-type Hfq effectively rescued the defect of hfq− strain in translation activation of rpoS. On the contrary, neither Y25A (Lane 2) nor F42S (Lane 1) mutant prominently increased GFPuv expression level as compared with hfq− strain carrying empty plasmid (lane 4). These observations are consistent with previous reports using different Hfq mutants (15,41). Clearly, both distal and proximal sides are important for Hfq in translation regulation. Mutant Hfq that cannot simultaneously bind both rpoS and DsrA in vitro is defective in translation activation of rpoS in vivo.
Figure 7.
RNA-binding sites in both distal and proximal sides are important for translation activation of rpoS facilitated by Hfq in vivo. (A) Schematic representation of the reporter construct for in vivo translation efficiency assays. Coding sequence of GFPuv proteins was fused to 5′ UTR of rpoS mRNA preceded by IPTG-inducible lac operator and T7 promoter. (B) GFPuv translation was detected by western blot using anti-GFP antibody. GroEL was detected as loading control. Deletion of hfq lowers GFPuv expression level, while wild-type HfqFL rescues translation activity. Brightness, contrast and gamma adjustments are applied to the whole image.
However, one might notice that there are still some inconsistencies between our results of FP and EMSA experiments (Figure 6 and Supplementary Figure S3) and in vivo translation assay (Figure 7). FP and EMSA results show that DsrA has decreased binding affinity with F42S mutation on the proximal side of Hfq, although the effect is not as drastic as Y25A mutation in the case of rpoS, which abolishes the binding. In vivo, however, Y25A mutant results in more rpoS expression compared with F42S mutant, which basically gives the same result as Hfq null mutant. It seems difficult to reconcile this disagreement between our in vitro and in vivo data. Interestingly, a recent work shows that DsrA accumulation is significantly reduced in F42A- and noticeably increased in Y25D-mutated Ec (42). Therefore, we speculate that F42S mutation might cause significant decrease of DsrA concentration in vivo, which in turn decreases the translation activity of rpoS mRNA. In contrast, increased DsrA accumulation in Y25A-mutated strain might compensate partly the loss in the translation activity, which is caused by disruption of interaction between Hfq and rpoS mRNA.
DISCUSSION
Possible role of transient ternary complex in promoting rpoS and DsrA annealing
Although many structural and functional studies have made tremendous advances in our understanding of the regulatory role of Hfq in facilitating base pairing between sRNA and target mRNA, the specific mechanisms by which Hfq engages sRNA and mRNA in the early encounter stage remain unclear. Two mutually non-exclusive mechanisms have been proposed to explain the above process: (i) Hfq may form ternary complex with two RNAs via co-binding to bring the RNA strands into proximity for optimal annealing. (ii) Hfq may bind one or both RNAs, and change its (or their) secondary (or tertiary) structure to facilitate base pairing. It has recently been demonstrated that the binding of Hfq to an (AAN)4 motif in 5′ UTR of rpoS is critical for regulation of rpoS by sRNAs (40,43). Binding of Hfq to this A-rich sequence may induce restructuring of the mRNA to promote base pairing with DsrA sRNA (19). Although supporting the second model, above research also indicated a role of possible intermediate ternary complex bridged by Hfq. Furthermore, a 1:1:1 ternary complex of a poly (A) stretch A18, a DsrA fragment DsrADII (nucleotides 23–60) and Hfq have been detected by mass spectroscopy using cross-linked sample. However, this ternary complex is unstable in solution (20). Seemingly, there are different ternary complexes that Hfq, DsrA and rpoS may form. The stable ternary complex is formed between DsrA-rpoS duplex and Hfq (Hfq only binds to one RNA, likely rpoS) (19). The unstable ternary complex is most likely formed via co-binding of sRNA and mRNA to Hfq, and this complex may exist as a transition state in the early encounter stage of sRNA and mRNA. To capture the structure of this transient ternary complex, we used the fragment AU6A and DsrAII of DsrA, which binds primarily to the proximal side of Hfq (8,15,17,18,23), and a stretch of rpoS 5′ UTR containing a short poly (A) A6 and an (AAN)4 element, rpoS-AA, which binds to the distal side of Hfq (43). We observed ternary complex bridged by HfqFL in EMSAs. Crystal structures of AU6A•Hfq•A7 ternary complex and Hfq•A7 complex showed that Hfq is capable of simultaneously binding to short poly (A) fragment and AU6A using its two distinct sides. In addition, NMR examination also indicated that Hfq can bind simultaneously to AU6A and an (AAN)3 fragment of rpoS in solution. Mutation of RNA-binding sites on either distal or proximal side of Hfq prevented formation of ternary complex. We further showed that the Hfq mutants, which cannot bridge ternary complex, are not efficient in rpoS translation activation at lower temperature in vivo. These results suggest that, via simultaneous binding to rpoS and DsrA, Hfq bridges a ternary complex, which is important for translation activation of rpoS by DsrA in vivo. The ternary complex we reported here very likely mimics the meta-stable transition state during Hfq-facilitated annealing of DsrA to rpoS where RNAs meet for subsequent base pairing (3).
Unstable nature of Hfq-bridged ternary complex may be due to the mutual effects between the binding of DsrAII and rpoS-AA on Hfq
Hfq binds to many RNAs tightly with nanomolar Kd values. At the same time, it is involved in many different sRNA-related regulatory processes, which require fast turnover among a large pool of binding RNAs. To reconcile this ‘strong-binding, high-turnover’ paradox, active cycling of Hfq by means of competition between RNAs for Hfq-binding sites has been proposed. The association of one RNA with Hfq may cause the replacement of the already-bound RNA (44). In addition, it has been shown in vivo that Hfq is sequestered by high-level transcription of sRNA or mRNA without base-pairing partners. However, when sRNA and mRNA are over-expressed in pairs, Hfq will not be sequestered, suggesting that duplex formation between sRNA and mRNA is coupled to Hfq dissociation from bound RNAs (45). The release of Hfq from RNA precedes duplex formation (46). Intriguingly, The Hfq-binding site of DsrA is partially overlapped with its base-pairing site with rpoS, suggesting a requirement of DsrA dissociation from Hfq during duplex formation (8). The ternary complex formed by co-binding of DsrA and rpoS fragments to Hfq that we observed likely forms the basis of an intermediate state during inter-molecular annealing (3). This intermediate ternary complex needs to be disrupted for effective base pairing. In our EMSAs, we observed the competition between rpoS-AA and DsrAII (Figure 1B and C). In addition, crystal structure comparison between AU6A•Hfq•A7 and AU6A•Hfq•ADP reveals that the binding sites of A7 molecules overlap with those of the 5′ adenosine of AU6A on the distal side of Hfq, indicating a possible competition between these two RNAs on this side. All our experimental evidences suggest a possibility that DsrA and rpoS will compete with each other for the binding to Hfq. Because the longer A-rich stretch [(AAN)4 and A6] on rpoS 5′ UTR very likely has higher affinity to the Hfq distal side than short A-rich segment preceding the AU6A site of DsrA, it is probable that, in vivo, the potential competition from rpoS acts to destabilize DsrA binding with Hfq. Furthermore, destabilization of DsrA-Hfq interactions may facilitate subsequent duplex formation between DsrA and rpoS. In summary, we propose that competition between the two RNAs might provide at least partly the driving force behind the unstable nature of the intermediate state we studied in this research, which definitely requires more detailed investigations.
Possible roles of lateral surface of Hfq in ternary complex formation
Besides the distal and the proximal RNA-binding sites, the lateral surface of Hfq may also play an important role in RNA binding. Several basic amino acid residues, R16, R17, R19 and K47, cluster near the outer rim of the ring-shaped Hfq hexamer, constituting a conserved positively charged surface. These residues have been reported to contribute to Hfq’s interactions with RNA and DNA (47,48). Recently it has been proposed that the lateral residues may bind to the sRNAs via their ‘body’ (16). In our previous research, we also observed that both the distal and proximal sides as well as the outer rim of the Hfq hexamer are involved in AU6A and Uex (nucleotides 23–35 of DsrA) binding in solution (23). Furthermore, the mutations of several residues, including R16, R17 and R19, were found to affect the translation efficiency of mRNA in vivo (42). Therefore, the positively charged lateral surface on Hfq hexamer might represent a new type of RNA-binding site apart from distal and proximal RNA-binding sites. In this research, we examined whether the mutations of these positively charged residues would also affect DsrAII•Hfq•rpoS-AA ternary complex formation (Supplementary Figure S6). Intriguingly, both the mutants of R16A/R17A (Supplementary Figure S6 lane 5) and R19A (Supplementary Figure S6 lane 6) fail to bridge ternary complex. Binary complex of Hfq with U-rich RNAs is also abolished by these mutations, indicating that the lateral site may be not very selective in RNA sequence. Because intact distal and proximal sides are also essential for ternary and corresponding binary complex formation (Figure 6D and E), it is likely that the positively charged lateral residues may act to enhance the binding affinities between Hfq and RNAs on both distal and proximal sides. Moreover, the competition for lateral surface may also exist between DsrA and rpoS, considering the lateral residues do not seem to be preferentially selective in binding RNA. Clearly, a more systemic investigation is needed for a better understanding of the roles of Hfq lateral sites in RNA binding.
RNA binding pattern on both distal and proximal side varies
The binding pattern of RNA on Hfq seems to differ depending on Hfq protein as well as RNA sequence. Three reported complex structures of Hfq bound to A-rich sequences showed interesting variations. In Ec, A7 binding pattern in AU6A•Hfq•A7 and Hfq•A7 structures is very similar to A15 binding in Hfq•A15 structure despite the difference in RNA length, exhibiting an A-R-N tripartite recognition motif (24). Interestingly in gram-positive bacteria, same A7 binds to Staphylococcus aureus (Sa) Hfq in a different A-L bipartite motif (25). An RNA tract (AG)3A, which is also seven nucleotides in length, binds to Bacillus subtilis (Bs) Hfq in a similar A-L motif (26). These structures indicate that A-rich tract recognition by distal side of Hfq is more sensitive to species of Hfq origin than to sequence feature of RNA. On proximal side, AU5G binds to Sa Hfq in a circular manner, with one nucleotide in the central pore (21). U6 bound to Salmonella typhimurium (St) Hfq showed a recognition mode of 3′-terminal poly-U pattern where all uridines were bound in the PUs (22). We have previously reported an AU6A•Hfq•ADP crystal structure in which AU6A bound to proximal side of Ec Hfq in a different manner: three uridines and one adenosine bound in PUs, while two uridines floated above the central pore. The 5′ adenosine bound to ‘R’ site on distal side of another Hfq hexamer (23). These structures show that variations in RNA sequences may lead to different recognition patterns on proximal side of Hfq, even in the same species. Interestingly, in the AU6A•Hfq•A7 structure we report here, only four uridines (bound in PU) were observed, indicating a destabilized binding of AU6A with Hfq. This is possibly caused by inaccessibility of ‘R’ sites in AU6A•Hfq•A7 structure, in which two A7 occupied all ‘R’ sites on distal side.
ACCESSION NUMBERS
PDB IDs of AU6A•Hfq•A7 and Hfq•A7 are 4HT8 and 4HT9, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–6.
ACKNOWLEDGEMENTS
The authors thank Prof. Baolin Sun, Ting Xue and Dr. Dan Yu for assistance in translation efficiency assays. Prof. Ke Ruan, Zhiyong Zhang, Jianye Zang, Changlin Tian, Dr. Fangming Wu, Zhenwei Song, Bin Wen, Pengzhi Wu, Jia Gao, Sai Li and Niamu for helpful discussions; Dr. Fudong Li, Yang Zou, Zhenhua Shao, Zhonghua Liu, Juncheng Wang, Chongyuan Wang, Minhao Wu, Yongxing He for assistance in reflection data collection; Dr. Yun-Xing Wang for kindly providing the plasmid of T7 RNA polymerase; F. Delaglio and A. Bax for providing the software NMRPipe; T. D. Goddard and D. Kneller for Sparky; and A. T. Brünger for CNS and W. L. DeLano for PyMol.
FUNDING
National Basic Research Program of China (973 Program) [2011CB911104, 2011CB966302]; Chinese National Natural Science Foundation [31270782, 30830031]; ‘Outstanding Technical Talent’ project of the Chinese Academy of Sciences. Funding for open access charge: National Basic Research Program of China (973 Program) [2011CB911104].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA: RNA interactions at multiple loci. Proc. Natl Acad. Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muffler A, Fischer D, HenggeAronis R. The RNA-binding protein HF-I, known as a host factor for phage Q beta RNA replication, is essential for rpoS translation in Escherichia coli. Gene Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 8.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat. Struct. Mol. Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- 10.Scofield DG, Lynch M. Evolutionary diversification of the Sm family of RNA-associated proteins. Mol. Biol. Evol. 2008;25:2255–2267. doi: 10.1093/molbev/msn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 12.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, Hajnsdorf E, Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 14.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Blasi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl Acad. Sci. USA. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Almeida Ribeiro E, Jr, Beich-Frandsen M, Konarev PV, Shang W, Vecerek B, Kontaxis G, Hammerle H, Peterlik H, Svergun DI, Blasi U, et al. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res. 2012;40:8072–8084. doi: 10.1093/nar/gks510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA. 2011;17:489–500. doi: 10.1261/rna.2452111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Wang L, Zou Y, Zhang J, Gong Q, Wu J, Shi Y. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 2011;25:2106–2117. doi: 10.1101/gad.16746011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19286–19291. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horstmann N, Orans J, Valentin-Hansen P, Shelburne SA, 3rd, Brennan RG. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012;40:11023–11035. doi: 10.1093/nar/gks809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–1867. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leslie AGW. In: Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography. 1992. Recent changes to the MOSFLM package for processing film and image plate data. 26. [Google Scholar]
- 31.Bailey S. The Ccp4 suite—programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011;30:2805–2816. doi: 10.1038/emboj.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. Plos One. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salim NN, Faner MA, Philip JA, Feig AL. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012;40:8021–8032. doi: 10.1093/nar/gks392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 2013 doi: 10.1016/j.jmb.2013.01.006. Jan 11 ( doi: 10.1016/j.jmb.2013.01.006; Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl Acad. Sci. USA. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 2011;39:5193–5202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun XG, Wartell RM. Escherichia coli Hfq binds A(18) and DsrA domain II with similar 2: 1 Hfq(6)/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 48.Updegrove TB, Correia JJ, Galletto R, Bujalowski W, Wartell RM. E. coli DNA associated with isolated Hfq interacts with Hfq's distal surface and C-terminal domain. Biochim. Biophys. Acta. 2010;1799:588–596. doi: 10.1016/j.bbagrm.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.